Abstract
The expression of certain bacterial genes is regulated in a cell-density dependent way, a phenomenon called quorum sensing. Both Gram-negative and Gram-positive bacteria use this type of communication, though the signal molecules (auto-inducers) used by them differ between both groups: Gram-negative bacteria use predominantly N-acyl homoserine lacton (AHL) molecules (autoinducer-1, AI-1) while Gram-positive bacteria use mainly peptides (autoinducer peptides, AIP or quorum sensing peptides). These quorum sensing molecules are not only involved in the inter-microbial communication, but can also possibly cross-talk directly or indirectly with their host. This review summarizes the currently applied analytical approaches for quorum sensing identification and quantification with additionally summarizing the experimentally found in vivo concentrations of these molecules in humans.
Keywords: quorum sensing peptides, analytical methods, reporter bacteria, biosensors, chromatography, microbiome
Introduction
The idea of bacteria being single isolated organisms has been outdated for many years. In the late 1960s, a cell-density dependent bioluminescence was observed in the marine symbiotic bacterium Vibrio fisheri (Nealson et al., 1970; Nealson and Hastings, 1979). This cell-density dependent regulation of gene expression is defined as quorum sensing and consists of at least four steps: (I) synthesis of signal molecules, called autoinducers, (II) excretion of the signal molecules, (III) at a certain threshold concentration, activation of a specific receptor and as a result (IV) activation or suppression of gene expression (Sifri, 2008). For example, with the increase of the number of Vibrio fisheri bacteria, the amount of autoinducer in the external environment reaches a certain level and triggers the production of the enzyme luciferase resulting in bioluminescence (Engebrecht and Silverman, 1984). The genes involved in quorum sensing, are responsible for activities that are only of use when performed by a large number of cells, for example: bioluminescence, antibiotic production, formation of biofilms, and production of virulence factors (Rutherford and Bassler, 2012).
Both Gram-negative and Gram-positive bacteria apply quorum sensing for communication, but they produce different auto-inducers. Gram-negative bacteria mainly depend on N-acyl homoserine lacton (AHL) molecules (autoinducer-1, AI-1) while Gram-positive bacteria use modified oligopeptides (autoinducer peptides, AIP) (Taga and Bassler, 2003). These peptides possess a large structural diversity and frequently undergo post-translational modifications (Sturme et al., 2002). A third type of autoinducers are boron-furan-derived signal molecules (autoinducer-2, AI-2) and are produced and detected by both Gram-negative and Gram-positive bacteria (Li and Nair, 2012). Besides these 3 main groups, there is also a fourth group of miscellaneous quorum sensing molecules (Barber et al., 1997; Flavier et al., 1997; Holden et al., 1999; Pesci et al., 1999; Higgins et al., 2007; Wei et al., 2011).
In this review, the focus will be on the production of quorum sensing molecules belonging to the first three groups, i.e., AHL molecules, AI-2 molecules and quorum sensing peptides.
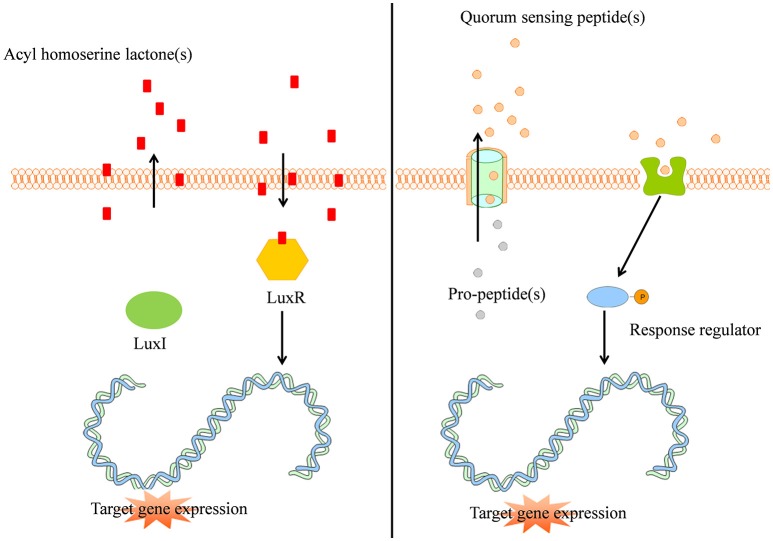
The quorum sensing mechanism of Gram-negative bacteria can be described using the example of Vibrio fisheri: an intracellular autoinducer synthase (LuxI) synthesizes AHL signal molecules by catalyzing a reaction between S-adenosylmethionine and an acyl carrier protein (Sifri, 2008; Rutherford and Bassler, 2012). Due to the small size and lipophilicity of AHL autoinducers, they readily pass the cell membrane by means of passive diffusion (Sifri, 2008). If the concentration of AHL is sufficiently high, the AHL autoinducer binds to the intracellular LuxR protein and provokes the LuxR DNA binding domain to reveal. Subsequently, the LuxR protein binds to DNA, causing activation of target gene transcription (Figure 1 left panel, Rutherford and Bassler, 2012). More than 100 Gram-negative bacterial species apply a LuxI/LuxR-type system with an autoinducer synthase (e.g., LuxI) and a transcriptional regulator (e.g., LuxR, Sifri, 2008).
Figure 1.
(Left panel) Typical Gram-negative quorum sensing mechanism. Acyl homoserine lactone molecules, synthesized by LuxI, passively pass the bacterial cell membrane and when a sufficient concentration is reached (threshold level) activate the intracellular LuxR which subsequently activates target gene expression in a coordinated way. (Right panel) Quorum sensing peptides are synthesized by the bacterial ribosomes as pro-peptidic proteins and undergo posttranslational modifications during excretion by active transport. The quorum sensing peptides bind membrane associated receptors which get autophosphorylated and activate intracellular response regulators via phosphor-transfer. These phosphorylated response regulators induce increased target gene expression.
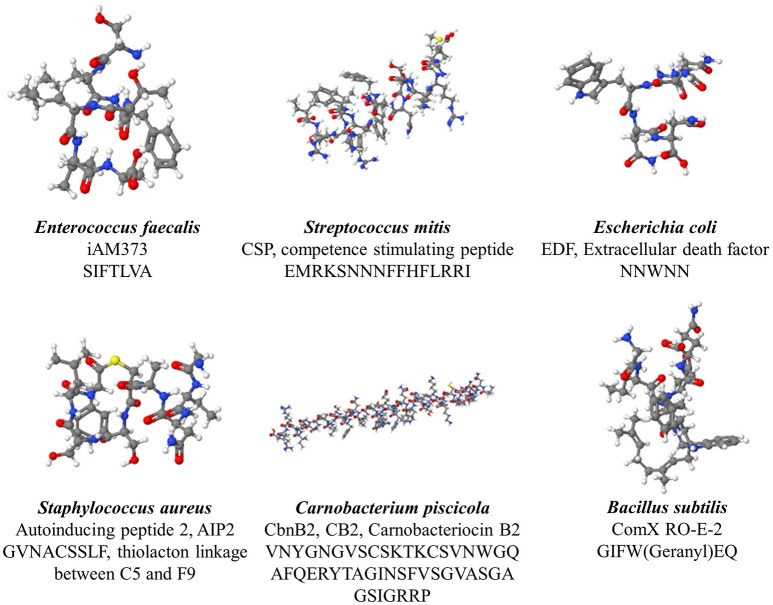
Gram-positive bacteria apply peptides as autoinducers for quorum sensing (Figure 1 right panel). Examples of this heterogeneous group of peptides are given in Figure 2. These peptides are synthesized by ribosomes as precursor peptides and undergo posttranslational modifications during excretion to become activated and stabilized (Sturme et al., 2002). In general, the secretion of the AIP is facilitated by a membrane associated ATP-binding cassette (ABC) transporter (Sturme et al., 2002). As the population density increases, the AIPs accumulate in the environment. When a certain threshold level is reached, binding of an AIP to a receptor initiates activation of the receptor kinase by phosphorylation on a conserved histidine residue (Bassler, 1999; Sturme et al., 2002). Subsequently, the activated receptor kinase transfers the phosphoryl group to a conserved aspartate residue of the intracellular response regulator, which in turn will be activated (Bassler, 1999). The activated response regulator influences the transcription of target genes, including the AIP genes, genes for the receptor kinase and response regulator and genes for the ABC transporter. Based on the species, the nomenclature of the quorum sensing mechanisms can be different, due to the involved genes and receptor(s). For example, Staphylococci species employ the agr-quorum sensing system, Streptococci species employ the ComX-quorum sensing system and Bacilli species use the Rap-quorum sensing system.
Figure 2.
Various quorum sensing peptides, illustrating the structural diversity observed among quorum sensing peptide.
The current mainstream opinion is that the AIPs only serve as inter-microbial communication molecules, but other biological actions are known and being explored as well, for example the lantibiotic nisin peptide of Lactococcus lactis, also has known antimicrobial properties (Sturme et al., 2002). The influence of quorum sensing peptides on human tumor progression has recently also been investigated (De Spiegeleer et al., 2015; Wynendaele et al., 2015a).
Quorum sensing molecules do not only serve as intra-species communication molecules. It has been demonstrated that certain AHLs are secreted and recognized by several species of Gram-negative bacteria, hereby potentially acting as “cross-talk” signals (interspecies communication). Another indication is the presence of AI-2 in both Gram-positive and Gram-negative bacteria. Co-culture systems of V. harveyi and E coli have indicated that AI-2 production by one species affects gene expression in the other (McNab and Lamont, 2003; Ryan and Dow, 2008; Pereira et al., 2013). For example, cell culture supernatant of P. aeruginosa is capable of mediating cell death of E. coli via a quorum sensing mediated process (Kumar et al., 2013).
Inter-kingdom communication
Inter-kingdom communication, due to co-evolution, between bacteria and eukaryotes is readily known (Lyte and Ernst, 1993). The knowledge of the inter-kingdom communication between bacteria and the human host has extended remarkably during recent years [reviewed in references Hughes and Sperandio, 2008; Karavolos et al., 2013; Clarke et al., 2014; O'Mahony et al., 2015; Kendall and Sperandio, 2016; Moos et al., 2016; Oleskin et al., 2016; Stilling et al., 2016]. For example, to modulate the expression of virulence factors, bacteria have shown the remarkable capacity to monitor neuroendocrine hormones produced by the host, e.g., adrenaline and noradrenaline (Lyte, 1993, 2004; Lyte and Ernst, 1993; Sperandio et al., 2003; Freestone et al., 2008; Karavolos et al., 2008, 2011; Pacheco and Sperandio, 2009; Spencer et al., 2010; Lyte et al., 2011). For example, Enterohaemorrhagic E. coli (EHEC) is able to modify its mobility and virulence expression in relationship to adrenaline and noradrenaline concentration (Rasko et al., 2008; Pacheco and Sperandio, 2009). Moreover, the mouse gut microbiome itself is capable of producing catecholamines, thus possessing the ability to interact with the host (Asano et al., 2012). The neurotransmitter γ-aminobutyric acid (GABA) produced by various Lactobacilli plays an important role in our brains (Barrett et al., 2012; Clarke et al., 2014). Another important group of messenger molecules in the gut-brain communication are the bacterial short chain fatty acids, fermentation products of bacterial metabolism as these molecules are capable of triggering peptide YY release (Holzer et al., 2012). Bacteria are also capable of sensing a variety of human peptide hormones, e.g., somatostatin (Yamashita et al., 1998) and gastrin (Chowers et al., 1999). Both bacteria and humans apply cyclic dipeptides for communication purposes and hence these molecules might be of extreme interest for the gut-brain communication axis [opinionated in reference (Belleza et al., 2014)]. It has also been demonstrated that some quorum sensing peptides are capable to penetrate the blood-brain barrier in a mouse model, without significant subsequent efflux from the brain (Wynendaele et al., 2015b). The Gram-negative quorum sensing molecules, i.e., N-acylhomoserinelactones (AHLs), easily pass eukaryotic cell membranes, due to their lipophilic nature (Fuqua et al., 1994).
Immunomodulation by odHL (N-3-oxo-dodecanoyl-L-homoserine lactone) has been reported (Pritchard, 2006a; Teplitski et al., 2011) and occurs in a TLR4-independent manner (Kravchenko et al., 2006). The activity of macrophages (Gomi et al., 2006; Kravchenko et al., 2006, 2008; Miyairi et al., 2006; Thomas et al., 2006), epithelial cells (Kravchenko et al., 2006, 2008; Jahoor et al., 2008; Cooley et al., 2010), mast cells (Li et al., 2009), fibroblasts (Kravchenko et al., 2006, 2008; Miyairi et al., 2006; Jahoor et al., 2008), T-lymphocytes (Wagner et al., 2007), B-lymphocytes (Telford et al., 1998a), and neutrophils (Zimmermann et al., 2006) is influenced by odHL. The outcome of the immunomodulatory properties of odHL are not straight-forward, as some studies suggest a proinflammatory response (Smith et al., 2001, 2002a,b; Vikström et al., 2005; Thomas et al., 2006; Jahoor et al., 2008; Mayer et al., 2011), whereas others point out an anti-inflammatory effect (Telford et al., 1998a; Chhabra et al., 2003; Hooi et al., 2004; Kravchenko et al., 2008; Skindersoe et al., 2009), thus facilitating persistent infection. As a rationale to elucidate this apparent discrepancy, a concentration-dependent effect has been proposed (Teplitski et al., 2011; Pritchard, 2006a). Besides odHL, P. aeruginosa also produces an aromatic quorum sensing molecule, i.e., 2-amino-acetophenon. This molecule contributes to the establishment of a chronic infection by damping the inflammatory immune response (Bandyopadhaya et al., 2012). A third quorum sensing molecule from P. aeruginosa, 2-heptyl-3-hydroxy-4(1H)-quinolone, has shown to inhibit T cell proliferation (Pritchard, 2006b), possibly via the IL-2 receptor pathway (Hooi et al., 2004). As a direct effect of its immunomodulating properties, odHL evokes IL-8 production on fibroblasts and bronchial epithelial cells, resulting in an accelerated apoptosis by mammalian macrophages and neutrophils (DiMango et al., 1995; Smith et al., 2002a; Tateda et al., 2003). Growth of human colorectal and prostate cancer cells is inhibited by odHL and analogs (Dolnick et al., 2005; Oliver et al., 2009), whereas down-modulation of STAT3 by odHL induces apoptosis in breast carcinoma cells (Li et al., 2004). Additionally, quorum sensing molecules produced by the Gram-negative P. aeruginosa in nosocomial urinary tract infection (UTI) might also be accountable for renal damage. As an opportunistic pathogen, P. aeruginosa especially affects immunocompromised patients and treatment is further hindered by its ability to form biofilms on urinary catheters (Gupta et al., 2013). Immunomodulation by these quorum sensing molecules can also have implications via the neuroendocrine-immune system axis, which is frequently involved in various diseases (Procaccini et al., 2014).
The clinical significance of the microbiome-host relationship is becoming increasingly apparent. Differences between the microbiome of healthy individuals and cancer patients have been related to susceptibility to cancer (Bultman, 2014), e.g., an increase in fecal E. coli has been associated with development of colon cancer (Wynendaele et al., 2015a). Strong indications have been found that changes in the microbiome are not only associated with tumorigenesis, but also directly contribute to tumorigenesis (Zackular et al., 2013; Baxter et al., 2014). The microbiome has been shown to alter the tumor-environment by influencing the host's neuroendocrine system (Erdman and Poutahidis, 2014). Helicobacter pylori is, besides causing gastro-duodenal ulcers, accountable for the majority of stomach cancers (Ernst and Gold, 2000). Moreover, the association between inflammatory bowel diseases, e.g., chronic ulcerative colitis and Chrohn's disease, and colon carcinoma is widely accepted (Balkwill and Mantovani, 2001). The microbiome is relevantly involved in alterations of the human immune system and host's metabolism (Thaiss et al., 2016). Quorum sensing molecules also affect human health via quorum sensing mediated biofilms (Kalia, 2013) which are associated with a wide array of infections (Beloin et al., 2014). These infections contribute to an increase in morbidity, mortality, and public costs (Haas et al., 2014). Alterations in the human microbiome have also been linked to several central nervous disorders, e.g., autism (Finegold et al., 2010; Tomova et al., 2015), depression (Naseribafrouei et al., 2014; Jiang et al., 2015), and schizophrenia (Castro-Nallar et al., 2015). The gut microbiome is likely associated with various conditions via the gut-brain axis, e.g., anxiety and altered neurochemistry (Cryan and O'Mahony, 2011) and increasing evidence points toward the influence of the microbiome in neuroendocrine diseases, e.g., as a mediator in diabetes mellitus via the cholinergic nervous system (Parekh et al., 2016). Additionally, aging has also been linked to alterations of the gut microbiome, which are, among others, related to immunosenescence and immune aging (De Spiegeleer et al., 2016; Mello et al., 2016).
Analytical techniques
A variety of analytical techniques have been reported in literature for the qualitative and quantitative analysis of quorum sensing molecules. Generally, these techniques can be considered as chromatographic/mass spectroscopic techniques or based on biosensor systems using (genetically modified) reporter bacteria and will be discussed in more detail in the following sections regarding the detection and quantification of quorum sensing molecules in human biological samples and bacterial cell culture broths.
Biosensor systems using reporter bacteria
In these systems, the presence of a compound of interest is qualitatively and/or quantitatively detected based on a signal produced by the bacterial reporter. The produced signal can be colorimetric, fluorimetric, bioluminescent, chemiluminescent, turbidimetric, colony forming units to name a few (van der Meer and Belkin, 2010).
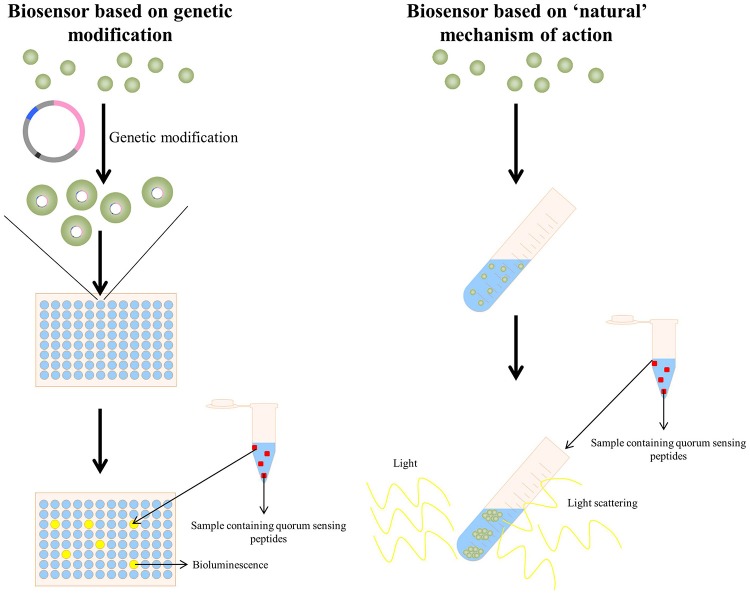
Regarding quorum sensing bacterial reporters, two approaches are currently being used, i.e., (I) those based on the “natural” provoked effect by the specific quorum sensing peptide and (II) by applying genetically modified bacteria acting as detecting strains (Figure 3). The clumping assay (Suzuki et al., 1984; Mori et al., 1988; Nakayama et al., 1995a) can be considered as a system based on the “natural” provoked effects of some quorum sensing peptides: the bacteria applied as reporter strain are wild-type bacteria and have not been genetically altered to obtain receptiveness toward the specific quorum sensing peptide(s). When certain enterococcal quorum sensing peptides are added to receptive Enterococci, these quorum sensing peptides provoke clumping of the Enterococci. Consecutively, the turbidity of the bacterial suspension enhances and provides a quantifiable result via turbidimetry. The second type of bacterial reporters consists of bacteria that have been genetically modified to produce a signal in the presence of the quorum sensing molecule of interest; this type of reporter bacteria is more elaborately explained in the following sections.
Figure 3.
(Left panel) A quorum sensing reporter strain based on genetic modification: bacteria are genetically modified to make them competent to detect the specific quorum sensing peptide. Upon addition of a sample containing the quorum sensing peptide, the activated quorum sensing peptide receptor triggers in this example bioluminescence, allowing detection and quantification of the quorum sensing peptide (La Rosa et al., 2015). (Right panel) A quorum sensing reporter strain based on the naturally observed quorum sensing mechanism of a quorum sensing peptide (in this example the clumping induction assay as observed with various E. faecalis quorum sensing peptides Suzuki et al., 1984; Mori et al., 1988; Nakayama et al., 1995a). A sample containing the quorum sensing peptide is added to a bacterial strain capable of detecting the quorum sensing peptide and the formation of clumps is observed via light scattering.
Acyl homoserine lactones
Detection of quorum sensing molecules is often performed by using quorum sensing reporter bacteria strains, of which most are sensitive to either autoinducer-1 (AHL's) or autoinducer-2 (Steindler and Venturi, 2007; Rai et al., 2015). As mentioned previously, these bacterial reporters can be based on non-genetically modified or genetically-modified bacteria.
Genetic modifications consist of altering the bacterial genome, in order to obtain a detectable signal in the presence of the quorum sensing molecule of interest. This genetic modification consists of coupling a suitable promoter to a desired reporter gene, and hence the desired detection signal. The detection signal is produced by a reporter protein, e.g., bioluminescence, chemiluminescence, fluorescence, colorimetric response, etc. Production of the reporter protein depends on activation of the promoter, which initiates transcription of the reporter gene.
Inherent to genetic modifications of the bacteria, is the need of selecting those bacteria that are successfully genetically modified [e.g., those bacterial cells that have acquired the plasmid(s)]. Selecting these correctly, genetically modified bacteria can be performed by for example antibiotic resistance genes (O'Connor et al., 2016).
Table 1 provides an overview of plasmid-based AHL reporter bacteria.
Table 1.
Overview of some plasmid-based AHL reporter strains (Steindler and Venturi, 2007; Rai et al., 2015; O'Connor et al., 2016).
| Plasmid | Bacterial strain | Analyte | Promoter | Reporter |
|---|---|---|---|---|
| pSB401 | E. coli | 3-oxo-C4- to 3-oxo-C14-HSL C4- to C12-HSL | luxI | luxCDABE |
| pSB1075 | E. coli | 3-oxo-C12- to 3-oxo-C16-HSL C12- to C16-HSL | lasI | luxCDABE |
| pECP61.5 | P. aeruginosa | C4-HSL | rhlA | lacZ |
| pKDT17 | E. coli | 3-oxo-C10- to 3-oxo-C12-HSL C10- to C12-HSL | lasB | lacZ |
| pCF218, pMV26 | A.tumefaciens | 3-oxo-C6- to 3-oxo-C12-HSL C4- to C12-HSL | traI | luxCDABE |
| pCF218, pCF372 | A.tumefaciens | 3-oxo-C4- to 3-oxo-C12-HSL C5- to C10-HSL | traI | lacZ |
| pSB406 | E. coli | 3-oxo-C4- to 3-oxo-C14-HSL C4- to C12-HSL | rhlI | luxCDABE |
| C. violaceum | 3-oxo-C6- to 3-oxo-C8-HSL C4- to C8-HSL | cviI | Violacein | |
| pAL105 | E. coli | 3-oxo-C12-HSL | lasI | luxCDABE |
| pAL101 | E. coli | C4-HSL | rhlI | luxCDABE |
| pSB536 | E. coli | C4-HSL | ahyI | luxCDABE |
| pSB403 | Broad host range | 3-oxo-C4- to 3-oxo-C14-HSL C4- to C12-HSL | luxI | luxCDABE |
| pHV2001 | E. coli | 3-oxo-C6- to 3-oxo-C8-HSL C6- to C8-HSL | luxI | luxCDABE |
| pZLR4 | A.tumefaciens | All 3-oxo-HSLs C6- to C14-HSL 3-OH-C6- to 3-OH-C10-HSL | traI | lacZ |
| pJZ384, pJZ410, pJZ372 | A.tumefaciens | 3-oxo-C4- to 3-oxo-C18-HSL C4- to C18-HSL | traI | lacZ |
| pSF105, pSF107 | P. fluorescens | 3-OH-C6-HSL C6-HSL 3-OH-C8-HSL | phzI | lacZ |
| pUCP18 | P. aeruginosa | 3-oxo-C12-HSL | rsaL | luxCDABE |
| pMS402 | P. aeruginosa | 3-oxo-C12-HSL | rsaL | luxCDABE |
| pUCGMAT1-4 | E. coli | 3-oxo-C6-HSL | ahlI | mcherry |
| pREC-FF | E. coli | 3-oxo-C6-HSL | luxI | cfp |
| M71LZ | P. aeruginosa lasI − | 3-oxo-C10 to 3-oxo-C12-HSL | lasI | lacZ |
| pAS-C8 | Broad host range | C8- to C10-HSL | cepI | gfp |
| pKR-C12 | Broad host range | 3-oxo-C10- to 3-oxo-C12-HSL | lasI | gfp |
| pJBA-132 | Broad host range | 3-oxo-C6-HSL C6- to C10-HSL | luxI | gfp |
Autoinducer-2
The Vibrio harveyi BB170 bioluminescence assay is an example of a reporter bacteria constructed by modifications of the bacterial genome to prevent production of and/or response to certain quorum sensing molecules. It is a frequently used reporter strain for quantification of autoinducer-2 molecules. Genetic modifications of the BB170 strain ensure it does not produce endogenous autoinducer-1 (AHL's) or autoinducer-2 and is insensitive for autoinducer-1. Hence, bioluminescence only occurs when autoinducer-2 is added, where the light intensity correlates with the amount of autoinducer-2 that is added (O'Connor et al., 2016).
Quorum sensing peptides
A variety of approaches are applied to proof the presence of, and in rare cases, eventually identify quorum sensing peptides (see Table 2). Currently, ~300 quorum sensing peptides and structural analogs have been published (Wynendaele et al., 2013). The majority of these peptides have been tested by the researchers on a system considered a reporter bacteria based biosensor system. While peptide structure elucidation was frequently performed by liquid chromatography-mass spectroscopy and/or Edman degradation. Analogs of the cognate quorum sensing peptides are also frequently evaluated using a specific biosensor.
Table 2.
Summary of quorum sensing peptide bio-sensor detection systems.
| Quorum sensing molecule (amino acid sequence) | Species | LOD | Operational principle |
|---|---|---|---|
| 21-CSP (SGSLSTFFRLFNRSFTQALGK) | S. mutans | Not specified | β-galactosidase assay, Bacteriocin production, Transformation assay (Syvitski et al., 2007; Tian et al., 2009) |
| Gelatinase Biosynthesis-Activating Pheromone (QNSPNIFGQWM, lacton linkage between S3 and M11) | E. faecalis | Not specified | Bioluminescence (La Rosa et al., 2015) |
| EntF (AGTKPQGKPASNLVECVFSLFKKCN) | E. faecium | 10 aM | Bacteriocin induction (Dunny et al., 1979) |
| cAM373 (AIFILAS) | E. faecalis | 50 pM | Microtiter dilution method (Mori et al., 1986a) |
| cAD1 (LFSLVLAG) | E. faecalis | 50 pM | Microtiter dilution method (Mori et al., 1984) |
| cCF10 (LVTLVFV) | E. faecalis | 25 pM | Clumping induction assay (Mori et al., 1988) |
| cPD1 (FLVMFLSG) | E. faecalis | 40 pM | Clumping induction assay (Suzuki et al., 1984) |
| cOB1 (VAVLVLGA) | E. faecalis | Not specified | Clumping induction assay (Nakayama et al., 1995a) |
| iPD1 (ALILTLVS) | E. faecalis | Not specified | Microtiter dilution method (Mori et al., 1987) |
| iAD1 (LFVVTLVG) | E. faecalis | Not specified | Microtiter dilution method (Mori et al., 1986b) |
| iAM373 (SIFTLVA) | E. faecalis | 200 pM | Clumping induction assay (Nakayama et al., 1995b) |
| iCF10 (AITLIFI) | E. faecalis | 10 nM | Inhibition of induced self-clumping (Nakayama et al., 1994) |
| CSP (DSRIRMGFDFSKLFGK) | S. constellatus, S. anginosus, S. thermophilus | 0.2 nM | Biofilm formation assay (Petersen et al., 2004) |
| CSP (DRRDPRGIIGIGKKLFG) | S. milleri | 200 ng/ml ≈ 105 nM | Transformation assay (Håvarstein et al., 1997) |
| CSP (SQKGVYASQRSFVPSWFRKIFRN) | S. gordonii | 200 ng/ml ≈ 72 nM | Transformation assay (Håvarstein et al., 1997) |
| CSP (EMRISRIILDFLFLRKK) | S. pneumoniae | 100 ng/ml ≈ 46 nM | Transformation assay (Pozzi et al., 1996) |
| CSP (EMRLSKFFRDFILQRKK) | S. pneumoniae | 100 ng/ml ≈ 45 nM | Transformation assay (Pozzi et al., 1996) |
| EDF (NNWNN) | E. coli | <100 ng/ml ≈ 150 nM | Colony forming units assay (Kolodkin-Gal et al., 2007) |
For example, two reporter strains in S. mutans GS5, capable of detecting the competence stimulating peptide (21-CSP) UA159sp4 have been described. The CSP-mediated quorum sensing sytem of S. mutans involves the genes comCDE, comAB, and comX. In brief, the CSP precursor, encoded by comC, is cleaved and exported to form CSP by an ABC-transporter encoded by comAB. Additionally, the comDE gene encodes a two-component signal transduction system that recognizes CSP and initiates a response. ComX encodes a sigma factor that regulates transcription of several genes needed for uptake of foreign DNA and the ComE protein is activated by autophosphorylation upon binding of CSP with the ComD histidine kinase receptor. Phosphorylated ComE activates target genes such as comCDE, comAB, and comX, thereby generating a positive feedback quorum sensing system (Syvitski et al., 2007). The reporter strain is composed of a comC mutant S. mutans GS5, resulting in a mutant strain which is unable to produce CSP itself, but still able to respond to exogenous CSP, named SMdC. Mutation of the comC gene is obtained by inserting a spectinomycin (Specr) resistance gene in the comC gene of S. mutans GS5. This mutant was used to develop two lacZ transcriptional reporter strains in which the lacZ gene is fused to the comDE promoter or the nlmAB promoter. To generate the first reporter strain, a vector pYH2 was formed by fusion of the comDE promoter with a promoterless lacZ gene in the Streptococcus—Escherichia coli shuttle vector pSL; subsequently, pYH2 was transformed into SMdC, resulting in the SMdC-pYH2 reporter strain. The second reporter strain is constructed by fusion of the nlmAB promoter with a promoter-lacking lacZ gene and transformation of this pOMZ47 plasmid into SMdC, resulting in the SMdC-PnlmAB reporter strain (Steindler and Venturi, 2007). Following addition of exogenous CSP (UA159sp), due to transcription of the lacZ gene, both SMdC-pYH2 and SMd-C-PnlmAB express β-galactosidase (Syvitski et al., 2007; Tian et al., 2009). Cleavage of O-nitrophenyl-β-D-galactoside by β-galactosidase renders O-nitrophenol, with the yellow color proportional to the enzyme activity and hence the quorum sensing peptide concentration (Syvitski et al., 2007).
The ability of CSP (0.2–100 nM) to induce competence in S. intermedius NCTC 11324 was tested: overnight cells of S. intermedius NCTC 11324 were diluted 1:200 and subsequently, quorum sensing peptide and a plasmid expressing erythromycin resistance genes were added. After incubation for 24 h, transformants were selected by growth on THB agar plates containing erythromycin. The amount of transformants/mL increased when increasing concentrations of CSP were added (Wynendaele et al., 2015a). Comparable transformation assays were performed by Håvarstein et al. (1997) with two other synthetic CSPs from Streptococcus milleri and from Streptococcus gordonii (Håvarstein et al., 1997) and by Pozzi et al. (1996).
The reporter strain for the Gram-negative E. coli quorum sensing peptide “extracellular death factor” (EDF) was described by Kolodkin-Gal et al. (2007). They studied the influence of the quorum sensing peptide on mazEF-mediated cell death in E. coli. The mazEF module encodes the stable toxin MazF, which exerts its toxic effects by cleavage of single-stranded mRNA at ACA sequences, and the labile antitoxin MazE, which counteracts the effect of MazF. The mazEF module is induced by stress. Stressful conditions that prevent transcription/translation of mazEF, such as inhibition of transcription and/or translation by antibiotics (e.g., rifampicin, chloramphenicol, spectinomycin) or DNA damage, reduces the level of the labile MazE, hereby reducing the counteracting effect on MazF. However, mazEF-mediated cell death only occurs in dense cultures since it requires the quorum sensing peptide EDF. Addition of rifampicin provokes mazEF-mediated cell death in dense cultures, but not in diluted cultures. The mazEF-mediated cell death in these diluted cultures could be restored by addition of supernatant of a dense culture. The bioassay for detection and quantification of EDF activity is based on this principle. Different dilutions of the supernatant of a dense culture of the E. coli strain MC4100relA+, containing EDF, were added to a diluted culture of MC4100relA+, after which stressful conditions (e.g., addition of chloramphenicol) were induced. The percentage of surviving colony forming units (CFUs) was determined after overnight incubation (Kolodkin-Gal et al., 2007).
Quorum sensing peptides can also be detected quantitatively (limit of detection 10 aM) by demonstrating a product of which formation is regulated by a quorum sensing peptide. Nilsen et al. (1998) studied the production of the bacteriocins enterocin A and enterocin B in Enterococcus faecium CTC492, regulated by the quorum sensing peptide EntF. Without EntF, a non-bacteriocin-producing culture (Bac–) of Enterococcus faecium CTC492 was obtained. Upon addition of induction factor EntF, production of enterocin A and enterocin B is induced and after 20–24 h incubation, bacteriocin activity is assayed by addition of sterilized supernatant to a culture of an indicator organism, e.g., Lactobacillus sakei NCDO 2714 (sensitive to both enterocin A and enterocin B), Pediococcus pentosaceus FBB 63 (sensitive to enterocin A) or Lactobacillus sakei FVM 148 (sensitive to enterocin B). Growth inhibition of these strains indicates presence of bacteriocins in the supernatant and thus, presence of induction factor EntF in the sample added to the non-bacteriocin-producing culture (Nilsen et al., 1998).
A different approach uses the phenomenon of plasmid transfer to demonstrate the presence of quorum sensing peptides. Genes located on plasmids and encoding for hemolysins, bacteriocins, and antibiotic resistance genes are transferable. Several Enterococcus faecalis strains are able to transfer plasmids, facilitated by sex pheromones (i.e., quorum sensing peptides) (Dunny et al., 1979). Certain quorum sensing peptides, excreted by recipient strains, induce donor strains, containing conjugative plasmids, to produce a proteinaceous substance on their surface. Thereby, mating aggregates are formed, facilitating plasmid transfer. These quorum sensing peptides are also called clumping-inducing agents (CIA) since they cause self-aggregation of donor cells when culture filtrate of recipients is added (Dunny et al., 1979). Mori et al. (1984, 1986a, 1988), Suzuki et al. (1984) and Nakayama et al. (1995a), isolated and determined the sequence of several of these CIAs. Once the recipient strain acquires the conjugative plasmid, the excretion of the specific CIA, corresponding with the acquired plasmid, stops and the strain becomes sensitive to exogenous quorum sensing peptides. However, production of other CIAs, specific for donor strains containing other conjugative plasmids, still continues (Ike et al., 1983). Quantification of CIA can be performed by means of a microtiter system. Serial dilutions of recipient strain filtrates (or sample) are added to responder cells (donor cells with the appropriate conjugative plasmid) and incubated for 90–120 min, after which clumping is observed in dilutions (samples) containing sufficient amount of CIA (quorum sensing peptide) (Dunny et al., 1979). The same principle can be applied to other quorum sensing peptides facilitating plasmid transfer in E. faecalis e.g., cAD1 (Mori et al., 1984), cCF10 (Mori et al., 1988), cPD1 (Suzuki et al., 1984), and cOB1 (Nakayama et al., 1995a). Quorum sensing peptides cOB1 and cAD1 are an exception to the assumption of one pheromone (cX) specifically activating plasmid transfer of its corresponding plasmid (pX) since both pOB1 and pY11 respond to cOB1 and both pBEM10 and pAD1 respond to cAD1 (Murray et al., 1988; Nakayama et al., 1995a).
The production stop of quorum sensing peptide, once the recipient strain acquires the corresponding plasmid, is due to the production of a quorum sensing inhibitor encoded on the plasmid, which inhibits the activity of its corresponding quorum sensing peptide (Mori et al., 1987). Inhibitory activity of iPD1 (Mori et al., 1986b, 1987) and iAM373 (Nakayama et al., 1995b) can be quantitatively measured by modifying the microtiter system described above. Using iAD1 as example, serially diluted sample solutions containing iAD1 are added to a mixture of responder cells OG1S(pAD1::Tn917) and cAD1. After incubation for 2–3 h, inhibition of cAD1-induced clumping of responder cells is observed (Mori et al., 1986b).
Chromatographic techniques
Liquid chromatography
A variety of liquid-chromatographic methods have been reported, especially for the N-acyl homoserine lactones (see Table 3) (Morin et al., 2003; Li et al., 2006; Fekete et al., 2007; Kumari et al., 2008; Ortori et al., 2011; Wang et al., 2011; Cataldi et al., 2013), including methods allowing the simultaneous analysis of various N-acyl homoserine lactones via a non-targeted LC-MS/MS method (Patel et al., 2016). Generally, these methods are capable of generating limits of detection (LOD) in the broad nM-range (median LOD of 2.3 nM for 19 N-acyl homoserine lactone standards; range: 0.6 nM–2838.9 nM), with some compounds owing a limit of detection around 0.6 nM (for C10-HSL) (Patel et al., 2016).
Table 3.
Summary of reported concentrations of quorum sensing molecules in different matrices.
| Quorum sensing molecule | Matrix | Detection method | Concentration |
|---|---|---|---|
| AUTOINDUCER-2 | |||
| AI-2 | S. oralis 34 in vitro biofilm | V. harveyi BB170 bioluminescence assay | 69–123 nM over 48 h (Rickard et al., 2008) |
| A. naeslundii T14V in vitro biofilm | V. harveyi BB170 bioluminescence assay | 153–382 nM over 48 h (Rickard et al., 2008) | |
| S. oralis 34 and A. naeslundii T14V in vitro dual-species biofilm | V. harveyi BB170 bioluminescence assay | 124–197 nM over 48 h (Rickard et al., 2008) | |
| S. oralis 34 luxS mutant and A. naeslundii T14V in vitro dual-species biofilm | V. harveyi BB170 bioluminescence assay | 122–140 nM over 48 h (Rickard et al., 2008) | |
| Saliva (n = 8) | HPLC-MS | 244–965 nM (mean = 526 nM) (Campagna et al., 2009) | |
| Saliva IBD patients (n = 3) | V. harveyi BB170 bioluminescence assay | 1.67–2.29 μM (mean = 2.05 μM) (Raut et al., 2013) | |
| Stool IBD patients (n = 2) | V. harveyi BB170 bioluminescence assay | 1.57–3.59 μM (mean = 2.76 μM) (Raut et al., 2013) | |
| Ileal washing IBD patients (n = 1) | V. harveyi BB170 bioluminescence assay | 2.29 μM (Raut et al., 2013) | |
| Saliva healthy volunteers (n = 3) | V. harveyi BB170 bioluminescence assay | 0.998–2.07 μM (mean = 1.39 μM) (Raut et al., 2013) | |
| AHL | |||
| 3-oxo-C12-HSL | Sputum samples (n = 47) (20 CF patients + 2 healthy volunteers) | LC-MS | 20.24–6833.20 nM (Struss et al., 2013) |
| P. aeruginosa in vitro biofilms (n = 4) | LC-MS | Mean = 0.95 ± 0.68 μM (Struss et al., 2013) | |
| Sputum CF patients (n = 38/59) | LC-MS/MS | LLOQ (not specified)–410 nM (Barr et al., 2015) | |
| Sputum CF patients (n = 10/12) | pKDT17 with lasB:lacZ fusion in E. coli | 0.92–21.20 nM (mean = 5.04 nM) (Marchesan et al., 2015) | |
| HHQ | Sputum CF patients (n = 35/59) | LC-MS/MS | LLOQ (not specified)–1066 nM (Barr et al., 2015) |
| Plasma CF patients (n = 40/56) | LC-MS/MS | 0.01 nM (i.e., LLOQ)–2.744 nM (Barr et al., 2015) | |
| Urine CF patients (n = 31/59) | LC-MS/MS | 0.02 nM (i.e., LLOQ)–4.028 nM (Barr et al., 2015) | |
| NHQ | Sputum CF patients (n = 43/59) | LC-MS/MS | LLOQ (not specified)–1563 nM (Barr et al., 2015) |
| Plasma CF patients (n = 25/56) | LC-MS/MS | 0.01 nM (i.e., LLOQ)–0.222 nM (Barr et al., 2015) | |
| Urine CF patients (n = 19/59) | LC-MS/MS | 0.01 nM (i.e., LLOQ)–0.347 nM (Barr et al., 2015) | |
| HQNO | Sputum CF patients (n = 47/59) | LC-MS/MS | LLOQ (not specified)–780 nM (Barr et al., 2015) |
| Plasma CF patients (n = 21/56) | LC-MS/MS | 0.03 nM (i.e., LLOQ)–0.904 nM (Barr et al., 2015) | |
| Urine CF patients (n = 48/59) | LC-MS/MS | 0.03 nM (i.e., LLOQ)–12.511 nM (Barr et al., 2015) | |
| NQNO | Sputum CF patients (n = 26/59) | LC-MS/MS | LLOQ (not specified)–1075 nM (Barr et al., 2015) |
| Plasma CF patients (n = 15/56) | LC-MS/MS | 0.04 nM (i.e., LLOQ)–0.705 nM (Barr et al., 2015) | |
| Urine CF patients (n = 16/59) | LC-MS/MS | 0.05 nM (i.e., LLOQ)–2.067 nM (Barr et al., 2015) | |
| C7-PQS | Sputum CF patients (n = 21/59) | LC-MS/MS | LLOQ (not specified)–873 nM (Barr et al., 2015) |
| Plasma CF patients (n = 8/56) | LC-MS/MS | 0.1 nM (i.e., LLOQ)–0.571 nM (Barr et al., 2015) | |
| C9-PQS | Sputum CF patients (n = 16/59) | LC-MS/MS | LLOQ (not specified)–4302 nM (Barr et al., 2015) |
| C4-HSL | Sputum CF patients (n = 18/59) | LC-MS/MS | LLOQ (not specified)–145 nM (Barr et al., 2015) |
| Sputum CF patients (n = 6/12) | pECP61.5 with rhlA:lacZ fusion in P. aeruginosa | 0.84–5.00 nM (mean = 1.04 nM) (Erickson et al., 2002) | |
| C10-HSL | Burkholderia cepacia JA-8 cell culture broth (n = 4) | LC-MS/MS | 260 nM (9% RSD) (Frommberger et al., 2004) |
| C8-HSL | Burkholderia cepacia JA-8 cell culture broth (n = 4) | LC-MS/MS | 180 nM (12% RSD) (Frommberger et al., 2004) |
Besides the closely related cyclodipeptides demonstrated qualitatively, the presence of quorum sensing peptides has not yet been demonstrated in vivo (Marchesan et al., 2015), except for one report (Darkoh et al., 2015) where the presence of a thiolacton peptide, without structural elucidation, is suggested in feces of patients suffering from C. difficile infection. Nevertheless, the presence of these peptides in cell culture broths has already been proven via reporter strains (see Table 2). Additionally, various authors have reported quorum sensing peptide levels in bacterial cell culture broths via chromatographic techniques, hence applying an orthogonal detection method compared to the biological reporter bacteria.
Additional to reporter bacteria based biosensors, several quorum sensing peptides have been qualitatively or quantitatively determined in bacterial cell culture broths (see Table 4). For example, the currently reported E. faecalis quorum sensing peptides have been quantified in cell culture broths. Based on the reports, the concentration of these quorum sensing peptides is in the broad picomolar range, though caution should be paid to the often elaborate sample preparation methods that have been applied, mostly intended for a qualitative approach.
Table 4.
Concentration of selected peptides in bacterial cell culture broths.
| Species | Quorum sensing peptide (sequence) | Strain | Concentration | Detection method |
|---|---|---|---|---|
| Enterococcus faecalis | cAM373 (AIFILAS) | E. faecalis JH2-2 (pAM351) | 4.4 μg/16 L = 374 pM | HPLC-UV/VIS (Mori et al., 1986a) |
| iPD1 (ALILTLVS) | E. faecalis JH2-2 (pAM351) | 6.3 μg/20 L = 378 pM | HPLC-UV/VIS (Mori et al., 1987) | |
| iCF10 (AITLIFI) | E. faecalis OG1RF(pMSP5011) | 15,360 pg/mL = 19,440 pM | Estimated via bio-assay via comparing with synthetic peptide (La Rosa et al., 2015) | |
| cOB1 (VAVLVLGA) | E. faecalis FA2-2 | 1.5 μg/15 L = 135 pM | HPLC-UV/VIS (Nakayama et al., 1995a) | |
| cAD1 (LFSLVLAG) | E. faecalis JH2-2 | 200 μg/300 L = 814 pM | HPLC-UV/VIS (Mori et al., 1984) | |
| iAD1 (LFVVTLVG) | E. faecalis FA2-2 (pAM727) | 20 μg/20 L = 787 pM | HPLC-UV/VIS (Mori et al., 1986b) | |
| cCF10 (LVTLVFV) | E. faecalis FA2-2 (pAM351) | 4.1 μg/60 L = 86 pM | HPLC-UV/VIS (concentration) FAB-MS (amino acid sequence confirmation) (Mori et al., 1988) | |
| iAM373 (SIFTLVA) | E. faecalisJSS2 (pAM377) | 3 μg/8 L = 500 pM | HPLC-UV/VIS (Nakayama et al., 1995b) |
Besides the presence of E. faecalis quorum sensing peptides in cell culture broths, other authors have developed chromatographic methods to demonstrate quorum sensing peptide presence of other species in their respective cell cultures (Junio et al., 2013; Todd et al., 2016). For example, the presence of Staphylococcus autoinducing peptide I was demonstrated using a ultra-high performance liquid chromatographic system coupled to a mass spectrophotometer in methicillin-resistant Staphylococcus aureus cell cultures with a limit of detection of 3.5 nM and a LOQ of 0.10 μM (Todd et al., 2016). Junio et al. (2013) demonstrated quantitatively (LOD 0.25 μM; LOQ 2.6 μM) the time- and strain dependent synthesis of AIP-1 in cell culture medium in the μM-magnitude, with concentrations as high as 13 ± 2 μM at 16 h incubation of the S. aureus LAC strain (Junio et al., 2013).
Additionally, the extracellular death factor quorum sensing peptide from E. coli has also been demonstrated in cell culture broths via a qualitative chromatographic method (Kolodkin-Gal et al., 2007).
Quorum sensing is not limited to bacteria, but eukaryotic yeasts have also shown to possess quorum sensing mechanisms. The C. albicans quorum sensing molecules farnesol and tyrosol are via a LC-MS/MS method quantifiable with limits of detection of respectively 15.2 and 3.0 nM (Gregus et al., 2010).
Gas chromatography
Via gas chromatography coupled to a mass spectrometer (GC-MS), an enantiomer-selective method was achieved for the analysis of C8-HSL (LOD: 0.1 mg/L ≈ 0.4 μM for L-C8-HSL), C10-HSL (LOD: 0.1 mg/L ≈ 0.4 μM for L-C10-HSL), and C12-HSL (LOD: 0.3 mg/L ≈ 1.1 μM for L-C12-HSL). By applying this method to Burkholderia cepacia LA3 cell culture supernatant, the authors demonstrated the in vitro production of D-C10-HSL (Malik et al., 2009). N-3-oxoacyl homoserine lactones were determined by derivatizing them into their pentafluorobenzyloxime derivatives with subsequent gas chromatography-mass spectrometry analysis. ODHL (biofilm: 3 ± 2 μM; effluent: 1 ± 0.1 nM), OdHL (biofilm: 632 ± 381 μM; effluent: 14 ± 3 nM), OtDHL (biofilm: 40 ± 15 μM; effluent: 1.5 ± 0.7 nM), and OOHL (effluent: 0.1 ± 0.1 nM) were by applying this method found in biofilms and/or effluent (Charlton et al., 2000). Cataldi et al. (2007) also reported a GC-MS method capable of detecting AHL's in the μM-range with an average LOD of 4.4 μM for the 6 investigated AHLs (Cataldi et al., 2007). The autoinducer-2, following silylation, has also proven to be analyzable by gas chromatography-mass spectrometry (LOD 5.3 nM and limit of quantification (LOQ) 16 nM) (Thiel et al., 2009).
Thin-layer chromatography
Besides liquid and gas chromatography, (high performance-) thin-layer chromatography has also been used in the analysis of certain quorum sensing molecules (Shaw et al., 1997; Bala et al., 2013). Shaw et al. (1997) achieved LODs ranging from 0.5 fmol to 300 pmol depending on the specific AHL (Shaw et al., 1997), whereas Bala et al. (2013) reported a lower limit of detection of 0.006 nmol/spot and a lower limit of quantification of 0.01 nmol/spot for PQS (Bala et al., 2013). A hybrid technique, combining bacterial reporters with thin-layer chromatography has been described (Andersen et al., 2001; Charlesworth et al., 2015) (LODs ranging from 0.012 to 1,710 ng depending on the specific AHL) (Charlesworth et al., 2015).
Capillary electrophoresis
Besides liquid and gas chromatography, some authors reported capillary electrophoresis as a suitable technique to quantify quorum sensing molecules. With partial filling micellar electrokinetic chromatography–electrospray ionization-ion trap mass spectrometry, two acyl homoserine lactone derivatives (i.e., N-octanoyl-L-homoserine lactone and N-decanoyl-L-homoserine lactone) molecules originating from Burkholderia cepacia could be qualitatively detected in bacterial cell culture broths down to a concentration of approximately 1 μM (Frommberger et al., 2003). The same authors were capable of determining quantitatively (LOQ 0.05 μg/ml) C8-HSL (0.26 μg/ml ≈ 1.1 μM), C10-HSL (1.93 μg/ml ≈ 7.6 μM), and C12-HSL (0.16 μg/ml ≈ 0.6 μM) in Burkholderia cepacia cell culture broths by means of capillary zone electrophoresis-mass spectrometry following hydrolysis of the homoserine lactones into their corresponding organic acids (Frommberger et al., 2005).
Several P. aeruginosa-quinolones are also detectable using capillary electrophoresis with LODs of 65 nM (PQS), 94 nM (HHQ), 61 nM (“Quinolone 1”) and 79 nM (“Quinolone 2”) (Zhou et al., 2012). Noteworthy is the eukaryotic quorum sensing molecule from C. albicans farnesol, which is also detectable via capillary electrophoresis. By applying this capillary electrophoresis method, the authors demonstrated the concentration of farnesol to be approximately 2 nM in a cell culture supernatant (Kubesová et al., 2010).
Miscellaneous analytical techniques
Cyclic voltammetry and amperometric detection by a bare boron-doped diamond electrode showed, however with selectivity limitations, to be a possible approach to detect 2-heptyl-3-hydroxy-4-quinolon from P. aeruginosa at approximately 30 μM in P. aeruginosa pqsL− mutant strains cell culture medium (Zhou et al., 2011).
Other authors also developed electrochemical biosensors for the detection of AHL's, achieving detection limits down to 2 pM via cyclic voltammetry with a gold microelectrode (Baldrich et al., 2011).
Zhang and Ye (2014) reported a Förster resonance energy transfer-based biosensor for the detection of N-(3-oxo-hexanoyl)-L-homoserine lactone. However, the reported method had a limit of detection as high as 100 μM (Zhang and Ye, 2014).
Enzyme-linked immunosorbent assay (ELISA) also proved a suitable approach for AHL-detection. Chen et al. (2010a) developed rat monoclonal antibodies achieving limits of detection of 31 nM concerning 3-oxo-C10-HSL and 476 nM regarding C8-HSL (Chen et al., 2010a). Additionally, Kaufmann et al. (2006) developed an antibody against 3-oxo-C12-HSL (Kaufmann et al., 2006) without aiming to develop an ELISA. However, this antibody could still be suitable for ELISA and has according to Chen et al. (2010a) a limit of detection of 15 nM (Chen et al., 2010a). Another ELISA method has been developed to monitor AHL production in B. cepacia LA3 cell cultures (Chen et al., 2010b).
Finally, surface-enhanced Raman spectroscopy also proved a feasible approach for AHL detection with a LOD estimated around 3 nM (Pearman et al., 2007).
In vivo concentration of quorum sensing molecules
Autoinducer-2
Streptococcus oralis, Streptococcus oralis luxS mutant, and Actinomyces naeslundii T14V each produce autoinducer-2 when they are grown in commercial media (Rickard et al., 2006). Rickard et al. (2006, 2008) investigated if each of these species also produce autoinducer-2 when they are part of single- or dual-species biofilms in saliva in an ex-vivo in-vitro design. Biofilms were grown within Sorbarods fed with 25% human saliva, after which the autoinducer-2 concentration in the effluent was determined by the Vibrio harveyi BB170 bioluminescence assay. Autoinducer-2 concentrations in a single-species biofilm of S. oralis 34 or A. naeslundii T14V varied between 69–123 and 153–382 nM over 48 h respectively. In the effluent from the S. oralis 34 luxS mutant biofilm, no autoinducer-2 could be detected. Autoinducer-2 concentrations in the effluent of dual-species biofilms containing S. oralis 34 and A. naeslundii T14V or S. oralis 34 luxS mutant and A. naeslundii T14V varied respectively between 124–197 and 122–140 nM over 48 h (Rickard et al., 2006, 2008).
Campagna et al. (2009) developed a HPLC-MS method (LOD in the low nM-range) for the detection of autoinducer-2 in human saliva. They asked 8 volunteers to donate approximately 1 ml of saliva and determined the concentration of autoinducer-2 in these samples. The average concentration was found to be 526 nM with individual values ranging from 244 to 965 nM (Campagna et al., 2009).
Autoinducer-2 concentrations in saliva, stool and intestinal samples from patients with irritable bowel disease (IBD), as well as in saliva samples from healthy volunteers, were determined using the Vibrio harveyi BB170 bioluminescence assay. Because of genetic modifications, the BB170 strain only emits light when autoinducer-2 is added. Dose-response curves were obtained using 50 nM–0.5 mM dilutions of synthetic autoinducer-2 in saliva or stool matrix. The autoinducer-2 concentration was quantified in stool (n = 2, 2.76 μM), ileal washing samples (n = 1; 2.29 μM) and saliva from patients with IBD (n = 3, 2.05 μM) and saliva from healthy volunteers (n = 3; 1.39 μM) (Raut et al., 2013).
An overview of the observed in vivo concentrations is given in Table 3.
Acyl homoserine lactones
Chronic lung disease in cystic fibrosis patients is frequently associated with Pseudomonas aeruginosa biofilm formation in the lungs (Singh et al., 2000). Quorum sensing in P. aeruginosa is mediated via N-3-oxododecanoyl homoserine lactone and N-butyryl homoserine lactone (Pesci et al., 1997). N-3-oxododecanoyl homoserine lactone is indispensable for the establishment of a persistent P. aeruginosa infection; therefore a LC-MS method for the quantitation of this quorum sensing molecule in sputum of cystic fibrosis patients has been developed (LLOQ of 10 nM for 3-oxo-C12-HSL) (Struss et al., 2013). In total, 47 samples originating from 20 cystic fibrosis and 2 healthy donors were collected. Concerning the cystic fibrosis patients, 9 patients donated more than 1 sample, resulting in 34 samples from these patients obtained in total at different time points. Cystic fibrosis patients were allocated into different groups depending on their disease state. Besides patient samples, they also investigated the N-3-oxododecanoyl homoserine lactone concentration in P. aeruginosa in vitro cultured biofilms (n = 4) and found a mean concentration of 0.95 ± 0.68 μM. Regarding the patient samples, 45 out of 47 samples contained N-3-oxododecanoyl homoserine lactone in quantifiable concentrations with concentrations ranging between 20 to > 1,000 nM; the highest measured concentration was approximately 6,900 nM (Struss et al., 2013). These results are potentially clinically significant as N-3-oxododecanoyl homoserine lactone in a concentration range of 0.1–100 μM has shown to alter the human immune response (Telford et al., 1998b).
Barr et al. (2015) determined the concentration of several quorum sensing molecules in sputum, plasma and urine of 60 cystic fibrosis patients with chronic Pseudomonas aeruginosa infection at the beginning of a pulmonary exacerbation (Barr et al., 2015). All samples were analyzed by liquid chromatography—tandem mass spectrometry (LC-MS/MS). Eight different quorum sensing molecules were observed in sputum: 2-heptyl-4-hydroxyquinoline (HHQ), 2-nonyl-4-hydroxyquinoline (NHQ), 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO), 2-nonyl-4-hydroxyquinoline-N-oxide (NQNO), 2-heptyl-3-hydroxy-4(1H)-quinolone (C7-PQS), 2-nonyl-3-hydroxy-4(1H)-quinolone (C9-PQS), N-3-oxododecanoyl-L-homoserine lactone (3-oxo-C12-HSL) and N-butanoyl-L-homoserine lactone (C4-HSL). Five of them were also detected in plasma (HHQ, NHQ, HQNO, NQNO, and C7-PQS) and four in urine samples (HHQ, NHQ, HQNO, NQNO). Concentrations in sputum, plasma, and urine varied respectively between lower limit of quantification (LLOQ) (not specified)–4,302 nM, LLOQ (i.e., 0.01 nM)–2.744 nM and LLOQ (i.e., 0.01 nM)–12.511 nM and do correlate with the disease state of cystic fibrosis (Barr et al., 2015).
The concentration of 3-oxo-C12-HSL and C4-HSL in sputum samples of 12 patients with cystic fibrosis has been assessed. In order to determine the 3-oxo-C12-HSL concentration, a pKDT17 plasmid in E. coli, which contains a copy of the lasR gene and a lasB-lacZ fusion, was used. Concentrations of C4-HSL were determined by means of a pECP61.5 plasmid in P. aeruginosa, which contains a copy of the rhlR gene and a rhlA-lacZ fusion. The measured β-galactosidase activity in both reporter bacteria correlates with the levels of respectively 3-oxo-C12-HSL and C4-HSL. The average concentrations of 3-oxo-C12-HSL and C4-HSL were found to be respectively 5.04 and 1.04 nM with individual values ranging from < LOD (not specified) to 21.2 nM and < LOD (not specified) to 5 nM. However, determination of the extraction efficiency by adding synthetic autoinducers to sputum samples that do not contain autoinducer showed only 80% recovery of 3-oxo-C12-HSL and 60–65% recovery of C4-HSL, indicating loss of autoinducer during extraction in sample preparation (Erickson et al., 2002).
A summary of the reported concentrations of quorum sensing molecules in different matrices, together with the method used for detection and the corresponding reference is given in Table 3. To our knowledge, quorum sensing peptides have not yet been quantitated nor identified in biological samples from human origin.
Discussion
Detection methods
A variety of analytical techniques is currently available for many quorum sensing molecules, mainly chromatographic methods and reporter bacteria based biosensors. However, caution must be paid when considering these different methods regarding their analytical characteristics like selectivity. The reporter bacteria are based on the interaction of the molecule of interest with a receptor. Hence, structural analogs can possibly also interact with the receptor and consecutively might impede a straight-forward quantitative or qualitative selective analysis of individual quorum sensing molecules, a rather general concern that is also valid for other analytical techniques. For example, applying ELISA as a quantitative analytical method, cannot exclude cross-reactivity toward other structurally analog quorum sensing molecules that are currently not yet discovered. Since these molecules currently might not yet have been identified, it is very difficult to test for cross-reactivity toward these molecules. This can be illustrated by a variety of quorum sensing peptides listed in the Quorumpeps database: various synthetic structural analogs of certain quorum sensing peptides have been tested toward the bacteria receptive for the cognate quorum sensing peptide. The reactivity of the receptors is not solely limited toward synthetic analogs: the receptor(s) have been proven to be activated by various naturally occurring structural analogs (e.g., the N-acyl homoserine lactones). This potential pitfall can be omitted by coupling the biosensor with an initial chromatographic separation by e.g., thin layer chromatography. On the other hand, mass spectroscopy, especially in its targeted modi, allows additional selectivity via quorum sensing molecule fragmentation. Another important method consideration for qualitative and/or quantitative analysis of samples is the achievable limit of detection and its related, always higher, limit of quantification. This analytical characteristic is depending on the analytical method as such, including the sample preparation steps and is also compound-dependent. The latter being illustrated by the large range of limits of detection reported for various methods. The bacteria reporter based biosensors have in some specific cases proven to achieve limits of detection and limits of quantification in the sub-pM range. Chromatography coupled to mass spectroscopy can also achieve limits of detection and quantification in the sub-nM or even below. Hence, the preferred method of detection and/or quantification mainly depends on the desired selectivity and sensitivity for a specific quorum sensing molecule or group of molecules.
In vivo quorum sensing molecule concentrations
The in vivo presence and concentration of quorum sensing peptides have been neglected, while the other classes of quorum sensing molecules (e.g., the N-acyl homoserine lactones) have attracted considerable attention. The concentration of these molecules has been demonstrated in both healthy and diseased subjects. Concentrations of AI-2 were found to be in the high nM–low μM range and AHL concentrations were found in a comparable concentration range.
The lack of information related to the large group of quorum sensing peptides in in vivo samples cannot be longer justified. Peptides play an important role in all aspects of the immune, endocrine, and neuronal systems, and hence, the biological roles of quorum sensing peptides as produced by our microbiome are expected to be elucidated in the near future. Moreover, taking into consideration the increasing evidence that our microbiome is involved in a variety of biological host processes, both in health as in disease, these quorum sensing peptides could act as biomarkers, similar to recent suggestions of using microbiota as biomarkers (Eloe-Fadrosh and Rasko, 2013). This can be illustrated by the reports of quorum sensing molecules in cystic fibrosis patients (Singh et al., 2000).
Conclusion
Quorum sensing molecules are a heterogeneous group of molecules currently only started to be explored in in vivo situations. For example, to our knowledge, the bacterial quorum sensing peptides have sparsely qualitatively or quantitatively detected in vivo within humans. Conversely, the N-acyl homoserine lactones do have attracted considerable scientific interest. They have been quantified in vivo at concentrations ranging in the broad nM-μM range. Quorum sensing peptides have, together with the other classes of quorum sensing molecules, been detected and quantified in bacterial cell culture broths. The concentration of these quorum sensing molecules is highly variable and in vitro observed concentrations can reach almost mM concentrations.
A variety of analytical techniques has been described in literature to investigate the major quorum sensing molecule classes. Generally, reporter bacteria based biosensors and chromatography, especially LC(-MS/MS) are most frequently applied for qualitative and quantitative purposes. Selectivity and sensitivity are key analytical characteristics when considering quorum sensing molecule identification and quantification. Concerning selectivity, reporter bacteria based biosensors will potentially be confronted with cross-reactivity, and hence potential false-positives, due to currently unknown quorum sensing molecules that might cross-react with the specific quorum sensing molecule receptor. However, these reporter bacteria biosensors are, in some specific cases, able to achieve very low limits of detection (e.g., pM range), sometimes outperforming the LC(-MS/MS) methods. Opposite, LC-MS/MS methods potentially show a higher degree of selectivity due to both the chromatographic separation and the molecule fragmentation patterns via mass spectroscopy. Hence, the preferred method of analysis does depend on the quorum sensing molecule(s) of interest and the objectives put forward.
Evidence of the role of the human microbiome, both in health and disease, is continuously increasing. Quorum sensing molecules are bacterial languages, and apart from their fundamental biological importance in pathophysiological conditions, they can serve as potential prognostic or diagnostic biomarkers. However, the vast potential field of quorum sensing molecule-biomarkers, especially quorum sensing peptides, have not yet received the attention they deserve.
Author contributions
FV and BD conceived the idea for this manuscript, while FV and SD wrote the manuscript, FV, SD, ND, YJ, EW, CV, and BD made substantial contributions to data interpretation and drafted/revised the manuscript. FV, SD, ND, YJ, EW, CV, and BD approved this manuscript and agree to be held accountable for all aspects of this manuscript.
Funding
FV is funded by the “Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen)” (Grant number 131356 to FV). ND is funded by “Fonds wetenschappelijk onderzoek-Vlaanderen (Fund Scientific Research-Flanders)” (Grant number 1S21017N to ND). IWT-Vlaanderen or Fonds wetenschappelijk onderzoek-Vlaanderen were not involved in any part of the study, and had no role in the conception, contributions to the consent nor in manuscript writing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer SOF and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review
References
- Andersen J. B., Heydorn A., Hentzer M., Eberl L., Geisenberger O., Christensen B. B., et al. (2001). gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67, 575–585. 10.1128/AEM.67.2.575-585.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y., Hiramoto T., Nishino R., Aiba Y., Yoshihara K., Koga Y., et al. (2012). Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G1288–G1295. 10.1152/ajpgi.00341.2012 [DOI] [PubMed] [Google Scholar]
- Bala A., Gupta R. K., Chhibber S., Harjai K. (2013). Detection and quantification of quinolone signalling molecule: a third quorum sensing molecule of Pseudomonas aeruginosa by high performance-thin layer chromatography. J. Chromatogr. B. 930, 30–35. 10.1016/j.jchromb.2013.04.027 [DOI] [PubMed] [Google Scholar]
- Baldrich E., Mu-oz F. X., García-Aljaro C. (2011). Electrochemical detection of quorum sensing signaling molecules by dual signal confirmation at microelectrode arrays. Anal. Chem. 83, 2097–2103. 10.1021/ac1028243 [DOI] [PubMed] [Google Scholar]
- Balkwill F., Mantovani A. (2001). Inflammation and cancer: back to Virchow? Lancet 357, 539–545. 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- Bandyopadhaya A., Kesarwani M., Que Y. A., He Y., Padfield K., Tompkins R., et al. (2012). The quorum sensing volatile molecule 2-amino-acetophenon modulates host immune responses in a manner that promotes life with unwanted guests. PLOS Pathog. 11:e1003024. 10.1371/journal.ppat.1003024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber C. E., Tang J. L., Feng J. X., Pan M. Q., Wilson T. J., Slater H., et al. (1997). A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. 10.1046/j.1365-2958.1997.3721736.x [DOI] [PubMed] [Google Scholar]
- Barr H. L., Halliday N., Cámara M., Barrett D. A., Williams P., Forrester D. L., et al. (2015). Pseudomonas aeruginosa quorum sensing molecules correlate with clinical status in cystic fibrosis. Eur. Respir. J. 46, 1046–1054. 10.1183/09031936.00225214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E., Ross R. P., O'Toole P. W., Fitzgerald G. F., Stanton C. (2012). Gamma-aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 113, 411–417. 10.1111/j.1365-2672.2012.05344.x [DOI] [PubMed] [Google Scholar]
- Bassler B. L. (1999). How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2, 582–587. 10.1016/S1369-5274(99)00025-9 [DOI] [PubMed] [Google Scholar]
- Baxter N. T., Zackular J. P., Chen G. Y., Schloss P. D. (2014). Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2:20. 10.1186/2049-2618-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleza I., Peirce M. J., Minelli A. (2014). Cyclic dipeptides: from bugs to brain. Trends Mol. Med. 20, 551–558. 10.1016/j.molmed.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Beloin C., Renard S., Ghigo J. M., Lebeaux D. (2014). Novel approaches to combat bacterial biofilms. Curr. Opin. Pharmacol. 18C, 61–68. 10.1016/j.coph.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Bultman S. J. (2014). Emerging roles of the microbiome in cancer. Carcinogenesis 35, 249–255. 10.1093/carcin/bgt392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna S. R., Gooding J. R., May A. L. (2009). Direct Quantitation of the quorum sensing signal, autoinducer-2, in clinically relevant samples by Liquid chromatography-tandem mass spectrometry. Anal. Chem. 81, 6374–6381. 10.1021/ac900824j [DOI] [PubMed] [Google Scholar]
- Castro-Nallar E., Bendall M. L., Pérez-Losada M., Sabuncyan S., Severance E. G., Dickerson F. B., et al. (2015). Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls. PeerJ 3:e1140. 10.7717/peerj.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi T. R. I., Bianco G., Fonseca J., Schmitt-Kopplin P. (2013). Perceiving the chemical language of Gram-negative bacteria: listening by high-resolution mass spectrometry. Anal. Bioanal. Chem. 405, 493–507. 10.1007/s00216-012-6371-2 [DOI] [PubMed] [Google Scholar]
- Cataldi T. R. I., Bianco G., Palazzo L., Quaranta V. (2007). Occurrence of N-acyl-L-homoserine lactones in extracts of some Gram-negative bacteria evaluated by gas chromatography–mass spectrometry. Anal. Biochem. 361, 226–235. 10.1016/j.ab.2006.11.037 [DOI] [PubMed] [Google Scholar]
- Charlesworth J., Kimyon O., Manefield M., Burns B. P. (2015). Detection and characterization of N-acyl-L-homoserine lactones using GFP-based biosensors in conjunction with thin-layer chromatography. J. Microbiol. Methods 118, 164–167. 10.1016/j.mimet.2015.09.012 [DOI] [PubMed] [Google Scholar]
- Charlton T. S., de Nys R., Netting A., Kumar N., Hentzer M., Givskov M., et al. (2000). A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography-mass spectrometry: application to a model bacterial biofilm. Environ. Microbiol. 2, 530–541. 10.1046/j.1462-2920.2000.00136.x [DOI] [PubMed] [Google Scholar]
- Chen X., Buddrus-Schiemann K., Rothballer M., Krämer P. M., Hartmann A. (2010b). Detection of quorum sensing molecules in Burkholderia cepacia culture supernatants with enzyme-linked immunosorbent assays. Anal. Bioanal. Chem. 398, 2669–2676. 10.1007/s00216-010-4045-5 [DOI] [PubMed] [Google Scholar]
- Chen X., Kremmer E., Gouzy M. F., Clausen E., Starke M., Wöllner K., et al. (2010a). Development and characterization of rat monoclonal antibodies for N-acylated homoserine lactones. Anal. Bioanal. Chem. 398, 2655–2667. 10.1007/s00216-010-4017-9 [DOI] [PubMed] [Google Scholar]
- Chhabra S. R., Harty C., Hooi D. S. W., Daykin M., Williams P., Telford G., et al. (2003). Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl-L-homoserine lactone as immune modulators. J. Med. Chem. 46, 97–104. 10.1021/jm020909n [DOI] [PubMed] [Google Scholar]
- Chowers M. Y., Keller N., Tal R., Barshack I., Lang R., Bar-Meir S., et al. (1999). Human gastrin: a Helicobacter pylori-specific growth factor. Gastroenterology 117, 1113–1118. 10.1016/S0016-5085(99)70396-3 [DOI] [PubMed] [Google Scholar]
- Clarke G., Stilling R. M., Kennedy P. J., Stanton C., Cryan J. F., Dinan T. G. (2014). Minireview: gut microbiota: the neglected endocrine organ. Mol. Endocrinol. 28, 1221–1238. 10.1210/me.2014-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley M. A., Whittall C., Rolph M. S. (2010). Pseudomonas signal molecule 3-oxo-C12-homoserine lactone interferes with binding of rosiglitazone to human PPARgamma. Microbes. Infect. 12, 231–237. 10.1016/j.micinf.2009.12.009 [DOI] [PubMed] [Google Scholar]
- Cryan J. F., O'Mahony S. M. (2011). The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol. Motil. 23, 187–192. 10.1111/j.1365-2982.2010.01664.x [DOI] [PubMed] [Google Scholar]
- Darkoh C., DuPont H. L., Norris S. J., Kaplan H. B. (2015). Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 6, e02569–e02514. 10.1128/mBio.02569-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Spiegeleer B., Verbeke F., D'Hondt M., Hendrix A., Van De Wiele C., Burvenich C., et al. (2015). The quorum sensing peptides PhrG, CSP and EDF promote angiogenesis and invasion of breast cancer cells in vitro. PLOS ONE 10:e0119471. 10.1371/journal.pone.0119471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Spiegeleer B., Wynendaele E., Bracke N., Veryser L., Taevernier L., Degroote A., et al. (2016). Regulatory development of geriatric medicines: to GIP or not to GIP? Ageing Res. Rev. 27, 23–36. 10.1016/j.arr.2016.02.004 [DOI] [PubMed] [Google Scholar]
- DiMango E., Zar H. J., Bryan R., Prince A. J. (1995). Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. Clin. Invest. 96, 2204–2210. 10.1172/JCI118275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnick R., Wu Q., Angelino N. J., Stephanie L. V., Chow K. C., Sufrin J. R., et al. (2005). Enhancement of 5-fluorouracil sensitivity by an rTS signaling mimic in H630 colon cancer cells. J. Cancer Res. 65, 5917–5924. 10.1158/0008-5472.CAN-05-0431 [DOI] [PubMed] [Google Scholar]
- Dunny G. M., Craig R. A., Carron R. L., Clewell D. B. (1979). Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2, 454–465. 10.1016/0147-619X(79)90029-5 [DOI] [PubMed] [Google Scholar]
- Eloe-Fadrosh E. A., Rasko D. A. (2013). The human microbiome: from symbiosis to pathogenesis. Annu. Rev. Med. 64, 145–163. 10.1146/annurev-med-010312-133513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. (1984). Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U.S.A. 81, 4154–4158. 10.1073/pnas.81.13.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S. E., Poutahidis T. (2014). The microbiome modulates the macroenvironment. Oncoimmunology 3:e28271. 10.4161/onci.28271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson D. L., Endersby R., Kirkham A., Stuber K., Vollman D. D., Rabin H. R., et al. (2002). Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 70, 1783–1790. 10.1128/IAI.70.4.1783-1790.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P. B., Gold B. D. (2000). The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and cancer. Annu. Rev. Microbiol. 54, 615–640. 10.1146/annurev.micro.54.1.615 [DOI] [PubMed] [Google Scholar]
- Fekete A., Frommberger M., Rothballer M., Li X., Englmann M., Fekete J., et al. (2007). Identification of bacterial N-acylhomoserine lactones (AHLs) with a combination of ultra-performance liquid chromatography (UPLC), ultra-high-resolution mass spectrometry, and in-situ biosensors. Anal. Bioanal. Chem. 387, 455–467. 10.1007/s00216-006-0970-8 [DOI] [PubMed] [Google Scholar]
- Finegold S. M., Dowd S. E., Gontcharova V., Liu C., Henley K. E., Wolcott R. D., et al. (2010). Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 16, 444–453. 10.1016/j.anaerobe.2010.06.008 [DOI] [PubMed] [Google Scholar]
- Flavier A. B., Clough S. J., Schell M. A., Denny T. P. (1997). Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26, 251–259. 10.1046/j.1365-2958.1997.5661945.x [DOI] [PubMed] [Google Scholar]
- Freestone P. E., Sandrini S. M., Haigh R. D., Lyte M. (2008). Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. 16, 55–64. 10.1016/j.tim.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Frommberger M., Hertkorn N., Englmann M., Jakoby S., Hartmann A., Kettrup A., et al. (2005). Analysis of N-acylhomoserine lactones after alkaline hydrolysis and anion-exchange solid-phase extraction by capillary zone electrophoresis-mass spectrometry. Electrophoresis 26, 1523–1532. 10.1002/elps.200410365 [DOI] [PubMed] [Google Scholar]
- Frommberger M., Schmitt-Kopplin P., Menzinger F., Albrecht V., Schmid M., Eberl L., et al. (2003). Analysis of N-acyl-L-homoserine lactones produced by Burkholderia cepacia with partial filling micellar electrikinetic chrormatgraphy – electrospray ionization-ion trap mass spectrometry. Electrophoresis 24, 3067–3074. 10.1002/elps.200305567 [DOI] [PubMed] [Google Scholar]
- Frommberger M., Schmitt-Kopplin P., Ping G., Frisch H., Schmid M., Zhang Y., et al. (2004). A simple and robust set-up for on-column sample preconcentration –nano-liquid chromatography – electrospray ionization mass spectrometry for the analysis of N-acylhomoserine lactones. Anal. Bioanal. Chem. 378, 1014–1020. 10.1007/s00216-003-2400-5 [DOI] [PubMed] [Google Scholar]
- Fuqua W. C., Winans S., Greensberg E. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K., Kikuchi T., Tokue Y., Fujimura S., Uehara A., Takada H., et al. (2006). Mouse and human cell activation by N-dodecanoyl-DL-homoserine lactone, a Chromobacterium violaceum auto-inducer. Infect. Immun. 74, 7029–7031. 10.1128/IAI.00038-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregus P., Vlcková H., Buchta V., Kestranek J., Krivcíková L., Nováková L. (2010). Ultra high performance liquid chromatography tandem mass spectrometry analysis of quorum-sensing molecules of Candida albicans. J. Pharm. Biomed. Anal. 53, 674–681. 10.1016/j.jpba.2010.05.029 [DOI] [PubMed] [Google Scholar]
- Gupta R. K., Chhibber S., Harjai K. (2013). Quorum sensing signal molecules cause renal tissue inflammation through local cytokine responses in experimental UTI caused by Pseudomonas aeruginosa. Immunobiology 218, 181–185. 10.1016/j.imbio.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Haas C. F., Eakin R. M., Konkle M. A., Blank R. (2014). Endotracheal tubes: old and new. Respir. Care 59, 933–952. 10.4187/respcare.02868 [DOI] [PubMed] [Google Scholar]
- Håvarstein L. S., Hakenbeck R., Gaustad P. (1997). Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179, 6589–6594. 10.1128/jb.179.21.6589-6594.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. A., Pomianek M. E., Kraml C. M., Taylor R. K., Semmelhack M. F., Bassler B. L. (2007). The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450, 883–886. 10.1038/nature06284 [DOI] [PubMed] [Google Scholar]
- Holden M. T., Chhabra S. R., de Nys R., Stead P., Bainton N. J., Hill P. J., et al. (1999). Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33, 1254–1266. [DOI] [PubMed] [Google Scholar]
- Holzer P., Reichmann F., Farzi A. (2012). Neuropeptide, Y, peptide YY and pancreatic polypeptide in the gut-brain axis. Neuropeptides 46, 261–274. 10.1016/j.npep.2012.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooi D. S. W., Bycroft B. W., Chhabra S. R., Williams P., Pritchard D. I. (2004). Differential immune modulatory activity of Pseudomonas aerigunosa quorum-sensing signal molecules. Infect. Immun. 72, 6463–6470. 10.1128/IAI.72.11.6463-6470.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. T., Sperandio V. (2008). Inter-kingdom signaling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6, 111–120. 10.1038/nrmicro1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike Y., Craig R. A., Yagi Y., Clewell D. B. (1983). Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. U.S.A. 80, 5369–5373. 10.1073/pnas.80.17.5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahoor A., Patel R. J., Bryan A., Do C., Krier J., Watters C., et al. (2008). Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J. Bacteriol. 190, 4408–4415. 10.1128/JB.01444-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Ling Z., Zhang Y., Mao H., Ma Z., Yin Y., et al. (2015). Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 48, 186–194. 10.1016/j.bbi.2015.03.016 [DOI] [PubMed] [Google Scholar]
- Junio H. A., Todd D. A., Ettefagh K. A., Ehrmann B. M., Kavanaugh J. S., Horswill A. R., et al. (2013). Quantitative analysis of autoinducing peptide I (AIP-I) from Staphylococcus aureus cultures using ultrahigh performance liquid chromatography–high resolving power mass spectrometry. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 930, 7–12. 10.1016/j.jchromb.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V. C. (2013). Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31, 224–245. 10.1016/j.biotechadv.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Karavolos M. H., Bulmer D. M., Spencer H., Rampioni G., Schmalen I., Baker S., et al. (2011). Salmonella typhi sense host neuroendocrine stress hormones and release the toxin haemolysin E. EMBO Rep. 12, 252–258. 10.1038/embor.2011.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavolos M. H., Spencer H., Bulmer D. M., Thompson A., Winzer K., Williams P., et al. (2008). Adrenaline modulates the global transcriptional profile of Salmonella revealing a role in the antimicrobial peptide and oxidative stress resistance responses. BMC Genomics 9:458. 10.1186/1471-2164-9-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavolos M. H., Winzer K., Williams P., Khan C. M. (2013). Pathogen espionage: multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 87, 455–465. 10.1111/mmi.12110 [DOI] [PubMed] [Google Scholar]
- Kaufmann G. F., Sartorio R., Lee S. H., Mee J. M., Altobell L. J. I. I. I., Kujawa D. P., et al. (2006). Antibody interference with N-acyl homoserine lactone-mediated bacterial quorum sensing. J. Am. Chem. Soc. 128, 2802–2803. 10.1021/ja0578698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. M., Sperandio V. (2016). What a dinner party! Mechanisms and functions of interkingdom signaling in host-pathogen associations. mBio 7:e01748-15. 10.1128/mBio.01748-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I., Hazan R., Gaathon A., Carmeli S., Engelberg-Kulka H. (2007). A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318, 652–654. 10.1126/science.1147248 [DOI] [PubMed] [Google Scholar]
- Kravchenko V. V., Kaufmann G. F., Mathison J. C., Scott D. A., Katz A. Z., Wood M. R., et al. (2006). N-(3-oxo-acyl)homoserine lactones signal cell activation through a mechanism distinct from the canonical pathogen-associated molecular pattern recognition receptor pathways. J. Biol. Chem. 281, 28822–28830. 10.1074/jbc.M606613200 [DOI] [PubMed] [Google Scholar]
- Kravchenko V. V., Kaufmann G. F., Mathison J. C., Scott D. A., Katz A. Z., Grauer D. C., et al. (2008). Modulation of gene expression via disruption of NF-kappaB signaling by a bacterial small molecule. Science 321, 259–263. 10.1126/science.1156499 [DOI] [PubMed] [Google Scholar]
- Kubesová A., Horká M., Ružička F., Slais K., Glatz Z. (2010). Separation of attogram terpenes by the capillary zone electrophoresis with fluorometric detection. J. Chromatogr. A 1217, 7288–7292. 10.1016/j.chroma.2010.09.038 [DOI] [PubMed] [Google Scholar]
- Kumar S., Kolodkin-Gal I., Engelberg-Kulka H. (2013). Novel quorum-sensing peptides mediating interspecies bacterial cell death. mBio 4, 1–12 10.1128/mBio.00314-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A., Pasini P., Daunert S. (2008). Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples. Anal. Bioanal. Chem. 391, 1619–1627. 10.1007/s00216-008-2002-3 [DOI] [PubMed] [Google Scholar]
- La Rosa S. L., Solheim M., Diep D. B., Nes I. F., Brede D. A. (2015). Bioluminescence based biosensors for quantitative detection of enterococcal peptide-pheromone activity reveal inter-strain telesensing in vivo during polymicrobial systemic infection. Sci. Rep. 5:8339. 10.1038/srep08339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang L., Ye L., Mao Y., Xie X., Xia C., et al. (2009). Influence of Pseudomonas aeruginosa quorum sensing signal molecule N-(3-oxododecanoyl) homoserine lactone on mast cells. Med. Microbiol. Immunol. 198, 113–121. 10.1007/s00430-009-0111-z [DOI] [PubMed] [Google Scholar]
- Li L., Hooi D., Chhabra S. R., Pritchard D., Shaw P. E. (2004). Bacterial N-acylhomoserine lactone-induced apoptosis in breast carcinoma cells correlated with down-modulation of STAT3. Oncogene 23, 4894–4902. 10.1038/sj.onc.1207612 [DOI] [PubMed] [Google Scholar]
- Li X., Fekete A., Englmann M., Götz C., Rothballer M., Frommberger M., et al. (2006). Development and application of a method for the analysis of N-acylhomoserine lactones by solid-phase extraction and ultra high pressure liquid chromatography. J. Chromatogr. A 1134, 186–193. 10.1016/j.chroma.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Li Z., Nair S. K. (2012). Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals? Protein Sci. 21, 1403–1417. 10.1002/pro.2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyte M. (1993). The role of microbial endocrinology in infectious disease. J. Endocrinol 137, 343–345. [DOI] [PubMed] [Google Scholar]
- Lyte M. (2004). Microbial endocrinology and infectious disease in the 21th century. Trends Microbiol. 12, 14–20. 10.1016/j.tim.2003.11.004 [DOI] [PubMed] [Google Scholar]
- Lyte M., Ernst S. (1993). Alpha and beta adrenergic receptor involvement in catecholamine-induced growth of Gram-negative bacteria. Biochem. Biophys. Res. Commun. 190, 447–452. 10.1006/bbrc.1993.1068 [DOI] [PubMed] [Google Scholar]
- Lyte M., Vulchanova L., Brown D. R. (2011). Stress at the intestinal surface: catecholamines and mucosa-bacteria interactions. Cell Tissue Res. 343, 23–32. 10.1007/s00441-010-1050-0 [DOI] [PubMed] [Google Scholar]
- Malik A. K., Fekete A., Gebefuegi I., Rothballer M., Schmitt-Kopplin P. (2009). Single drop microextraction of homoserine lactones based quorum sensing signal molecules, and the separation of their enantiomers using gas chromatography mass spectrometry in the presence of biological matrices. Microchim. Acta 166, 101–107. 10.1007/s00604-009-0183-x [DOI] [Google Scholar]
- Marchesan J. T., Morelli T., Moss K., Barros S. P., Ward M., Jenkins W., et al. (2015). Association of synergistetes and cyclodipeptides with periodontitis. J. Dent. Res. 94, 1425–1431. 10.1177/0022034515594779 [DOI] [PubMed] [Google Scholar]
- Mayer M. L., Sheridan J. A., Blohmke C. J., Turvey S. E., Hancock R. E. W. (2011). The Pseudomonas autoinducer 3O-C12 homoserine lactone provokes hyperinflammatory responses from cystic fibrosis airway epithelial cells. PLOS ONE 6:e16246. 10.1371/journal.pone.0016246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab R., Lamont R. J. (2003). Microbial dinner-party conversations: the role of LuxS in interspecies communication. J. Med. Microbiol. 52, 541–545. 10.1099/jmm.0.05128-0 [DOI] [PubMed] [Google Scholar]
- Mello A. M., Paroni G., Daragjata J., Pilotto A. (2016). Gastrointestinal microbiota and their contribution to healthy aging. Dig. Dis. 34, 194–201. 10.1159/000443350 [DOI] [PubMed] [Google Scholar]
- Miyairi S., Tateda K., Fuse E. T., Ueda C., Saito H., Takabatake T., et al. (2006). Immunization with 3-oxododecanoyl-L-homoserine lactone-protein conjugate protects mice from lethal Pseudomonas aeruginosa lung infection. J. Med. Microbiol. 55, 1381–1387. 10.1099/jmm.0.46658-0 [DOI] [PubMed] [Google Scholar]
- Moos W. H., Faller D. V., Harpp D. N., Kanara I., Pernokas J., Powers W. R., et al. (2016). Microbiota and neurological disorders: a gut feeling. Biores Open Access. 5, 137–145. 10.1089/biores.2016.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Isogai A., Sakagami Y., Fujino M., Kitada C., Clewell D. B., et al. (1986b). Isolation and structure of the Streptococcus-faecalis sex-pheromone inhibitor, iAD1, that is excreted by the donor strain harboring plasmid pAD1. Agric. Biol. Chem. 50, 539–541. 10.1271/bbb1961.50.539 [DOI] [Google Scholar]
- Mori M., Sakagami Y., Ishii Y., Isogai A., Kitada C., Fujino M., et al. (1988). Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 263, 14574–14578. [PubMed] [Google Scholar]
- Mori M., Sakagami Y., Narita M., Isogai A., Fujino M., Kitada C., et al. (1984). Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett. 178, 97–100. 10.1016/0014-5793(84)81248-X [DOI] [PubMed] [Google Scholar]
- Mori M., Tanaka H., Sakagami Y., Isogai A., Fujino M., Kitada C., et al. (1986a). Isolation and structure of the Streptococcus faecalis sex pheromone, cAM373. FEBS Lett. 206, 69–72. 10.1016/0014-5793(86)81342-4 [DOI] [PubMed] [Google Scholar]
- Mori M., Tanaka H., Sakagami Y., Isogai A., Kitada C., Clewell D. B., et al. (1987). Isolation and structure of the sex pheromone inhibitor, iPD1, excreted by Streptococcus faecalis donor strains harboring plasmid pPD1. J. Bacteriol. 169, 1747–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin D., Grasland B., Vallée-Réhel K., Dufau C., Haras D. (2003). On-line high-performance liquid chromatography–mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 1002, 79–92. 10.1016/S0021-9673(03)00730-1 [DOI] [PubMed] [Google Scholar]
- Murray B. E., An F. Y., Clewell D. B. (1988). Plasmids and pheromone response of the beta-lactamase producer Streptococcus (Enterococcus) faecalis HH22. Antimicrob. Agents Chemother. 32, 547–551. 10.1128/AAC.32.4.547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Abe Y., Ono Y., Isogai A., Suzuki A. (1995a). Isolation and structure of the Enterococcus faecalis sex pheromone, cOB1, that induces conjugal transfer of the hemolysin/bacteriocin plasmids, pOB1 and pYI1. Biosci. Biotechnol. Biochem. 59, 703–705. 10.1271/bbb.59.703 [DOI] [PubMed] [Google Scholar]
- Nakayama J., Ono Y., Suzuki A. (1995b). Isolation and structure of the sex pheromone inhibitor, iAM373, of Enterococcus faecalis. Biosci. Biotechnol. Biochem. 59, 1358–1359. 10.1271/bbb.59.1358 [DOI] [PubMed] [Google Scholar]
- Nakayama J., Ruhfel R. E., Dunny G. M., Isogai A., Suzuki A. J. (1994). The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J. Bacteriol. 176, 7405–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseribafrouei A., Hestad K., Avershina E., Sekelja M., Linløkken A., Wilson R., et al. (2014). Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil. 26, 1155–1162. 10.1111/nmo.12378 [DOI] [PubMed] [Google Scholar]
- Nealson K. H., Platt T., Hastings J. W. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K., Hastings J. W. (1979). Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T., Nes I. F., Holo H. (1998). An exported inducer peptide regulates bacteriocin production in Enterococcus faecium CTC492. J. Bacteriol. 180, 1848–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor G., Knecht L. D., Salgado N., Strobel S., Pasini P., Daunert S. (2016). Whole-cell biosensors as tools for the detection of quorum-sensing molecules: uses in diagnostics and the investigation of the quorum-sensing mechanism. Adv. Biochem. Eng. Biotechnol. 154, 181–200. 10.1007/10_2015_337 [DOI] [PubMed] [Google Scholar]
- Oleskin A. V., El'-registan G. I., Shenderov B. A. (2016). Role of neuromediators in the functioning of the human microbiota: “business talks” among microorganisms and the microbiota-host dialogue. Microbiology 85, 1–22. 10.1134/S0026261716010082 [DOI] [PubMed] [Google Scholar]
- Oliver C. M., Schaefer A. L., Greenberg E. P., Sufrin J. R. (2009). Microwave synthesis and evaluation of phenacylhomoserine lactones as anticancer compounds that minimally activate quorum sensing pathways in Pseudomonas aeruginosa. J. Med. Chem. 52, 1569–1575. 10.1021/jm8015377 [DOI] [PubMed] [Google Scholar]
- O'Mahony S. M., Clarke G., Borre Y. E., Dinan T. G., Cryan J. F. (2015). Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 277, 32–48. 10.1016/j.bbr.2014.07.027 [DOI] [PubMed] [Google Scholar]
- Ortori C. A., Dubern J. F., Chhabra S. R., Cámara M., Hardie K., Williams P., et al. (2011). Simultaneous quantitative profiling of N-acyl-l-homoserine lactone and 2-alkyl-4(1H)-quinolone families of quorum-sensing signaling molecules using LC-MS/MS. Anal. Bioanal. Chem. 399, 839–850. 10.1007/s00216-010-4341-0 [DOI] [PubMed] [Google Scholar]
- Pacheco A. R., Sperandio V. (2009). Inter-kingdom signaling: chemical language between bacteria and host. Curr. Opin. Microbiol. 12, 192–198. 10.1016/j.mib.2009.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh P. J., Nayi V. R., Johnson D. A., Vinik A. I. (2016). The Role of gut microflora and the cholinergic anti-inflammatory neuroendocrine system in diabetes mellitus. Front. Endocrinol. 7:55. 10.3389/fendo.2016.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N. M., Moore J. D., Blackwell H. E., Amador-Noguez D. (2016). Identification of unanticipated and novel N-acyl L-homoserine lactones (AHLs) using a sensitive non-targeted LC-MS/MS method. PLOS ONE 11:e0163469. 10.1371/journal.pone.0163469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearman W. F., Lawrence-Snyder M., Angel S. M., Decho A. W. (2007). Surface-enhanced raman spectroscopy for in situ measurements of signaling molecules (autoinducers) relevant to bacteria quorum sensing. Appl. Spectrosc. 61, 1295–1300. 10.1366/000370207783292244 [DOI] [PubMed] [Google Scholar]
- Pereira C. S., Thompson J. A., Xavier K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. 10.1111/j.1574-6976.2012.00345.x [DOI] [PubMed] [Google Scholar]
- Pesci E. C., Milbank J. B. J., Pearson J. P., McKnight S., Kende A. S., Greenberg E. P., et al. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96, 11229–11234. 10.1073/pnas.96.20.11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci E. C., Pearson J. P., Seed P. C., Iglewski B. H. (1997). Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179, 3127–3132. 10.1128/jb.179.10.3127-3132.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen F. C., Pecharki D., Scheie A. A. (2004). Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J. Bacteriol. 186, 6327–6331. 10.1128/JB.186.18.6327-6331.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi G., Masala L., Lannelli F., Manganelli R., Havarstein L. S., Piccoli L., et al. (1996). Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178, 6087–6090. 10.1128/jb.178.20.6087-6090.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard D. (2006a). Immune modulation by Pseudomonas aeruginosa quorum-signal molecules. Int. J. Med. Microbiol. 296, 111–1166. 10.1016/j.ijmm.2006.01.037 [DOI] [PubMed] [Google Scholar]
- Pritchard Q. I. (2006b). Immune modulation by Pseudomonas aeruginosa quorum-sensing signal molecules. Int. J. Med. Microbiol. 296, 111–116. 10.1016/j.ijmm.2006.01.037 [DOI] [PubMed] [Google Scholar]
- Procaccini C., Pucino V., De Rosa V., Marone G., Matarese G. (2014). Neuro-endocrine networks controlling immune system in health and disease. Front. Immunol. 5:143. 10.3389/fimmu.2014.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai N., Rai R., Venkatesh K. V. (2015). Quorum Sensing Biosensors, in Quorum Sensing vs. Quorum Quenching: A Battle with No End in Sight, ed Kalia V. C. (New Delhi: Springer; ), 173–183. [Google Scholar]
- Rasko D. A., Moreira C. G., Li D. R., Reading N. C., Ritchie J. M., Waldor M. K., et al. (2008). Targeting QsecC signaling and virulence for antibiotic development. Science 321, 1078–1080. 10.1126/science.1160354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raut N., Pasini P., Daunert S. (2013). Deciphering bacterial universal language by detecting the quorum sensing signal, autoinducer-2, with a whole-cell sensing system. Anal. Chem. 85, 9604–9609. 10.1021/ac401776k [DOI] [PubMed] [Google Scholar]
- Rickard A. H., Campagna S. R., Kolenbrander P. E. (2008). Autoinducer-2 is produced in saliva-fed flow conditions relevant to natural oral biofilms. J. Appl. Microbiol. 105, 2096–2103. 10.1111/j.1365-2672.2008.03910.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard A. H., Palmer R. J., Jr., Blehert D. S., Campagna S. R., Semmelhack M. F., Egland P. G., et al. (2006). Autoinducer 2, a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60, 1446–1456. 10.1111/j.1365-2958.2006.05202.x [DOI] [PubMed] [Google Scholar]
- Rutherford S. T., Bassler B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, 1–25. 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P., Dow J. M. (2008). Diffusible signals and interspecies communication in bacteria. Microbiology 154, 1845–1858. 10.1099/mic.0.2008/017871-0 [DOI] [PubMed] [Google Scholar]
- Shaw P. D., Ping G., Daly S. L., Cha C., Cronan J. E., Jr., Rinehart K. L., et al. (1997). Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. U.S.A. 94, 6036–6041. 10.1073/pnas.94.12.6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifri C. D. (2008). Quorum sensing: bacteria talk sense. Clin. Infect. Dis. 47, 1070–1076. 10.1086/592072 [DOI] [PubMed] [Google Scholar]
- Singh P. K., Schaefer A. L., Parsek M. R., Moninger T. O., Welsh M. J., Greenberg E. P. (2000). Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764. 10.1038/35037627 [DOI] [PubMed] [Google Scholar]
- Skindersoe M. E., Zeuthen L. H., Brix S., Fink L. N., Lazenby J., Whittall C., et al. (2009). Pseudomonas aeruginosa quorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol. Med. Microbiol. 55, 335–345. 10.1111/j.1574-695X.2008.00533.x [DOI] [PubMed] [Google Scholar]
- Smith R. S., Fedyk E. R., Springer T. A., Mukaida N., Iglewski B. H., Phipps R. P. (2001). IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappa B and activator protein-2. J. Immunol. 167, 366–374. 10.4049/jimmunol.167.1.366 [DOI] [PubMed] [Google Scholar]
- Smith R. S., Harris S. G., Phipps R., Iglewski B. (2002a). The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 184, 1132–1139. 10.1128/jb.184.4.1132-1139.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. S., Kelly R., Iglewski B. H., Phipps R. P. (2002b). The Pseudomonas autoinducer N-(3-oxododecanoyl)homoserine lactone induces cyclooxygenase-2 and prostaglandin E2 production in human lung fibroblasts: implications for inflammation. J. Immunol. 169, 2636–2642. 10.4049/jimmunol.169.5.2636 [DOI] [PubMed] [Google Scholar]
- Spencer H., Karavolos M. H., Bulmer D. M., Aldridge P., Chhabra S. R., Winzer K., et al. (2010). Genome-wide transposon mutagenesis identifies a role for host neuroendocrine stress hormones in regulating the expression of virulence genes in T. typhimurium. J. Bacteriol. 192, 714–724. 10.1128/JB.01329-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V., Torres A. G., Jarvis B., Nataro J. P., Kaper J. B. (2003). Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U.S.A. 100, 8951–8956. 10.1073/pnas.1537100100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler L., Venturi V. (2007). Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. Fems Microbiol. Lett. 266, 1–9. 10.1111/j.1574-6968.2006.00501.x [DOI] [PubMed] [Google Scholar]
- Stilling R. M., van de Wouw M., Clarke G., Stanton C., Dinan T. G., Cryan J. F. (2016). The neuropharmacology of butyrate: the bread and butter of the microbiota-gut-brain axis. Neurochem. Int. 99, 110–132. 10.1016/j.neuint.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Struss A. K., Nunes A., Waalen J., Lowery C. A., Pullanikat P., Denery J. R., et al. (2013). Toward implementation of quorum sensing autoinducers as biomarkers for infectious disease states. Anal. Chem. 85, 3355–3362. 10.1021/ac400032a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturme M. H. J., Kleerebezem M., Nakayama J., Akkermans A. D. L., Vaughan E. E., de Vos W. M. (2002). Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 81, 233–243. 10.1023/A:1020522919555 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Mori M., Sakagami Y., Isogai A., Fujino M., Kitada C., et al. (1984). Isolation and structure of bacterial sex pheromone, cPD1. Science 226, 849–850. 10.1126/science.6436978 [DOI] [PubMed] [Google Scholar]
- Syvitski R. T., Tian X. L., Sampara K., Salman A., Lee S. F., Jakeman D. L., et al. (2007). Structure-activity analysis of quorum-sensing signaling peptides from Streptococcus mutans. J. Bacteriol. 189, 1441–1450. 10.1128/JB.00832-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga M. E., Bassler B. L. (2003). Chemical communication among bacteria. Proc. Natl. Acad. Sci. U.S.A. 100, 14549–14554. 10.1073/pnas.1934514100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda K., Ishii Y., Horikawa M., Matsumoto T., Miyairi S., Pechere J. C., et al. (2003). The Pseudomonas aeruginosa autoinducer N-3-dodecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect. Immun. 71, 5785–5793. 10.1128/IAI.71.10.5785-5793.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford G., Wheeler D., Williams P., Tomkins P. T., Appleby P., Sewell H., et al. (1998a). The Pseudomonas aeruginosa Quorum-sensing signal molecule N-(3-oxadodecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect. Immun. 66, 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford G., Wheeler D., Williams P., Tomkins P. T., Appleby P., Sewell H., et al. (1998b). The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-1-homoserine lactone has immunomodulatory activity. Infect. Immun. 66, 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M., Mathesius U., Rumbaugh K. P. (2011). Perception and degradation of N-acyl homoserine lactone quorum sensing signals by mammalian and plant cells. Chem. Rev. 111, 100–116. 10.1021/cr100045m [DOI] [PubMed] [Google Scholar]
- Thaiss C. A., Zmora N., Levy M., Elinav E. (2016). The microbiome and innate immunity. Nature 535, 65–74. 10.1038/nature18847 [DOI] [PubMed] [Google Scholar]
- Thiel V., Vilchez R., Sztajer H., Wagner-Döbler I., Schulz S. (2009). Identification, quantification, and determination of the absolute configuration of the bacterial quorum-sensing signal autoinducer-2 by gas chromatography–mass spectrometry. ChemBioChem 10, 479–485. 10.1002/cbic.200800606 [DOI] [PubMed] [Google Scholar]
- Thomas G. L., Böhner C. M., Williams H. E., Walsh C. M., Ladlow M., Welch M., et al. (2006). Immunomodulatory effects of Pseudomonas aeruginosa quorum sensing small molecule probes on mammalian macrophages. Mol. Biosyst. 2, 132–137. 10.1039/B517248A [DOI] [PubMed] [Google Scholar]
- Tian X., Syvitski R. T., Liu T., Livingstone N., Jakeman D. L., Li Y. H. (2009). A method for structure-activity analysis of quorum-sensing signaling peptides from naturally transformable Streptococci. Biol. Proced. Online 11, 207–226. 10.1007/s12575-009-9009-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd D. A., Zich D. B., Ettefagh K. A., Kavanaugh J. S., Horswill A. R., Cech N. B. (2016). Hybrid Quadrupole-Orbitrap mass spectrometry for quantitative measurement of quorum sensing inhibition. J. Microbiol. Methods 127, 89–94. 10.1016/j.mimet.2016.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomova A., Husarova V., Lakatosova S., Bakos J., Vlkova B., Babinska K., et al. (2015). Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 138, 179–187. 10.1016/j.physbeh.2014.10.033 [DOI] [PubMed] [Google Scholar]
- van der Meer J. R., Belkin S. (2010). Where microbiology meets microengineering: desing and applications of reporter bacteria. Nat. Rev. Microbiol. 8, 511–522. 10.1038/nrmicro2392 [DOI] [PubMed] [Google Scholar]
- Vikström E., Magnusson K. E., Pivoriünas A. (2005). The Pseudomonas aerigunosa quorum-sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone stimulates phagocytic activity in human macrophages through the p38 MAPK pathway. Microbes Infect. 7, 1512–1518. 10.1016/j.micinf.2005.05.012 [DOI] [PubMed] [Google Scholar]
- Wagner C., Zimmermann S., Brenner-Weiss G., Hug F., Prior B., Obst U., et al. (2007). The quorum-sensing molecule N-3-oxododecanoyl homoserine lactone (3OC12-HSL) enhances the host defence by activating human polymorphonuclear neutrophils (PMN). Anal. Bioanal. Chem. 387, 481–487. 10.1007/s00216-006-0698-5 [DOI] [PubMed] [Google Scholar]
- Wang J., Quan C., Wang X., Zhao P., Fan S. (2011). Extraction, purification and identification of bacterial signal molecules based on N-acyl homoserine lactones. Microb. Biotechnol. 4, 479–490. 10.1111/j.1751-7915.2010.00197.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Perez L. J., Ng W. L., Semmelhack M. F., Bassler B. L. (2011). Mechanism of Vibrio cholerae autoinducer-1 biosynthesis. ACS Chem. Biol. 6, 356–365. 10.1021/cb1003652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynendaele E., Bronselaer A., Nielandt J., D'Hondt M., Stalmans S., Bracke N., et al. (2013). Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides. Nucleic Acids Res. 41, D655–D659. 10.1093/nar/gks1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynendaele E., Verbeke F., D'Hondt M., Hendrix A., Van De Wiele C., Burvenich C., et al. (2015a). Crosstalk between the microbiome and cancer cells by quorum sensing peptides. Peptides 64, 40–48. 10.1016/j.peptides.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Wynendaele E., Verbeke F., Stalmans S., Gevaert B., Janssens Y., Van De Wiele C., et al. (2015b). Quorum sensing peptides selectively penetrate the blood-brain barrier. PLoS ONE 10:e0142071. 10.1371/journal.pone.0142071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K., Kaneko H., Yamamoto S., Konagaya T., Kusugami K., Mitsuma T. (1998). Inhibitory effect of somatostatin on Helicobacter pylori proliferation in vitro. Gastroenterology 115, 1123–1130. 10.1016/S0016-5085(98)70083-6 [DOI] [PubMed] [Google Scholar]
- Zackular J. P., Baxter N. T., Iverson K. D., Sadler W. D., Petrosino J. F., Chen G. Y., et al. (2013). The gut microbiome modulates colon tumorigenesis. mBio 4, e00692–e00613. 10.1128/mBio.00692-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Ye B. C. (2014). Real-time measurement of quorum-sensing signal autoinducer 3OC6HSL by a FRET-based nanosensor. Bioprocess Biosyst. Eng. 37, 849–855. 10.1007/s00449-013-1055-7 [DOI] [PubMed] [Google Scholar]
- Zhou L., Glennon J. D., Luong J. H., Reen F. J., O'Gara F., McSweeney C., et al. (2011). Detection of the Pseudomonas Quinolone Signal (PQS) by cyclic voltammetry and amperometry using a boron doped diamond electrode. Chem. Commun. 47, 10347–10349. 10.1039/c1cc13997e [DOI] [PubMed] [Google Scholar]
- Zhou L., Reen F. J., O'Gara F., McSweeney C. M., Clarke S. L., Glennon J. D., et al. (2012). Analysis of pseudomonas quinolone signal and other bacterial signalling molecules using capillaries coated with highly charged polyelectrolyte monolayers and boron doped diamond electrode. J. Chromatogr. A 1251, 169–175. 10.1016/j.chroma.2012.06.064 [DOI] [PubMed] [Google Scholar]
- Zimmermann S., Wagner C., Müller W., Brenner-Weiss G., Hug F., Prior B., et al. (2006). Induction of neutrophil chemotaxis by the quorum-sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone. Infect. Immun. 74, 5687–5692. 10.1128/iai.01940-05 [DOI] [PMC free article] [PubMed] [Google Scholar]