Abstract
CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli emerged recently in Cameroon. CTX-M-15 was encoded by two different multiresistance plasmids, of which one carried an ISEcp1-blaCTX-M-15 element flanked by a 5-bp target site duplication and inserted within a Tn2-derived sequence. A truncated form of this element in the second plasmid was identified.
Extended-spectrum β-lactamase (ESBL)-positive enterobacteria are frequently isolated in hospitals in Cameroon. Up to 1999, SHV-12 and SHV-2a were the dominant ESBLs (J. Gangoue-Pieboji, B. Bedenic, S. Koula-Shiro, et al., Program Abstr. 9th Int. Congr. Infect. Dis., abstr. 15419, 2000). In a PCR-based screening for bla types applied to enterobacteria collected during July and August 2002 in Yaounde Central Hospital, it was found that 14 out of 17 ESBL-positive isolates produced SHV ESBLs, confirming previous findings. The remaining isolates (one Klebsiella pneumoniae isolate and two Escherichia coli isolates), however, were blaCTX-M positive. CTX-M is a rapidly growing family of ESBLs that preferentially hydrolyze cefotaxime. The blaCTX-M genes are commonly found in plasmids carried by enterobacteria. CTX-M ESBLs have been reported worldwide, the highest prevalence being observed in Latin America, Eastern Europe, and the Far East (3, 16). We report here on the emergence of CTX-M producers also in Cameroon.
The three clinical isolates studied (K. pneumoniae YC-17 and E. coli YC-5b and YC-14) had been derived from patients with urinary tract infection acquired during hospitalization. The isolates were resistant to amoxicillin, amoxicillin-clavulanate, piperacillin, cefotaxime, ceftazidime, cefepime, and aztreonam, as determined by the agar dilution method. Activity of cefotaxime and ceftazidime was restored by clavulanic acid. MICs of piperacillin-tazobactam, cefoxitin, and imipenem were within the susceptibility range. Isolates were also resistant to various non-β-lactam antibiotics by a disk diffusion assay (Table 1).
TABLE 1.
Antibiotic susceptibility of CTX-M-15-producing strains
| Strain | MICs (μg/ml) ofa:
|
Other resistance markersb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | PIP | TZP | FOX | CTX | CTX+ | CAZ | CAZ+ | ATM | FEP | IMI | ||
| E. coli YC-14 | ≥256 | 32 | ≥256 | 32 | 8 | ≥256 | 0.5 | 32 | 1 | 64 | 64 | 0.25 | Gm, Tb, Sul, Tmp, Cm |
| E. coli K-12(pYC-14) | ≥256 | 32 | ≥256 | 16 | 8 | ≥256 | 0.25 | 32 | 0.5 | 32 | 32 | 0.12 | Gm, Tb |
| K. pneumoniae YC-17 | ≥256 | 64 | ≥256 | 32 | 16 | ≥256 | 1 | 128 | 2 | 128 | 128 | 0.5 | Gm, Tb, Sul, Tmp, Cm |
| E. coli K-12(pYC-17) | ≥256 | 32 | ≥256 | 16 | 8 | ≥256 | 0.25 | 32 | 0.5 | 32 | 32 | 0.12 | Gm, Tb |
| E. coli YC-5b | ≥256 | 32 | ≥256 | 32 | 16 | ≥256 | 0.5 | 64 | 1 | 64 | 64 | 0.12 | Gm, Tb, Sul, Tmp |
| E. coli DH5α(pYC-5b) | ≥256 | 32 | ≥256 | 8 | 4 | ≥256 | 0.12 | 32 | 0.5 | 32 | 16 | ≤0.06 | Gm, Tb, Sul, Tmp |
| E. coli K-12 | 4 | 2 | 1 | 1 | 4 | ≤0.06 | —c | 0.25 | — | ≤0.06 | ≤0.06 | ≤0.06 | |
| E. coli DH5α | 2 | 2 | 1 | 1 | 4 | ≤0.06 | — | 0.12 | — | ≤0.06 | ≤0.06 | ≤0.06 | |
AMX, amoxicillin; AMC, amoxicillin-clavulanic acid (2:1); PIP, piperacillin; TZP, piperacillin plus tazobactam (4 μg/ml); Fox, cefoxitin; Ctx, cefotaxime; CTX+, cefotaxime plus clavulanic acid (4 μg/ml); CAZ, ceftazidime; CAZ+, ceftazidime plus clavulanic acid (4 μg/ml); ATM, aztreonam; IMI, imipenem.
Gm, gentamicin; Tb, tobramycin; Sul, sulfonamides; Tmp, trimethoprim; Cm, chloramphenicol.
—, not done.
β-Lactamases were extracted by ultrasonic treatment and characterized by isoelectric focusing. Isolates produced β-lactamases with apparent isoelectric points (pIs) equal to 7.3 and 8.8. E. coli YC-5b produced an additional β-lactamase focusing at 5.4. Isolates were positive in a PCR specific for blaCTX-M-3-related genes (6). Sequencing the PCR products showed 100% homology with blaCTX-M-15 (accession no. AY044436) (6). CTX-M-15 corresponded to the β-lactamase with a pI of 8.8. Also, by PCR with blaTEM- and blaOXA-specific primers (1, 15) and the sequencing of the amplicons the β-lactamases with pIs of 7.3 and 5.4 were identified as OXA-30 and TEM-1. Therefore, oxyimino-β-lactam resistance was mainly due to CTX-M-15.
In conjugation experiments performed in liquid media E. coli YC-14 and K. pneumoniae YC-17 transferred resistance to oxyimino-β-lactams and aminoglycosides to an E. coli K-12 host (Table 1). Plasmid analysis indicated transfer of 90-kb plasmids that produced similar PstI restriction patterns. Additionally, in both preparations, PstI fragments equal in size (5.3 kb) hybridized with a digoxigenin-labeled blaCTX-M-15 probe, suggesting spread of a single plasmid (pYC-14). E. coli YC-5b harbored a 50-kb plasmid (pYC-5b) that was used to transform E. coli DH5α. Transformants exhibited the resistance phenotype of E. coli YC-5b (Table 1). The PstI-generated restriction pattern of pYC-5b was different from that of pYC-14. Hybridization of the blaCTX-M-15 probe occurred on a 3.4-kb PstI fragment of pYC-5b. Isoelectric focusing and PCR experiments showed that pYC-5b and pYC-14 coded also for the penicillinases produced by the respective clinical isolates.
Plasmids pYC-5b and pYC-14 were partially digested with Sau3A, and the fragments were ligated into pBCSK(+) (Stratagene). Recombinant plasmids were used to transform E. coli DH5a. Selection was performed in media containing either cefotaxime or ampicillin. Colony hybridization with a blaCTX-M probe was also applied to facilitate selection. Nucleotide sequences of overlapping fragments were determined with an ABI 377 sequencer (Applied Biosystems).
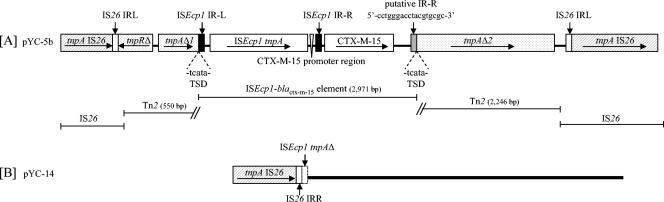
In pYC-5b, an ISEcpI insertion sequence, comprising an intact tnpA gene and two 30-bp imperfect inverted repeats (IRL and IRR) characteristic of this element (accession no. AJ242809) (9), was located 48 bp upstream of blaCTX-M-15. The promoter driving blaCTX-M transcription was identified within the 3′ noncoding sequence of ISEcpI (13). An 18-bp sequence corresponding to the external part of IRR of ISEcpI (putative IRR) was found 373 bp downstream of blaCTX-M-15. The intervening 373-bp sequence had 55% homology with the respective chromosomal region of Kluyvera cryocrescens (from nucleotide [nt] 3304 to 3677 in the sequence with accession no. AY026417) (4). The ISEcpI-blaCTX-M-15-containing sequence was flanked by 5-bp direct repeats and inserted within tnpA (tnpAΔ1, 214 nt from the 5′ end; tnpAΔ2, 2,246 nt) of a Tn2-derived sequence. The latter also contained part of the respective tnpR (tnpRΔ; 173 nt from the 5′ end) and was flanked by directly repeated IS26 elements (Fig. 1A). The truncated forms of transposase and resolvase of Tn2 were, most likely, not functional. A homologous segment, extending from the 3′ end of the tnpA gene of ISEcp1 up to the IS26 of the right end, was carried by the self-transferable plasmid pYC-14. This sequence was preceded by IS26 (Fig. 1B).
FIG. 1.
Schematic representation of the ISEcp1-blaCTX-M-15-containing sequences in plasmids pYC-5b (A) and pYC-14 (B). Inverted repeat sequences (IR) and target site duplications (TSD) are shown. Arrows indicate direction of transcription. The thick line (B) denotes homology with the sequence in panel A.
Since its first description in 2001, CTX-M-15 has been identified in multiple locations in Asia and Europe (2, 5-8, 10-12, 17). This study documents for the first time the emergence of CTX-M-15-producing enterobacteria in an African country. CTX-M-15 differs from CTX-M-3 by an Asp-240→Gly substitution that increases activity against ceftazidime (14). The enhanced substrate spectrum of CTX-M-15 is probably a factor contributing to its spread.
ISEcp1-like sequences have been associated with various blaCTX-M genes of the three major evolutionary groups (3). The presence of a 5-bp duplication at the boundaries of the ISEcp1-blaCTX-M-15 element and the resemblance of its right end to the IRR of ISEcp are indicative of transposition. Similar sequence characteristics in the recently described ISEcp1B-blaCTX-M-19 element led to the hypothesis that ISEcp1 mediates a regular transposition process (13). However, the putative IRRs of these elements had less than 60% homology with the corresponding region of IRR and also differ from each other by 9 nt (50% homology). Therefore, the possibility for a one-ended transposition mechanism cannot be definitely excluded (P. D. Stapleton, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1457, 1999).
Geographical and temporal clusters of identical blaCTX-M genes carried by apparently different plasmids have also been reported in previous studies (reviewed in reference 3). Notably, the sequence homology of the CTX-M-encoding loci in pYC-5b and pYC-14 extends beyond ISEcpI-blaCTX-M-15, including parts of the Tn2 flanking segments. Recently, Lartigue et al. described plasmids carrying ISEcp1-blaCTX-M-15 elements inserted within tnpA of a Tn2-like transposon harbored by E. coli isolates from France and India (8). Furthermore, a GenBank search revealed a plasmid from E. coli isolated in Canada (pC15-1a) that also contained a Tn2-inserted ISEcp1-blaCTX-M-15 (from nt 17077 to 23482 in the sequence with accession no. AY458016 [M. R. Mulvey et al., unpublished data]). This sequence was homologous to that found in pYC-5b except that it lacked the left-hand IS26. Also, in silico restriction analysis of pC15-1a indicated patterns different from that of pYC-5b. Since ISEcp1 does not exhibit marked target site selectivity, it can be hypothesized either that the CTX-M-15-encoding plasmids discussed here diverged from an ancestral ISEcp1-blaCTX-M-carrying plasmid or that the ISEcp1-blaCTX-M-15 sequence was independently acquired as part of a larger mobile element.
Nucleotide sequence accession numbers.
The described sequences have been assigned GenBank accession numbers AY604721 and AY604722.
REFERENCES
- 1.Arlet, G., G. Brami, D. Decre, A. Flippo, O. Gaillot, P. H. Lagrange, and A. Philippon. 1995. Molecular characterisation by PCR-restriction fragment length polymorphism of TEM β-lactamases. FEMS Microbiol. Lett. 134:203-208. [DOI] [PubMed] [Google Scholar]
- 2.Baraniak, A., J. Fiett, W. Hryniewicz, P. Nordmann, and M. Gniadkowski. 2002. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum-β-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 50:393-396. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edelstein, M., M. Pimkin, I. Palagin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum-β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 7.Lartigue, M. F., L. Poirel, C. Heritier, V. Tolun, and P. Nordmann. 2003. First description of CTX-M-15-producing Klebsiella pneumoniae in Turkey. J. Antimicrob. Chemother. 52:315-316. [DOI] [PubMed] [Google Scholar]
- 8.Lartigue, M. F., L. Poirel, and P. Nordmann. 2004. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol. Lett. 234:201-207. [DOI] [PubMed] [Google Scholar]
- 9.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mushtaq, S., N. Woodford, N. Potz, and D. M. Livermore. 2003. Detection of CTX-M-15 extended-spectrum-β-lactamase in the United Kingdom. J. Antimicrob. Chemother. 52:528-529. [DOI] [PubMed] [Google Scholar]
- 11.Neuwirth, C., E. Siebor, M. Pruneaux, M. Zarnayova, C. Simonin, J. P. Kisterman, and R. Labia. 2003. First isolation of CTX-M15-producing Escherichia coli from two French patients. J. Antimicrob. Chemother. 51:471-473. [DOI] [PubMed] [Google Scholar]
- 12.Pagani, L., E. Dell'Amico, R. Migliavacca, M. M. D'Andrea, E. Giacobone, G. Amicosante, E. Romero, and G. M. Rossolini. 2003. Multiple CTX-M-type extended-spectrum-β-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J. Clin. Microbiol. 41:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel, L., J.-W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum-β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 15.Siu, L. K., J. Y. C. Lo, K. Y. Yuen, P. Y. Chau, M. H. Ng, and P. L. Ho. 2000. β-Lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like β-lactamase, OXA-30. Antimicrob. Agents Chemother. 44:2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTXM-type beta-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 17.Yu, W.-L., K.-C. Cheng, L.-T. Wu, M. A. Pfaller, P. L. Winokur, and R. N. Jones. 2004. Emergence of two Klebsiella pneumoniae isolates harboring plasmid-mediated CTX-M-15 β-lactamase in Taiwan. Antimicrob. Agents Chemother. 48:362-363. [DOI] [PMC free article] [PubMed] [Google Scholar]