Abstract
Extended-spectrum β-lactamases (ESBLs) emerge by point mutation from non-extended-spectrum precursors. The aims of this study were to reveal the basis for variations in resistance levels found in a collection of 21 Klebsiella pneumoniae clinical isolates from Brisbane, Australia. Previous studies have shown that 20 of these isolates possess blaSHV-11, blaSHV-2a, and/or blaSHV-12, and there is an association between the copy numbers of the ESBL-encoding genes and resistance levels. In this study, a real-time PCR method for interrogating the polymorphic sites at codons 238 and 240 was developed, and this confirmed the relationship between mutant gene copy numbers and resistance levels. The blaSHV promoter region was cloned from one of the ESBL-expressing isolates, and this showed that blaSHV genes exist downstream of two different promoters within this single isolate. These promoters have both been reported previously, and they differ by virtue of the presence or absence of an IS26 insertion. The blaSHV copy numbers in cis with the different promoters were measured, and the copy number of the IS26 promoter was correlated with resistance levels. Cloning and analysis of PCR products showed that different blaSHV variants existed in cis with individual promoters in individual isolates but that mutant genes were more abundant downstream of the IS26 promoter. There were no ESBL-positive isolates without this promoter. It was concluded that blaSHV in cis with the IS26 promoter is located on an amplifiable replicon, and the presence of the IS26 insertion may facilitate the acquisition of an ESBL-positive phenotype.
SHV β-lactamases are prevalent in gram-negative bacteria (2). SHV-1 can hydrolyze penicillin and cephalosporins but not expanded-spectrum antibiotics such as oxyimino cephalosporins and monobactams. The introduction and widespread use of expanded-spectrum antibiotics has led to the appearance of extended spectrum β-lactamases (ESBLs) which are able to hydrolyze these compounds (1).
Numerous variants of the SHV and TEM enzymes have been reported (www.lahey.org/studies/webt.stm). Variation is exclusively in the form of amino acid substitutions (11, 13). Jacoby (7) has assembled data from studies on the effects of substitutions on activity. It appears that conversion of a non-ESBL SHV enzyme to an ESBL is nearly always caused by a G238S substitution, while a further extension of spectrum and increased enzyme activity can be conferred by an E240K substitution. Although many other substitutions have been reported, it appears that for the SHV family sites 238 and 240 are the most important for acquiring ESBL activity.
Previously, we have reported the study of a collection of 21 Klebsiella pneumoniae isolates that included 13 isolates that expressed SHV ESBLs (6). The SHV β-lactamase genes identified in this collection encoded SHV-1 and SHV-11 (non-ESBL), SHV-2a (ESBL with a G238S mutation), and SHV-12 (ESBL with G238S and E240K mutations). SHV-11, SHV-2a, and SHV-12 differ from SHV-1, SHV-2, and SHV-5, respectively, at codon 35, where there is an L35Q substitution. Limited cloning of PCR products and isoelectric focusing suggested that most if not all of the isolates contain multiple SHV β-lactamase-encoding genes. A single-base extension method for interrogating the polymorphic sites in codons 238 and 240 was developed, and this revealed a strong correlation between the level of resistance to expanded-spectrum β-lactam antibiotics and the relative copy numbers of the different blaSHV alleles (6).
The coexistence of different blaSHV alleles within individual isolates is of considerable practical significance because, as has been noted by others (3, 12, 19), it raises questions about the validity of surveys in which blaSHV genes are amplified from genome extracts and the primary PCR product is identified by sequencing. Genes with both the codon 238 and codon 240 mutations would be expected to be dominant (as opposed to recessive) over single-mutation or unmutated genes with respect to resistance to expanded spectrum β-lactams. It is easy to envisage a situation in which an ESBL-encoding mutant gene is in a small minority in comparison to a non-ESBL-encoding homolog and yet confers the clinically relevant resistance phenotype. Therefore, the first part of this study was designed to confirm the results of the minisequencing-based allele copy number measurements by using a newly developed real-time PCR assay for allele copy number ratios and also by carrying out extensive cloning and sequencing of blaSHV-derived PCR products. In the second part of this study, we characterized the blaSHV promoters and investigated whether different promoters could coexist in single isolates, whether homologous recombination equilibrates blaSHV across different replicons, which promoters are associated with copy number variation, and whether our results are consistent with previous studies of the epidemiology of the SHV-11,2a,12 family. In the course of this work we developed a suite of real-time PCR-based methods for directly determining blaSHV copy numbers, and these will likely prove to be useful in future studies of the natural history of SHV ESBLs.
MATERIALS AND METHODS
Bacterial isolates.
The 21 isolates of K. pneumoniae used in this study were isolated at the Princess Alexandra Hospital in Brisbane, Australia, and have been previously characterized (6, 17). All strains were cultured in Luria-Bertani (LB) broth and stored in cryovials with 12% glycerol at −80°C.
DNA extraction.
DNA was extracted from 2.5-ml cultures grown overnight in LB broth. For each isolate, 1 ml of culture was centrifuged for 2 min at room temperature. The supernatant was discarded, and the pellet was washed twice in TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]). The pellet was then resuspended in 500 μl of TE buffer and boiled for 20 min. The lysed cells were then centrifuged for 2 min, and the supernatant was removed and stored at −20°C.
Detection of mutations in bla genes by kinetic PCR.
All reactions were performed with an ABI Prism 7000 real-time PCR device (Applied Biosystems). Allele-specific PCR primers were designed to interrogate single-nucleotide polymorphisms (SNPs) within the blaSHV gene. PCR was performed in 96-well optical plates (Applied Biosystems) with a total reaction volume of 20 μl. SYBR-Green 1 2× master mix (Applied Biosystems) was used for all reactions, with 5 pmol of common primer, 5 pmol of either the wild-type-specific primer or mutant-specific primer, and 1 μl of bacterial cell lysate. The thermocycling parameters were as follows: an initial denaturation step of 95°C for 60 s, 40 cycles of denaturation at 94°C for 15 s, primer annealing at 60°C for 20 s, and extension and optical read at 72°C for 30 s. The point at which the fluorescence reaches a defined threshold defines a cycle number, called the CT value. The threshold for each reaction plate was set in the logarithmic phase of amplification.
The sequence and annealing sites of allele-specific primers used are shown in Table 1. All primers were designed by using Primer Express version 2.0 from Applied Biosystems and have a theoretical melting temperature of 60°C. The codon 238 SNP was interrogated by using Shv238mt and Shv238wt as the allele-specific primers and Shv238 reverse as the common primer. The codon 240 SNP was interrogated by using Shv240mt and Shv240wt as the allele-specific primers and Shv240 reverse as the common primer.
TABLE 1.
Primers for kinetic and relative quantitation PCR.
| Primer name | Gene | Annealing region | Primer sequence (5′→3′)a |
|---|---|---|---|
| 16sAllBactF | 16s rRNA | (+) 1297/1318 | TCCATGAAGTCGGAATCGCTAG |
| 16sAllBactR | 16s rRNA | (−) 1374/1392 | CACTCCCATGGTGTGACGG |
| SHV-F | blaSHV | (+) 310/327 | TCAGCGAAAAACACCTTG |
| Pr-F | pNSF-1b | (+) −107/−82 | GATGAAGGAAAAAAGAGGAATTGTGA |
| SHVquantF | blaSHV | (+) 730/748 | TGCTTGGCCCGAATAACAA |
| SHVquantR | blaSHV | (−) 766/786 | GCGTATCCCGCAGATAAATCA |
| Pr::IS26-F | pMPA2ac | (+) −158/−141 | CCGGCCTTTGAATGGGTT |
| Prquant-R | blaSHV | (−) −3/24 | TAATACACAGGCGAATATAACGCATAAC |
| Shv238mt | blaSHV | (+) 680/699 | CGCCGATAAGACCGGAGCTA |
| Shv238wt | blaSHV | (+) 680/699 | CGCCGATAAGACCGGAGCTG |
| Shv238reverse | blaSHV | (−) 770/781 | CGGCGTATCCCGCAGATAA |
| Shv240mt | blaSHV | (−) 702/716 | GCGCGCACCCCGCTT |
| Shv240wt | blaSHV | (−) 702/716 | GCGCGCACCCCGCTC |
| Shv240reverse | blaSHV | (+) 663/679 | CCGGCGGGCTGGTTTAT |
| Shv35mt | blaSHV | (+) 70/91 | AGCCGCTTGAGCAAATTAAACA |
| Shv35wt | blaSHV | (+) 70/91 | AGCCGCTTGAGCAAATTAAACT |
| Shv35reverse | blaSHV | (−) 174/194 | CATCATGGGAAAGCGTTCATC |
Determination of sequence flanking the blaSHV gene in isolate I1.
A gene library of isolate I1 was constructed by using the λ EMBL3 Packagene system (Promega) in accordance with the manufacturer's instructions. Resultant plaques were transferred to nitrocellulose filters for screening with nucleic acid probes. Digoxigenin (DIG)-labeled blaSHV PCR product was generated by using primer set SHV-F/Shv238reverse (Table 1) with DIG-labeled deoxynucleoside triphosphate mix (Roche). Southern hybridizations with DIG-labeled PCR product were then performed by using the DIG Easy Hyb kit (Roche), anti-DIG antibody (Roche), and CDP-star (Roche) according to the manufacturer's instructions. Hybridizing plaques were selected, and lysate preparations were made by the liquid culture method (16). Recombinant λ DNA was isolated by a miniprep procedure according to the instructions for the λ EMBL Packagene system (Promega). Purified recombinant λ was restriction digested with SalI, BamHI, and EcoRI and subjected to double digestion with SalI-BamHI, SalI-EcoRI, and BamHI/EcoRI. Restriction digests were subject to gel electrophoresis with a 0.7% agarose gel and Tris-borate-EDTA buffer (40 V for 20 h). The gels were then transferred to nylon membranes (16) and probed with DIG-labeled blaSHV. Restriction fragments that contained sequence complementary to the blaSHV probe and were of suitable size for cloning and sequencing were cloned into appropriately digested pBlueScriptII. White colonies were selected and grown overnight in LB-ampicillin broth, and the recombinant plasmid was purified by the alkaline lysis plasmid miniprep procedure (16). The recombinant plasmid inserts were then sequenced by using primer-walking methods. The complete insert sequences were compared to all known sequences by using the BLASTn program (National Center for Biotechnology Information).
Cloning and analysis of blaSHV genes and gene fragments.
PCR amplicons were generated from isolates B2, K2, J3, D1, L1, A1, J2, and I1 by using primer sets that amplify all blaSHV variants (SHV-F/Shv238reverse) and primer sequences that amplify the 5′ end of blaSHV together with upstream flanking sequence (Pr-F/Shv238reverse and Pr::IS26-F/Shv238reverse) (Table 1). Amplification products were cloned by using the pGEM-T Easy plasmid kit (Promega) and JM109 high-efficiency electrocompetent Escherichia coli cells prepared as described by Sambrook and Russell (16). Twenty clones were selected for each strain, and plasmid miniprep isolations were carried out for use in subsequent sequencing and real-time PCR interrogation analysis. Plasmid miniprep solutions diluted 400-fold were used as templates for analysis of the codon 238 and codon 240 polymorphic sites. This was done by kinetic PCR with the method described above.
Sequence determination.
Cloned plasmid DNA was purified by using QIAquick miniprep kits (QIAGEN), and plasmid DNA was quantified by UV spectrophotometry. Plasmid DNA (200 to 500 ng) was sequenced by using 3.2 pmol of the appropriate primer. Sequencing was performed at the Australian Genome Research Facility, University of Queensland, Brisbane, Australia.
Relative quantitation by real-time PCR.
Real-time PCR was used to determine the interisolate relative copy numbers of total blaSHV and also blaSHV in cis with different flanking sequences. The comparative CT method (ABI PRISM 7000 sequence detection system user bulletin 2; Applied Biosystems) was used with the 16S RNA-encoding gene as the standard. All measurements were carried out in triplicate. The procedure was validated by making a twofold dilution of the genomic preparation from isolate I1 and determining that for all four primer sets the slopes of CT versus log dilution fell between −0.1 and +0.1.
While the comparative CT method can be used to estimate the absolute ratio between the copy numbers of the same target in different isolates, it cannot be used to determine the absolute copy number ratio of different targets. The approach taken to estimating this parameter was to assume that the total amount of blaSHV in any given isolate is equal to the amount of blaSHV in cis with flanking sequence 1 plus the amount of blaSHV in cis with flanking sequence 2 (see Results and Discussion). The relative copy number data from different isolates were then used to generate simultaneous equations, the solution of which provided an estimation of the absolute copy number ratios of the different targets.
The primers used are shown in Table 1 and are fully specified in Results and Discussion.
Typing by PFGE.
The isolates were grown on Mueller-Hinton II agar at 37°C for 24 h. A single colony from each isolate was then selected and grown in 3 ml of Mueller-Hinton II broth at 37°C for 20 h with gentle agitation. The control strain used for pulsed-field gel electrophoresis (PFGE) experiments was K. pneumoniae ATCC 13883. DNA was extracted from isolates, digested with XbaI, and subjected to PFGE with the GenePath group 6 reagent kit (Bio-Rad) according to the manufacturer's instructions.
RESULTS AND DISCUSSION
Interrogation of the codon 238 and 240 polymorphic sites using kinetic PCR.
The first aim of this study was to develop a single-step real-time PCR method for interrogating the blaSHV codon 238 and 240 polymorphisms. This was done to determine if the minisequencing-based relative copy number determinations by Howard et al. (6) could be confirmed and to facilitate experiments designed to elucidate the molecular basis for the allele copy number variations. The approach used was allele-specific real-time PCR, which is also known as kinetic PCR (5). This exploits the reduced extension frequency from a mismatched 3′ end of a PCR primer. It is more inherently robust than conventional, end-point-analyzed allele-specific PCR because it depends upon a difference between the allele-specific reactions with respect to the thermocycle number at which amplimer is detectable (the ΔCT) rather than a difference between the amounts of amplimer in the allele-specific reactions at the reaction end points. With equal copy numbers of the two alleles, each amplification product should reach a detectable level at the same CT value. If an unequal amount of an allele is present, the difference in the cycle number (or ΔCT value) between the two amplification reactions provides a measure of relative gene dosages of each allele.
All 21 isolates were subjected to three real-time assays with each allele-specific primer set, and the ΔCT values and their standard deviations were calculated (Table 2). It is clear that the isolates may be divided into distinct clusters. There is a discontinuity in the codon 238 results that separates the double-disk synergy test (DDST)-negative isolates (ΔCT of <−4.14) from the DDST-positive isolates (ΔCT of >−0.65). A second discontinuity in the codon 240 results separates the high-resistance (i.e., high drug MIC) DDST-positive isolates (ΔCT of >−2.46) from the low-resistance DDST-positive and DDST-negative isolates (ΔCT of <−5.91). In the case of the high-resistance isolates (L1, J2, C1, J1, G1, L1, and H1), there was a clear trend for the higher ΔCT values to be associated with the highest resistance, although the codon 240 ΔCT for isolate C1 is unexpectedly high. Despite the result for isolate C1, the correlation coefficient for these isolates for the log MIC of aztreonam (chosen because of the greatest range) versus the sum of the codon 238 and codon 240 ΔCT values is 0.73. The results are entirely consistent with the previously reported minisequencing data (5) and confirm that relationship between blaSHV-12 copy numbers and resistance levels.
TABLE 2.
Kinetic PCR results
| Isolatea | Phenotypic datab
|
Kinetic PCRc at codon:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DDST | MIC (μg/ml)
|
238
|
240
|
|||||||||
| ATM | CAZ | CTX |
CT
|
ΔCT
|
CT
|
ΔCT
|
||||||
| Mutant | Wild type | Mean | SD | Mutant | Wild type | Mean | SD | |||||
| B2 | − | 0.03 | 0.13 | 0.03 | 22.0 | 16.0 | −4.9 | 1.22 | 25.1 | 17.7 | −6.6 | 0.82 |
| K2 | − | 0.03 | 0.13 | 0.03 | 21.7 | 14.5 | −7.0 | 0.33 | 23.8 | 13.1 | −10.4 | 0.55 |
| M1 | − | 0.03 | 0.13 | 0.03 | 24.1 | 18.2 | −6.8 | 0.71 | 28.7 | 20.1 | −7.3 | 2.67 |
| K1 | − | 0.06 | 0.50 | 0.06 | 21.8 | 15.1 | −6.7 | 0.27 | 24.1 | 17.8 | −5.9 | 0.52 |
| J5 | − | 0.06 | 0.50 | 0.13 | 24.6 | 16.1 | −8.0 | 1.68 | 22.3 | 13.1 | −9.1 | 0.39 |
| J3 | − | 0.13 | 0.50 | 0.13 | 22.6 | 17.5 | −4.1 | 1.16 | 21.0 | 14.4 | −6.5 | 0.78 |
| L2 | − | 0.13 | 0.25 | 0.25 | 26.4 | 17.2 | −8.3 | 1.30 | 24.1 | 15.0 | −8.5 | 1.01 |
| J4 | − | 0.25 | 2.00 | 0.13 | 21.0 | 15.1 | −6.0 | 0.25 | 21.3 | 11.7 | −9.3 | 0.31 |
| F2 | + | 0.25 | 0.5 | 0.5 | 16.4 | 17.9 | 1.3 | 0.50 | 24.7 | 13.6 | −10.6 | 0.75 |
| A1 | + | 0.5 | 1 | 1 | 15.2 | 16.7 | 1.4 | 0.31 | 23.3 | 12.9 | −10.0 | 0.58 |
| E1 | + | 0.5 | 1 | 1 | 15.9 | 19.2 | 2.2 | 1.23 | 24.8 | 13.7 | −9.7 | 1.39 |
| B1 | + | 1 | 1 | 1 | 16.3 | 17.8 | 1.3 | 0.20 | 23.1 | 14.1 | −8.8 | 0.42 |
| F1 | + | 1 | 1 | 1 | 15.2 | 16.6 | 1.4 | 0.28 | 22.0 | 13.3 | −9.0 | 0.76 |
| D1 | + | 2 | 4 | 4 | 14.2 | 17.0 | 2.9 | 0.71 | 22.4 | 12.5 | −9.8 | 1.05 |
| L1 | + | 32 | 32 | 1 | 15.3 | 16.4 | 1.5 | 0.46 | 15.0 | 13.7 | −0.9 | 0.43 |
| J2 | + | 64 | 32 | 2 | 15.9 | 16.0 | 0.2 | 0.38 | 14.7 | 13.0 | −1.8 | 0.49 |
| C1 | + | 64 | 64 | 4 | 16.1 | 16.0 | −0.7 | 0.48 | 14.9 | 18.4 | 4.1 | 0.68 |
| J1 | + | 64 | 32 | 4 | 16.6 | 16.4 | −0.2 | 0.22 | 15.5 | 14.0 | −2.5 | 1.48 |
| G1 | + | 128 | 128 | 16 | 12.9 | 15.2 | 2.7 | 0.37 | 11.7 | 14.5 | 2.8 | 0.19 |
| I1 | + | 128 | 128 | 16 | 13.0 | 15.4 | 2.3 | 0.59 | 11.7 | 14.7 | 3.4 | 0.28 |
| H1 | + | 128 | 128 | 128 | 11.6 | 14.3 | 2.7 | 0.47 | 10.6 | 14.8 | 3.9 | 0.45 |
Designations are as in reference 6.
The DDST and MIC data are from reference 17. The highest concentration tested when the isolates were characterized (17) was 128 μg/ml, so some MICs may be higher than this. ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime.
Shown are data from single replicates of CT measurements from each isolate and the mean ΔCT and associated standard deviation from all replicates.
Direct determination of allele copy number ratios by analysis of cloned blaSHV PCR products.
In order to provide additional confirmation that the kinetic PCR procedure provided a true indication of the blaSHV allelic dosage within an isolate, we carried out cloning and analysis of blaSHV PCR products from isolates B2, K2, J3, A1, D1, J2, L1, and I1. blaSHV internal sequences spanning codons 238 and 240 were amplified from total cellular DNA. The amplified material was ligated to pGEM-T and transformed into E. coli JM109. Initially, inserts in five clones derived from each isolate in the total blaSHV PCR group were sequenced and also subjected to the kinetic PCR assays. Both methods clearly identified the same bases at the codon 238 and 240 polymorphic sites, and it was concluded that the real-time PCR procedure was suitable for screening of more clones. Fifteen more clones from each isolate were analyzed by this method.
The results of clone interrogation are shown in Table 3. These are consistent with the data in Table 2. Isolates B2, K2, and J3 are DDST negative and, as expected, possessed no mutant alleles. Isolate A1 is one of the lowest-resistance DDST-positive isolates and, as expected, had a high proportion of wild-type alleles and no double-mutant alleles. Isolate D1 has a slightly higher level of resistance than A1, and, consistent with this, a lower proportion of wild-type clones were recovered. Interestingly, this isolate also yielded a small number of double-mutation clones, despite having an intermediate resistance phenotype. Isolate J2 is at the low-resistance end of the group of isolates that have previously been shown to possess the double mutation. This isolate did not yield any clones of the single-mutation allele. Rather, the majority of clones were wild type and a substantial minority were of the double-mutation allele. Although this result was somewhat unexpected, it is consistent with both the high-resistance phenotype and also the significantly negative ΔCT value obtained from the interrogation of the codon 238 mutation site (Table 4). Isolate L1 has a resistance phenotype similar to that of J2. The distribution of clones is also similar, with the only differences being a slightly lower proportion of wild-type alleles and the presence of a small number of single-mutation alleles. Finally, isolate I1 has one of the highest-resistance phenotypes. As expected, the majority of the clones recovered were of the double-mutation allele. These data confirmed that there is a correlation between relative allele copy numbers and resistance phenotypes. In addition, they reveal a complex pattern of variation from isolate to isolate and suggest that different mixes of alleles can, on occasion, give rise to similar resistance levels.
TABLE 3.
blaSHV allele distributions found in a diverse subset of isolates, as determined by analysis of cloned PCR products
| Isolate | % blaSHV allelesa
|
||
|---|---|---|---|
| 238-G, 240-E | 238-S, 240-E | 238-S, 240-K | |
| B2 | 100 | ||
| K2 | 100 | ||
| J3 | 100 | ||
| D1 | 25 | 65 | 10 |
| L1 | 60 | 5 | 35 |
| A1 | 50 | 50 | |
| J2 | 80 | 20 | |
| I1 | 10 | 25 | 65 |
The deduced gene products are shown.
TABLE 4.
Relative quantitations of total blaSHV and blaSHV in cis with the different promoters and PFGE
| Isolate | DDST | MIC (μg/ml)a
|
Mean copy number (range)
|
Pulsotype | ||||
|---|---|---|---|---|---|---|---|---|
| ATM | CAZ | CTX | blaSHV | pr::IS26-blaSHV | pr-blaSHV | |||
| B2 | − | 0.03 | 0.13 | 0.03 | 0.396 (0.356-0.440) | Nilb | 0.082 (0.075-0.090) | D |
| K2 | − | 0.03 | 0.13 | 0.03 | 1.00 (0.929-1.076) | 0.897 (0.783-1.028) | 0.102 (0.089-0.117) | A |
| M1 | − | 0.03 | 0.13 | 0.03 | 0.444 (0.360-0.548) | Nil | 0.128 (0.102-0.161) | C |
| K1 | − | 0.06 | 0.5 | 0.06 | 0.331 (0.302-0.363) | Nil | 0.178 (0.150-0.211) | B |
| J5 | − | 0.06 | 0.5 | 0.13 | 0.696 (0.627-0.773) | 0.417 (0.395-0.441) | 0.175 (0.163-0.188) | A4 |
| J3 | − | 0.13 | 0.5 | 0.13 | 0.289 (0.258-0.324) | Nil | 0.197 (0.161-0.242) | A2 |
| L2 | − | 0.13 | 0.25 | 0.25 | 0.395 (0.303-0.515) | Nil | 0.124 (0.101-1.53) | A5 |
| J4 | − | 0.25 | 2 | 0.13 | 1.875 (1.657-2.122) | 1.986 (1.642-2.402) | 0.072 (0.053-0.96) | A2 |
| F2 | + | 0.25 | 0.5 | 0.5 | 0.785 (0.643-0.958) | 0.313 (0.294-0.332) | 0.099 (0.094-0.105) | A |
| A1 | + | 0.5 | 1 | 1 | 0.794 (0.716-0.879) | 0.433 (0.391-0.480) | 0.189 (0.160-0.224) | A |
| E1 | + | 0.5 | 1 | 1 | 0.664 (0.570-0.774) | 0.613 (0.503-0.746) | 0.197 (0.138-0.282) | A6 |
| B1 | + | 1 | 1 | 1 | 0.655 (0.547-0.785) | 0.427 (0.384-0.475) | 0.150 (0.132-0.172) | A5 |
| F1 | + | 1 | 1 | 1 | 0.707 (0.661-0.757) | 0.369 (0.307-0.445) | 0.084 (0.068-0.105) | A |
| D1 | + | 2 | 4 | 4 | 1.701 (1.556-1.860) | 1.721 (1.490-1.987) | 0.085 (0.069-0.104) | A1 |
| L1 | + | 32 | 32 | 1 | 0.809 (0.689-0.949) | 0.319 (0.254-0.400) | 0.087 (0.079-0.096) | A |
| J2 | + | 64 | 32 | 2 | 1.692 (1.552-1.848) | 1.277 (1.064-1.533) | 0.060 (0.018-0.206) | A5 |
| C1 | + | 64 | 64 | 4 | 0.835 (0.752-0.927) | 0.613 (0.508-0.739) | 0.133 (0.114-0.155) | B |
| J1 | + | 64 | 32 | 4 | 0.968 (0.793-1.182) | 1.32 (1.16-1.51) | 0.124 (0.092-0.167) | A3 |
| G1 | + | 128 | 128 | 16 | 2.955 (2.498-3.496) | 3.854 (3.418-4.347) | 0.158 (0.122-0.206) | A |
| I1 | + | 128 | 128 | 16 | 4.616 (4.203-5.070) | 5.241 (4.863-5.648) | 0.093 (0.086-0.102) | A |
| H1 | + | 128 | 128 | 128 | 8.244 (7.628-8.909) | 8.111 (6.776-9.709) | 0.124 (0.102-0.150) | A1 |
ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime.
On occasion, amplimer appeared very late in the cycling protocol, i.e., at least seven cycles later than for the isolates with measurable pr::IS26-blaSHV. This was not reproducible (i.e., on most occasions the PCRs were entirely negative) and was probably due to contamination.
Characterization of sequences flanking blaSHV in isolate I1.
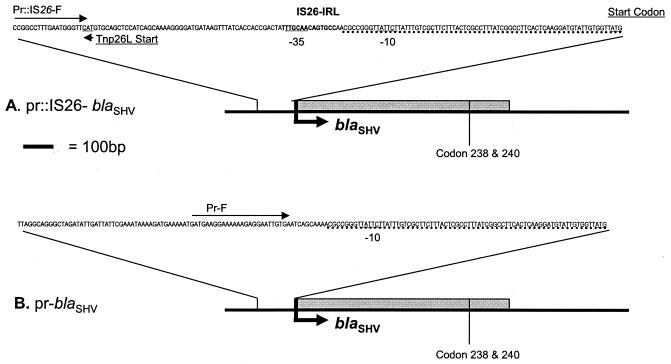
To further understand the molecular basis for the coexistence of different blaSHV alleles, sequences upstream from blaSHV were cloned and characterized. Isolate I1 was selected, as this isolate is DDST positive and highly resistant. To avoid prejudging the nature of the upstream sequences, a cloning-based approach (as opposed to PCR) was used. A λEMBL3 library of partially Sau3A-cleaved DNA was constructed and screened by plaque hybridization with a blaSHV-derived probe. Two hybridizing plaques, λI1-3 and λI1-9, were isolated, and their genomes were purified. Restriction analysis revealed that the inserts were 22 to 24 kb in size. There was no discernible overlap; i.e., there was no commonality of restriction fragments. Southern hybridization indicated that both inserts carry blaSHV and confirmed that the two inserts represent different genetic environments for blaSHV. Hybridizing fragments were subcloned and sequenced and were found to be identical to the two previously described blaSHV promoter regions. The cloned fragment from I1-9 was found to contain an IS26 element insertion next to the −10 upstream region, as has been reported for plasmid pMPA2a (10, 14). The cloned fragment from I1-3 did not contain an IS26 element and was homologous to the prototype SHV-1 upstream region (14, 15). In the interests of clarity, we have termed the two promoter regions in cis with blaSHV, pr::IS26-blaSHV and pr-blaSHV (Fig. 1). The genomes of an additional eight hybridizing phage were subjected to restriction analysis, and they were all identical to λI1-3.
FIG. 1.
pr::IS26-blaSHV (A) and pr-blaSHV (B). Identical sequences in the promoter regions are underlined with dotted lines. The pr::IS26-blaSHV cassette contains the IS26 element inserted next to the −10 region (15). The pr-blaSHV cassette does not contain IS26 element and is identical to the prototype SHV-1 upstream region (15).
Interisolate relative copy numbers of blashv variants.
The results described to this point show that the level of resistance is a function of the gene dosage of blaSHV mutant alleles. However, given that we had found that blaSHV genes can exist in two different genetic environments in individual isolates, it was of interest to know whether the genetic environment dictates the likelihood that a blaSHV gene will confer a high level of resistance. Accordingly, the strategy for the remainder of this study was to use real-time PCR to determine the interisolate relative copy numbers of total blaSHV and also blaSHV in cis with the two different promoters and to use cloning and analysis of PCR products to determine the distributions of blaSHV alleles located in cis with the different promoters in a subset of isolates.
The real-time PCR primers used are shown in Table 1. The 16S RNA-encoding genes were amplified by using primers 16sAllBactF and 16sAllBactR. Total blaSHV was measured by using SHVquant and SHVquantR. pr::IS26-blaSHV was measured by using Pr::IS26-F and Prquant-R. pr-blaSHV was measured by using Pr-F and Prquant-R.
In order to meaningfully measure relative copy numbers by using real-time PCR, several normalization steps were required. In summary, the 16S RNA-encoding gene was chosen as the reference point to control for variability in the genome extraction efficiencies, and isolate K2 was selected to be the calibrator isolate. The quantity of each target in each isolate was expressed as the difference in ΔCT between the 16S RNA gene and the target. The interisolate copy number ratios were then calculated from the difference between these ΔCT values for the different isolates. At this point the data were normalized such that the quantity of each target in the calibrator isolate was 1. To provide an approximate indication of the relative copy numbers of different targets within individual isolates, the data were analyzed further. This was accomplished by solving simultaneous equations derived from the data from isolates K2 and H1, which were chosen because they spanned a wide range of pr::IS26-blaSHV copy numbers. These equations stated that, for both of these isolates, the copy number of the total blaSHV equaled the copy number of pr-blaSHV plus pr::IS26-blaSHV. The solution to these equations was that in isolate K2 there are 8.8 times more copies of pr::IS26-blaSHV than there are of pr-blaSHV. This value was then used to normalize the copy numbers of the other targets in all of the other isolates, and this is what is shown in Table 4.
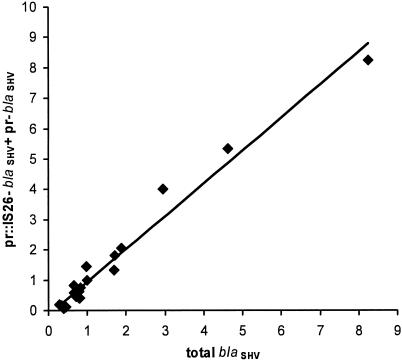
It would be expected that the pr-blaSHV copy number plus the pr::IS26-blaSHV copy number equals the total blaSHV copy number for all of the isolates. If this were not the case, then either the copy number determinations did not provide meaningful results and/or blaSHV exists in cis with an undiscovered flanking sequence. Therefore, the correlation between total blaSHV and pr-blaSHV plus pr::IS26-blaSHV was determined (Fig. 2). A linear relationship exists between these two independently determined parameters; the slope of the line approximates to 1, and the intercept is close to the origin. This demonstrates that the interisolate copy number variations are not experimental artifacts and essentially rules out the possibility that the blaSHV gene exists with an undetected different upstream sequence in any of these isolates.
FIG. 2.
Linear regression analysis for blaSHV relative quantitation measurements.
It can be seen in Table 4 that pr-blaSHV displays very little copy number variation and is present in all isolates, while pr::IS26-blaSHV is not universally present and displays considerable copy number variation. Also, the isolates with the highest resistance levels possessed the highest copy numbers of pr::IS26-blaSHV. These data confirm the relationship between copy number and resistance levels and suggest that pr::IS26-blaSHV is plasmid borne while pr-blaSHV is located either on the chromosome or on a plasmid with very tight copy number control.
PFGE analysis of isolates.
Interpretation of DNA restriction patterns generated by PFGE was performed as described by Tenover et al. (18). There was an apparent association between pulsotype A and the pr::IS26-blaSHV cassette. All except one of the non-pulsotype A isolates (C1) were negative for this cassette, while all except two of the pulsotype A isolates (J3 and L2) carried this cassette (Table 4). The presence of pr::IS26-blaSHV in the pulsotype B isolate C1 suggests that it is present on a transmissible plasmid. However, without more detailed analyses we cannot exclude the possibility of pr::IS26-blaSHV being present on different plasmids in different isolates.
Direct determination of the allele copy number ratios of pr-blaSHV and pr::IS26-blaSHV.
In order to determine whether homologous recombination has resulted in the blaSHV alleles located on different replicons being in equilibrium within individual isolates, PCR products representing blaSHV in cis with the different upstream sequences were cloned and analyzed. A representative subset of isolates was used for this experiment, and the results are shown in Table 5. It is clear that in all isolates, pr-blaSHV is predominantly but not exclusively wild type at codons 238 and 240, while the relative copy numbers for the allelic variants of pr::IS26-blaSHV vary greatly. It may therefore be concluded that the alleles are not in equilibrium within individual isolates and that pr::IS26-blaSHV has the dominant effect on the resistance phenotype, particularly in the highly resistant isolates. Also of interest was the finding that all sequences analyzed possessed the L35Q substitution that differentiates SHV-1/2/5 from SHV-11/2a/12, irrespective of whether they were derived from pr-blaSHV or pr::IS26-blaSHV.
TABLE 5.
blaSHV allele distributions found in cis with the different promoters in a diverse subset of isolates, as determined by analysis of the cloned PCR products
| Isolate | % pr::IS26-blaSHV allelesa
|
% pr-blaSHV allelesa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 35-L | 35-Q | 238-G, 240-E | 238-S, 240-E | 238-S, 240-K | 35-L | 35-Q | 238-G, 240-E | 238-S, 240-E | 238-S, 240-K | |
| B2 | NAb | NA | NA | NA | NA | 100 | 95 | 5 | ||
| K2 | 100 | 100 | 100 | 100 | ||||||
| J3 | NA | NA | NA | NA | NA | 100 | 95 | 5 | ||
| D1 | 100 | 5 | 90 | 5 | 100 | 95 | 5 | |||
| L1 | 100 | 5 | 95 | 100 | 95 | 5 | ||||
| A1 | 100 | 100 | 100 | 95 | 5 | |||||
| J2 | 100 | 70 | 30 | 100 | 100 | |||||
| I1 | 100 | 0 | 5 | 95 | 100 | 85 | 15 | |||
The deduced gene products are shown.
NA, not applicable because this promoter is absent from this isolate.
The proportions of the different alleles are consistent with the resistance phenotypes. The DDST-negative isolates B2 and K2 have very few mutant alleles, the highly resistant J2 and L1 have high proportions of the double-mutant allele, and the other intermediate-resistance isolates have intermediate allelic contents. These data confirm that resistance levels are very much a function of the copy numbers of mutant alleles. The data also emphasise the generality of the coexistence of different alleles in these isolates and clearly demonstrate that, despite the effects of homologous recombination, the probability that an individual blaSHV gene is mutated is a function of the replicon on which the gene is located.
An unexpected aspect of these results was the detection of unmutated pr::IS26-blaSHV, most particularly in isolates K2 and J2. This observation means that it is very unlikely that a single plasmid with a mutated blaSHV was introduced into southeast Queensland or the Princess Alexandra Hospital and then underwent a dissemination process. Rather, the simplest explanation is that a plasmid containing blaSHV-11 was extant, and mutated derivatives of this were selected on one or more occasions. If this is the case, then the low proportion of mutant alleles of pr-blaSHV suggests that if an isolate has both pr::IS26-blaSHV and pr-blaSHV and is subject to selection for ESBL expression, then mutations in pr::IS26-blaSHV are more likely to be evolutionarily fixed than mutations in pr-blaSHV. A possible explanation for this is that a multicopy plasmid location and/or the presence of IS26 in the promoter increase expression and thus increase the impact upon the resistance phenotype of the mutations that confer ESBL activity. It is consistent with this hypothesis that all of the ESBL-positive isolates in this study possessed pr::IS26-blaSHV.
Our results are similar to findings from studies on the distribution and genetics of SHV-2a and SHV-12 in Korea (8, 9). These enzymes are the dominant ESBL SHV variants found in that country, and it appears that their genes invariably possess the promoter with the IS26 insertion (9). Those authors suggested that the SHV-2a and SHV-12 enzymes arose from an SHV-11 precursor whose gene also possessed the promoter with the IS26 insertion. Our study has provided direct evidence in support of this model, because we have detected blaSHV-11 in cis with the IS26 promoter and have shown that it is present in both ESBL-positive and ESBL-negative isolates. However, the isolate-to-isolate variations in copy number of blaSHV-11 in cis with the IS26 promoter do not support the conjecture of Kim et al. (9) that the blaSHV-11 is chromosomally located and distinct from the plasmid-located blaSHV-2a and blaSHV-12. Enterobacteriaceae expressing SHV-12 have also been found to be very widespread in Italy (12). It would be most interesting to determine whether instances of wide dissemination of SHV-12 are always associated with IS26 insertions in the promoter.
Kim et al. (9) also postulated that the acquisition of the point mutation that confers the L35Q substitution that defines SHV-11, SHV-2a, and SHV-12 was coincident with the acquisition of the IS26 insertion in the promoter. Our study has demonstrated that this is not the case, as all cloned PCR products that were fully sequenced contained the mutation, irrespective of whether there was an IS26 insertion in the promoter.
In conclusion, we have used a detailed study of a small collection of geographically and temporally linked K. pneumoniae clinical isolates to illuminate aspects of the natural history of blaSHV genes. This has revealed that a mix of alleles in individual isolates is the norm rather than the exception and that there is a strong relationship between high levels of resistance and a high copy number of blaSHV-12. The most likely mechanism for the acquisition of resistance in this collection is the dissemination of a plasmid carrying blaSHV-11 followed by one or more rounds of selection for the ESBL-positive mutants. The apparent ability of a gene cassette consisting of IS26 and blaSHV-11 to facilitate the appearance of resistance in response to selection, even in the presence of a blaSHV gene with a different promoter, may be of clinical significance. We are currently investigating the relationship between the presence of this cassette and frequencies of mutation to an ESBL-positive phenotype.
Acknowledgments
This work was supported by the Australian Federal Government Program for Cooperative Research Centres.
REFERENCES
- 1.Blazquez, J., M. I. Morosini, M. C. Negri, and F. Baquero. 2000. Selection of naturally occurring extended-spectrum TEM β-lactamase variants by fluctuating β-lactam pressure. Antimicrob. Agents Chemother. 44:2182-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantu, C., W. Huang, and T. Palzkill. 1996. Selection and characterization of amino acid substitutions at residues 238-240 of TEM-1 β-lactamase with altered substrate specificity for aztreonam and ceftazidime. J. Biol. Chem. 271:22538-22545. [DOI] [PubMed] [Google Scholar]
- 3.Essack, S. Y., L. M. Hall, D. G. Pillay, M. L. McFadyen, and D. M. Livermore. 2001. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum β-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fortineau, N., T. Naas, O. Gaillot, and P. Nordmann. 2001. SHV-type extended-spectrum β-lactamase in a Shigella flexneri clinical isolate. J. Antimicrob. Chemother. 47:685-688. [DOI] [PubMed] [Google Scholar]
- 5.Germer, S., M. J. Holland, and R. Higuchi. 2000. High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res. 10:258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard, C., A. van Daal, G. Kelly, J. Schooneveldt, G. Nimmo, and P. M. Giffard. 2002. Identification and minisequencing-based discrimination of SHV β-lactamases in nosocomial infection-associated Klebsiella pneumoniae in Brisbane, Australia. Antimicrob. Agents Chemother. 46:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby, G. A. 1994. Genetics of extended-spectrum β-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 13(Suppl. 1):S2-S11. [DOI] [PubMed] [Google Scholar]
- 8.Kim, J., Y. Kwon, H. Pai, J. W. Kim, and D. T. Cho. 1998. Survey of Klebsiella pneumoniae strains producing extended-spectrum β-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J. Clin. Microbiol. 36:1446-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, J., H. S. Shin, S. Y. Seol, and D. T. Cho. 2002. Relationship between blaSHV-12 and blaSHV-2a in Korea. J. Antimicrob. Chemother. 49:261-267. [DOI] [PubMed] [Google Scholar]
- 10.Nuesch-Inderbinen, M. T., H. Hachler, and F. H. Kayser. 1995. New system based on site-directed mutagenesis for highly accurate comparison of resistance levels conferred by SHV β-lactamases. Antimicrob. Agents Chemother. 39:1726-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peixe, L. V., J. C. Sousa, J. C. Perez-Diaz, and F. Baquero. 1997. A bla(TEM-1b)-derived TEM-6 beta-lactamase: a case of convergent evolution. Antimicrob. Agents Chemother. 41:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perilli, M., E. Dell'Amico, B. Segatore, M. R. de Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitout, J. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, E. S. Moland, and C. C. Sanders. 1998. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob Agents Chemother. 42:1350-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podbielski, A., J. Schonling, B. Melzer, and K. Warnatz. 1991. Molecular cloning and nucleotide sequence of a new plasmid-coded Klebsiella pneumoniae beta-lactamase gene (SHV-2a) responsible for high-level cefotaxime resistance. Zbl. Bakteriol. 275:369-373. [DOI] [PubMed] [Google Scholar]
- 15.Podbielski, A., J. Schonling, B. Melzer, and G. Haase. 1991. Different promoters of SHV-2 and SHV-2a β-lactamase lead to diverse levels of cefotaxime resistance in their bacterial producers. J. Gen. Microbiol. 137:569-578. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press., Cold Spring Harbor, N.Y.
- 17.Schooneveldt, J. M., G. R. Nimmo, and P. Giffard. 1998. Detection and characterisation of extended spectrum β-lactamases in Klebsiella pneumoniae causing nosocomial infection. Pathology 30:164-168. [DOI] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, Y., N. Bhachech, P. A. Bradford, B. D. Jett, D. F. Sahm, and K. Bush. 1998. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli isolates producing TEM-10 and TEM-43 beta-lactamases from St. Louis, Missouri. Antimicrob. Agents Chemother. 42:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]