Abstract
A novel trimethoprim resistance gene, designated dfrA20, was detected on the 11-kb plasmid pCCK154 from Pasteurella multocida. The dfrA20 gene codes for a dihydrofolate reductase of 169 amino acids. Sequence comparisons revealed that the DfrA20 protein differed distinctly from all dihydrofolate reductases known so far.
Trimethoprim (TMP) competitively inhibits the enzyme dihydrofolate reductase, which is responsible for the reduction of dihydrofolate to tetrahydrofolate (6, 19). Bacterial resistance to trimethoprim can be intrinsic or acquired. Intrinsic resistance by either permeability barriers, folate auxotrophy, or dihydrofolate reductases with low affinity for TMP have been detected in various bacterial pathogens, including Pseudomonas aeruginosa, Clostridium spp., Brucella spp., Bacteroides spp., and Enterococcus spp. (15). Different types of acquired TMP resistance, including mutations in the promoter region or the dihydrofolate reductase structural gene, have been reviewed by Sköld (19). The most widespread TMP resistance mechanism, namely, the replacement of a TMP-sensitive dihydrofolate reductase by a plasmid-, transposon-, or cassette-borne TMP-resistant dihydrofolate reductase, causes high-level TMP resistance in various bacteria (6, 19). Up to now, more than 25 different TMP resistance-mediating dihydrofolate reductase (dfr) genes, subdivided on the basis of their structure into major types 1 and 2 (14), which nowadays are referred to as dfrA and dfrB (16), have been identified (17, 19). Although trimethoprim resistance is widespread among bacterial pathogens from human and animal sources, previous attempts to identify TMP resistance genes in bacteria of the genus Pasteurella have failed (3). It was assumed that bacteria of the genera Pasteurella and Mannheimia may carry dfrA or dfrB genes different from those previously identified in other gram-negative bacteria (8). In the present study, we describe a novel dfr gene, designated dfrA20, from Pasteurella multocida.
P. multocida strain GB154 was obtained from the nasal swab specimen of a calf suffering from pneumonia. Antimicrobial susceptibility testing (13) revealed that the strain was resistant to ampicillin (MIC, 32 μg/ml), sulfamethoxazole (MIC, 1,024 μg/ml), and trimethoprim (MIC, ≥128 μg/ml). Strain GB154 harbored a plasmid of 11 kb, designated pCCK154, which upon transformation into Escherichia coli JM109 (Stratagene, Amsterdam, The Netherlands) and electrotransformation into the plasmid-free and antibiotic-susceptible P. multocida field isolate B130 (9) proved to mediate resistance to sulfamethoxazole and trimethoprim. MICs, determined by broth macrodilution (13), were 1,024 μg of sulfamethoxazole/ml and ≥128 μg of trimethoprim/ml in both recipient strains. PCR assays for the most frequently detected dfrA and dfrB genes of gram-negative bacteria as described by Frech et al. (4) confirmed that none of these genes was present on plasmid pCCK154. However, a sul2 gene, coding for a type II dihydropteroate synthase, was identified on this plasmid by PCR (4, 10). A restriction map of plasmid pCCK154 was constructed and served as a basis for cloning experiments. A ca. 1.9-kb PstI fragment was found to mediate trimethoprim resistance. This fragment plus another 0.7 kb upstream located on an EcoRV fragment of 5.6 kb and 1.2 kb downstream located on an EcoRV fragment of 5.4 kb were sequenced by primer walking (Fig. 1).
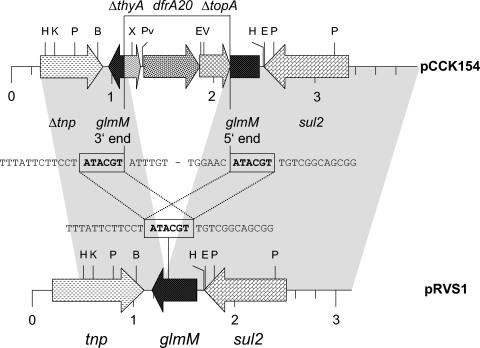
FIG. 1.
Comparison of the dfrA20-sul2 area of plasmid pCCK154 from P. multocida with the corresponding region of plasmid pRVS1. The arrows indicate the extents of the genes tnp (transposase), glmM (phosphoglucosamine mutase), sul2 (sulfonamide-resistant dihydropteroate synthase), dfrA20 (TMP-resistant dihydrofolate reductase), thyA (thymidylate synthase), and topA (type IA topoisomerase), with the arrowheads showing the directions of transcription. The prefix Δ means that the gene is truncated. The regions of homology between pCCK154 and pRVS1 are marked by grey shading. The sequences at the junctions of pRVS1-homologous and -nonhomologous parts in pCCK154 are shown in comparison to the corresponding pRVS1 sequence between the two maps. The 6-bp direct repeats are boxed. Restriction endonuclease cleavage sites are abbreviated as follows: B (BamHI), E (EcoRI), EV (EcoRV), H (HpaI), K (KpnI), P (PstI), Pv (PvuII), and X (XbaI).
Sequence analysis of the ca. 3.8-kb segment revealed two stretches of 99% sequence identity between pCCK154 and the Vibrio salmonicida plasmid pRVS1 (GenBank accession number AJ289135). These homologous areas were from bases 293 to 1100 and 2179 to 3891 in the pCCK154 sequence (Fig. 1). The 1,078-bp sequence between these two pRVS1-homologous regions contained the novel TMP resistance gene. The dfrA20 gene was bracketed by the 3′ end of a thyA-like gene whose deduced amino acid sequence showed 82% identity to the terminal 57 amino acids of a thymidylate synthase from Cytophaga hutchinsonii (accession number ZP00117986) and the 5′ end of a topA-like gene whose deduced amino acid sequence revealed 68% identity to the initial 119 amino acids of a type IA topoisomerase from C. hutchinsonii (accession number ZP00118579). The organization of a thymidylate synthase gene and a dihydrofolate reductase gene in the same operon has been observed in a number of bacteria, including, among others, Bacillus subtilis (7), Bordetella bronchiseptica, Bordetella parapertussis, and Bordetella pertussis (accession numbers NC 002927 to NC 002929). Both enzymes are essential for DNA synthesis in that thymidylate synthase catalyzes the transfer of a methyl group from N5,N10-methylentetrahydrofolate to deoxyuridylate, thereby generating deoxythymidylate and 7,8-dihydrofolate, whereas dihydrofolate reductase then converts 7,8-dihydrofolate to tetrahydrofolate, a precursor of N5,N10-methylentetrahydrofolate. At both junctions of pRVS1-homologous and -nonhomologous sequences in pCCK154, the 6-bp direct repeat ATACGT was detected (Fig. 1). The sequence ATACGT is part of the phosphoglucosamine mutase gene glmM, which was disrupted in pCCK154 by the integration of the dfrA20-containing 1,078-bp segment (Fig. 1). Whether these 6-bp direct repeats represent relics of a transposable element that had been involved in the integration of the dfrA20-containing segment remains to be clarified. Bases 1 to 292 in the pCCK154 sequence did not reveal significant homology to sequences deposited in the databases.
The sul2 gene was located in close proximity to the glmM gene in pCCK154 (Fig. 1). Its reading frame codes for a protein of 289 amino acids and thus represents the largest type II dihydropteroate synthase enzyme known to date. Previous studies showed that mutations in the terminal part of the sul2 reading frame might cause an extension of the reading frame without having an impact on the functioning of the enzyme (1, 10). In the present case, the loss of a single A at position 793 within the sul2 reading frame caused a frameshift mutation which led to the substitution of 6 codons and extended the reading frame by 18 codons compared to sul2 from pRVS1. The high MIC of sulfamethoxazole indicates that these alterations in the C terminus had no negative impact on sulfonamide resistance.
The dfrA20 gene is—to the best of our knowledge—the first TMP resistance gene detected in P. multocida. Analysis of the flanking regions did not reveal structures resembling gene cassettes (16). The deduced DfrA20 protein sequence consists of 169 amino acids and thus is in the same size range (152 to 189 amino acids) as most of the bacterial DfrA proteins known so far. Phylogenetic analysis showed that DfrA20 from P. multocida represents a separate branch in the phylogenetic tree and clusters with Dfr proteins from gram-positive bacteria such as Staphylococcus, Bacillus, and Listeria (Fig. 2). Identities to the known DfrA proteins determined on the basis of a multisequence alignment varied between 20.0 and 37.7%, with the highest levels of identity to the Dfr proteins being found for Bacillus subtilis (37.7%) (7) and Staphylococcus haemolyticus (37.0%) (2). This observation confirmed that DfrA20 is only distantly related to other DfrA proteins. In addition to the DfrA proteins shown in Fig. 2, there are numerous reading frames which have been identified during whole-genome sequencing and are assumed to code for dihydrofolate reductases. Their functional annotation, however, was usually based only on Conserved Domain Database and Clusters of Orthologous Groups assignments, not on experimental proof of their role in TMP resistance.
FIG. 2.
Phylogenetic tree of the DfrA proteins involved in TMP resistance. Branch lengths are scaled according to amino acid exchanges observed in a multisequence alignment. The number at each major branch point refers to the percentage of times that a particular node was found in 10,000 bootstrap replications. The bacterial source and the GenBank accession number are given for each DfrA protein. For a number of DfrA proteins, e.g., DfrA1, several closely related sequences from different bacterial sources have been deposited in the databases. To reduce the complexity of this phylogenetic tree, only one representative for each type of DfrA protein was chosen. For this, the cutoff was set at ≥95% amino acid identity. According to the classification of the known Dfr proteins (also referred to as DHFR proteins in several database entries) into the two classes A and B (14, 16), all class A proteins for which numerals (arabic or roman) have been used in the database entries were indicated as DfrA protein followed by the respective arabic numeral. For the Dfr proteins from gram-positive bacteria which have not yet been assigned a number, only the bacterial source and the database accession number are given. DfrB proteins, which differ distinctly from DfrA proteins by their size and structure, have been excluded from this phylogenetic analysis. V. cholerae, Vibrio cholerae; A. salmonicida, Aeromonas salmonicida; P. mirabilis, Proteus mirabilis; S. Typhimurium, Salmonella enterica serovar Typhimurium; M. profunda, Moritella profunda; L. monocytogenes, Listeria monocytogenes; S. epidermidis, Staphylococcus epidermidis; S. aureus, Staphylococcus aureus.
In contrast to the situation with tetracycline resistance genes (11) and macrolide-lincosamide-streptogramin B resistance genes (18), there is still no accepted nomenclature for the dfrA genes. Hall and Partridge addressed this problem and suggested that numbers should be assigned in the order of the database entries (5). Therefore, we tentatively designated the new dfrA gene from P. multocida as dfrA20. Since dfrA20 is located on a small plasmid that also carries the sulfonamide resistance gene sul2, there is potential for this gene to be disseminated horizontally but also to be coselected by the use of sulfonamides. The origin of the gene dfrA20 remains to be answered. However, a lower GC content (35%) of the 1,078-bp segment containing dfrA20 than of the whole genome of P. multocida strain Pm70 (41%) (12) suggested that dfrA20 is most likely not an indigenous P. multocida gene.
Nucleotide sequence accession number.
The sequence of a 3,891-bp segment of plasmid pCCK154 has been deposited in the EMBL database under accession number AJ605332.
Acknowledgments
We thank Vera Nöding for excellent technical assistance.
REFERENCES
- 1.Chang, Y. F., D. P. Ma, H. Q. Bai, R. Young, D. K. Struck, S. J. Shin, and D. H. Lein. 1992. Characterization of plasmids with antimicrobial resistant genes in Pasteurella haemolytica. DNA Seq. 3:89-97. [DOI] [PubMed] [Google Scholar]
- 2.Dale, G. E., H. Langen, M. G. P. Page, R. L. Then, and D. Stüber. 1995. Cloning and characterization of a novel, plasmid-encoded trimethoprim-resistant dihydrofolate reductase from Staphylococcus haemolyticus MUR313. Antimicrob. Agents Chemother. 39:1920-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escande, F., G. Gerbaud, J.-L. Martel, and P. Courvalin. 1991. Resistance to trimethoprim and 2,4-diamino-6,7-diisopropyl-pteridine (O/129) in Pasteurella haemolytica. Vet. Microbiol. 26:107-114. [DOI] [PubMed] [Google Scholar]
- 4.Frech, G., C. Kehrenberg, and S. Schwarz. 2003. Resistance phenotypes and genotypes of multiresistant Salmonella enterica subsp. enterica serovar Typhimurium var. Copenhagen isolates from animal sources. J. Antimicrob. Chemother. 51:180-182. [DOI] [PubMed] [Google Scholar]
- 5.Hall, R., and S. Partridge. 2003. Unambiguous numbering of antibiotic resistance genes. Antimicrob. Agents Chemother. 47:3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huovinen, P., L. Sundström, G. Swedberg, and O. Sköld. 1995. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 39:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwakura, M., M. Kawata, K. Tsuda, and T. Tanaka. 1988. Nucleotide sequence of the thymidylate synthase B and dihydrofolate reductase genes contained in one Bacillus subtilis operon. Gene 64:9-20. [DOI] [PubMed] [Google Scholar]
- 8.Kehrenberg, C., G. Schulze-Tanzil, J.-L. Martel, E. Chaslus-Dancla, and S. Schwarz. 2001. Antimicrobial resistance in Pasteurella and Mannheimia: epidemiology and genetic basis. Vet. Res. 32:323-339. [DOI] [PubMed] [Google Scholar]
- 9.Kehrenberg, C., and S. Schwarz. 2001. Molecular analysis of tetracycline resistance in Pasteurella aerogenes. Antimicrob. Agents Chemother. 45:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kehrenberg, C., and S. Schwarz. 2001. Occurrence and linkage of genes coding for resistance to sulfonamides, streptomycin and chloramphenicol in bacteria of the genera Pasteurella and Mannheimia. FEMS Microbiol. Lett. 205:283-290. [DOI] [PubMed] [Google Scholar]
- 11.Levy, S. B., L. M. McMurray, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Pattishall, K. H., J. Acar, J. J. Burchall, F. W. Goldstein, and R. J. Harvey. 1977. Two distinct types of trimethoprim-resistant dihydrofolate reductase specified by R-plasmids of different compatibility groups. J. Biol. Chem. 252:2319-2323. [PubMed] [Google Scholar]
- 15.Quintiliani, R., Jr., D. F. Sahm, and P. Courvalin. 1999. Mechanisms of resistance to antimicrobial agents, p. 1505-1525. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 16.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 17.Roberts, M. C. 2002. Resistance to tetracycline, macrolide-lincosamide-streptogramin, trimethoprim, and sulfonamide drug classes. Mol. Biotechnol. 20:261-283. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sköld, O. 2001. Resistance to trimethoprim and sulfonamides. Vet. Res. 32:261-273. [DOI] [PubMed] [Google Scholar]