Abstract
After screening marine actinomycetes isolated from sediment samples collected from the Arctic fjord Kongsfjorden for potential anticancer activity, an isolate identified as Streptomyces artemisiae MCCB 248 exhibited promising results against the NCI-H460 human lung cancer cell line. H460 cells treated with the ethyl acetate extract of strain MCCB 248 and stained with Hoechst 33342 showed clear signs of apoptosis, including shrinkage of the cell nucleus, DNA fragmentation and chromatin condensation. Further to this treated cells showed indications of early apoptotic cell death, including a significant proportion of Annexin V positive staining and evidence of DNA damage as observed in the TUNEL assay. Amplified PKS 1 and NRPS genes involved in secondary metabolite production showed only 82% similarity to known biosynthetic genes of Streptomyces, indicating the likely production of a novel secondary metabolite in this extract. Additionally, chemical dereplication efforts using LC–MS/MS molecular networking suggested the presence of a series of undescribed tetraene polyols. Taken together, these results revealed that this Arctic S. artemisiae strain MCCB 248 is a promising candidate for natural products drug discovery and genome mining for potential anticancer agents.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0610-3) contains supplementary material, which is available to authorized users.
Keywords: Actinomycetes, Streptomyces, Anticancer, Natural products, Apoptosis
Introduction
Microorganisms have been the source of many chemotherapeutics used in cancer treatment, and amongst them, actinomycetes are the most promising source organisms. Actinomycetes account for about 45% of all microbial secondary metabolites, of which 7600 (80%) are produced by Streptomyces (Bérdy 2005). Over the years, the rate of discovery of new bioactive compounds has reduced, but the rediscovery of known compounds has increased (Fenical et al. 1999). Hence there is a need for bioprospecting of unexplored or underexplored habitats for unique and rare microorganisms, as these can be expected to yield a higher percentage of novel metabolites with desirable bioactivities (Bredholt et al. 2008; Pathom-aree et al. 2006). The Arctic region remains one of the least well-explored geographical locations on Earth for novel bioactive metabolites. Herein, we report on the isolation and screening of actinomycetes from sediment collected from an Arctic fjord and discovery of the production of potential anticancer natural products.
Materials and methods
Isolation of actinomycetes
Sediment samples were collected using a van Veen grab from varying depths (50–250 m) in the Arctic Fjord ‘Kongsfjorden’ located in Ny-Ålesund, Svalbard in, July 2012. The samples were serially diluted in ice-cold sterile seawater and plated on Actinomycetes Isolation Agar (AIA) (Himedia, India) supplemented with 0.2 mg/L gentamycin, 0.25 mg/L cycloheximide, and 0.1 mg/L amphotericin B. These were incubated at 20 °C for 4 weeks at which time characteristic actinobacterial colonies were isolated and their extracts screened for anticancer activity as described below.
Fermentation and crude extract preparation
The isolated strains of actinomycetes were mass cultured for the production of their secondary metabolites. Briefly, a loop full of actinomycete spores was inoculated into a seawater-based seed medium (beef extract 3 g/L, peptone 5 g/L). The flasks were incubated at 20 °C for 48 h on a rotary shaker set at 150 rpm. Subsequently, these were transferred to seawater-based fermentation medium (Yang et al. 2013) (soybean meal 3 g/L, yeast extract 3 g/L, proline 1 g/L, beef extract 3 g/L, glycerol 6 mL/L, K2HPO4 0.5 g/L, MgSO4·7H2O 0.5 g/L, FeSO4·7H2O 0.5 g/L, CaCO3 2 g/L, pH 7.4) and incubated at 20 °C for 10 days on a rotary shaker at 150 rpm. The liquid cultures were centrifuged at 6500 g for 5 min, and the supernatant was extracted three times with an equal volume of ethyl acetate. The extract was concentrated under reduced vacuum on a rotary evaporator at 40 °C, and the dry residue (~10 mg) was re-dissolved in 1 mL DMSO for cytotoxicity screening against NCI-H460 non-small cell human lung cancer (NSCLC) cells as well as for chemical dereplication efforts.
Screening for potential anticancer activity using NCI-H460 cells in vitro
Cytotoxicity evaluation of the crude extracts was performed using a sulforhodamine B (SRB) (Sigma, USA) colorimetric assay on 96-well culture plates (Skehan et al. 1990). The NCI-H460 cell line, obtained from the National Centre for Cell Science (NCCS, Pune, India), was maintained in RPMI-1640 (Himedia, India) supplemented with 10% fetal bovine serum (FBS) (Himedia, India) at 37 °C in an incubator with 5% carbon dioxide. Aliquots of 190 µL cell suspension at a density of 1.9 × 104 cells/well were pipetted into 96-well micro titer plates. The crude extracts were diluted to 1 mg/mL with sterile deionized water, and then 10 µL of each were added to each well to achieve a final concentration of 50 µg/mL. Control wells were composed of 190 µL cell suspension plus 10 µL of 10% DMSO. All assays were performed in triplicate. The plates were incubated at 37 °C in a CO2 (5%) incubator for 72 h, and fixed with 100 µL ice-cold 30% trichloroacetic acid (TCA) and incubated at 4 °C for another 1 h. The plates were gently washed four times and air-dried at room temperature. To each well, 100 µL of 0.057% (wt/vol) sulforhodamine B (SRB) prepared in deionized water with 1% acetic acid was added to each well and incubated at room temperature for 30 min. Unbound stain was removed by washing with 1% acetic acid and then the plates were air dried. To dissolve the cell bound dye, 200 µL of 10 mM Tri base solution (pH 10.5) was added to each well and the plate was shaken on a gyratory shaker for 10 min. Optical density (OD) was read at 510 nm in a microplate reader (Tecan, Switzerland). Percentage of cell-growth inhibition (GI) was calculated according to the following equation (Vichai and Kirtikara 2006):
Based on the results of a primary screening effort, a potent actinomycete isolate, MCCB 248, was selected for further studies. IC50 values were determined to the NCI-H460 human lung cancer cell line as well as to a cell line derived from normal epithelial kidney cells from the African green monkey (BS-C-1). Briefly, two fold serial dilution of the crude extract of the MCCB 248 isolate was prepared to obtain concentrations ranging from 1 mg/mL to 15.625 µg/mL in 10% DMSO; these were added to the various cell lines as described above. IC50 values were determined based on probit analysis (Brownlee et al. 1952).
Molecular identification of MCCB 248 by 16S rRNA Gene Analysis
The 16S rRNA gene from isolate MCCB 248 was PCR-amplified, sequenced, and a phylogenetic tree was constructed. Briefly, DNA was extracted from a 48 h broth culture of strain MCCB using the PureLink Genomic DNA mini kit (Invitrogen, USA). PCR amplification of the 16S rRNA was performed using universal primers 16S1 (GAGTTTGATCCTGGCTCA) and 16S2 (ACGGCTACCTTGTTACGACTT). The amplified PCR product was purified by Exosap (Affymetrix, USA) and sequenced. Nucleotide sequence data were analyzed using BLAST and deposited in NCBI GenBank. The nucleotide sequences were aligned using Clustal W on MEGA 6 (Tamura et al. 2013). The 16S rRNA sequences of closely related actinomycetes were retrieved from GenBank, and their similarity to the present isolate was assessed at the nucleotide level. A phylogenetic tree was constructed using the neighbor-joining method with boot strap values based on 1000 replicates.
Amplification of biosynthetic genes encoding for secondary metabolites
PCR based screening for polyketide synthases (PKS I & PKS II), nonribosomal peptide synthetases (NRPS), aminodeoxyisochorismate synthase (phzE), and cytochrome P450 hydroxylase (CYP) genes was performed using the degenerate primers reported previously (Izumikawa 2003; Wawrik et al. 2005; Ayuso-Sacido and Genilloud 2005; Lee et al. 2006; Schneemann et al. 2011). The composition of the reaction mixture for all PCR amplifications was: 0.4 µL template, 5 µL 2× EmeraldAmp GT PCR Master Mix (Takara Bio Inc., Japan), and 0.5 µL each of the primers (10×).
PCR conditions were as described below.
PKS 1 (10 µL): 2 min at 95 °C, followed by 25 cycles of 40 s at 95 °C, 40 s at 62 °C and 45 s at 72 °C, followed by a 5 min extension at 72 °C.
PKS 2 (10 µL): 5 min at 95 °C, followed by 40 cycles of 1 min at 95 °C, 30 s at 62 °C and 1 min at 72 °C, followed by a 10 min extension at 72 °C.
NRPS (10 µL): 5 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 2 min at 59 °C and 4 min at 72 °C, followed by a 10-min extension at 72 °C (Yuan et al. 2014).
phzE (10µL): 2 min at 94 °C, followed by 36 cycles of 1 min at 94 °C, 1 min at 54.7 °C and 2 min at 72 °C, followed by a 7-min extension at 72 °C (Yuan et al. 2014).
CYP (10µL): 5 min at 96 °C, followed by 45 cycles of 1 min at 96 °C, 30 s at 60 °C and 45 s at 72 °C, followed by a 5-min extension at 72 °C (Yuan et al. 2014).
Similarity searching for the gene sequences was performed using BLASTX against the NCBI GenBank. For phylogenetic analysis, the nucleic acid sequences were translated to protein sequences and aligned with other similar biosynthetic proteins in the NCBI database using the Clustal W program on MEGA 6 (Tamura et al. 2013). A phylogenetic tree of the corresponding biosynthetic genes was constructed by the neighbor-joining method with boot strap values based on 1000 replications.
Chemical dereplication efforts by LC-PDA-MS/MS and molecular networking
An aliquot (1 mg) of the fermentation culture extract was prepared for chemical analysis by filtration over GracePure C18-Max 100 mg/1 mL SPE cartridges (Grace Technologies, USA). Compounds were eluted from the column using acetonitrile and methanol to produce a final volume of 1 mL. This material was taken for nominal mass resolution LC-PDA-MS/MS analysis on a Thermo Finnigan system with a Surveyor PDA Plus Detector, Autosampler Plus, and LC Pump Plus coupled to an LCQ Advantage Plus mass spectrometer (Thermo Fisher Scientific, USA), and with a Phenomenex Kinetex 150 mm × 10 mm × 5 µm C18 analytical column installed (Phenomenex, USA). The UV–Vis spectrum from 200 to 600 nm and positive mode ESI mass spectrum from m/z 190–2000 were recorded, and the mass spectrometer was configured for an automated sample-dependent MS/MS scan. Data were analyzed both manually and by mass spectrometric molecular networking using the Global Natural Products Social Molecular Networking (GNPS) (Wang et al. 2016). This was performed in duplicate using both the native library searching and analog detection search modes. Additionally, for compound dereplication masses of major components of the mixture were searched manually using MarinLit and the Dictionary of Natural Products databases.
Evaluation of apoptosis by Hoechst 33342 staining
To evaluate morphological changes induced by the Streptomyces sp. MCCB 248 metabolites, NCI-H460 cells were stained with Hoechst 33342 and observed for characteristics of apoptosis using a fluorescence microscope (Zhang et al. 2007). Briefly, NCI-H460 cells, seeded in a chamber slide having 1.9 × 104 cells per well, were treated with the culture extract at its IC50 value. After 24 h of treatment, the culture medium was removed and cells were washed twice with PBS and stained with DNA specific Hoechst 33342 dye (Sigma Chemicals, USA) (2 μg/mL in PBS) for 10 min at 37 °C. At the end of staining, cells were observed under a fluorescence microscope for apoptotic features. Doxorubicin (Sigma, USA) was used as a positive control and DMSO was used as a negative control.
Evaluation of apoptosis by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
To detect cells undergoing apoptosis, in situ labeling of the 3′ OH end of the DNA fragments generated during apoptosis was performed using the In Situ Cell Death Detection Kit (TUNEL) (Roche Diagnostics, Switzerland) according to the manufacturer’s instructions. Briefly, NCI-H460 cells in their log phase were treated with the culture extract at its IC50 value and incubated for 24 h. Subsequently, culture medium was removed and cells were washed with PBS, air-dried and fixed with 4% paraformaldehyde (in PBS) for 1 h at 25 °C. Fixed cells were washed 3 times with PBS and incubated with permeabilization solution (1% Triton-X in 1% sodium citrate) for 2 min at 4 °C. As a positive control, NCI-H460 cells were fixed, permeabilized and treated with recombinant DNAase I (New England Biolabs, USA) for 10 min to induce DNA strand nicks. Both control and treated cells were washed twice with PBS, and then 50 µL of TUNEL reaction mixture was added to each well. The cells were incubated for 1 h at 37 °C in a humidified dark chamber and subsequently washed three times with PBS and observed under a fluorescence microscope.
Annexin V–propidium iodide (PI) double staining
Externalization of membrane phosphatidylserine (PS) is an early event occurring in cells undergoing apoptosis, and can be visualized with FITC labeled Annexin V staining. Annexin V is a Ca2+ dependent phospholipid-binding protein that has high affinity for PS, and binds to cells with exposed PS. To confirm apoptosis induction by Streptomyces sp. MCCB 248 metabolites, Annexin-V-FLOUS/Propidium iodide (PI) staining was performed as per the manufacturer’s instructions (Roche Diagnostics, Switzerland). Briefly, cells grown in chamber slides (Millicell EZ slide, Millipore Corporation, US) were treated with the MCCB 248 extract at its IC50 value. After adding the extract, cells were observed and counted under a fluorescent microscope for apoptotic features at regular intervals of treatment (0, 6, 12 and 24 h). A minimum of four different microscopic fields was counted and the average calculated and expressed as a percentage of the total cell population. Cells treated with 10% DMSO were used as a negative control.
Results
Isolation of actinomycetes and screening for anticancer activity
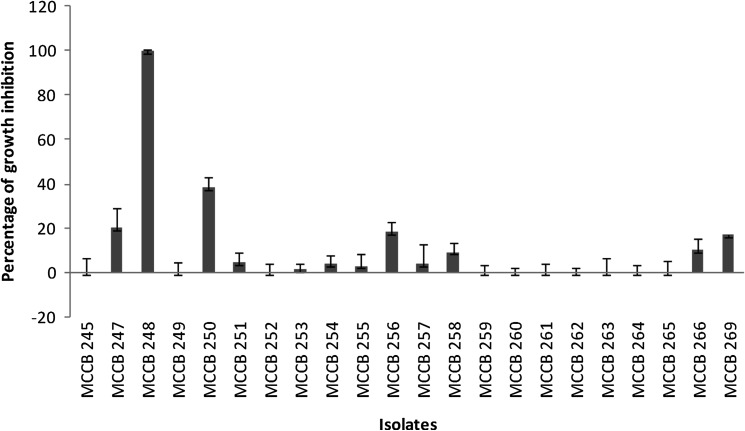
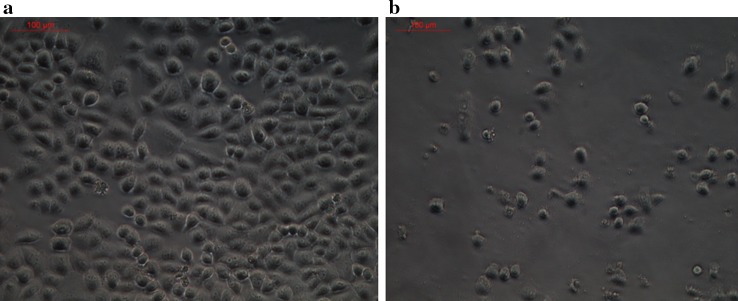
A total of 22 morphologically distinct actinobacterial isolates were obtained from the Arctic sediment samples. Preliminary screening for anticancer activity of the extracts of these isolates resulted in the identification of one isolate, designated as MCCB 248 (Fig. 1), as possessing the most potent growth inhibition against NCI-H460 cells (Fig. 2). Upon further study, the IC50 of the MCCB 248 crude extract was found to be 8.1 and 8.2 µg/mL in NCI-H460 human non-small cell lung cancer cells and non-cancerous BS-C-1 African green monkey kidney cells, respectively. During this preliminary screening effort, it was observed that cells exposed to this extract were less confluent after 24 h of incubation, and attached to the substratum with pronounced shrinkage (Fig. 3).
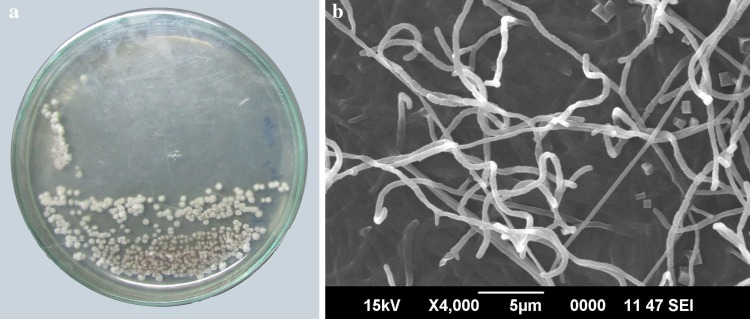
Fig. 1.
a Colony morphology and b scanning electron micrograph of aerial mycelia of S. artemisia strain MCCB 248 after incubation for 14 days on Nutrient agar at 28 °C
Fig. 2.
Percentage growth inhibition by various actinomycete crude extracts on NCI-H460 cell lines at 50 µg/mL (normalized to a control without extract)
Fig. 3.
Phase contrast microscope image (20×) of NCI-H460 cells; a control cells, b treated cells with S. artemisiae MCCB 248 extract at its IC50 value for 24 h
Molecular identification of the isolate MCCB 248
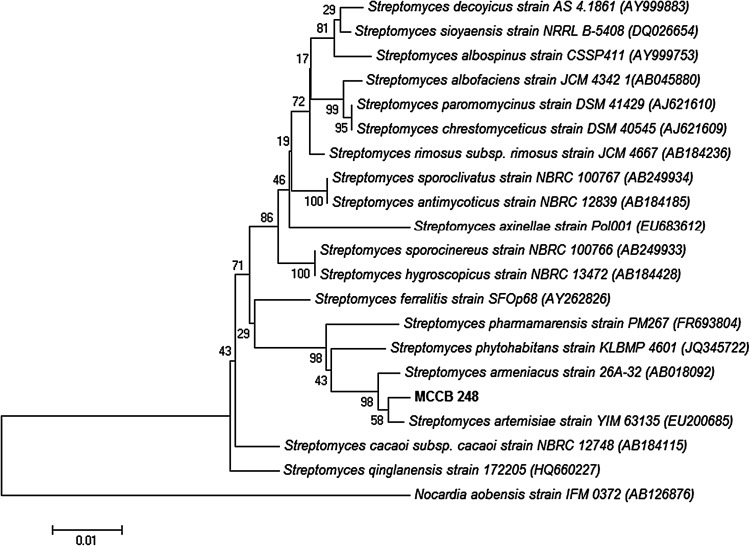
A 16S rRNA gene sequence consisting of 1350 bp from the isolate MCCB248 identified it by BLAST analysis as belonging to the genus Streptomyces, having highest sequence similarity (99%) to the type strain Streptomyces artemisiae YIM 63135. The latter strain was isolated from surface-sterilized tissue of Artemisia annua L., collected in Yunnan Province, southwest China,and described by (Zhao et al. 2010). Moreover, phylogenetic analysis using the neighbor-joining method positioned it in a distinct clade along with Streptomyces artemisiae YIM 63135 (Fig. 4). Accordingly, the isolate MCCB 248 was identified as Streptomyces artemisiae, and hence designated as Streptomyces artemisiae MCCB 248 (GenBank accession number KP313874).
Fig. 4.
Neighbour-joining phylogenetic dendrogram based on 16S rRNA gene sequences showing relationships between the isolated S. artemisiae MCCB 248 and related taxa
Detection and phylogenetic analysis of biosynthetic genes in Streptomyces artemisiae MCCB248
After screening for the presence of genes involved in the biosynsthetic pathways, such as type I polyketide synthase (PKS I), type II polyketide synthase (PKS II), nonribosomal peptide synthetase (NRPS), aminodeoxyisochorismate synthase (phzE), and cytochrome P450 hydroxylase (CYP), Streptomyces artemisiae MCCB 248 was found to have type I polyketide synthase (PKS I) (GenBank accession number KT251042) and nonribosomal peptide synthase (NRPS) (KT277491) genes. After NCBI GenBank BLASTX searching, the PKS 1 nucleotide sequence had its closest match with Streptomyces sp. ID05-A0179 (83%). Phylogenetic analysis of the S. artemisiae MCCB248 PKS 1 using translated nucleic acid sequences revealed that it formed a separate cluster along with Streptomyces himastatinicus ATCC 53653 and Streptomyces aurantiacus JA4570 (Supplementary material). Blast analysis of the NRPS sequence from S. artemisiae MCCB248 showed that it had closest similarity (83%) with one from Streptomyces sp. SCAU5132. Phylogenetic analysis of this NRPS using the deduced amino acid sequence positioned S. artemisiae MCCB 248 as a distinct clade in the tree (Supplementary material).
Chemical dereplication efforts by LC-PDA-MS/MS and molecular networking
The crude extract of S. artemisiae MCCB 248 was first subjected to a C18 SPE clean-up step, and then analyzed by positive ionization LC–MS/MS analysis. Four major chemical components were determined to be present in the crude extract. These four molecules had mass spectra indicating protonated mass ions of m/z 761, 827, 879, and 913, and appeared to possess a common UV chromophore (ν = 312 nm, broad absorption from 280 to 340 nm, and ν = 240 nm). These MS/MS spectra did not match with any library entry upon molecular networking. However, they clustered together in a small molecular family, and upon library comparison with the analog search option, were suggested to be structurally related to a polyhydroxy macrolide, such as bastimolide A (Shao et al. 2015). A number of minor metabolites were also detected in the chromatogram by MS or DAD UV, but were not analyzed further due to their low abundance.
DNA damage and induced apoptosis
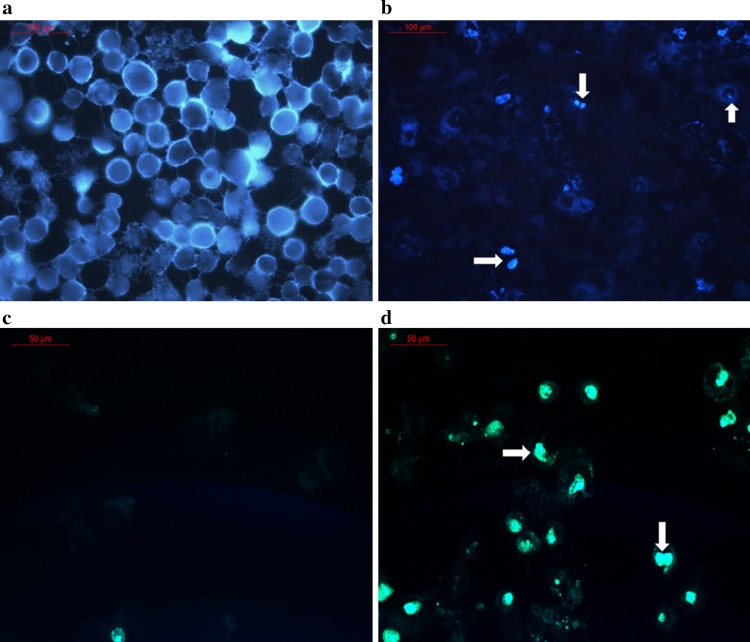
NCI-H460 cells exposed to the S. artemisiae MCCB248 extract exhibited a characteristic apoptotic morphology, such as shrinkage of cell nuclei, chromatin condensation and nuclear fragmentation (Fig. 5a, b), which indicated that one or more components of this extract induced apoptosis. This was further in evidence using a TUNEL assay, which demonstrated condensed TUNEL positive chromatin within the cell nuclei (Fig. 5c, d) when treated with the S. artemisiae MCCB 248 extract. DNase I treated cells were used as a positive control.
Fig. 5.
Fluorescent microscope image (20×) of Hoechst 33342 stained H-460 cells; a control cells, and b S. artemisiae MCCB 248 extract treated cells. Also shown are fluorescent microscope images (40×) of the TUNEL assay of c control cells, and d S. artemisiae MCCB 248 extract treated cells for 24 h
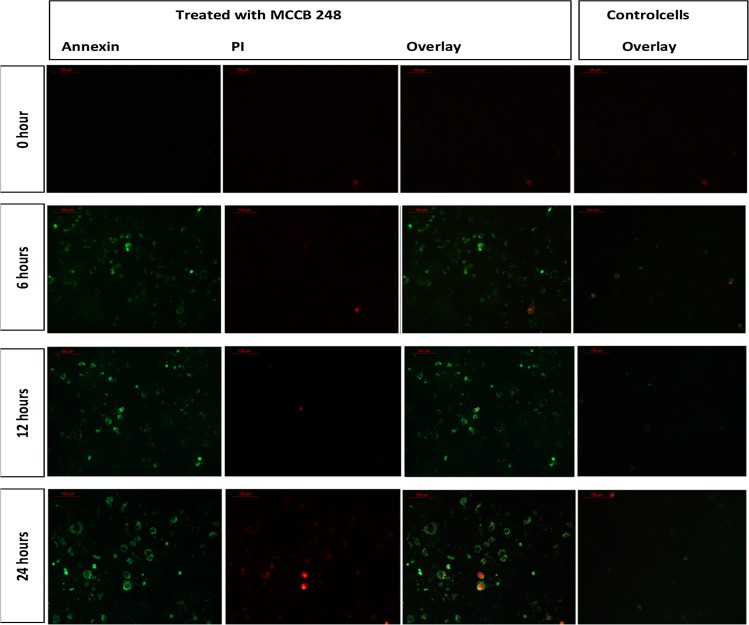
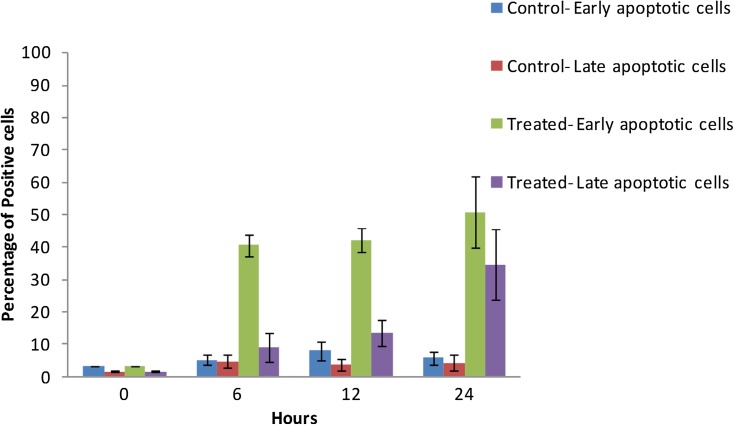
In Annexin V-PI assays, a time-dependent increase in Annexin V positive cells was observed in cells treated with S. artemisiae MCCB 248 extract, implying that there was an increased PS translocation to the outer leaflet of the plasma membrane. At the beginning of treatment (0 h), a majority of the cells were negative for both Annexin V and propidium iodide (PI) staining. After 6 h of treatment, 40% of the cells were Annexin V positive and only 9% of cells were both Annexin V and PI positive. A similar observation was observed after 12 h (42% Annexin V and 13% both Annexin V and PI positive reaction), which was indicative of the early stages of apoptosis. After 24 h, 51% of cells were Annexin V positive and more cells had become positive for PI (34%), indicative of the later stages of apoptosis and signified more dead cells (Figs. 6, 7).
Fig. 6.
Fluorescent microscope image (20×) of Annexin-V/PI double-staining assay. After treating with S. artemisiae MCCB 248 extract, cells were stained with Annexin V-FITC and propidium iodide and analyzed after 0, 6, 12 and 24 h of treatment
Fig. 7.
Percentage of H-460 cells showing early and late apoptotic cell death at 0, 6, 12, 24 h after exposure to the extract of S. artemisiae MCCB 248 (data shown are mean of four independent observations and its standard deviation)
Discussion
Actinomycetales are excellent sources of novel bioactive metabolites with potential pharmaceutical applications (Lam 2006). Actinomycetes contribute over half of the known bioactive secondary metabolites, especially in the classes of antibiotics, antitumor agents, enzymes and immunosuppressive agents (Bérdy 2005). With improved sampling and culture techniques, the isolation of new groups of actinomycetes are being made from sediments collected from even the deepest parts of the oceans (Kwon et al. 2006; Pathom-aree et al. 2006). Such efforts are yielding such chemically rich genera as Salinispora (Feling et al. 2003) and Marinispora (Kwon et al. 2006).
In the present work, 22 different actinomycetes were isolated and cultured from sediment samples collected from the Arctic fjord, Kongsfjorden. Among these was S. artemisiae MCCB248, the extract of which exhibited cytotoxicity with IC50 values of 8.1 µg/mL on NCI-H460 cells and 8.2 µg/mL on BS-C-1 cells. The biosynthetic potential of this microorganism for the production of secondary metabolites was evaluated by detecting the presence of genes involved in secondary metabolite production. The presence of PKS 1 and NRPS genes in S. artemisiae MCCB 248 suggests the possibility that it can produce bioactive secondary metabolites belonging to these two classes of natural products, or a hybrid of both. Relatively low sequence similarity of the PKS and NRPS gene sequences with those available in GenBank indicates the possibility that novel compounds are produced by S. artemisiae MCCB 248. By phylogenetic analysis of PKS 1 using the deduced amino acid sequence, it formed a separate branch along with Streptomyces himastatinicus ATCC 53653 and Streptomyces aurantiacus.
In the present study, H460 human non-small cell lung cancer cells treated with the Streptomyces sp. MCCB 248 extract showed nuclear condensation and fragmentation as evident from Hoechst 33342 staining. This is similar to what is observed with the known anticancer agent doxorubicin (Xin et al. 2012; Zhang et al. 2010). A significant number of anticancer agents display cytotoxicity by damaging DNA, leading to apoptosis (Johnstone et al. 2002). The morphological hallmark of apoptotic cell death includes shrinkage of the cell and nucleus as well as condensation of nuclear chromatin (Saraste 2000), as was observed with application of the S. artemisiae MCCB248 extract.
The DNA fragmentation observed in the TUNEL assay confirmed apoptotic cell death in cells treated with the S. artemisiae MCCB248 extract. Further, a time-dependent increase in Annexin V positive cells implied more cells had undergone a flip-flop of PS to the outer leaflet of the plasma membrane, another indicator of apoptosis. These results demonstrated that S. artemisiae MCCB 248 extract induced apoptotic cell death in NCI-H460 lung cancer cells.
The HPLC–PDA-MS/MS chemical analysis and dereplication efforts for the crude extract of S. artemisiae MCCB 248 suggested some key molecular features associated with the major components of this mixture. The MS and MS/MS spectra of the major four metabolites in the extract showed repeated losses of 18 mass units, which is typical of polyols that show neutral ion losses of water. Furthermore, the UV–Vis spectrum associated with these compounds was suggestive of a tetraene moiety in each, due to the typical absorption band from 280 to 340 nm with a clear maxima at 312 nm (Supplementary material). Taken together, these data suggest the presence of tetraene polyols in the extract, which would be in-line with the production of PKS or hybrid PKS natural products by similar actinomycetes such as the novonestmycins, separacenes, bahamaolides and marinisporolides (Kwon et al. 2009; Kim et al. 2012; Bae et al. 2013; Wan et al. 2015). However, the parent masses observed in this study were not found to correlate with any known molecules of this class in the MarinLit or Dictionary of Natural Products databases. Additionally, the molecular networking performed using GNPS failed to yield any matches against spectra present in the available MS/MS library databases for the four major metabolites present in the extract, but did suggest that these compounds could be analogs of the reported polyhydroxy macrolide natural product known as bastimolide A (Shao et al. 2015). On the other hand, of the lower abundance molecules in the extract, it was possible to identify several known diketopiperazines.
In conclusion, we have isolated and identified the actinomycete Streptomyces artemisiae MCCB 248 from an Arctic fjord that showed promising cancer cell toxicity to NCI-H460 cells in vitro. From the cell-based assays, it could be concluded that this activity likely resulted from the induction of apoptotic pathways leading to cell death. Chemical analysis, dereplication efforts and biosynthetic gene comparisons for the isolate suggested that this organism produces potentially novel bioactive secondary metabolites. Such a promising finding warrants further study, including the purification and characterization of the active compounds as well as description of their biosynthetic gene cluster(s).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to acknowledge University Grants Commission, New Delhi for the funding (major project Grant; F. No 41-1152/2012 (SR) dated 26 July 2012 and Indo—US 21st Century Knowledge Initiative; F. No. 194-1/2009(IC) dated 23rd July 2015) and the Director, ESSO-NCAOR, and Ministry of Earth Sciences, Government of India, for providing the logistic support for field access to the Arctic samples. CBN acknowledges and appreciates financial support provided by a postdoctoral fellowship from a U.S. NCI/NIH Training Program in the Biochemistry of Growth Regulation and Oncogenesis (T32 CA009523). DM would like to thank Cochin University of Science and Technology and Kerala State Council for Science, Technology and Environment, Govt. of Kerala, India for providing fellowship funding.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0610-3) contains supplementary material, which is available to authorized users.
References
- Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol. 2005;49:10–24. doi: 10.1007/s00248-004-0249-6. [DOI] [PubMed] [Google Scholar]
- Bae M, Kim H, Shin Y, et al. Separacenes A-D, novel polyene polyols from the marine actinomycete, streptomyces sp. Mar Drugs. 2013;11:2882–2893. doi: 10.3390/md11082882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Bredholt H, Fjærvik E, Johnsen G, Zotchev SB. Actinomycetes from sediments in the Trondheim fjord, Norway: diversity and biological activity. Mar Drugs. 2008;6:12–24. doi: 10.3390/md6010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee KA, Finney DJ, Tattersfield F. Probit analysis: a statistical treatment of the sigmoid response curve. J Am Stat Assoc. 1952;47:687. doi: 10.2307/2280787. [DOI] [Google Scholar]
- Feling RH, Buchanan GO, Mincer TJ, et al. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew Chemie Int Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- Fenical W, Baden D, Burg M, et al. Marine derived pharmaceuticals and related bioactive compounds. In: Fenical W, et al., editors. From Monsoons to Microbes Underst. Ocean. Role Hum. Heal. Washington, DC: National Academies Press; 1999. pp. 71–86. [Google Scholar]
- Izumikawa M. Cloning of modular type I polyketide synthase genes from salinomycin producing strain of streptomyces albus. Bioorg Med Chem. 2003;11:3401–3405. doi: 10.1016/S0968-0896(03)00337-7. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108:153–164. doi: 10.1016/S0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- Kim DG, Moon K, Kim SH, et al. Bahamaolides A and B, antifungal polyene polyol macrolides from the marine actinomycete streptomyces sp. J Nat Prod. 2012;75:959–967. doi: 10.1021/np3001915. [DOI] [PubMed] [Google Scholar]
- Kwon HC, Kauffman CA, Jensen PR, Fenical W. Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J Am Chem Soc. 2006;128:1622–1632. doi: 10.1021/ja0558948. [DOI] [PubMed] [Google Scholar]
- Kwon HC, Kauffman CA, Jensen PR, Fenical W. Marinisporolides, polyene–polyol macrolides from a marine actinomycete of the new genus Marinispora. J Org Chem. 2009;74:675–684. doi: 10.1021/jo801944d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KS. Discovery of novel metabolites from marine actinomycetes. Curr Opin Microbiol. 2006;9:245–251. doi: 10.1016/j.mib.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Lee M, Myeong J, Park H, et al. Isolation and partial characterization of a cryptic polyene gene cluster in Pseudonocardia autotrophica. J Ind Microbiol Biotechnol. 2006;33:84–87. doi: 10.1007/s10295-005-0018-7. [DOI] [PubMed] [Google Scholar]
- Pathom-aree W, Stach JEM, Ward AC, et al. Diversity of actinomycetes isolated from challenger deep sediment (10,898 m) from the mariana trench. Extremophiles. 2006;10:181–189. doi: 10.1007/s00792-005-0482-z. [DOI] [PubMed] [Google Scholar]
- Saraste A. Morphologic and biochemical hallmarks of apoptosis. Cardiovasc Res. 2000;45:528–537. doi: 10.1016/S0008-6363(99)00384-3. [DOI] [PubMed] [Google Scholar]
- Schneemann I, Wiese J, Kunz AL, Imhoff JF. Genetic approach for the fast discovery of phenazine producing bacteria. Mar Drugs. 2011;9:772–789. doi: 10.3390/md9050772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao CL, Linington RG, Balunas MJ, et al. Bastimolide A, a potent antimalarial polyhydroxy macrolide from the marine Cyanobacterium Okeania hirsuta. J Org Chem. 2015;80:7849–7855. doi: 10.1021/acs.joc.5b01264. [DOI] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- Wan Z, Fang W, Shi L, et al. Novonestmycins A and B, two new 32-membered bioactive macrolides from Streptomyces phytohabitans HBERC-20821. J Antibiot (Tokyo) 2015;68:185–190. doi: 10.1038/ja.2014.123. [DOI] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan VV, et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrik B, Kerkhof L, Zylstra GJ, et al. Identification of unique type ii polyketide synthase genes in soil identification of unique type II polyketide synthase genes in soil. Appl Environ Microbiol. 2005;71:2232–2238. doi: 10.1128/AEM.71.5.2232-2238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin W, Ye X, Yu S, et al. New capoamycin-type antibiotics and polyene acids from marine Streptomyces fradiae PTZ0025. Mar Drugs. 2012;10:2388–2402. doi: 10.3390/md10112388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-W, Zhang G-Y, Ying J-X, et al. Isolation, characterization, and bioactivity evaluation of 3-((6-methylpyrazin-2-yl)methyl)-1H-indole, a new alkaloid from a deep-sea-derived actinomycete Serinicoccus profundi sp. nov. Mar Drugs. 2013;11:33–39. doi: 10.3390/md11010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Yu Y, Li H, et al. Phylogenetic diversity and biological activity of actinobacteria isolated from the Chukchi shelf marine sediments in the arctic ocean. Mar Drugs. 2014;12:1281–1297. doi: 10.3390/md12031281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Wu HY, Xia XK, et al. Anthracenedione derivative 1403P-3 induces apoptosis in KB and KBv200 cells via reactive oxygen species-independent mitochondrial pathway and death receptor pathway. Cancer Biol Ther. 2007;6:1413–1421. doi: 10.4161/cbt.6.9.4543. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tao L, Liang Y, et al. Anthracenedione derivatives as anticancer agents isolated from secondary metabolites of the mangrove endophytic fungi. Mar Drugs. 2010;8:1469–1481. doi: 10.3390/md8041469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G-Z, Li J, Qin S, et al. Streptomyces artemisiae sp. nov., isolated from surface-sterilized tissue of Artemisia annua L. Int J Syst Evol Microbiol. 2010;60:27–32. doi: 10.1099/ijs.0.011965-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.