Abstract
We developed a kinetic, 96-well turbidimetric procedure that is capable of testing the antimicrobial properties of six human α-defensins concurrently on a single microplate. The defensins were prepared by solid-phase peptide synthesis and tested against gram-positive bacteria (Staphylococcus aureus and Bacillus cereus) and gram-negative bacteria (Enterobacter aerogenes and Escherichia coli). Analysis of the growth curves provided virtual lethal doses (vLDs) equivalent to conventional 50% lethal doses (LD50s), LD90s, LD99s, and LD99.9s obtained from colony counts. On the basis of their respective vLD90s and vLD99s, the relative potencies of human myeloid α-defensins against S. aureus were HNP2 > HNP1 > HNP3 > HNP4. In contrast, their relative potencies against E. coli and E. aerogenes were HNP4 > HNP2 > HNP1 = HNP3. HD5 was as effective as HNP2 against S. aureus and as effective as HNP4 against the gram-negative bacteria in our panel. HD6 showed little or no activity against any of the bacteria in our panel, including B. cereus, which was highly susceptible to the other five α-defensins. The assay described provides a quantitative, precise, and economical way to study the antimicrobial activities of host-defense peptides. Its use has clarified the relative potencies of human α-defensins and raised intriguing questions about the in vivo function(s) of HD6.
Antimicrobial peptides, such as α-defensins, are believed to play substantial roles in the innate host defense against bacterial, fungal, and viral pathogens (3, 5, 24). Four of these peptides (HNP1 to HNP4) were initially isolated from human leukocytes (13). HNP1 to HNP3 differ only at the N-terminal position, while the other sequences are more diverse. Human defensin 5 (HD5) and HD6, which are synthesized in and secreted by intestinal Paneth cells, were discovered through genomic studies (2, 7, 8). Because native HNP1, HNP2, and HNP3 are easily purified from leukocytes, they have been widely studied. As the other native α-defensin peptides have been recovered in amounts that are small (HNP4), smaller (HD5), or nil (HD6), considerably less is known about their properties.
A recent synthesis procedure has made all six human α-defensins available for in vitro analysis (22, 23). These advances are especially significant for the characterization of HNP4, HD5, and HD6. To study the six α-defensins described in this report and to facilitate future studies of selectively modified defensins that we hope to perform in the future, we developed a facile way to assay their antimicrobial properties quantitatively.
Broth microdilution methods for the testing of antibiotics are traditionally conducted in 96-well plates, ideally according to guidelines approved by the National Center for Clinical Laboratory Standards (NCCLS) (15). Although such methods are simple to perform, their inherent precision is limited by the use of serial dilutions rather than a continuous calibration curve. Colony counting procedures are considerably more labor intensive to set up and analyze. We refer to the alternative procedure described below as “virtual colony counting” and to the CFU values that it provides as “CFUv.” The virtual colony counting assay eliminates the multiple dilutions, spreading, and colony counting steps required in actual colony counting assays. It provides data comparable to colony counts from efforts commensurate with those needed to set up broth microdilution assays.
Since the scientific underpinnings of the virtual colony count method are fairly obvious, it is surprising that it has not been applied previously. Brewster (1a) applied its principles to formulate a microplate procedure to enumerate bacterial concentrations as low as ∼10 CFU/ml. His assay, as well as the one described here, is based on the fact that the time required for the turbidity (optical density) of an inoculated well to reach an arbitrary threshold value allows the number of viable bacteria in that inoculum to be estimated from an appropriate growth calibration curve (1a). Because Brewster's procedure (1a), our assay, and traditional colony counting procedures all enumerate only viable cells, in principle, all should be applicable to a wide variety of physiologically disparate strains.
MATERIALS AND METHODS
α-Defensins.
All six human α-defensins were synthesized by procedures based upon an optimized Boc chemistry protocol developed by Kent and colleagues (9, 17), as described in detail elsewhere (22, 23). Disulfide mapping of HNP1 to HNP3 (23) and the solution of the X-ray crystal structures of HNP4, HD5, and HD6 (Jacek Lubkowski, personal communication) unequivocally demonstrate correct folding and disulfide connectivity. The concentrations of the pure final products were determined spectroscopically by using molar extinction coefficients at 280 nm calculated by the algorithm of Pace et al. (15a), which takes into account amino acid variations.
Bacteria.
Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923, S. aureus ATCC 29213, and Enterobacter aerogenes ATCC 13048 were from Becton-Dickinson (Sparks, Md.); Bacillus cereus ATCC 10876 and E. coli ATCC 8739 were from MicroBioLogics (St. Cloud, Minn.) and Remel (Lenexa, Kans.), respectively. Mueller-Hinton (MH) broth (MHB) or MH II Agar (Becton Dickinson) was prepared in deionized water. A Millipore Milli-Q deionization system was used to prepare water for the buffers and peptides. Sterile, polystyrene, flat-bottom, 96-well tissue culture plates (3595; Costar) were used. Growth kinetics in the 96-well microplates was monitored (as turbidity) with a computer-controlled Vmax plate reader (Molecular Devices, Sunnyvale, Calif.) running Softmax Pro software (version 3.1) and a 650-nm filter. Growth of the seed cultures was monitored on a Beckman DU-640 spectrophotometer by measuring the optical density at 650 nm (OD650) of 1-ml samples.
Conversion of turbidity to number of CFU per milliliter.
E. coli ATCC 25922 was grown overnight in 2 ml of MHB. An aliquot of the overnight culture was inoculated 1/100 into 25 ml of MHB and incubated with shaking (250 rpm) at 37°C. When this culture reached an OD650 of 0.5, dilutions were plated in triplicate on MH agar, and colonies were counted the next day, yielding a conversion factor of 5 × 108 CFU/ml per OD650 unit. For convenience, turbidity, not the initial cell concentration, was held constant among the six strains, such that the cell “concentration” quantities of the other five strains corresponded to the concentration that E. coli ATCC 25922 would have produced at the same OD650. Moreover, turbidity is a more appropriate basis of comparison than CFU, since turbidity should correlate more closely with the amount of cytoplasmic membrane present. Membranes are the substrates of the lethal activity of defensins (3). If the number of CFU had been held constant, the result would have been a variation in the amount of plasma membrane roughly proportional to the variation in the surface-to-volume ratios among these six strains. To emphasize this distinction, the notation C′0 for the initial cell concentration has been introduced in place of the C0 defined by Brewster (1a) in the calculations below.
Growth of seed cultures.
Single colonies from MH agar plates were inoculated into 2 ml of PMH (MHB with 5 mM sodium phosphate buffer [pH 7.4]). After overnight growth at 37°C, 250 μl of this culture was added to 25 ml of PMH in a 250-ml capped Erlenmeyer flask and grown at 37°C with shaking (250 rpm) to an OD650 of 0.45 to 0.55.
Calibration.
One milliliter of seed culture was added to 1.5 ml of PMH to provide a suspension with ∼108 CFU/ml. A 10-fold dilution series of this suspension ranging from 107 to 10−1 CFU/ml was made in PMH in sextuplicate. The final volume per well was 200 μl. The last well in each row was a negative control that contained 200 μl of uninoculated PMH. The 60 internal wells (rows B to G and columns 2 to 11) contained the dilution series. The 36 edge wells (rows A and H and columns 1 and 12) contained 200 μl of uninoculated PMH to minimize evaporation from the internal wells and to control for contamination.
Bactericidal assay.
A nine-step, twofold dilution series of synthetic human α-defensins was prepared in 10 mM sodium phosphate (pH 7.4) to provide concentrations ranging from 512 to 2.0 μg/ml. The final volume of the peptide solutions placed in each well was 50 μl. The last (10th) well in each row was a negative control that contained 50 μl of buffer. Cultures were serially diluted 125-fold in three steps (2.5-fold, then 5-fold, and then 10-fold) in 10 mM sodium phosphate (pH 7.4). For E. coli ATCC 25922, this resulted in 2 × 106 CFU/ml in 10 mM sodium phosphate (pH 7.4). Fifty microliters was added to each of the 60 internal wells, resulting in a cell concentration of 106 CFU/ml for E. coli ATCC 25922. This addition resulted in final defensin concentrations that ranged from 256 to 1.0 μg/ml. The edge wells remained empty. The plate was placed in a Molecular Devices Vmax plate reader in a warm (37°C) room with shaking for 30 s initially and then for 3 s every 5 min thereafter. The purpose of placing the 96-well plate in the plate reader during this defensin incubation was to subject the cells to the identical condition to which they were subjected during the subsequent outgrowth and to shake the plate during the incubation to ensure proper mixing. Over the next 2 h readings were taken every 5 min by using the 650-nm filter. Then, 100 μl of double-strength (2×) MHB was added to each of the 60 internal wells. C′0 was 5 × 105 CFUv/ml for control wells (zero killing) after the last 2× MHB addition. Two hundred microliters of a 3:1 mixture of MHB and sodium phosphate buffer was added to each of the 36 edge wells as a contamination check and to retard evaporation of the internal wells. The plate was again shaken for 30 s initially and then for 3 s every 5 min thereafter. Readings were taken at 650 nm every 5 min over the next 12 h. Each experiment was repeated three times on separate days by starting with overnight cultures inoculated from three different colonies.
Data processing.
The OD measurements from the 12-h plate reader run were imported into Microsoft Excel software and corrected by subtracting each well's initial reading from the subsequent data for that well. The times at which each curve crossed the threshold change in OD650 (ΔOD650) of 0.02 absorbance units were plotted against log(C′0) to generate a calibration curve that related the threshold times to C′0. The rate of survival was calculated as the number of CFUv of defensin-treated cells/number of CFUv of control cells. The virtual 50% lethal dose (vLD50), vLD90, vLD99, and vLD99.9 were reported as the defensin concentrations that resulted in survival rates of 0.5, 0.1, 0.01, and 10−3, respectively. The raw data were plotted, and the y axis was rescaled to show only the region near the threshold ΔOD650, which allows visual assessment of the point where the threshold crossed each growth curve.
RESULTS
Calibration curves.
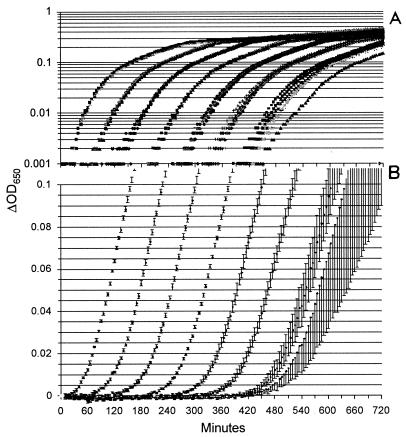
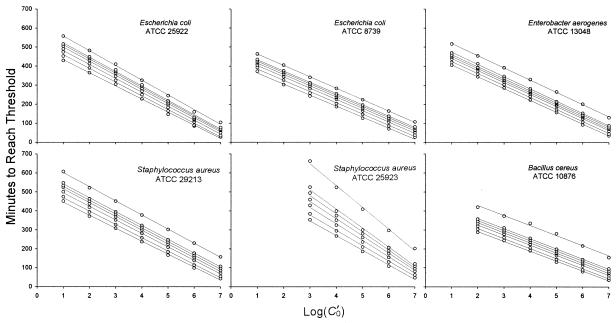
The data used to construct a calibration curve for S. aureus ATCC 29213 are shown in Fig. 1. Consistent with the laws of probability and the small volume (200 μl) per well, only two of six wells became turbid when they were inoculated with a dilution that nominally contained 1 CFUv/ml, and none became turbid when they were inoculated with 0.1 CFUv/ml. The standard deviation for the sextuplicate wells inoculated with 1 or 10 CFUv/ml was greater than that for the wells inoculated with higher concentrations. Again, this result is consistent with expectations based on probability. All six standard curve experiments yielded parallel curves that could be used to calculate the number of CFUv over at least 5 orders of magnitude (Fig. 2). We always excluded the points at 100 (1 CFUv/ml) from linear regression analysis and excluded the point at 101 (10 CFUv/ml) when it was noticeably off-line. Linear regression (r2) values ranged from 0.9946 to 0.9998 at a threshold ΔOD650 of 0.02. Similar slopes, y intercepts, and r2 values were obtained at other thresholds, and these ranged from 0.005 to 0.1 (Table 1).
FIG. 1.
S. aureus ATCC 29213 calibration growth kinetics. Tenfold dilutions yielded C′0 values of 107 (far left) through 100 (far right). (A) Log-normal plot of the raw growth kinetics data; (B) averages for each set of sextuplicate growth curves; the rightmost (C′0 = 100) series is the average for duplicate growth curves. Error bars represent standard deviations.
FIG. 2.
Calibration curves calculated from the threshold times of the curves shown in Fig. 1B (or the equivalent plot for the other five strains) at ΔOD650s of 0.005, 0.01, 0.02, 0.03, 0.04, 0.05, and 0.1 (from bottom to top). The value of 0.02 was used to calibrate the growth of all six strains. In practice, any of these ΔOD650 values could be used for calibration.
TABLE 1.
Slopes, y intercepts, and r2 values of the regression lines shown in Fig. 2
| Strain | ΔOD650 | Slope | y-intercept | r2 |
|---|---|---|---|---|
| E. coli ATCC 8739 | 0.005 | −57.8 | 421.3 | 0.9970 |
| 0.01 | −59.1 | 444.1 | 0.9979 | |
| 0.02 | −59.2 | 462.6 | 0.9997 | |
| 0.03 | −59.3 | 475.1 | 0.9998 | |
| 0.04 | −59.2 | 484.7 | 0.9997 | |
| 0.05 | −59.2 | 492.9 | 0.9998 | |
| 0.1 | −59.6 | 522.6 | 0.9998 | |
| E. coli ATCC 25922 | 0.005 | −68.7 | 501.7 | 0.9978 |
| 0.01 | −71.9 | 531.7 | 0.9975 | |
| 0.02 | −73.1 | 555.6 | 0.9973 | |
| 0.03 | −74.1 | 574.4 | 0.9967 | |
| 0.04 | −75.0 | 587.9 | 0.9971 | |
| 0.05 | −75.5 | 598.3 | 0.9972 | |
| 0.1 | −77.3 | 635.7 | 0.9982 | |
| E. aerogenes ATCC 13048 | 0.005 | −62.2 | 469.1 | 0.9997 |
| 0.01 | −62.8 | 486.6 | 0.9995 | |
| 0.02 | −62.8 | 502.3 | 0.9998 | |
| 0.03 | −63.3 | 514.3 | 0.9999 | |
| 0.04 | −63.1 | 522.7 | 0.9997 | |
| 0.05 | −64.2 | 537.7 | 0.9997 | |
| 0.1 | −64.1 | 583.3 | 0.9996 | |
| S. aureus ATCC 25923 | 0.005 | −77.1 | 587.4 | 0.9956 |
| 0.01 | −81.0 | 631.4 | 0.9941 | |
| 0.02 | −86.6 | 688.6 | 0.9946 | |
| 0.03 | −90.6 | 732.1 | 0.9962 | |
| 0.04 | −95.5 | 767.1 | 0.9955 | |
| 0.05 | −100.0 | 822.3 | 0.9972 | |
| 0.1 | −114.0 | 1,006.1 | 0.9966 | |
| S. aureus ATCC 29213 | 0.005 | −68.5 | 513.6 | 0.9988 |
| 0.01 | −70.3 | 539.1 | 0.9991 | |
| 0.02 | −71.2 | 566.1 | 0.9994 | |
| 0.03 | −72.6 | 588.9 | 0.9992 | |
| 0.04 | −72.5 | 600.7 | 0.9996 | |
| 0.05 | −73.0 | 615.1 | 0.9995 | |
| 0.1 | −74.4 | 675.7 | 0.9996 | |
| B. cereus ATCC 10876 | 0.005 | −51.8 | 394.3 | 0.9979 |
| 0.01 | −53.1 | 414.1 | 0.9978 | |
| 0.02 | −53.1 | 431.5 | 0.9985 | |
| 0.03 | −53.3 | 444.9 | 0.9985 | |
| 0.04 | −53.4 | 456.5 | 0.9990 | |
| 0.05 | −53.4 | 467.2 | 0.9991 | |
| 0.1 | −53.0 | 535.0 | 0.9917 |
Survival curves.
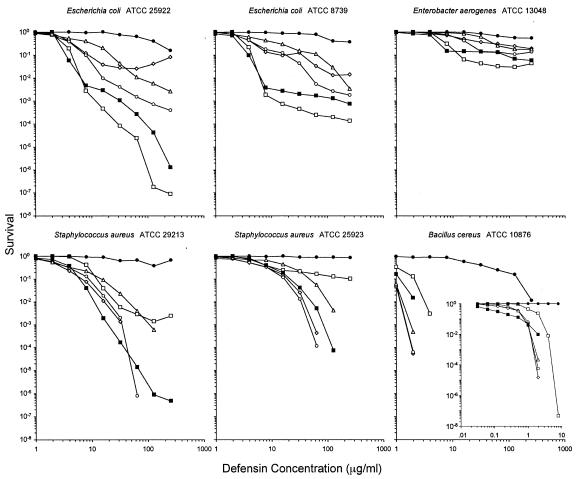
Figure 3 plots the rate of survival (number of CFUv) against the defensin concentration on a log-log scale. Zero values cannot be plotted on such scales, and some values were mathematically below a single surviving cell given the input number of CFUv, indicating that a lag time must have occurred. Most curves had a concave-down shape over the full range of defensin concentrations, but a few deviated from this with greater than expected rates of survival, which was especially notable when E. coli ATCC 25922 was exposed to HNP1 at concentrations greater than 64 μg/ml. We noted the same effect when we performed conventional colony counting experiments with this strain and HNP1 (data not shown).
FIG. 3.
Survival curves. Each curve is the mean of triplicate experiments. Open markers are human neutrophil defensins (⋄, HNP1; ○, HNP2; ▵, HNP3; □, HNP4). Filled symbols are human intestinal defensins (▪, HD5; •, HD6). Points scored as zero survival could not be plotted. All strains were exposed to defensins at concentrations that varied twofold from 1 to 256 μg/ml. In addition, B. cereus was assayed against a twofold dilution series of 0.031 to 8 μg/ml (inset).
Bactericidal concentrations.
The concentrations of each of the six defensins that killed 50, 90, 99, or 99.9% of each of the six strains are presented in Table 2. All six strains were tested with defensin concentrations that varied in twofold steps from 1 to 256 μg/ml. This range was sufficient to determine the vLDs for five of the six strains. B. cereus, however, was more susceptible to all six human α-defensins than any of the other strains, such that some quantities were determined by repeating the analysis with defensin concentrations of 0.031 to 8 μg/ml. The potency of HD5 at a concentration of 0.031 μg/ml resulted in greater than 50% killing of B. cereus in two of the three replicates.
TABLE 2.
Antimicrobial activities of human α-defensins against six strains
| Test organism | vLD50 (μg/ml [mean ± SEM])a
|
vLD90 (μg/ml [mean ± SEM])a
|
vLD99 (μg/ml [mean ± SEM])a
|
vLD99.9 (μg/ml [mean ± SEM])a
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HNP-1 | HNP-2 | HNP-3 | HNP-4 | HD5 | HD6 | HNP-1 | HNP-2 | HNP-3 | HNP-4 | HD5 | HD6 | HNP-1 | HNP-2 | HNP-3 | HNP-4 | HD5 | HD6 | HNP-1 | HNP-2 | HNP-3 | HNP-4 | HD5 | HD6 | |
| E. coli ATCC 8739 | 3.6 ± 0.3 | 3.5 ± 0.7 | 6.2 ± 0.9 | 3.3 ± 0.4 | 2.5 ± 0.3 | >256 | 14 ± 4.3 | 20 ± 7.3 | 61 ± 25 | 4.9 ± 1.0 | 4.5 ± 0.9 | >256 | 106 ± 4.8b | 49 ± 11 | 163 ± 41 | 7.5 ± 0.3 | 5.9 ± 1.2 | >256 | ||||||
| E. coli ATCC 25922 | 3.7 ± 0.4 | 3.5 ± 0.6 | 5.9 ± 2.1 | 3.0 ± 0.7 | 2.1 ± 0.9 | 103 ± 14 | 9.4 ± 0.9 | 8.4 ± 1.3 | 23 ± 4.2 | 4.8 ± 1.1 | 3.6 ± 0.8 | >256 | >256 | 19 ± 4.4 | 93 ± 43 | 7.0 ± 0.5 | 5.3 ± 1.3 | >256 | ||||||
| E. aerogenes ATCC 13048 | 10 ± 0.5b | 16 ± 4.0 | 41 ± 9.2 | 6.6 ± 0.2 | 5.5 ± 0.5 | 156 ± 11b | 70 ± 35b | 26 ± 1.7b | 82 ± 19b | 12 ± 0.8b | 7.6 ± 0.04b | >256 | >256 | >256 | >256 | 12 ± 0.8b | >256 | >256 | ||||||
| S. aureus ATCC 25923 | 4.2 ± 1.0 | 4.2 ± 0.9 | 13 ± 2.1 | 7.3 ± 1.4 | 6.3 ± 0.5 | >256 | 21 ± 0.8 | 18 ± 1.2 | 52 ± 5.9 | >256 | 24 ± 1.1 | >256 | 48 ± 3.1 | 37 ± 6.0 | 117 ± 9.9 | >256 | 58 ± 0.7 | >256 | ||||||
| S. aureus ATCC 29213 | 2.1 ± 0.3 | 2.4 ± 0.3 | 2.2 ± 0.4 | 7.2 ± 0.2 | 3.2 ± 0.3 | >256 | 7.0 ± 0.4 | 9.4 ± 1.1 | 16 ± 2.1 | 14 ± 0.2 | 7.0 ± 0.4 | >256 | 17 ± 0.7 | 22 ± 1.0 | 46 ± 8.8 | 29 ± 0.3 | 13 ± 1.1 | >256 | ||||||
| B. cereus ATCC 10876 | 0.22 ± 0.03 | 0.22 ± 0.04 | 0.37 ± 0.08 | 0.87 ± 0.08 | <0.31 | 25 ± 13 | 0.82 ± 0.09 | 0.79 ± 0.09 | 0.74 ± 0.2 | 2.5 ± 0.5 | 0.44 ± 0.1 | 100 ± 49 | 1.9 ± 0.04 | 1.8 ± 0.1 | 1.6 ± 0.3 | 5.1 ± 1.1 | 2.3 ± 0.8 | 240 ± 7.9 | 3.9 ± 0.03 | 3.9 ± 0.02 | 4.1 ± 0.13 | 12 ± 1.8 | 7.5 ± 0.2 | >256 |
The values are for three replicates of each experiment unless indicated otherwise.
The value for one of the three replicates was >256 μg/ml; in these cases, the standard error of the mean is reported for n = 2. When one of the three replicates was determinable and the other two were >256 μg/ml, a value of >256 μg/ml was entered.
Strain selectivities of human α-defensins.
vLD values (Table 2) and survival plots (Fig. 3) showed that high concentrations of HNP1 were notably less effective against E. coli (two strains) and E. aerogenes than against S. aureus (two strains) and B. cereus. In contrast, the five highest concentrations of HNP4 exhibited the greatest killing of all three gram-negative strains; yet, except for HD6, HNP4 was the least potent against all three gram-positive strains at the two highest concentrations. HNP1 and HNP2 showed almost indistinguishable activities against the gram-positive strains, but HNP2 appeared to be more potent than HNP1 against the gram-negative strains. Other defensins showed no clear strain selectivity. HNP3 was consistently less active than HNP2 against all six strains. HD5 showed a relatively high level of activity against all six strains, although its activity was the highest of the six defensins only at intermediate concentrations against S. aureus ATCC 29213 and only at low concentrations against B. cereus. HD6 was active against B. cereus and both E. coli strains, but its activity was the lowest against all six strains at all concentrations. Similar results were obtained for the pairs of E. coli and S. aureus strains. The curves for E. coli varied slightly for HNP1 to HNP3, which intersected between different concentrations, but the orders of points on this plot were not meaningfully different at 4, 8, 16, and 256 μg/ml for all six defensins (Fig. 3). The two S. aureus strains also yielded sets of curves that were materially congruent.
DISCUSSION
α-Defensins are found in various leukocytes and in the Paneth cells of the small intestines of many mammals. Although human neutrophils contain four α-defensins, HNP1 to HNP3 are collectively about 60 times more abundant than HNP4 (6). Largely because it has been difficult to obtain HNP4 from natural sources, this defensin has been studied less than HNP1 to HNP3. Wilde et al. (21) used size-exclusion and reverse-phase high-performance liquid chromatography to purify native HNP4 from human neutrophils and then tested its antimicrobial activity. They asserted, “compared to a mixture of the other human defensins, HNP4 was found to be approximately 100 times more potent against E. coli and four times more potent against both Streptococcus faecalis and Candida albicans” (21). Our study provides partial support to their findings with E. coli. The relative potencies of two peptides, e.g., HNP4 and HNP1, can be estimated from the ratio of their vLDs. When we used the mean vLD90s shown in Table 2 for E. coli ATCC 8739, these ratios were 3 for HNP1, 4 for HNP2, and 13 for HNP3, signifying that HNP4 is approximately 3- to 13-fold more potent on a weight basis. If we used the mean vLD90 ratios for the other E. coli strain, ATCC 25922, the ratios were 2 for HNP1 and HNP2 and 5 for HNP3. Had we chosen to use the vLD99s for E. coli ATCC 8739, they would have returned ratios that were two- to fourfold higher than the vLD90 ratios: 15 for HNP1 and 23 for HNP2 and HNP3.
Human neutrophils may have compensated for the lower intrinsic potencies of HNP1 to HNP3 against gram-negative bacteria by producing these peptides at a 60-fold greater abundance. The greater intrinsic potency of HNP4 against gram-negative bacteria evidently came with a price, since it was less potent than HNP1 and HNP2 against the gram-positive test organisms in our panel. In contrast, HD5 was as active as (or more so than) HNP4 against gram-negative bacteria, and it was almost as effective as HNP2 against the gram-positive organisms.
Perhaps the greatest surprise from our study was the exceedingly poor performance of HD6 against our bacterial test panel. Indeed, the relative inactivity of HD6 against bacteria initially led us to question its raison d'être. However, subsequent data from our group indicated that HD6 can inhibit infection of peripheral blood mononuclear cells by human immunodeficiency virus type 1 as effectively as HNP1 to HNP4 (unpublished data), suggesting that HD6 may have evolved specificity for viruses at the expense of its antibacterial activity. In the future, HD6 (and other intestinal defensins) deserve to be tested for their activities against anaerobes, fungi, protozoans, and helminths and for other properties within the purview of defensins, e.g., chemotaxis, adjuvanticity, and enhancement of wound healing (11, 12, 18).
The conspicuous diversity of the survival curves shown in Fig. 3 was also unexpected. Most defensins tested against gram-positive strains, including all six defensins against B. cereus, consistently produced concave-down dose-response relationships that follow simple exponential killing (14). Each curve that appears entirely concave down, as plotted on the log-log scale of Fig. 3, is linear on a lognormal plot (data not shown). This type of result would suggest that neither saturation by increasing defensin concentrations nor resistance to defensins influences the activity measured. By contrast, HNP4 produced bimodal curves against both S. aureus strains, consistent with a small (<1:100) subpopulation of organisms that were phenotypically resistant to defensins rather than genotypically resistant (1, 10), especially since we used cloned organisms in each experiment. Similarly, bimodal curves were observed with HNP4 and HD5 against gram-negative strains E. coli ATCC 8739 and E. aerogenes. With the exception of the scant activity of HD6, none of the defensins tested against gram-negative strains produced strictly exponential survival curves. When deviations from a linear lognormal survival curve exist, only the most linear portions of the curve should be used to calculate relative potency. The human α-defensins are considerably less potent than antimicrobial peptides, such as ovine SMAP-29, porcine protegrins, or the horseshoe crab's tachyplesins. Although the vLD90s and vLD99s of human defensins are reliable, vLD99.9s could be calculated for only one organism in our test panel. When more potent antimicrobial peptides (e.g., SMAP-29, protegrins, and tachyplesins) are assayed, reliable vLD99.9s can also be obtained.
With these caveats, the virtual colony counting assay described in this report can produce results comparable to those of the NCCLS-approved colony counting procedures with far less effort. Our virtual constants were so named to distinguish them from any existing published constants while making their meaning readily understandable to workers in various fields of study. We do not intend to imply that our vLDs have been rigorously verified to correlate with LDs determined by other methods. However, our findings do appear to correlate well with those produced by colony counting. As expected, the mean vLD50s reported in Table 2 were slightly greater than the LD50s reported previously (22) in a colony counting study of the same preparations of these six defensins against E. coli ATCC 25922 and S. aureus ATCC 29213 with 3-h defensin exposure times. Colony counting data comparable to the remainder of the data in Table 2 have yet to be reported, but our results agree generally with what is known about these peptides. For instance, in our study HNP3 was less active than HNP1 and HNP2 against all six strains, which agrees with the findings from studies that used a variety of methods (4, 16, 19). Furthermore, the calibration data shown in Table 1 and Fig. 2 satisfied each of the assumptions underlying the use of growth kinetic curves to calculate C′0, as specified by Brewster (1a), i.e., the curves were parallel and evenly spaced, with similar slopes for all thresholds and excellent linear regression r2 values. Any error added by using this calibration procedure to enumerate small concentrations of bacteria is insignificant. The growth rates in the experimental wells with the same strain were similar, but they were not as close to identical as those for the strain in the calibration growth curves (Fig. 1) or the control wells. Threshold time variations were minimized by averaging the results of experiments performed on 3 days and by choosing a low threshold ΔOD650 of 0.02.
Several technical details were crucial in the development of the virtual colony counting assay. A potential drawback of using 96-well plates for a 14-h experiment is that moisture can condense on the inner surface of the lid. Brewster (1a) applied detergent to the insides of 96-well lids before their use to deter the accumulation of hanging droplets. We found this step to be unnecessary, perhaps because we did the experiments in a 37°C room or used a different instrument for the measurements. Evaporation of liquid from the edge wells caused us to limit the collection of experimental data to the internal 60 wells, none of which decreased in volume by more than 3% when the volumes were measured after overnight experiments. Antimicrobial activity must be antagonized in order to allow cells to grow exponentially after the addition of twice-concentrated media. Inhibition of defensin activity was likely due to the carbohydrates present in the growth medium. MH II powder is composed of Casamino Acids; 14% (wt/wt) beef extract, which contains a variety of carbohydrates; and 7% (wt/wt) starch. Defensins are known to bind to various polysaccharides, including protein glycosyl moieties (20), cellulose, and dextran sulfate, with high affinities. Virtual colony counting could be adapted to measure the activities of agents other than defensins if their antimicrobial activities are also strongly inhibited by 2× MHB itself or if a peptide binder, such as dextran sulfate, that does not slow bacterial growth is added to the MHB. Virtual colony counting is also adaptable to address hypotheses involving high salt concentrations, various divalent cation concentrations, or physiological conditions by supplementing the low-salt 10 mM sodium phosphate incubation buffer as desired.
Our virtual colony counting procedure allows the convenient, concurrent comparison of the antibacterial specificities of the human α-defensins. In addition, libraries of mutated defensins could be screened at one (or more) critical concentration(s) to deduce sequence-to-activity relationships, test mechanistic hypotheses, or identify candidate therapies that can be used to combat pathogenic bacteria. The assay may also have potential as a general method for determination of cationic peptide antibacterial activity.
Acknowledgments
We thank Jacek Lubkowski of the National Cancer Institute, Frederick, Md., for communicating the results of crystallographic studies; Fiorenza Cocchi for conducting anti-human immunodeficiency virus assays; and Rob Powell for valuable technical advice.
This work was supported by National Institutes of Health grants AI056264 and AI058939 (to W.L.).
REFERENCES
- 1.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 1a.Brewster, J. D. 2003. A simple micro-growth assay for enumerating bacteria. J. Microbiol. Methods 53:77-86. [DOI] [PubMed] [Google Scholar]
- 2.Cunliffe, R. N. 2003. Alpha-defensins in the gastrointestinal tract. Mol. Immunol. 40:463-467. [DOI] [PubMed] [Google Scholar]
- 3.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 4.Ganz, T., M. E. Selsted, D. Szklarek, S. S. Harwig, K. Daher, D. F. Bainton, and R. I. Lehrer. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 6.Harwig, S. S., A. S. Park, and R. I. Lehrer. 1992. Characterization of defensin precursors in mature human neutrophils. Blood 79:1532-1537. [PubMed] [Google Scholar]
- 7.Jones, D. E., and C. L. Bevins. 1993. Defensin-6 mRNA in human Paneth cells: implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 315:187-192. [DOI] [PubMed] [Google Scholar]
- 8.Jones, D. E., and C. L. Bevins. 1992. Paneth cells of the human small intestine express an antimicrobial peptide gene. J. Biol. Chem. 267:23216-23225. [PubMed] [Google Scholar]
- 9.Kent, S. B. 1988. Chemical synthesis of peptides and proteins. Annu. Rev. Biochem. 57:957-989. [DOI] [PubMed] [Google Scholar]
- 10.Koch, A. L. 1996. Similarities and differences of individual bacteria within a clone, p. 1640-1651. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 11.Kopp, E., and R. Medzhitov. 2002. Skin antibiotics get in the loop. Nat. Med. 8:1359-1360. [DOI] [PubMed] [Google Scholar]
- 12.Lehrer, R. I., and T. Ganz. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14:96-102. [DOI] [PubMed] [Google Scholar]
- 13.Lehrer, R. I., A. K. Lichtenstein, and T. Ganz. 1993. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu. Rev. Immunol. 11:105-128. [DOI] [PubMed] [Google Scholar]
- 14.Luria, S. E., and R. Latarjet. 1947. Ultraviolet irradiation of bacteriophage during intracellular growth. J. Bacteriol. 53:149-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NCCLS. 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A. NCCLS, Wayne, Pa.
- 15a.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raj, P. A., K. J. Antonyraj, and T. Karunakaran. 2000. Large-scale synthesis and functional elements for the antimicrobial activity of defensins. Biochem. J. 347(Pt. 3):633-641. [PMC free article] [PubMed] [Google Scholar]
- 17.Schnolzer, M., P. Alewood, A. Jones, D. Alewood, and S. B. Kent. 1992. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int. J. Pept. Protein Res. 40:180-193. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen, O. E., J. B. Cowland, K. Theilgaard-Monch, L. Liu, T. Ganz, and N. Borregaard. 2003. Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J. Immunol. 170:5583-5589. [DOI] [PubMed] [Google Scholar]
- 19.Takemura, H., M. Kaku, S. Kohno, Y. Hirakata, H. Tanaka, R. Yoshida, K. Tomono, H. Koga, A. Wada, T. Hirayama, and S. Kamihira. 1996. Evaluation of susceptibility of gram-positive and -negative bacteria to human defensins by using radial diffusion assay. Antimicrob. Agents Chemother. 40:2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, W., A. M. Cole, T. Hong, A. J. Waring, and R. I. Lehrer. 2003. Retrocyclin, an antiretroviral theta-defensin, is a lectin. J. Immunol. 170:4708-4716. [DOI] [PubMed] [Google Scholar]
- 21.Wilde, C. G., J. E. Griffith, M. N. Marra, J. L. Snable, and R. W. Scott. 1989. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J. Biol. Chem. 264:11200-11203. [PubMed] [Google Scholar]
- 22.Wu, Z., B. Ericksen, K. Tucker, J. Lubkowski, and W. Lu. 2004. High-yield synthesis and characterization of human alpha defensins 4-6. J. Pept. Res. 64:118-125. [DOI] [PubMed] [Google Scholar]
- 23.Wu, Z., R. Powell, and W. Lu. 2003. Productive folding of human neutrophil alpha-defensins in vitro without the pro-peptide. J. Am. Chem. Soc. 125:2402-2403. [DOI] [PubMed] [Google Scholar]
- 24.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]