Abstract
A survey of emm gene sequences and an analysis of the pulsed-field electrophoretic profiles of 30 Streptococcus pyogenes isolates with reduced susceptibilities to ciprofloxacin detected the prevalence of isolates with emm type 6 and considerable genetic diversity among isolates. The mechanism of ciprofloxacin resistance in these isolates was based on point mutations in topoisomerase IV subunit C encoded by parC, mainly replacement of serine-79 by alanine.
Streptococcus pyogenes is the etiologic agent of a wide range of human infections, including streptococcal sore throat, skin and soft tissue infections, and the postinfectious syndromes of glomerulonephritis and acute rheumatic fever. Penicillin remains the drug of choice for the treatment of these infections because S. pyogenes remains susceptible to this antibiotic despite its intensive use. By contrast, an increasing frequency of S. pyogenes isolates that are resistant to macrolides, probably due to the increasing use of these antibiotics for the treatment of other bacterial respiratory pathogens, i.e., Streptococcus pneumoniae (5), has been reported in different countries (2, 7, 13). Similarly, the increasing use of fluoroquinolones, due to their excellent activities against some bacterial pathogens, has led to the emergence of S. pyogenes isolates with reduced susceptibilities to these antibiotics (4, 15). Increased resistance of S. pyogenes to ciprofloxacin has been reported in Spain at the highest rates ever published (10). Using emm gene typing and pulsed-field electrophoresis, we studied whether those resistant isolates represent only a few clonal types or a genetically unrelated set of strains that have emerged through the antibiotic's selective pressure.
Bacterial strains.
S. pyogenes isolates included in this study belong to the SAUCE II surveillance collection and were collected between November 1998 and October 1999 from patients with acute pharyngitis in 17 different hospitals selected on the bases of population and geographical location in Spain (10). Among 70 S. pyogenes isolates with reduced susceptibilities to ciprofloxacin (for MICs of ≥4 μg/ml, no NCCLS susceptibility breakpoint was available) collected during this period, a random sample of 30 isolates was selected for emm-typing studies. All but 6 of these 30 isolates were from pediatric samples. An additional 32 pharyngeal isolates were also selected from isolates susceptible to ciprofloxacin (MIC ≤ 1 μg/ml) so as to match the resistant strains in terms of location, age group, and temporal proximity during the same surveillance period.
emm gene typing.
The emm gene types of the S. pyogenes isolates were determined by amplification and sequencing of the emm genes as described by Beall et al. (3). Lysates of the S. pyogenes isolates were prepared with mutanolysin as described previously (1). Primers GASM1 and GASM2 were used in PCRs carried out according to a method described previously (3). PCR products were sequenced with primer GASM1 with a dye terminator mix (Perkin Elmer, Applied Biosystems, Madrid, Spain) and were subjected to automated sequence analysis on a model 377 DNA sequencer (Perkin Elmer, Applied Biosystems). DNA sequences were subjected to homology searches against the bacterial DNA database with BLASTN. Sequences were given GenBank emm designations based on previously described criteria (3). We found a strong association between emm type 6 and resistance to ciprofloxacin. Thus, 19 of the ciprofloxacin-resistant isolates (63.3%) exhibited emm type 6, while only 1 ciprofloxacin-susceptible isolate (3.1%) exhibited this emm type. The remaining resistant isolates exhibited other emm types (Table 1). emm type distribution among susceptible isolates was highly heterogeneous. Up to 12 different emm types accounted for 100% of the susceptible isolates (data not shown).
TABLE 1.
Characteristics of pharyngeal ciprofloxacin-resistant S. pyogenes isolates collected in Spain (1998 to 1999)a
| No. of isolates | emm type | PFGE pattern | QRDR mutation(s) |
|---|---|---|---|
| 9 | 6 | A | Ser79Ala |
| 3 | 6 | D | Ser79Ala |
| 2 | 6 | B | Ser79Ala |
| 2 | 6 | H | Ser79Ala |
| 1 | 6 | G | Ser79Ala |
| 1 | 6 | E | Ser79Ala |
| 1 | 6 | N | Ser79Ala |
| 2 | 73 | M | Ser79Ala |
| 1 | 28 | R | Ser79Phe, Asp91Asn |
| 1 | 28 | C | Asp91Asn |
| 1 | 75 | J | Ser79Phe, Asp91Asn |
| 1 | 75 | L | Ser79Ala |
| 1 | 22 | F | Asp91Asn |
| 1 | 78 | I | Asp91Asn |
| 1 | 3 | K | Ser79Ala |
| 1 | st1815 | B | Ser79Ala |
| 1 | 12 | B | Ser79Ala |
A MIC of ≥4 μg/ml was used as the ciprofloxacin resistance breakpoint. No NCCLS breakpoint was available.
PFGE.
Our emm-typing results suggest that ciprofloxacin-resistant S. pyogenes isolates could be genetically related, since a high percentage of them exhibited emm type 6. To test this hypothesis, we analyzed by pulsed-field gel electrophoresis (PFGE) the genomic DNA of all the ciprofloxacin-resistant S. pyogenes isolates. Total DNA was prepared, digested with SfiI, and resolved by PFGE as previously described (2). Differences in banding patterns were documented by visual examination and indexed by capital lettering. Interpretation of restriction fragments was performed in accordance with recent consensus publications (14).
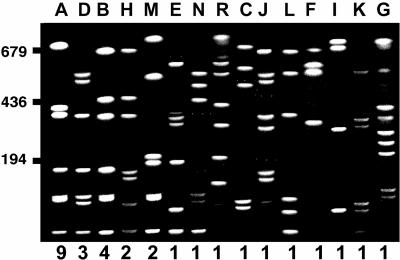
Results of PFGE analysis are shown in Fig. 1. Nine emm type 6 isolates presented PFGE pattern A, confirming our hypothesis that these isolates were genetically related. However, the remaining 10 emm type 6 isolates exhibited six different PFGE patterns. Moreover, the remaining ciprofloxacin-resistant isolates, which exhibited different emm types, also presented different PFGE patterns (Table 1). These findings suggest that ciprofloxacin-resistant S. pyogenes strains are the products of independent mutational events selected by the drugs from among a diverse population of S. pyogenes strains, presumably during treatment of other bacterial pathogens with these antibiotics. However, certain clones (of emm type 6 and PFGE pattern A) are preferentially selected and disseminated. Interestingly, eight of the nine isolates of this prevalent clone were also resistant to erythromycin (data not shown). The mechanism of resistance to erythromycin of these eight isolates is based on the presence of an efflux pump encoded by mefA as previously described (2). Moreover, all these isolates were collected from different patients in the same city. Thus, it is more probable that this specific clone was preferentially selected due to the use of macrolides than due to the use of quinolones, since all isolates were from children, in whom quinolone consumption, if any, is extremely low.
FIG. 1.
Representative PFGE types of ciprofloxacin-resistant S. pyogenes isolated between 1998 and 1999 in Spain. S. pyogenes genomic DNA was isolated, restricted with SfiI, and analyzed by PFGE. Letters above the lanes refer to PFGE types, and the numbers below the lanes refer to the number of isolates of each type. Molecular size markers (in kilobases) are indicated on the left.
Mechanisms of ciprofloxacin resistance in S. pyogenes.
The MICs of the antibiotics were determined by the microdilution method according to NCCLS guidelines. To eliminate the possibility that ciprofloxacin resistance was due to the presence of an active efflux pump, we determined the ciprofloxacin MICs in the presence of the efflux pump inhibitor reserpine (10 μg/ml). The ciprofloxacin MICs remained unchanged in the presence of reserpine, indicating that ciprofloxacin resistance in S. pyogenes was not due to the expulsion of the antimicrobial drug from the intracellular compartment by an efflux pump. Reduced susceptibility of S. pyogenes to ciprofloxacin has been associated with point mutations in either gyrA or parC (15). Those point mutations are located along the quinolone resistance-determining regions (QRDRs) of both genes (15). We investigated whether the ciprofloxacin-resistant isolates included in this study presented point mutations in the QRDRs of gyrA and parC. For this purpose, genomic DNA purified as described above was used as the template for PCR amplification of a 614-bp fragment of gyrA and a 520-bp fragment of parC containing the QRDRs of both genes with the pairs of primers described by Yan et al. (15). PCR products were sequenced on both strands, and sequences were compared with S. pyogenes gyrA and parC sequences from ciprofloxacin-resistant and -susceptible isolates previously described (15).
All ciprofloxacin-susceptible isolates demonstrated nucleotide sequences for the QRDRs of both gyrA and parC that were identical to those of the previously described susceptible strain ATCC 700294 (GenBank accession number AF220946) (data not shown). In contrast, mutations were identified only in parC, not in gyrA, in all the ciprofloxacin-resistant isolates. Specifically, two point mutations within the QRDR were identified in parC, with codon TCC (Ser-79; S. pyogenes coordinates) being replaced by TTC (Phe) or GCC (Ala) and with GAT (Asp-91) being replaced by AAT (Asn). Almost all ciprofloxacin-resistant isolates (25) showed only the replacement of Ser-79 by Ala, since the most prevalent ciprofloxacin-resistant clone (of emm type 6 and PFGE type A) contained this point mutation (Table 1). We found three isolates with a mutation in which Asp-91 was replaced by Asn and two isolates with this mutation together with a replacement of Ser-79 by Phe (Table 1).
Our results are consistent with previous reports which demonstrated that resistance to fluoroquinolones usually results from alterations in the QRDRs of either gyrA or parC or of both genes. This observation has been reported for S. pneumoniae (8, 9) and S. pyogenes; Yan et al. demonstrated that one isolate of S. pyogenes that was resistant to high levels of quinolones showed mutations in both gyrA and parC (15). Two recent studies conducted in the United States (12) and Germany (11) have confirmed the results reported by Yan et al. and identified S. pyogenes isolates with high levels of resistance to fluoroquinolones with mutations in both genes gyrA and parC. In these isolates, some mutations observed in parC were identical to those identified in our study, such as the replacement of Ser-79 by Phe (11, 12) and the replacement of Asp-91 by Asn (11). Recently, Alonso et al. showed that reduced susceptibility to ciprofloxacin is mainly associated with single mutations preferentially in parC, in which Ser-79 is replaced by Ala (R. Alonso, M. Galimand, and P. Courvalin, Letter, Antimicrob. Agents Chemother. 46:3686-3687, 2002). In our study, nearly all the resistant isolates exhibited levels of resistance to ciprofloxacin (MIC = 4 μg/ml) similar to those reported by Alonso et al., and consistently, almost all the isolates showed the same mutation, Ser79Ala in parC.
The fact that quinolone resistance in S. pneumoniae arises through mutations of parC before changes in gyrA (6), together with the experimental observation that high levels of resistance to quinolones in S. pyogenes are associated with mutations in both gyrA and parC (15), suggests that the ciprofloxacin-resistant isolates identified in Spain are good candidates to become highly resistant to fluoroquinolones.
In summary, since the year 1988, in which ciprofloxacin was introduced into therapeutic practice in Spain, several clones of S. pyogenes have developed mutational alterations of key topoisomerases and exhibit reduced susceptibilities to ciprofloxacin. Certain clones may be selected and further disseminated, particularly if they combine resistance to macrolides and fluoroquinolones.
Acknowledgments
We thank C. Vidal for DNA sequencing and A. Oliver for technical assistance with PFGE.
This study was supported by a grant from GlaxoSmithKline, Madrid, Spain.
Members of the Spanish Surveillance Group for Respiratory Pathogens are as follows: F. Marco and T. Jiménez de Anta, Hospital Clinic i Provincial, Barcelona; C. Fernández-Mazarrasa, Hospital Marqués de Valdecilla, Santander; C. García-Riestra, B. Regueiro, A. Jato, and M. Prieto, Hospital Clínico Universitario, Santiago de Compostela; M. Casal and A. Ibarra, Hospital Reina Sofía, Córdoba; M. de la Rosa, Hospital Virgen de las Nieves, Granada; C. de la Torre and A. Gené, Hospital San Joan de Deus, Barcelona; C. García, Hospital Clínico Universitario, Zaragoza; E. Perea and L. Martínez, Hospital Virgen de la Macarena, Seville; J. Nogueira, Hospital Dr. Peset, Valencia; E. Pérez-Trallero and J. Larruskain, Hospital Donostia, San Sebastián; I. Trujillano, Hospital Clínico Universitario, Salamanca; J. Ruiz and E. Simarro, Hospital Virgen de la Arrixaca, Murcia; E. Cercenado, Hospital Gregorio Marañón, Madrid; A. M. Martín and F. Cañas, Hospital Insular, Las Palmas; J. Barrón and L. López, Hospital de Cruces, Bilbao; A. García, S. García, and M. Güeni, Hospital La Paz, Madrid; D. Romero and M. González, Hospital Nuestra Señora de Alarcos, Ciudad Real; A. Fenoll and J. Casal, Instituto Carlos III, Madrid; J. J. Granizo, Fundación Jiménez Díaz, Madrid; J. García-de-Lomas, C. Gimeno, and E. Esteban, Instituto Valenciano de Microbiología, Valencia; and R. Dal-Ré, GlaxoSmithKline S.A., Madrid, Spain.
REFERENCES
- 1.Albertí, S., C. D. Ashbaugh, and M. R. Wessels. 1998. Structure of the has operon promoter and regulation of hyaluronic acid capsule expression in group A Streptococcus. Mol. Microbiol. 28:343-353. [DOI] [PubMed] [Google Scholar]
- 2.Albertí, S., C. García-Rey, M. A. Domínguez, L. Aguilar, E. Cercenado, M. Gobernado, A. García-Perea, and Spanish Surveillance Group for Respiratory Pathogens. 2003. Survey of emm gene sequences from pharyngeal Streptococcus pyogenes isolates collected in Spain and their relationship with erythromycin susceptibility. J. Clin. Microbiol. 41:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall, B., R. Facklam, and T. Thompson. 1995. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondeau, J. M. 1999. A review of the comparative in-vitro activities of 12 antimicrobial agents with a focus on five new respiratory quinolones. J. Antimicrob. Chemother. 43(Suppl. B):1-11. [DOI] [PubMed] [Google Scholar]
- 5.Granizo, J. J., L. Aguilar, R. Casal, R. Dal-Ré, and F. Baquero. 2000. Streptococcus pyogenes resistance to erythromycin in relation to macrolide consumption in Spain (1986-1997). J. Antimicrob. Chemother. 46:959-964. [DOI] [PubMed] [Google Scholar]
- 6.Janoir, C., V. Zeller, M.-D. Kitzis, N. J. Moreau, and L. Gutmann. 1996. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob. Agents Chemother. 40:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin, J. M., M. Green, K. A. Barbadora, and E. R. Wald. 2002. Erythromycin-resistant group A streptococci in schoolchildren in Pittsburgh. N. Engl. J. Med. 346:1200-1206. [DOI] [PubMed] [Google Scholar]
- 8.Muñoz, R., and A. G. De La Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan, X.-S., and L. M. Fisher. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Trallero, E., C. Fernández-Mazarrasa, C. García-Rey, E. Bouza, L. Aguilar, J. García-de-Lomas, F. Baquero, and The Spanish Surveillance Group for Respiratory Pathogens. 2001. Antimicrobial susceptibilities of 1,684 Streptococcus pneumoniae and 2,039 Streptococcus pyogenes isolates and their ecological relationships: results of a 1-year (1998-1999) multicenter surveillance study in Spain. Antimicrob. Agents Chemother. 45:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinert, R. R., R. Lutticken, and A. Al-Lahham. 2004. High-level fluoroquinolone resistance in a clinical Streptococcus pyogenes isolate in Germany. Clin. Microbiol. Infect. 10:659-662. [DOI] [PubMed] [Google Scholar]
- 12.Richter, S. S., D. J. Diekema, K. P. Heilmann, L. S. Almer, V. D. Shortridge, Z. R. Flamm, and G. V. Doern. 2003. Fluoroquinolone resistance in Streptococcus pyogenes. Clin. Infect. Dis. 36:380-383. [DOI] [PubMed] [Google Scholar]
- 13.Seppala, H. A., A. Nissinen, H. Jarvinen, S. Houvinen, T. Henriksson, and E. Herva. 1992. Resistance to erythromycin in group A streptococci. N. Engl. J. Med. 326:292-297. [DOI] [PubMed] [Google Scholar]
- 14.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan, S. S., M. L. Fox, S. M. Holland, F. Stock, V. J. Gill, and D. P. Fedorko. 2000. Resistance to multiple fluoroquinolones in a clinical isolate of Streptococcus pyogenes: identification of gyrA and parC and specification of point mutations associated with resistance. Antimicrob. Agents Chemother. 44:3196-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]