Abstract
Bacterial elongation factor Tu (EF-Tu) and EF-Ts are interacting proteins involved in polypeptide chain elongation in protein biosynthesis. A novel scintillation proximity assay for the detection of inhibitors of EF-Tu and EF-Ts, as well as the interaction between them, was developed and used in a high-throughput screen of a chemical library. Several compounds from a variety of chemical series with inhibitory properties were identified, including certain indole dipeptides, benzimidazole amidines, 2-arylbenzimidazoles, N-substituted imidazoles, and N-substituted guanidines. The in vitro activities of these compounds were confirmed in a coupled bacterial transcription-translation assay. Several indole dipeptides were identified as inhibitors of bacterial translation, with compound 2 exhibiting a 50% inhibitory concentration of 14 μM and an MIC for S. aureus ATCC 29213 of 5.6 μg/ml. Structure-activity relationship studies around the dipeptidic indoles generated additional analogs with low micromolar MICs for both gram-negative and gram-positive bacteria. To assess the specificity of antibacterial action, these compounds were evaluated in a metabolic labeling assay with Staphylococcus aureus. Inhibition of translation, as well as limited effects on other macromolecular pathways for some of the analogs studied, indicated a possible contribution from a non-target-based antibacterial mechanism of action.
Emerging bacterial resistance to antimicrobial agents, such as resistance to methicillin and vancomycin in Staphylococcus aureus, is a major public health problem that requires the discovery of new antibiotics with novel mechanisms of action to combat these organisms. Oxazolidinones are the only new class of antibacterial agents discovered and commercially launched within the last 30 years. Elongation factor Ts (EF-Ts) and EF-Tu are interacting proteins involved in extending the nascent polypeptide chain during the elongation stage of bacterial translation (18). During polypeptide chain synthesis, the complex of GTP and EF-Tu (EF-Tu*GTP) triggers the binding of an aminoacyl-tRNA to the acceptor site of the ribosome (3, 15). EF-Ts subsequently binds to EF-Tu, which promotes the release of GDP from EF-Tu after GTP hydrolysis, resulting in regeneration of the active form of EF-Tu (10). The essentialities of both proteins and their scant homologies to their eukaryotic counterparts make these components of the translation machinery attractive targets for drug discovery.
In the present work we describe the development of a high-throughput in vitro assay that monitors the activity of the EF-Tu*GTP regeneration process. Using this assay, we screened a large collection of compounds, resulting in the identification of several new translation inhibitors. A new chemical class of inhibitors, indole dipeptides, was identified as having both in vitro and in vivo activities. Structure-activity relationship studies with this series led to the identification of compounds with broad-spectrum antibacterial activity.
MATERIALS AND METHODS
Reagents.
Antibiotics were purchased from Sigma (St. Louis, Mo.). Taq polymerase was obtained from Perkin-Elmer Life Sciences (Boston, Mass.). Restriction enzymes, components of the Escherichia coli S30 Extract System for Circular DNA, and the Steady-Glo luciferase assay system were purchased from Promega (Madison, Wis.). Expression vector pET28a was obtained from Novagen (Madison, Wis.). Minimal essential medium (MEM) amino acid solution, MEM nonessential amino acids, and the TA cloning kit, which included the cloning vector pCR2.1, were purchased from Invitrogen (Carlsbad, Calif.). Primers were ordered from Integrated DNA Technologies (Caralville, Iowa). [3H]GDP, [3H]UTP, and [3H]leucine were purchased from NEN Life Science Products (Boston, Mass.). [3H]TTP and [3H]N-acetyl-d-glucosamine were obtained from Amersham (Piscataway, N.J.). Isopropyl-β-d-thiogalactopyranoside (IPTG) was purchased from U.S. Biochemicals (Cleveland, Ohio). All inhibitors were synthesized at Johnson and Johnson Pharmaceutical Research and Development, L.L.C., and were dissolved in 100% dimethyl sulfoxide (DMSO) to achieve a stock concentration of 20 mM. Multiscreen filter plates (N4550) were purchased from Millipore (Bedford, Mass.). The 96-well flat-bottom sterile plates (655076) used in the in vitro bacterial transcription-translation assay were purchased from Greiner (Frickenhausen, Germany). The 96-well round-bottom plates (3787) used for MIC determinations were purchased from Corning (Corning, N.Y.). Ni-coated flash plates were purchased from Perkin-Elmer Life Sciences.
Bacterial strains.
Haemophilus influenzae ATCC 49724, Streptococcus pneumoniae ATCC 49619, S. aureus ATCC 29213, E. coli ATCC 25922, and Enterococcus faecalis ATCC 29212 were used for susceptibility testing, along with E. coli tolC CGSC 5634, which was purchased from the E. coli Genetic Stock Center at Yale University. Strain BL21(DE3) was purchased from Novagen.
MICs.
MICs were determined by the broth microdilution method according to the guidelines of the National Committee for Clinical Laboratory Standards (12).
Cloning, overexpression, and purification of elongation factors.
The tufA gene encoding EF-Tu was amplified from E. coli strain K-12 by PCR with high-fidelity Taq polymerase and primers 5′-CCCCCGGATCCATGTCTAAAGAAAAATTTGAACGTAC-3′ and 5′-CCCCCCTCGAGGCCCAGAACTTTAGCAACAACGCC-3′, which contain BamHI and XhoI sites. The tufA fragment was cloned into the pCR2.1 vector with TA cloning kits that allow the direct ligation of Taq-amplified PCR products with 3′ overhangs. After transformation, the plasmids were purified from colonies resistant to kanamycin and were screened for the presence of the insert of the correct size. After sequence confirmation, the plasmid construct with the tufA insert was digested with BamHI and XhoI, and the resulting restriction fragment was ligated into the pET28a vector in order to attach a six-histidine tag. The resulting plasmid was transformed into strain BL21(DE3), and the sequence was confirmed and used for protein overexpression.
Primers 5′-TATACATATGGCTGAAATTACCGCATCCCTGG-3′ and 5′-ATACTCGAGAGACTGCTTGGACATCGCAGC-3′, which contain NdeI and XhoI sites, were used to PCR amplify the tsf gene encoding EF-Ts from E. coli strain K-12 by using high-fidelity Taq polymerase. The tsf fragment was cloned into the pCR2.1 vector with TA cloning kits. After transformation, plasmids were purified from colonies resistant to kanamycin and were screened for the presence of the insert of the correct size. After sequence confirmation, the plasmid construct with the tsf insert was digested with NdeI and XhoI, and the resulting restriction fragment was ligated into the pET28a vector in order to attach a six-histidine tag. The resulting plasmid was transformed into strain BL21(DE3), and the sequence was confirmed and used for protein overexpression. Overexpression was achieved at 37°C by induction for 3 h with 0.2 and 1.0 mM IPTG for EF-Tu and EF-Ts, respectively. EF-Tu was used without any further purification. EF-Ts was purified as described previously (17).
EF-Tu*EF-Ts binding assay.
The reaction mixture, which contained 40 pmol of EF-Tu, 50 mM Tris-HCl (pH 7.8), 100 mM KCl, 5 mM MgCl2, and 0.28 μM [3H]GDP (1μCi) in a total volume of 90 μl, was preincubated on Ni-coated flash plates to allow EF-Tu immobilization. Complex formation between EF-Tu and EF-Ts was assayed by monitoring the reduction in the radioactive signal upon addition of 30 μl of the second solution, which contained 7 pmol of EF-Ts, to the preformed EF-Tu*[3H]GDP complex, as described previously (6). The assay was validated with kirromycin, a known inhibitor of EF-Ts binding to EF-Tu (16). The reaction mixture was incubated for 60 min at room temperature, followed by aspiration of the liquid from the wells. Typically, reaction wells lacking EF-Ts produced a signal in the range of 20,000 to 30,000 cpm, whereas the complete reaction caused an approximately 10-fold reduction in the signal. High-throughput screening was performed on an Allegro robotic system (Zymark, Hopkinton, Mass.). The reaction components were added as described above, with the addition of a 5-μl aliquot of the compounds to be tested to columns 2 through 11 of each plate before the addition of EF-Tu*[3H]GDP. The final compound concentration in each well was 16 μM in 1.2% DMSO. A control plate containing kirromycin was inserted in every 35th plate for quality control purposes. Column 12 of each plate contained an identical aliquot of 30% DMSO to equalize the DMSO concentration in the control wells. Plates were read with a TopCount reader (read time, 30 s/well). It was found that counting within a 4-h period allowed detection of a stable signal (data not shown). Raw data from the TopCount reader were analyzed for percent inhibition by using Activity Base software (ID Business Solutions, Guildford, United Kingdom).
In vitro bacterial transcription-translation assay.
The enzymatic components of the translation machinery, which consisted of 1.1 μl of the E. coli S30 extract, 1.6 μl of premixture, 0.16 μl of a 5 mM amino acid mixture, and 4.14 μl of water, were added to a template mixture that contained 0.4 μg of pBest Luc circular DNA in 1.5 μl of water and 1.5 μl of compound in 5% DMSO in a total volume of 10 μl. The level of inhibition of the transcription-translation reaction with 0.75% DMSO was ∼20%, which did not exceed the standard variation of the assay. The reaction mixture was incubated for 40 min at room temperature in 96-well flat-bottom Greiner plates, and the formation of luciferase was measured by adding 30 μl of Steady-Glo luciferase reagent and reading on a TopCount reader. The assay was validated with kirromycin, rifampin, and chloramphenicol, as described previously (5).
Macromolecular synthesis.
To examine the effects of the experimental compounds on macromolecular biosynthetic pathways, DNA, RNA, protein, and cell wall biosyntheses were monitored by incorporation of [3H]TTP, [3H]UTP, [3H]leucine, and [3H]N-acetylglucosamine, respectively. Exponentially grown S. aureus was incubated in thiamine-nicotinic acid minimal essential medium, which contained 10 mM NaH2PO4, 12 mM K2SO4, 6 mM MgCl2, 16 mM (NH4)2SO4, 24 mM NaCl, 20 mg of thiamine per liter, 20 mg of nicotinic acid per liter, 0.5% glucose, 100 μM each amino acid except leucine in the presence of the appropriate tritiated precursors, and the test compound at a concentration of one-half the MIC for 30 min. The incorporation reaction was stopped with ice-cold 100% ethanol, and the macromolecules were precipitated with 10% trichloroacetic acid and collected on filter membranes. The membranes were washed with ethanol and allowed to dry completely, and the incorporation of radioactivity was read on the TopCount reader (Packard). Tetracycline, rifampin, ciprofloxacin, and vancomycin were used as control inhibitors.
RESULTS AND DISCUSSION
EF-Tu and EF-Ts are ideal targets for the development of novel antibacterial agents, as both proteins are essential and are highly conserved among bacteria and EF-Ts lacks homology with its eukaryotic counterparts; in addition, eukaryotic EF1α functions by a different mechanism than EF-Tu (9, 16). In addition, high-resolution crystallographic data for EF-Tu in complex with several known inhibitors are available (1-2, 4, 6-8, 11, 13-15). To identify inhibitors of these elongation factors, we have developed a novel high-throughput screening scintillation proximity assay based on the competition between EF-Ts and tritiated GDP for EF-Tu immobilized on a Ni-coated flash plate. By using this assay, the members of a chemical library were screened at 16 μM. Inhibitors belonging to four different chemical series were identified and confirmed (Fig. 1). A group of nine indole dipeptide analogs inhibited EF-Tu and EF-Ts binding, suggesting that these compounds were not providing false-positive results. An indole dipeptide, shown in Fig. 1, had an MIC of 2 μg/ml for S. aureus. Less potent compounds in the high throughput screening binding assay, 2-arylbenzimidazoles, an N-substituted imidazole, and an N-substituted guanidine, displayed activities against S. aureus, with MICs of 2, 8, and 0.5 μg/ml, respectively.
FIG. 1.
Structures of EF-Tu and EF-Ts inhibitors.
An in vitro transcription-translation reaction, which monitors the activities of both elongation factors, was used as a secondary functional assay. In addition, the activities of these compounds against several microorganisms were determined.
Two- and three-dimensional similarity and substructure searches were conducted on the hits that demonstrated both in vitro and antibacterial activities in order to identify structurally similar compounds that were not part of the original screening set. Through this process 76 additional benzimidazole analogs were identified. Upon further testing, only three compounds from this series were inhibitory in the in vitro transcription-translation assay, and none had antibacterial activity. The series of N-substituted guanidines, comprising 10 compounds, did not produce a viable lead compound that would possess both in vitro and antibacterial activities.
Among the remaining chemical classes, we selected the indole dipeptide series for further development, as it was the only structural series that both inhibited in vitro transcription-translation and displayed activity against gram-negative and gram-positive bacteria. Structure-activity relationship studies were conducted with this series to define the pharmacophore of the molecule responsible for biological activity. These analogs were screened for their inhibition of in vitro transcription-translation and activities against several pathogens. Eighty-two analogs showed inhibitory activity in the in vitro transcription-translation assay. A majority of the analogs that inhibited in vitro transcription-translation also displayed antibacterial activity.
Representatives of the indole dipeptide class, of which five of seven analogs inhibited protein biosynthesis, are shown in Table 1. Compounds 1 and 2 inhibited in vitro transcription-translation with 50% inhibitory concentrations (IC50s) of 15.3 and 14 μM, respectively. Compound 3, which contains a diaminobutyric acid moiety, inhibited in vitro transcription-translation with an IC50 of 34.7 μM. Compounds 4 and 6, in which the C-terminal amino acid residue was replaced by an aminoalkyl moiety, were less potent, with IC50s ≥50 μΜ. Indole dipeptides with activities against gram-positive strains E. faecalis, S. pneumoniae, and S. aureus and gram-negative strains E. coli, a tolC E. coli efflux pump mutant, and H. influenzae are illustrated in Table 2. The MICs of compounds 1 and 2 for the gram-positive species S. aureus and the gram-negative species H. influenzae were 6.3 μM. Compound 4 displayed weak activity against E. coli, with an MIC of 50 μM. A wide range of antibacterial activities was observed for each of the analogs tested. A phylogenetic analysis did not reveal any correlation between the sequence homology of the elongation factors and the observed variations in MICs for the species. Therefore, we concluded that the differences in activities against various microorganisms are more likely to be due to differential cellular permeability or efflux.
TABLE 1.
Inhibition of in vitro transcription-translation pathway by indole dipeptide analogsa
MW, molecular weight; boldface elements of the molecular structures indicate an indole or benzimidazole-2-one scaffold.
TABLE 2.
Activities of indole dipeptides against gram-positive and gram-negative bacteria
| Compound no. | MIC (μM)
|
Mol wt | |||||
|---|---|---|---|---|---|---|---|
| E. faecalis | S. pneumoniae | S. aureus | H. influenzae | E. coli tolC | E. coli | ||
| 1 | 25 | 50 | 6.3 | 6.3 | 3.1 | >200 | 1,132.49 |
| 2 | 25 | 25 | 6.3 | 6.3 | 6.3 | >200 | 885.01 |
| 3 | 25 | 50 | 12.5 | 50 | 6.3 | 100 | 1,049.81 |
| 4 | 25 | 25 | 12.5 | >200 | 6.3 | 50 | 881.67 |
| 5 | >200 | 50 | 12.5 | 6.3 | 6.3 | >200 | 1,203.57 |
| 6 | 200 | 100 | 25 | 50 | 25 | >200 | 900.05 |
| 7 | 25 | 25 | 25 | >400 | 25 | >200 | 767.37 |
It is noteworthy that all seven compounds contain a structurally similar fused heterocycle, either an indole or benzimidazole-2-one scaffold, highlighted in boldface in Table 1, suggesting that this portion of the molecule may be essential for biological activity. Preliminary analysis of this series, however, suggests that even the heterocycle can be modified significantly without a loss of activity. For example, the indole heterocycle may be replaced by a benzimidazole-2-one, as in the case of compound 3, and the amide substituent at the 6 position of the indole may be replaced by an amide moiety (2, 7) or a sulfonamide moiety (data not shown).
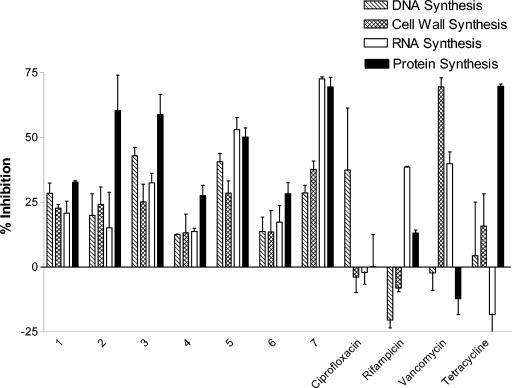
To filter out compounds with nonspecific antimicrobial activity, selected analogs that inhibited bacterial translation in vitro were evaluated for their intracellular mechanisms of action in a metabolic labeling assay. The activities of the compounds against the major macromolecular biosynthetic pathways, DNA, RNA, protein, and cell wall biosyntheses, were tested (Fig. 2). Activity was monitored by incorporation of the corresponding radioactive precursors, as described in Materials and Methods. Standard antibiotics were tested concurrently with the experimental compounds. All measurements were carried out in duplicate. In addition to the expected inhibition of the corresponding pathways by the control antibiotics, we found that tetracycline upregulated transcription, rifampin inhibited translation to some extent and activated DNA synthesis and cell wall synthesis to a limited extent, and vancomycin had a reproducible effect on RNA synthesis.
FIG. 2.
Effects of indole dipeptides on inhibition of macromolecular biosynthetic pathways. S. aureus 29213 was grown exponentially in minimal essential medium, without leucine, in the presence of compounds at a concentration of one-half the MIC and individual 3H-labeled precursors for 30 min.
Preferential inhibition of protein biosynthesis in S. aureus by compound 2 was observed, as illustrated in Fig. 2. Like tetracycline, compound 2 inhibited DNA synthesis and cell wall synthesis to some degree. Unlike tetracycline, transcription was not activated but was inhibited, albeit to a lesser extent than translation. Although compounds 3 and 4 inhibited other pathways, they were comparatively selective for translation. While compounds 5 and 7 inhibited translation, transcription was also affected to a similar degree. Compounds 1 and 6 inhibited all four pathways. Unlike the control antibiotics, the test compounds did not upregulate any of the pathways. These results indicate that potential nonspecific mechanisms contribute to the antibacterial effect, along with the specific inhibition of translation, for some of the compounds studied. Despite the inhibition of in vitro transcription-translation by the majority of the indole dipeptide analogs, one cannot rule out the possibility that the antimicrobial activity of this chemical class is due to a nonspecific mechanism of action. Thus, additional mechanism-of-action studies are required to elucidate the precise molecular target of these novel inhibitors. One of the ways to ultimately confirm the drug target would be to select for mutations that confer resistance to the novel antimicrobial agent, thus identifying the site of its action. There is no guarantee, though, that this approach would always work, as exemplified by studies of the mechanism of action of tetracycline, a translation inhibitor that was widely used in clinics for decades without knowledge of its molecular target. Only cocrystallization of this antibiotic with the 30S ribosome, a breakthrough that happened very recently, has finally provided compelling evidence of its site of action.
The identification of indole dipeptides represents a successful early attempt to find small-molecule inhibitors of translation elongation factors. Further chemical exploration of this class is needed to improve the potency and specificity of these translation inhibitors.
REFERENCES
- 1.Abel, K., M. D. Yoder, R. Hilgenfeld, and F. Jurnak. 1996. An alpha to beta conformational switch in EF-Tu. Structure 4:1153-1159. [DOI] [PubMed] [Google Scholar]
- 2.Berchtold, H., L. Reshetnikova, C. O. A. Reiser, N. K. Schirmer, M. Sprinzl, and R. Hilgenfeld. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365:126-132. [DOI] [PubMed] [Google Scholar]
- 3.Gordon, J. 1968. A stepwise reaction yielding a complex between a supernatant fraction from E. coli, guanosine 5′-triphosphate, and aminoacyl-sRNA. Proc. Natl. Acad. Sci. USA 59:179-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heffron, S. E., and F. Jurnak. 2000. Structure of an EF-Tu complex with a thiazolyl peptide antibiotic determined at 2.35 Å resolution: atomic basis for GE2270A inhibition of EF-Tu. Biochemistry 39:37-45. [DOI] [PubMed] [Google Scholar]
- 5.Kariv, I., H. Cao, P. D. Marvil, E. V. Bobkova, Y. E. Bukhtiyarov, Y. P. Yan, U. Patel, L. Coudurier, K. R. Oldenburg, and T. D. Y. Chung. 2001. Identification of the inhibitors of bacterial transcription/translation machinery utilizing the miniaturized 1536-well format screen. J. Biomol. Screen. 6:233-243. [DOI] [PubMed] [Google Scholar]
- 6.Kawashima, T., C. Berthet-Colominas, M. Wulff, S. Cusack, and R. Leberman. 1996. The structure of the Escherichia coli EF-Tu/EF-Ts complex at 2.5 Å resolution. Nature 379:511-518. [DOI] [PubMed] [Google Scholar]
- 7.Kjeldgaard, M., and J. Nyborg. 1992. Refined structure of elongation factor EF-Tu from Escherichia coli. Mol. Biol. 223:721-742. [DOI] [PubMed] [Google Scholar]
- 8.Kjeldgaard, M., P. Nissen, S. Thirup, and J. Nyborg. 1993. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1:35-50. [DOI] [PubMed] [Google Scholar]
- 9.Knudsen, C., H. Wieden, and M. V. Rodnina. 1991. The importance of structural transitions of the switch II region for the functions of EF-Tu on the ribosome. J. Biol. Chem. 276:22183-22190. [DOI] [PubMed] [Google Scholar]
- 10.Lucas-Lenard, J., and F. Lipmann. 1966. Separation of three microbial amino acid polymerization factors. Proc. Natl. Acad. Sci. USA 55:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mesters, J. R., L. A. H. Zeef, R. Hilgenfeld, J. M. de Graat, B. Kraal, and L. Bosch. 1994. The structural and functional basis for the kirromycin resistance of mutant EF-Tu species in Escherichia coli. EMBO J. 13:4877-4885. [DOI] [PMC free article] [PubMed]
- 12.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Nissen, P., M. Kjeldgaard, S. Thirup, G. Polekhina, L. Reshetnikova, B. Clark, and J. Nyborg. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464-1472. [DOI] [PubMed] [Google Scholar]
- 14.Polekhina, G., S. Thirup, M. Kjeldgaard, P. Nissen, C. Lippman, and J. Nyborg. 1996. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure 4:1141-1151. [DOI] [PubMed] [Google Scholar]
- 15.Ravel, J. M., R. L. Shorey, C. W. Garner, R. C. Dawkins, and W. Shive. 1969. The role of an aminoacyl-tRNA-GTP-protein complex in polypeptide synthesis. Cold Spring Harbor Symp. Quant. Biol. 34:321-330. [DOI] [PubMed] [Google Scholar]
- 16.Vogeley, L., G. F. Palm, J. R. Mesters, and R. Hilgenfeld. 2001. Conformational change of EF-Tu induced by antibiotic binding. J. Biol. Chem. 276:17149-17155. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, Y., J. Tao, M. Zhou, Q. Meng, L. Zhang, L. Shen, R. Klein, and D. L. Miller. 1997. Elongation factor Ts of Chlamydia trachomatis: structure of the gene and properties of the protein. Arch. Biochem. Biophys. 344:42-52. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, Y., N. Yu, and L. L. Spremulli. 1998. Functional analysis of the roles of residues in E. coli EF-Ts in the interaction with EF-Tu. J. Biol. Chem. 273:4556-4562. [DOI] [PubMed] [Google Scholar]