Two major groups of acquired β-lactamases have emerged in Pseudomonas aeruginosa: Ambler class A extended-spectrum β-lactamases (ESBLs) and class B metallo-β-lactamases (MBLs) (7, 9). MBLs are an expanding group of carbapenemases that includes the VIM family. To date, multiple allelic variants, namely, VIM-1 to VIM-10 (http://www.lahey.org/studies), have been described in Europe, Asia, and the Americas, VIM-2 being the most ubiquitous enzyme by far (7, 8; R. E. Mendes, M. Castanheira, P. Garcia, M. Guzman, M. A. Toleman, T. R. Walsh, and R. N. Jones, Letter, Antimicrob. Agents Chemother. 48:1433-1434, 2004). Additionally, five types of ESBLs have been detected in P. aeruginosa: TEM, SHV, PER, VEB, and IBC/GES (9). Association between VIM and ESBLs still appears to be a rare event in P. aeruginosa and was reported only for VIM-2 with either PER-1 or IBC-2 (3; J. D. Docquier, J. D., F. Luzzaro, G. Amicosante, A. Toniolo, and G. M. Rossolini, Letter, Emerg. Infect. Dis. 7:910-911, 2001). MBL- or ESBL-producing P. aeruginosa isolates have not yet been reported in Argentina, while the coexistence of VIM with GES-1 in a single clinical isolate has not been documented anywhere.
In November 2002, after 8 days of meropenem treatment (120 mg/kg of body weight/day), P. aeruginosa M5109 was recovered as the sole isolate from the catheter of a 7-month-old patient at the Hospital de Niños “Ricardo Gutierrez.” The isolate was confirmed to be P. aeruginosa by using the API20NE system (bioMérieux, Marcy l'Etoile, France) and displayed uncommonly high levels of carbapenem resistance and synergism between imipenem- and zinc chelator-containing disks. Based on the antibiotype profile, colistin was added to the meropenem treatment, and the patient was discharged alive.
By susceptibility analysis carried out by agar dilution according to NCCLS guidelines (6), M5109 showed resistance to ticarcillin (MIC, 1,024 μg/ml), piperacillin (MIC, >1,024 μg/ml), piperacillin-tazobactam (MIC, 512 μg/ml), cefotaxime (MIC, 512 μg/ml), ceftazidime (MIC, 256 μg/ml), cefepime (MIC, 64 μg/ml), aztreonam (MIC, 32 μg/ml), imipenem (MIC, 512 μg/ml), meropenem (MIC, 128 μg/ml), amikacin (MIC, 256 μg/ml), gentamicin (MIC, >512 μg/ml), and ciprofloxacin (MIC, 32 μg/ml) but not to colistin (MIC, 0.5 μg/ml). The addition of 0.4 mM EDTA produced a ≥32-fold decrease in the carbapenem MICs. A carbapenemase was detected by a microbiological assay (4). Attempts to transfer ceftazidime or imipenem resistance by biparental conjugation (4) to P. aeruginosa ATCC 27853 were unsuccessful.
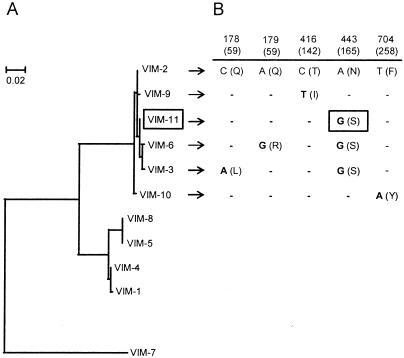
PCR screening of bla genes followed by DNA sequencing revealed the presence of blaTEM-1-like (6), blaOXA-2-like (1), blaGES-1-like (primers used were GES-F [5′-GAAAAAGCAGCTCAGATCG] and GES-R [5′-CAACAACCCAATCTTTAGGA]), and blaVIM-2-like (primers used were VIM-F and VIM-R [5]) (amplicons of 500, 480, 580, and 261 bp, respectively). Amplification for other ESBL (blaCTX-M-2, blaPER, and blaSHV [4]) or MBL (blaSPM-1 and blaIMP [unpublished data]) genes was negative. Since all the blaVIM genes and most of the blaGES genes presently reported are integron located (7, 9), we performed PCRs combining the primer 5′-CS, directed against the 5′-conserved segment of class 1 integrons (4), with either VIM-R or GES-R, rendering amplicons of 475 and 819 bp, respectively. Thus, both blaVIM-2-like and blaGES-1-like were found as the first cassettes of different class 1 integrons in M5109. For further sequencing of these genes, additional reverse primers (VIM-Rb [5′-TGTTATGCCGCATCTGCCTG] and GES-Rc [5′-TCAACTATTTGTCCGTGCTC], respectively) were designed based on sequence alignments of highly related genes (Clustal X; ftp://ftp-igbmc.u-strasbg.fr/pub/). The 5′-CS-VIM-Rb amplicon (948 bp) contained an 801-bp-long open reading frame carrying the new blaVIM-11 allele, which differs from blaVIM-2 in a unique, nonsynonymous mutation and from blaVIM-3 or blaVIM-6 in another nonsilent change. Therefore, it may be considered an evolutionary intermediate between blaVIM-2 and blaVIM-3/6 (Fig. 1). The 1,008-bp-long 5′-CS-GES-Rc amplicon harbored an open reading frame of 864 bp that showed a unique silent change (C→T) at nucleotide position 591 relative to the blaGES-1 sequence.
FIG. 1.
Relatedness of blaVIM genes. (A) Neighbor-joining tree of all the following blaVIM genes reported to date (with GenBank accession numbers given in parentheses): blaVIM-1 (Y18050), blaVIM-2 (AF191564), blaVIM-3 (AF300454), blaVIM-4 (AY135661), blaVIM-5 (AY144612), blaVIM-6 (AY165025), blaVIM-7 (AJ536835), blaVIM-8 (AY524987), blaVIM-9 (AY524988), blaVIM-10 (AY524989), and blaVIM-11 (AY605049). (B) Substitutions in genes highly related to blaVIM-2. The nucleotide positions of the indicated mutations (relative to the blaVIM-2 sequence) are shown. Amino acid changes and their positions based on the numbering system described by Galleni et al. (2) are indicated in parentheses.
By isoelectric focusing (4), only three bands at pI 5.4 (TEM-1), 5.8 (ESBL activity), and 7.85 (OXA-2-like) were visualized in M5109. A carbapenemase activity, inhibited in situ by EDTA (30 mM), was also detected at pI 5.8. Therefore, the coexistence of two β-lactamases at pI 5.8 (GES-1 and VIM-11) is proposed.
This is the first report of an MBL-mediated carbapenem-resistant and ESBL-producing P. aeruginosa isolate in Argentina and the first description anywhere of the coexistence of a VIM variant with GES-1 in a single strain. Our results, together with previous findings (M. Castanheira, R. E. Mendes, T. R. Walsh, A. C. Gales, and R. N. Jones, Letter, Antimicrob. Agents Chemother. 48:2344-2345, 2004; R. E. Mendes et al., letter, 2004), indicate that both VIM- and GES-producing P. aeruginosa strains have already become established in Latin America. The emergence of these enzymes in P. aeruginosa constitutes a public health concern in Argentina which requires efficient detection and brisk intervention to preserve antibiotic options.
REFERENCES
- 1.Bert, F., C. Branger, and N. Lambert-Zechovsky. 2002. Identification of PSE and OXA β-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J. Antimicrob. Chemother. 50:11-18. [DOI] [PubMed] [Google Scholar]
- 2.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, J.-M. Frère, and the Metallo-β-Lactamase Working Group. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mavroidi, A., E. Tzelepi, A. Tsakris, V. Miriagou, D. Sofianou, and L. S. Tzouvelekis. 2001. An integron-associated β-lactamase (IBC-2) from Pseudomonas aeruginosa is a variant of the extended-spectrum β-lactamase IBC-1. J. Antimicrob. Chemother. 48:627-630. [DOI] [PubMed] [Google Scholar]
- 4.Melano, R., A. Corso, A. Petroni, D. Centrón, B. Orman, A. Pereyra, N. Moreno, and M. Galas. 2003. Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum β-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J. Antimicrob. Chemother. 52:36-42. [DOI] [PubMed] [Google Scholar]
- 5.Miriagou, V., E. Tzelepi, D. Gianneli, and L. S. Tzouvelekis. 2003. Escherichia coli with a self-transferable, multiresistant plasmid coding for metallo-β-lactamase VIM-1. Antimicrob. Agents Chemother. 47:395-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 7.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 8.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weldhagen, G. F., L. Poirel, and P. Nordmann. 2003. Ambler class A extended-spectrum β-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob. Agents Chemother. 47:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]