Abstract
High-level fluoroquinolone (FQ) resistance in Salmonella enterica serovar Typhimurium phage type DT204 has been previously shown to be essentially due to both multiple target gene mutations and active efflux by the AcrAB-TolC efflux system. In this study we show that in intermediatly resistant acrB-inactivated serovar Typhimurium DT204 mutants, high-level resistance to FQs can be restored on in vitro selection with FQs. In each FQ- resistant mutant selected from serovar Typhimurium DT204 acrB mutant strains, an insertion sequence (IS1 or IS10) was found integrated upstream of the acrEF operon, coding for AcrEF, an efflux pump highly homologous to AcrAB. In one of the strains, transposition of IS1 caused partial deletion of acrS, the putative local repressor gene of the acrEF operon. Sequence analysis showed that both IS1 and IS10 elements contain putative promoter sequences that might alter the expression of adjacent acrEF genes. Indeed, reverse transcription-PCR experiments showed an 8- to 10-fold increase in expression of acrF in these insertional mutants, relative to their respective parental strain, which correlated well with the resistance levels observed to FQs and other unrelated drugs. It is noteworthy that AcrEF did not contribute to the intrinsic drug resistance of serovar Typhimurium, since acrF deletion in wild-type strains did not result in any increase in drug susceptibility. Moreover, deletion of acrS did not cause any acrF overexpression or any decrease in drug susceptibility, suggesting that acrEF overexpression is mediated solely by the IS1 and IS10 promoter sequences and not by inactivity of AcrS. Southern blot experiments showed that the number of chromosomal IS1 and IS10 elements in the serovar Typhimurium DT204 genome was about 5 and 15 respectively. None were detected in epidemic serovar Typhimurium DT104 strains or in the serovar Typhimurium reference strain LT2. Carrying IS1 and/or IS10 elements in their chromosome may thus be a selective advantage for serovar Typhimurium DT204 strains as opposed to DT104 strains for which no high-level FQ resistance nor insertional mutations were found. Taken together, the results of the present study indicate that the IS1- or IS10- activated AcrEF efflux pump may relay AcrAB in serovar Typhimurium, and underline the importance of transposable elements in the acquisition of FQ and multidrug resistance.
Salmonella enterica serovar Typhimurium is a food-borne pathogen and is an etiologic agent of gastrointestinal infections. Among the different phage types causing salmonellosis in humans and animals, the epidemic S. enterica serovar Typhimurium phage type DT104 emerged during the 1990s with stable resistance to the most common antibiotic families (ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline) (10). Although human gastroenteritis is not usually treated with antibiotics, severe cases in adults are treated with fluoroquinolones (FQs) such as ciprofloxacin. Emerging resistance to these antibiotics in Salmonella spp. has been found in both humans and animals and is thus a potentially serious public health problem (9, 45). However, high-level FQ resistance (MIC of ciprofloxacin, ≥32 μg/ml) has only rarely been described. The first reported example was the emergence and probable clonal spread of S. enterica serovar Typhimurium variant Copenhagen of phage type DT204 (hereafter referred to as serovar Typhimurium DT204) in limited areas in Europe (3, 17, 18, 20, 30). These strains were isolated mainly between 1991 and 1995 from animals and humans and were highly resistant to ciprofloxacin (MIC, 32 μg/ml). More recently, such high-level ciprofloxacin resistance has been found in other S. enterica serovars such as Choleraesuis (7) and Schwarzengrund (42). Recent reports also suggest that such high-level resistance is reemerging in serovar Typhimurium in different parts of the world (6, 19, 26, 39).
Quinolone resistance in Salmonella has been attributed to point mutations in the gyrA gene, coding for the A subunit of DNA gyrase, the primary target of quinolones. These mutations cluster in a region termed the quinolone resistance-determining region (QRDR). Amino acid changes at Ser83 (to Phe, Tyr, or Ala) or at Asp87 (to Gly, Asn, or Tyr) are the most frequently observed changes in nalidixic acid-resistant strains (9, 45). Double mutations at both residues 83 and 87 have been identified in clinical isolates of serovar Typhimurium DT204 showing high-level resistance to FQs, together with one mutation leading to the amino acid change Ser464Phe in the QRDR of gyrB encoding the B subunit of DNA gyrase. More recently, these high-level FQ-resistant isolates have been shown to carry a mutation leading to the amino acid change Ser80Ile in the QRDR of parC coding for the ParC subunit of topoisomerase IV, the secondary target of quinolones (2, 3, 14, 15, 17, 18).
FQ resistance in serovar Typhimurium has also been attributed to active efflux mechanisms (9, 13, 45, 46), and we have recently reported that the AcrAB-TolC efflux system is an important mechanism of high-level resistance to FQs in serovar Typhimurium DT204 (2, 3). High-level resistance to FQs in Salmonella is thus essentially explained by the combination of two major resistance mechanisms, i.e., multiple target gene mutations and active efflux mediated by AcrAB-TolC (2).
In this study, participation of other potential mechanisms in high-level resistance to FQs was assessed by in vitro selection experiments of FQ-resistant mutants of serovar Typhimurium DT104 and DT204 isolates and their respective acrB-inactivated mutants. The serovar Typhimurium DT204 strains 102SA00 and 902SA92 used in this study were previously characterized and contain a double mutation in gyrA leading to amino acid changes Ser83Tyr and Asp87Asn, one mutation in gyrB leading to the amino acid change Ser464Phe and one mutation in parC leading to the amino acid change Ser80Ile (3). The serovar Typhimurium DT104 strains BN10055 and 543SA98 used contain a single mutation in gyrA, leading to the amino acid change Ser83Tyr or Ser83Phe, respectively (1). It has been shown that another multidrug efflux system, AcrEF in Escherichia coli, which is highly homologous to AcrAB, was recruited to restore FQ and solvent resistance in acrB-inactivated mutants (21, 25). The mechanism responsible for the activation of expression of acrEF was the integration of insertion sequences (IS). Many IS elements are present in the genomes of bacteria and have been shown, in some cases, to alter the expression of neighboring genes (12). We investigated the role of such integrational elements in the recruitment of the putative efflux pump AcrEF in high-level FQ-resistant mutants derived from acrB mutants of serovar Typhimurium DT204 selected in vitro with the FQs enrofloxacin and marbofloxacin, currently used in veterinary medicine.
MATERIALS AND METHODS
Bacterial strains.
FQ-resistant mutants were selected with multidrug-resistant serovar Typhimurium DT104 strains BN10055 and 543SA98 and DT204 strains 902SA92 and 102SA00. Strain BN10055 was isolated from cattle in France in 1997; strains 902SA92 and 543SA98 were isolated from cattle in Belgium in 1992 and 1998, respectively; and strain 102SA00 was isolated from animal feed imported to Belgium from China in 2000 (3, 20). The selection was also performed with acrB::kan mutants of these strains (3). Additional strains used in this study were susceptible serovar Typhimurium DT104 strain S92/1495 isolated from cattle in Scotland in 1992 (4); susceptible serovar Typhimurium reference strain LT2, whose genome has completely been sequenced (36); and E. coli cloning strain TG1. All strains were cultivated at 37°C in Luria-Bertani (LB) or brain heart infusion medium. Mutants carrying the kan gene were grown in the presence of kanamycin (Fluka), at 50 μg/ml. Transformants carrying the pRS415 vector were grown in the presence of ampicillin (Sigma, St. Louis, Mo.) at 100 μg/ml. The strains used are listed in Table 1.
TABLE 1.
S. enterica serovar Typhimurium strains studied
| Parental strains and derived mutants | Phage type | IS upstream of acrEF | MICa (μg/ml) of:
|
||||
|---|---|---|---|---|---|---|---|
| Marbofloxacin | Enrofloxacin | Ciprofloxacin | Florfenicol | Erythromycin | |||
| S/921495 | DT104 | 0.06 [≤0.015] | 0.125 [≤0.015] | 0.03 [≤0.015] | 8 [≤0.5] | 200 [≤0.78] | |
| S/921495acrB::kan | ≤0.015 [≤0.015] | ≤0.015 [≤0.015] | ≤0.015 [≤0.015] | 2 [≤0.5] | 6.25 [≤0.78] | ||
| S/921495ΔacrF::kan | 0.03 [≤0.015] | 0.03 [≤0.015] | ≤0.015 [≤0.015] | 4 [1] | 100 [≤0.78] | ||
| S/921495ΔacrS::kan | 0.03 [≤0.015] | 0.06 [≤0.015] | ≤0.015 [≤0.015] | 4 [1] | 100 [≤0.78] | ||
| 102SA00 | DT204 | 32 [2] | 64 [2] | 32 [8] | 16 [1] | 200 [≤0.78] | |
| 102SA00 M1b | 64 [4] | 512 [2] | 256 [8] | 32 [1] | 400 [≤0.78] | ||
| 102SA00acrB::kan | 2 [≤0.015] | 2 [≤0.015] | 2 [≤0.015] | ≤0.5 [≤0.5] | 3.12 [≤0.78] | ||
| 102SA00acrB::kan M1b | IS10 | 16 [2] | 64 [2] | 32 [8] | 8 [1] | 100 [≤0.78] | |
| 102SA00acrB::kan M2b | IS1 | 16 [2] | 32 [2] | 32 [8] | 8 [1] | 200 [≤0.78] | |
| 102SA00acrB::kan M3b | IS1 | 16 [2] | 32 [2] | 32 [8] | 8 [1] | 200 [≤0.78] | |
| 102SA00ΔacrF::kan | 32 [2] | 128 [2] | 64 [8] | 16 [1] | 400 [≤0.78] | ||
| 102SA00ΔacrS::kan | 32 [2] | 128 [2] | 64 [8] | 16 [1] | 200 [≤0.78] | ||
| 902SA92 | DT204 | 32 [4] | 64 [2] | 32 [4] | 32 [≤0.5] | 400 [≤0.78] | |
| 902SA92 M1b | 64 [2] | 512 [2] | 128 [4] | 64 [≤0.5] | 400 [≤0.78] | ||
| 902SA92acrB::kan | 2 [≤0.015] | 2 [≤0.015] | 2 [≤0.015] | ≤0.5 [≤0.5] | 6.25 [≤0.78] | ||
| 902SA92acrB::kan M1b | IS10 | 16 [2] | 64 [2] | 32 [8] | 8 [1] | 200 [≤0.78] | |
| 902SA92acrB::kan M2b | IS1 | 16 [2] | 32 [4] | 32 [4] | 8 [1] | 200 [≤0.78] | |
| 902SA92ΔacrF::kan | 16 [2] | 128 [2] | 32 [8] | 16 [1] | 400 [≤0.78] | ||
| 902SA92ΔacrS::kan | 32 [2] | 64 [1] | 32 [8] | 16 [1] | 400 [≤0.78] | ||
MICs in the presence of 80 μg of PAβN are shown in brackets.
Mutants selected in vitro using enrofloxacin or marbofloxacin.
Drug susceptibility determination.
The MICs of enrofloxacin (Vetoquinol, Lure, France), marbofloxacin (Vetoquinol), ciprofloxacin (Bayer AG, Leverkusen, Germany), florfenicol (Shering-Plough Animal Health, Kenilworth, N.J.), and erythromycin (Fluka, Steiheim, Germany) were determined by the standard agar doubling dilution method on Mueller-Hinton medium with inocula of 104 CFU per spot. The MIC was defined as the lowest concentration of the drug that completely inhibited visible growth after an incubation of 18 h at 37°C. The following MIC breakpoints (c and C μg/ml), defined by the Comité de l'Antibiogramme de la Société Française de Microbiologie (37), were used to classify strains as susceptible (MIC ≤ c), intermediate (c < MIC < C), or resistant (MIC > C): enrofloxacin, ciprofloxacin, and marbofloxacin, 1 and 2; florfenicol, 8 and 16; and erythromycin, 1 and 4. MICs of these antibiotics were also determined in the presence of 80 μg of the efflux pump inhibitor Phe-Arg-β-naphthylamide (PAβN) (Sigma-Aldrich, Steinheim, Germany) per ml (29, 49).
Selection of FQ-resistant mutants.
Strains were grown overnight, and 100 μl of the inoculum (109 CFU/ml) was plated onto LB-agar plates containing concentrations of enrofloxacin or marbofloxacin from one to eight times the MIC of the parental strain. Selection was performed with 0, 20, or 80 μg of PAβN per ml. Plates were incubated for 24 to 48 h at 37°C, and singly resistant colonies were purified on selective LB-agar plates containing the same FQ concentration as that used for the selection. The frequency with which mutants were selected was calculated as the ratio of the number of bacteria growing at a defined FQ concentration divided by the number of bacteria in the original inoculum.
PCR amplification and sequencing of QRDR regions of gyrA, gyrB, parC, and parE genes.
The sequences of the primers used in the PCR amplifications of the QRDR of gyrA, gyrB, parC, and parE are shown in Table 2. PCR and nucleotide sequencing were performed as described previously (3).
TABLE 2.
Primers used for PCRs
| Target region | Primer | Nucleotide positiona | Oligonucleotide sequence (5′ to 3′) | Annealing temp (°C) |
|---|---|---|---|---|
| Detection of QRDR mutations | ||||
| gyrA | stgyrA1 | 2376240 | TGTCCGAGATGGCCTGAAGC | 55 |
| stgyrA12 | 2375771 | CGTTGATGACTTCCGTCAG | ||
| gyrB | stgyrB6 | 4039392 | AACGGTCTGCTCATCAGAAAGG | 58 |
| stgyrB7 | 4040102 | GAAATGACCCGCCGTAAAGG | ||
| parC | stparC1 | 3339205 | ATGAGCGATATGGCAGAGCG | 53 |
| stparC2 | 3338793 | TGACCGAGTTCGCTTAACAG | ||
| parE | stparE1 | 3344622 | GACCGAGCTGTTCCTTGTGG | 52 |
| stparE2 | 3344130 | GCGTAACTGCATCGGGTTCA | ||
| Detection of IS | ||||
| acrRA | acrR1 | 533463 | CAGTGGTTCCGTTTTTAGTG | 58 |
| acrR2 | 534455 | ACAGAATAGCGACACAGAAA | ||
| acrSE | acrsD | 3559826 | TGGCGAAAGCGTTAAATCTG | 58 |
| acrsG | 3561164 | CAGCGCAGCAGAGAATATGA | ||
| emrRA | emrG | 2961079 | TGTCGTTACTATATCGGCTG | 58 |
| emrD | 2962150 | CTGCTTGCTGTTAATCATCA | ||
| Screening of IS1 and IS10 | ||||
| IS1-acrE | IS1D | TCAGTAAGTTGGCAGCATCA | 58 | |
| acrsG | 3561164 | CAGCGCAGCAGAGAATATGA | ||
| IS10-acrE | IS10D | TCGCTTTGGTTGGCAGGTTA | 58 | |
| acrsG | 3561164 | CAGCGCAGCAGAGAATATGA | ||
| Construction of IS1 and IS10 probes | ||||
| IS1 | IS1-11 | GATAATGCCCGATGACTTTG | 55 | |
| IS1-12 | TGATGGTGTTTTTGAGGTGC | |||
| IS10 | IS10-11 | CCGTAGGCAGGACTTTTCAA | 55 | |
| IS10-12 | CGACTTATGGTATTGCGAGC | |||
| Construction of deletion mutants | ||||
| acrF-kan | acrFH1-P1 | 3564560 | CGATAAAGTCGTTAACGTAATAGCCGCCCAACGCCG | 52 |
| CGGAGATGGTTTCG-GTGTAGGCTGGAGCTGCTTC | ||||
| acrFH2-P2 | 3562888 | CAGTTAAAAGTACAGAACGACCAGATTGCGGCAGGC | ||
| CAACTGGGCGGCAC-CATATGAATATCCTCCTTAG | ||||
| acrS-kan | acrSH1-P1 | 3560688 | GCGAAGAAAACGAAGGCGGATGCGCTTAAAACGCG | |
| GCAACATTTGATTGA-GTGTAGGCTGGAGCTGCTTC | ||||
| acrSH2-P2 | 3560031 | GGCTTCTTCCGCCTGTTGTTCATTTGGCATTAATTGC | 52 | |
| CTCACACTGCCAT-CATATGAATATCCTCCTTAG | ||||
| RT-PCR expression analysis | ||||
| acrF | acrFP1 | 3562338 | TGGCGCACTGGCAATAATGCAA | 63 |
| acrFP2 | 3562623 | AAAGGCGTGGCGAGCTGCAATTTA | ||
| acrS | acrSRP1 | 3560663 | TTAAAACGCGGCAACATTTGA | 58 |
| acrSP2 | 3560495 | GCTGTAACCAAACCTCATTAAACA | ||
| gyrB | gyrBP1 | 4041269 | TGTCGAATTCTTATGACTCCTCCA | 60 |
| gyrBP2 | 4041121 | TCGTCGATAGCGTTATCTACCA | ||
| Construction of fusions plasmids | ||||
| p415PacrE | EcoRIpacrE102-F | 3560761 | CTAGGAATTCTCATGTCCTTTTTAGTCAGACGA | 50 |
| BamHIpacrE102-R | 3561081 | CTAGGGATCCAGCTCGTTATTCACCGGATA | ||
| p415PIS1 | EcoRIpIS1-F | CTAGGAATTCCCATTCATGGCCATATCAAT | ||
| p415PIS10 | EcoRIpIS10-F | CTAGGAATTCAACATTGCTCTGAAAGCGGG | 50 | |
| BamHIpacrE102-2R | 3561087 | CTAGGGATCCAGCTCGTTATTCACCGGATA |
Detection of IS upstream of acrA, acrE, and emrA.
The presence of IS in the regions covering acrR to acrA, acrS to acrE, and emrR to emrA of the respective acrRAB, acrSEF, and emrRAB efflux loci was assessed by PCR. The sequences of the primers used are shown in Table 2. PCR was performed with 0.2 μM each primer, 200 μM each deoxynucleoside triphosphate (dNTP), 1× Taq buffer, and 1.25 U of Taq DNA polymerase. A single colony was used as template DNA. After a 5-min denaturation at 95°C, amplification was performed for 30 cycles of 1 min at 95°C, 2 min at 58°C, and 3 min at 72°C, with a final extension of 10 min at 72°C. The presence of IS was detected by an increase in the size of the PCR amplicon relative to that expected for the respective parental strain. Three representative PCR products of different sizes longer than expected were sent for nucleotide sequencing to Genome Express (Meylan, France), and the IS were identified by homology searches using BLASTN (www.ncbi.nlm.nih.gov/BLAST).
Screening of IS1 or IS10 in the acrSE intergenic region.
PCRs were performed to screen the presence of an IS1 or an IS10 element in the acrSE intergenic region of all the selected mutants carrying an IS. IS1- and IS10-specific primers were used together with acrsG (Table 2). A single colony was used as template DNA, with 0.2 μM each primer, 200 μM each dNTP, 1× Taq buffer, and 0.6 U of Taq DNA polymerase. The PCR conditions were as follows: 5 min at 95°C, 30 cycles of 1 min at 95°C, 2 min at 58°C, and 3 min at 72°C, and a final extension of 10 min at 72°C.
RNA extraction and DNase treatment.
Overnight cultures were diluted 1/100 in LB medium and grown to the mid-logarithmic phase (optical density at 600 nm [OD600] = 0.5) at 37°C with shaking, Aliquots of cultures (4 ml) were pelleted by centrifugation at 14,000 × g for 10 min and lysed with 1 ml of TRIzol (Invitrogen, Cergy-Pontoise, France). Homogenized samples were incubated for 5 min at 30°C, shaken with 0.2 ml of chloroform, and centrifuged for 15 min at 12,000 × g. RNA, in the aqueous phase, was precipitated with 0.5 ml of isopropyl alcohol. After 10 min of incubation at room temperature, samples were centrifuged for 10 min at 12,000 × g. The RNA pellet was washed with 1 ml of 75% ethanol, air dried for 5 to 10 min, and dissolved in 25 μl of RNase-free water at 55°C. The total RNA concentration was estimated by an OD260 measurement. A 1-μg portion of total RNA was mixed with 1 U of DNase 1 (amplification grade; Invitrogen) in 1× DNase I buffer for 15 min at room temperature in a volume of 10 μl. The DNase was inactivated with 1 μl of 25 mM EDTA followed by 10 min at 65°C.
RT-PCR.
Reverse transcription-PCR (RT-PCR) was used to assess expression of acrF and acrS. Total RNA (1 μg), dNTPs (500 μM each), and 50 ng of random hexamers (Promega, Madison, Wis.) were incubated for 5 min at 65°C, chilled on ice, and then reverse transcribed in a volume of 20 μl containing 0.01 M dithiothreitol, 40 U of RNaseOUT RNase inhibitor (Invitrogen, Cergy-Pontoise, France), 200 U of Superscript II reverse transcriptase (Invitrogen), and 1× first-strand buffer for 50 min at 42°C and then 15 min at 70°C. Generated cDNA was incubated for 20 min at 37°C with 1 μl of RNase A (500 μg/ml; Qbiogene) and stored at −20°C until use. Differences in acrF and acrS gene expression were estimated by PCR, using target-specific primers acrFP1 and acrFP2 for acrF and envRP1 and envRP2 for acrS (Table 2). Total cDNA (1 μl) was amplified in a 20-μl final volume containing 0.5 μM each target specific primer, 250 μM each dNTP. 1× Taq buffer, and 0.5 U of Taq DNA. Amplifications were performed with an initial step of 3 min at 95°C followed by 35 cycles of 20 s at 95°C, 20 s at the annealing temperature, and 20 s at 72°C. Constitutive expression of gyrB assessed in the same cDNA preparation was used as a control, using primers gyrBP1 and gyrBP2 (Table 2). PCR products were detected on a 1.5% agarose gel containing ethidium bromide, and the level of gene overexpression was estimated by comparison of the band intensities relative to twofold serial dilutions of cDNAs.
Promoter strength determination.
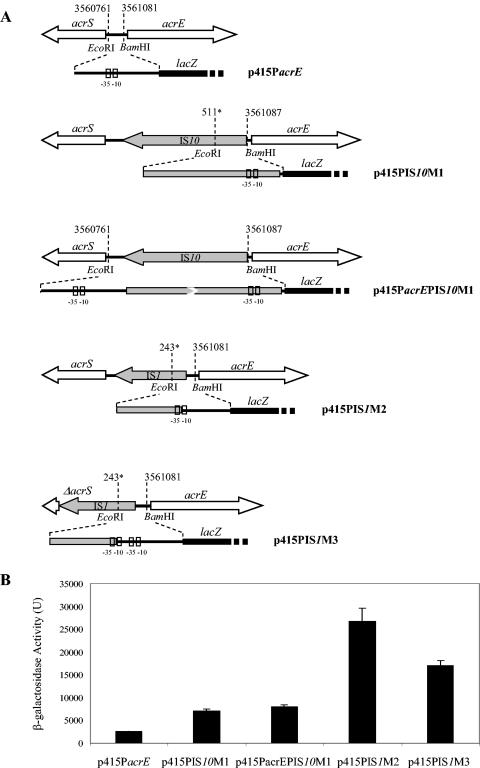
Fusions of the lacZ gene with different promoters for acrEF were constructed on plasmid pRS415 (52) (see Fig. 3). The promoters created after insertion of an IS upstream of acrEF, and also the wild-type promoter for acrEF used as control, were amplified by PCR using primers EcoRIpacrE102-F plus BamHIpacrE102-R for 102SA00, EcoRIpIS10-F plus BamHIpacrE102-2R or EcoRIpacrE102-F plus BamHIpacrE102-2R for 102SA00acrB::kan M1, and EcoRIpIS1-F plus BamHIpacrE102-R for both 102SA00acrB::kan M2 and 102SA00acrB::kan M3 (Table 2). PCR was performed on 200 ng of genomic DNA in a volume of 100 μl with 0.4 μM each primer, 200 μM each dNTP, 1× buffer, and 1 U of DyNazyme EXT polymerase (Finnzymes, Espoo, Finland). After denaturation for 3 min at 94°C, amplification was performed for 30 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C, with a final 10-min extension at 72°C. PCR products of 320 bp (strain 102SA00), 531 bp or 1,668 bp (strain 102SA00acrB::kan M1), 428 bp (strain 102SA00acrB::kan M2), and 477 bp (strain 102SA00acrB::kan M3) were purified with a Qiaquick spin PCR purification kit (Qiagen S.A.). PCR products and plasmid pRS415 were digested with EcoRI and BamHI (Qbiogene) and ligated overnight with T4 DNA ligase (Promega). Ligation products were electroporated in E. coli TG1 cells. Transformants carrying plasmids p415PacrE, p415PIS10-M1, p415PacrEPIS10-M1, p415PIS1-M2, and p415PIS1-M3 (see Fig. 3) were selected on LB plates containing 100 μg of ampicillin per ml and confirmed by PCR. Overnight cultures of selected transformants were diluted 1/50 in A medium [K2HPO4, 10.5 g/liter; KH2PO4, 4.5 g/liter; (NH4)2SO4 1 g/liter; sodium citrate · 2H2O, 0.5 g/liter] containing 100 μg of ampicillin per ml 0.4% glucose (Prolabo, Paris, France), 1 μg of vitamin B1 (Sigma) per ml, and 1 mM MgSO4 and grown to the mid-logarithmic phase (OD600 = 0.4) at 37°C with shaking. Cultures were cooled on ice for 20 min, and the cells were immediately permeabilized by mixing 1 ml of culture with 0.9 ml of Z buffer (1 mM MgSO4 · 7H2O, 0.6 M Na2HPO4 · 7H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, 50 mM β-mercaptoethanol), 2 drops of chloroform, and 1 drop of 0.1% sodium dodecyl sulfate. Samples were equilibrated at 28°C. At zero time, 0.2 ml of o-nitrophenyl-β-d-galactopyranoside at 4 mg/ml (Sigma) was added. The time was recorded, and the reaction was stopped by adding 0.5 ml of 1 M Na2CO3 after a sufficient yellow color has developed. The OD420 and OD550 were recorded for each sample. The activity of β-galactosidase was determined in Miller units, using the formula Units = 1,000 × [OD420 − (1.75 × OD550)]/(T × V × OD600), with OD420 and OD550 being read from the reaction mixture, OD600 being read from the culture before the assay, T being the time of the reaction in minutes, and V being the volume of culture in milliliters (38).
FIG. 3.
(A) Schematic representation of fusions of lacZ with different acrEF promoters flanked by EcoRI and BamHI restriction sites and cloned in pRS415 (52). Position numbers are relative to GenBank accession number NC_003197. Numbers followed by a star are relative to the IS1 and IS10 nucleotide sequences. The −35 and −10 hexamers of the respective acrEF promoters are represented by small boxes. (B) Specific β-galactosidase activity is represented in Miller units (38) as the mean of the results of three experiments; standard deviations are shown by thin bars.
Construction of acrS and acrF deletion mutants.
The gene inactivation method of Datsenko and Wanner (11) was used to create ΔacrS::kan and ΔacrF::kan mutants of susceptible serovar Typhimurium DT104 strain S92/1495 as described previously (2). Plasmid pKD4 carrying the kan gene was used as the plasmid template. The 50 nucleotides homologous to the gene to be inactivated and extended to pKD4 specific primers P1 and P2 (11) are listed in Table 2. Both mutations were then introduced into FQ-resistant serovar Typhimurium DT204 strains 102SA00 and 902SA92 by transduction using phage P22 as described previously (2, 3, 50). The resulting ΔacrS::kan and ΔacrF::kan mutants were selected on LB plates containing 50 μg of kanamycin per ml. Replacement of the target gene with the kan resistance gene was confirmed by PCR.
Southern blot analysis.
The number of IS elements was assessed by Southern blotting. Chromosomal and plasmid DNAs were extracted using the QIAamp DNA mini kit and the Qiagen plasmid mini kit (Qiagen, Courtaboeuf, France), respectively. Both DNAs were digested with EcoRV (one restriction site in IS10, none in IS1) or Mlu1 (one restriction site in IS1, none in IS10) (Qbiogene). Restriction fragments were separated by electrophoresis on a 0.8% agarose gel, depurinated with 0.25 N HCl for 20 min, denatured with 0.5 N NaOH-1.5 M NaCl for 20 min, neutralized with 1 M Tris (pH 7.4)-1.5 M NaCl for 20 min, and transferred to positively charged nylon membranes (Roche), using a 20× standard sodium citrate solution (SSC) (Quantum Appligene), for 1 h. Blots were washed in 2X SSC solution, and DNA fragments were cross-linked to the membrane at 120°C for 20 min. Prehybridization was carried out at 42°C in ECL gold hybridization buffer containing 5 M NaCl and 5% of blocking reagent (Amersham, Orsay, France). Hybridization was carried out overnight in prehybridization solution containing 2 ng of denatured probes per ml. Probes consisted of IS1 and IS10 internal PCR products, using primers listed in Table 2 and labeled by the ECL direct nucleic acid labeling system (Amersham, Orsay, France). The blots were washed in a urea (360 g/liter)-SDS (4 g/liter)-5× SSC buffer at 42°C for 20 min and then in 2× SSC at room temperature. Southern blots were developed using the ECL peroxidase detection system (Amersham), and revealed in a dark room on ECL Hyperfilms (Amersham).
RESULTS
Restoration of high-level resistance to FQs and other drugs by selection on FQs.
The in vitro selection of FQ-resistant S. enterica serovar Typhimurium strains was achieved with one single passage using increasing concentrations of the two FQs enrofloxacin and marbofloxacin from one to eight times the MIC of the parental strain (Table 3). High-level FQ-resistant mutants of the two serovar Typhimurium DT204 strains 902SA92 and 102SA00, with FQ MICs ranging from 64 to 512 μg/ml, i.e., from two to eight times the MIC for the parental strains (Table 1), were selected with frequencies ranging from 10−7 to 10−9 (Table 3). The frequency of selection decreased by about 1/10 with the addition of 20 μg of PAβN per ml, an efflux pump inhibitor previously shown to potentiate the activity of FQs in E. coli, Pseudomonas aeruginosa, and serovar Typhimurium (3, 29, 35, 49), and decreased below 10−9 when 80 μg of PAβN per ml was added. No such high-level FQ-resistant mutants were obtained when the selection was performed with the two serovar Typhimurium DT104 strains BN10055 and 543SA98 (data not shown). However, high-level FQ-resistant mutants were also selected from the acrB::kan mutants of serovar Typhimurium DT204 strains 902SA92 and 102SA00. These selected mutants showed FQ MICs ranging from 16 to 128 μg/ml, i.e., from 8 to 64 times the MIC for their parental acrB::kan strain (Table 1). The frequencies with which these strains were selected ranged from 10−7 to 10−9, and this value decreased to 10−9 or even below when 20 or 80 μg of PAβN per ml, respectively, was added to the selection medium (Table 3). FQ MICs for these selected mutants decreased from 4- to 32-fold in the presence of 80 μg of PAβN per ml, suggesting that the efflux-mediated mechanism plays a role in their high-level FQ resistance phenotype (Table 1). This was further supported by the restoration of the multiple antibiotic resistance phenotype in these selected clones, since they showed additional restoration of resistance to unrelated antibiotics such as florfenicol and erythromycin (Table 1). Moreover, if strains 102SA00 and 902SA92 both contain a double mutation in gyrA, one mutation in gyrB, and one mutation in parC, no additional mutations were found in the QRDRs of gyrA, gyrB, parC, and parE of these high- level FQ-resistant selected mutants (data not shown). Thus, these results showed that high-level FQ-resistant mutants were easily selectable from serovar Typhimurium DT204 acrB::kan mutant strains and that another efflux mechanism(s) from that mediated by AcrAB-TolC was probably involved. Therefore, we further investigated, for these high-level FQ-resistant mutants, the activity of other putative efflux pumps reported for E. coli to participate in FQ and multidrug resistance, in particular AcrEF and EmrAB (23, 27, 40).
TABLE 3.
Frequency of selection of FQ-resistant mutants of S. enterica serovar Typhimuriuma
| Parental strain | FQ used for selection | MIC (μg/ml) | Antibiotic strengh | Frequency of selection
|
||
|---|---|---|---|---|---|---|
| − PAβN | + PAβN (20 μg/ml) | + PAβN (80 μg/ml) | ||||
| 102SA00 | Marbofloxacin | 32 | 4× MIC | 1.8 × 10−8 | 8.3 × 10−8 | <10−9 |
| 8× MIC | <10−9 | <10−9 | <10−9 | |||
| Enrofloxacin | 64 | 4× MIC | 3.15 × 10−6 | 2 × 10−6 | <10−9 | |
| 8× MIC | 1.5 × 10−7 | 5.9 × 10−8 | <10−9 | |||
| 102SA00acrB::kan | Marbofloxacin | 2 | 4× MIC | 1.2 × 10−8 | 5.2 × 10−9 | <10−9 |
| 8× MIC | 1.7 × 10−9 | <10−9 | <10−9 | |||
| Enrofloxacin | 2 | 4× MIC | 8.2 × 10−8 | 5.3 × 10−8 | <10−9 | |
| 8× MIC | 4.4 × 10−9 | 2.6 × 10−9 | <10−9 | |||
| 902SA92 | Marbofloxacin | 32 | 2× MIC | 6.8 × 10−9 | <10−9 | <10−9 |
| 4× MIC | <10−9 | <10−9 | <10−9 | |||
| 8× MIC | <10−9 | <10−9 | <10−9 | |||
| Enrofloxacin | 64 | 2× MIC | >10−6 | 5.8 × 10−7 | <10−9 | |
| 4× MIC | 3.7 × 10−7 | <10−9 | <10−9 | |||
| 8× MIC | <10−9 | <10−9 | <10−9 | |||
| 902SA92acrB::kan | Marbofloxacin | 2 | 4× MIC | 6.3 × 10−9 | 1.6 × 10−9 | <10−9 |
| 8× MIC | <10−9 | <10−9 | <10−9 | |||
| Enrofloxacin | 2 | 4× MIC | 8 × 10−8 | 2.8 × 10−8 | <10−9 | |
| 8× MIC | 5 × 10−8 | 1 × 10−9 | <10−9 | |||
Mutants were selected on plates containing either marbofloxacin or enrofloxacin at concentrations from one to eight times the MIC. Selection was performed without (−PAβN) or with (+PAβN) 20 or 80 μg of the efflux pump inhibitor PAβN per ml.
Identification of IS1 and IS10 elements upstream of acrEF.
For all the FQ-resistant mutants selected in vitro, the presence of any IS in the regulatory sequences of acrAB, acrEF and emrRAB was sought by PCR. PCR products larger than expected from the wild-type background, i.e., larger than 992, 1,338 and 1,071 bp, respectively, according to the nucleotide sequences of serovar Typhimurium strain LT2 (36), were indicative of the presence of IS. None were detected in the regions covering acrR to acrA and emrR to emrA for all the FQ-selected mutants (data not shown). However, acrS to acrE region PCR products larger than expected were obtained for all the FQ-selected mutants derived from strains 902SA92acrB::kan and 102SA00acrB::kan. Indeed, amplicon sizes were about 1,400 bp (1 strain), 2,100 bp (9 strains), or 2,700 bp (11 strains) long, instead of the 1,338 bp expected for the wild-type strains. It is interesting that no such insertions were found in the acrS to acrE region of the mutants selected from wild-type serovar Typhimurium DT204 strains (20 strains tested) or from wild-type or acrB::kan mutant strains of serovar Typhimurium DT104 strains (14 strains tested) (data not shown).
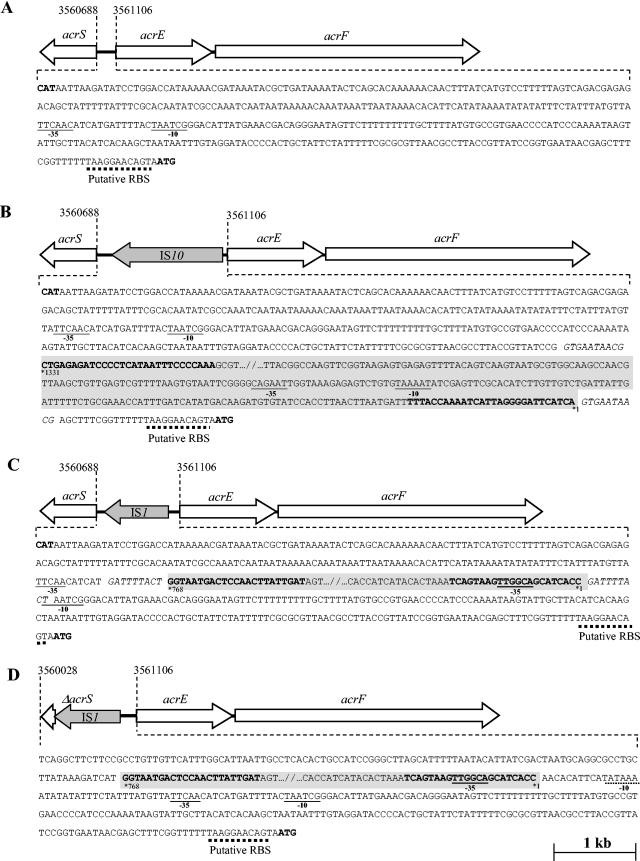
To identify the nature of these insertions, the acrS to acrE region of three representative mutants selected from strain 102SA00acrB::kan (M1, M2, and M3, with amplicon sizes of 2,633, 2,062 and 1,351 bp, respectively) was sequenced. Nucleotide sequencing identified either an IS1 or an IS10 element integrated upstream of acrE for the three mutants. IS10 was 98% identical to that identified in plasmid pHCM1 of S. enterica serovar Typhi strain CT18, and IS1 was 100% identical to that identified in the chromosome of the same bacterium (GenBank accession number NC_003198) (44). The three following situations were observed (Fig. 1). In mutant M1, an IS10 element of 1,331 bp was found inserted at position 3561078 relative to the strain LT2 genome sequence (36), separating acrE from its normal promoter. IS10 carries a 1,209-bp open reading frame encoding a transposase, comprises two imperfect inverted terminal repeats about 29 bp long (IRR and IRL), and is flanked by a 10-bp duplication of the target sequence GTGAATAACG (33). Both ends of IS10 carry entire promoter sequences named pOUT and OUTIIp (8, 34, 51). Our hypothesis was that the −35 and −10 sequences located at the left end of IS10 and previously described as pOUT could provide an alternative promoter for acrEF (Fig. 1). In mutant M2, an IS1 element of 768 bp was found inserted at position 3560906 relative to the strain LT2 genome sequence, disrupting the promoter of acrE. IS1 contains two adjacent genes called insA and insB, both required for IS1 transposition and IS1-mediated plasmid cointegration, comprises two imperfect terminal repeats about 23 bp long (IRR and IRL), and is flanked by a 9-bp duplication of the target sequence GATTTTACT (22, 41). Both ends of IS1 contain outward-facing −35 regions, which can form hybrid promoters with the existing −10 regions when transposed at the proper distance on target DNA (Fig. 1) (47). Our hypothesis was that the new −35 sequence located in the terminal IRL of IS1, together with the acrEF original −10 hexamer, could also provide an alternative promoter for acrEF. Finally, the situation was quite different for mutant M3, where an IS1 element was also found integrated upstream of the acrE gene but together with a deletion of most of acrS, the putative local repressor gene of acrEF. Indeed, 706 bp between positions 3560140 and 3560847 relative to the strain LT2 genome sequence was deleted and replaced by the IS1 sequence. In this case, the original acrEF promoter was still intact but the optimal 18 bp between the −35 hexamer located at the end of IS1 and a cryptic −10 hexamer on the target sequence was very likely to provide a new promoter for acrEF (Fig. 1). It is interesting that the IS1-mediated deletion in this mutant also resulted in the lack of any functional AcrS protein, a protein supposed to downregulate acrEF expression (21, 32, 43).
FIG.1.
Schematic representation of IS1 and IS10 integrated upstream of acrEF and organization of new promoter sequences. (A) In S. enterica serovar Typhimurium strain LT2, acrS and acrEF are transcribed divergently, as shown by the open arrows. The putative −35 and −10 promoter regions of acrEF are underlined, and the putative ribosome binding site (RBS) is shown by the dotted line (24). The positions of the first codons of acrS and acrE are indicated as in GenBank accession number NC_003197. (B) In mutant 102SA00acrB::kan M1, IS10 inserted upstream of acrEF is represented by a grey background. It is not represented in full, as shown by the slash bars. The insertion separates the original acrEF promoter from acrEF itself, and new putative −35 and −10 promoter regions located at the left end of IS10, providing an alternative promoter for acrEF, pOUT, are indicated (8, 51). IS10 carries two imperfect inverted terminal repeats about 29 bp long (IRR and IRL), shown in bold type, and is bracketed by a 10-bp duplication of the target sequence, shown in italics (33). (C) In mutant 102SA00acrB::kan M2, IS1 is inserted upstream of acrEF, separating the normal −35 and −10 promoter regions of acrEF. IS1 also carries two imperfect inverted terminal repeats about 23 bp long, shown in bold type, and is bracketed by a 9- bp duplication of the target sequence, shown in italics (22, 41). A new putative promoter sequence created by the −35 hexamer located in the terminal IRL of IS1 (47) and the original −10 hexamer is indicated. (D) In mutant 102SA00acrB::kan M3. IS1 is inserted upstream of acrEF, together with a deletion of 706 bp including the majority of acrS. The original acrEF promoter is intact, but another putative promoter created by the −35 hexamer located in the terminal IRL of IS1 (47) and a cryptic −10 hexamer located in the target sequence (underlined with a dotted line) are indicated.
Once the IS elements were identified in these three representative revertants, we further screened all the other serovar Typhimurium DT204 acrB::kan mutants selected on FQs for the presence of either an IS1 or an IS10 element upstream of acrE. Locus-specific primers together with IS1- or IS10-specific primers were used in PCR experiments (Table 2). The results showed that for 13 revertants selected from strain 102SA00acrB::kan, 8 carried an IS1 element and 5 carried an IS10 element in the acrSE region. Similarly, for 10 revertants selected from strain 902SA92acrB::kan, 2 carried an IS1 element and 6 carried an IS10 element in the acrSE region (data not shown). Based on these findings, the integration of either an IS1 or an IS10 element upstream of acrE appeared to be quite common in the high-level FQ-resistant mutants selected from serovar Typhiumrium DT204 strains in which acrB was inactivated.
The consequence of this IS1 or IS10 integration led us to consider the creation of new promoters for acrEF as the most attractive hypothesis (22, 34, 51). However, acrS and acrEF are transcribed divergently (36) and the integration of such IS elements could then also affect (i) acrS transcription, (ii) specific binding of the putative acrEF local repressor AcrS, or (iii) the presence of any functional AcrS protein in the particular case of mutant M3.
Activation of acrEF overexpression.
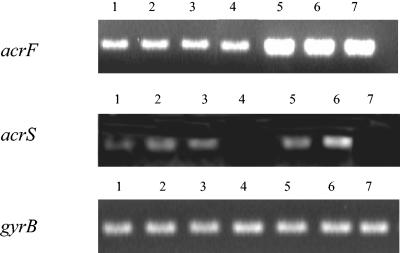
We investigated whether IS1 and IS10 insertions affected the expression of acrEF, The transcription level of acrF was estimated by RT-PCR for the three revertants 102SA00acrB::kan M1, M2, and M3 (Fig. 2). Comparing the acrF transcription levels of these revertants to those of the parental strain 102SA00 and its acrB::kan mutant, an 8- to 10-fold increase in the transcription of acrF was observed. It correlated well with the enhanced FQ MICs for these FQ-selected revertants compared to their respective parental acrB::kan strain (Table 1). Interpretation of these data was correlated by reference to a negative control in RT-PCR experiments, i.e., gyrB, whose transcription was not affected by the treatments used in this study. Indeed, the same band density of gyrB transcription was observed for the seven strains tested. We also investigated whether IS1 and IS10 affected the expression of the outward-facing acrS gene. The results showed that except for revertant 102SA00acrB::kan M3 in wich acrS is mostly deleted, the transcription level of acrS was approximately the same for the five strains tested. Thus, these results strongly suggested that the integration of either an IS1 or an IS10 element in the acrSE region caused an approximately 8- to 10-fold enhancement of the expression of acrEF in the revertants selected from the serovar Typhimurium DT204 acrB::kan mutants. Apparently it did not affect the expression of the outward-facing acrS gene.
FIG. 2.
RT-PCR expression analysis of the efflux pump gene acrF, its local repressor acrS, and the standard gene gyrB in strains S92/1495 (lane 1). 102SA00 (lane 2), 102SA00 acrB::kan (lane 3), 102SA00ΔacrS::kan (lane 4), 102SA00acrB::kan M1 (lane 5), 102SA00acrB::kan M2 (lane 6), and 102SA00acrB::kan M3 (lane 7).
Comparative strength of new promoter sequences.
To confirm that IS1 or IS10 created new promoters for acrEF in revertants 102SA00acrB::kan M1, M2, and M3, fusions of putative promoters with the lacZ gene were constructed in plasmid pRS415 (Fig. 3A). The β-galactosidase activity of constructs p415PIS10-M1, p415PacrEPIS10-M1, p415PIS1-M2, and p415PIS1-M3 was assayed and compared to the β-galactosidase activity obtained when the wild-type promoter for acrEF was fused to lacZ (p415PacrE). The activities were 2,621 ± 22, 7,033 ± 426, 7,995 ± 432, 26,643 ± 2,971, and 17,007 ± 1,049 Miller units for plasmids p415PacrE, p415PIS10-M1, p415PacrEPIS10-M1, p415PIS1-M2, and p415PIS1-M3, respectively (Fig. 3B). This corresponded to a 2.7-, 3-, 10.1-, and 6.4-fold increase in the expression of β-galactosidase for plasmids p415PIS10-M1, p415PacrEPIS10-M1, p415PIS1-M2, and p415PIS1-M3, respectively, compared to wild-type p415PacrE. These results confirmed that the new promoters created after IS1 or IS10 integration in mutants 102SA00acrB::kan M1, M2, and M3 were, at least in part, responsible for the increase in expression of the downstream genes acrEF.
Role of acrSEF in FQ and multidrug resistance.
The acrF and acrS genes were deleted by method of Datsenko and Wanner (11) in one serovar Typhimurium DT104 (S92/1495) and two serovar Typhimurium DT204 (102SA00 and 902SA92) strains to determine their respective role in multidrug resistance. MICs of the FQs marbofloxacin, enrofloxacin, ciprofloxacin, and the unrelated drugs florfenicol and erythromycin, known to be substrates of RND pumps, were determined in the presence or absence of 80 μg of the efflux pump inhibitor PAβN per ml (Table 1). This efflux pump inhibitor was previously shown to decrease the FQ and florfenicol MICs for high-level FQ-resistant serovar Typhimurium DT204 strains up to 32-fold (3). Inactivation of either acrB or tolC also resulted in an up to 32-fold decrease of the FQ MICs for these strains (2, 3). In marked contrast, we did not find any significant differences in FQ, florfenicol, or erythromycin MICs between wild-type strains and their ΔacrF::kan or ΔacrS::kan mutants (Table 1). These results showed that neither the AcrEF efflux pump nor the AcrS putative local repressor seemed to play any role in multidrug resistance in a wild-type background.
We also investigated the role of AcrS in acrEF expression. RT-PCR experiments with wild-type serovar Typhimurium DT204 strain 102SA00 and its ΔacrS::kan mutant showed that deletion of acrS did not result in enhanced expression of acrF and therefore AcrS did not appear to affect acrF transcription (Fig. 2). Thus, and in contrast to what has previously been suggested (21, 32, 43), AcrS does not seem to act as a local repressor for acrEF. This hypothesis is also supported by the lack of any increase in MICs for ΔacrS::kan mutants compared to wild-type strains (Table 1).
Occurrence of IS1 and IS10 elements in the serovar Typhimurium chromosome.
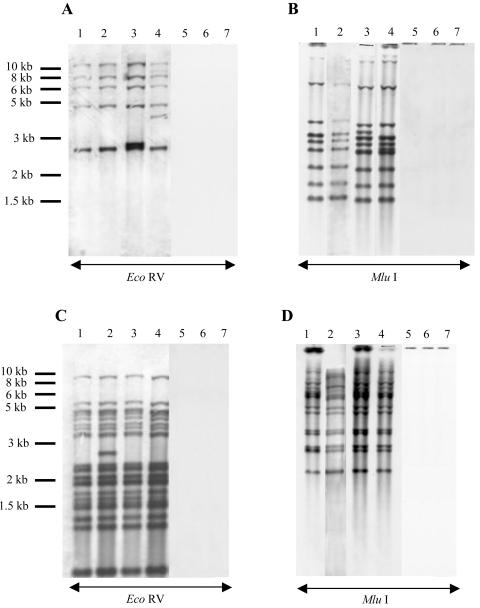
Neither IS1 nor IS10 sequences are present in the genome of serovar Typhimurium reference strain LT2 (36). Since we found such sequences in the revertants selected from serovar Typhimurium DT204 acrB::kan strains, the last aim of this study was to assess the occurrence of IS1 and IS10 in serovar Typhimurium phage types DT104 and DT204. Southern blot experiments were done on chromosomal or plasmid DNAs digested either with EcoRV (one restriction site in IS10, none in IS1) or Mlu1 (one restriction site in IS1, none in IS10). Internal IS1 and IS10 specific probes were used. The hybridization patterns of genomic DNAs are shown in Fig. 4. According to the number of bands detected, we could thus identify around 5 copies of IS1 and 15 copies of IS10 in the chromosome of the serovar Typhimurium DT204 strain 102SA00, with one additional copy of IS10 in 102SA00acrB::kan M1 and one additional copy of IS1 in both 102SA00acrB::kan M2 and M3. Similar results were obtained with the other serovar Typhimurium DT204 strain 902SA92 and two IS1- and IS10-carrying revertants (902SA92 acrB::kan M1 and M2 [Table 1]) (data not shown).
FIG. 4.
Southern blot hybridization analysis of chromosomal DNAs with either IS1 (A and B) or IS10 (C and D) probes. Genomic DNAs were digested with either EcoRV (one restriction site in IS10, none in IS1) or MluI (one restriction site in IS1, none in IS10). The following strains were tested: S. enterica serovar Typhimurium DT204 strains 102SA00 (lane 1), 102SA00acrB::kan M1 (lane 2), 102SA00acrB::kan M2 (lane 3), and 102SA00acrB::kan M3 (lane 4), serovar Typhimurium DT104 strains BN10055 (lane 5), and 543SA98 (lane 6), and serovar Typhimurium reference strain LT2 (lane 7).
IS1 or IS10 elements were not detected in serovar Typhimurium DT104 strains BN10055 and 543SA98 or, as expected, in the serovar Typhimurium reference strain LT2. They were also not detected on plasmid DNAs, whether from the serovar Typhimurium DT104, DT204, or LT2 strains (data not shown). These results showed that only serovar Typhimurium phage type DT204 carries IS1 and IS10 elements, with 5 and 15 chromosomal copies, respectively.
DISCUSSION
As shown in this study, the use of the efflux pump inhibitor PAβN considerably reduced the frequencies with which resistant mutants were selected, whether selection was performed with enrofloxacin or marbofloxacin. This strongly supported the role of an efflux-mediated mechanism in the resistance phenotype of the selected mutants. It had already been shown that active efflux mediated by AcrAB-TolC played a major role in high-level FQ-resistance in S. enterica serovar Typhimurium DT204 (2, 3), together with multiple target gene mutations in gyrA, gyrB, and parC (2, 3, 9). The most interesting observation of the present study was that high-level FQ-resistant mutants could be selected from the serovar Typhimurium DT204 strains in which the acrB gene was inactivated. For these selected mutants, the initial FQ resistance levels observed for the wild-type strains were restored and could therefore be considered “revertants”. Moreover, for these revertants, the levels of resistance to FQs and other unrelated drugs such as florfenicol and erythromycin were very sensitive to the efflux pump inhibitor PAβN whereas it was confirmed that the acrB gene was kept inactivated. Therefore, we investigated the participation of other efflux pumps in their resistance phenotype.
According to previous reports on characterization of FQ efflux mechanisms in E. coli, the multidrug efflux pumps AcrEF and EmrAB, present in the genome of the serovar Typhimurium LT2 strain, appeared good candidates for this reversion phenomenon (23, 27, 32, 36). Indeed, AcrF, like AcrB, belongs to the resistance nodulation division family of transporters, which recognize a wide variety of substrates including FQs (23, 31, 32). ClustalW alignment with sequences from the serovar Typhimurium strain LT2 genome showed that AcrE and AcrF have 66 and 80% amino acid sequence identity to AcrA and AcrB, respectively. The situation is similar in E. coli, where AcrE and AcrF have 65 and 77% amino acid sequence identity to AcrA and AcrB, respectively (31). In a similar manner to acrR and acrAB, acrE and acrF are organized in an operon, with a putative repressor gene acrS located immediately upstream of acrEF and transcribed divergently (31) (Fig. 1). EmrB belongs to the major facilitator superfamily of transporters that also recognizes a wide variety of substrates including quinolones (27). The emrAB genes encoding the pump are also organized in an operon together with emrR, a repressor gene located upstream of emrA (28). EmrB does not have significant identity to AcrB (8%), but, as with AcrF, its role in multidrug resistance in E. coli has previously been described (27, 40). It also therefore seemed a good candidate to study.
Depending on the strains, we identified IS1 or IS10 elements integrated upstream of acrEF, leading to overexpression of acrF. We did not find any insertional mutations in the region covering emrR to emrA or in the region covering acrR to acrA in all strains studied including wild-type strains and acrB::kan mutants. RT-PCR experiments showed that the IS1 or IS10 transposition resulted in an approximately 8- to 10-fold increase in acrF transcription that correlated well with the enhanced FQ, florfenicol, and erythromycin MICs observed for the in vitro FQ-selected revertants. The RT-PCR results also showed that except for revertant 102SA00acrB::kan M3, in which acrS was mostly deleted, the transcription level of acrS was not affected by IS1 or IS10. Moreover, no additional mutations in the QRDR regions of gyrA, gyrB, parC, and parE were detected in the revertants. Therefore, we assumed that restoration of high-level resistance to FQs in these revertants was essentially due to activation of overexpression of acrEF by IS1 or IS10. Since it is only indirect evidence, several complementation experiments were attempted to ensure that the IS elements inserted upstream of acrEF were the only cause of these high-level of FQ resistance, but they failed to do so. Mainly, two previous studies with E. coli are in accord with our findings with serovar Typhimurum, supporting the functional complementation of acrB mutations by acrEF activation with IS elements. The first showed enhanced efflux in acrAB knockout strains selected in vitro with FQs due to enhanced expression of acrEF associated with insertion of IS2 upstream of acrEF (21). The other showed that the solvent hypersensitivity of acrB mutants was suppressed by insertion of IS1 or IS2 elements upstream of acrEF (25). In addition we showed that, as in E. coli (23, 32, 53), the AcrEF efflux pump did not play any intrinsic role in resistance to FQs and other unrelated drugs in serovar Typhimurium. Indeed, deletion of acrF in wild-type serovar Typhimurium did not result in increased susceptibility to FQs, florfenicol, or erythromycin. Another interesting finding in this study was that the putative repressor AcrS does not appear to have any effect on acrEF expression, although ClustalW alignment with sequences from the serovar Typhimurium LT2 genome showed that it has 35% amino acid sequence identity to AcrR, the local repressor of acrAB. It thus ruled out the hypothesis in which overexpression of acrEF, following integration of IS1 or IS10, was due to a lack of functional AcrS and confirmed that IS1 and IS10 integration directly affect the transcription of acrEF.
The mechanism by which acrEF overexpression was activated in the FQ-selected revertants is interesting since it always appeared to be related to IS1 or IS10 transposition upstream of acrEF. Indeed, for the three representative revertants, M1, M2, and M3, analyzed, IS1 or IS10 integration created new promoter sequences for acrEF, according to the consensus sequences described by Hawley and McClure (16), and to the outfacing IS1 and IS10 promoter sequences previously reported (34, 47, 51). In mutant M1, the integration of an IS10 element separated the normal acrEF promoter from acrEF itself but supplied a new promoter, pOUT, located at its terminal end (51). In mutant M2, the integration of an IS1 element disrupted the original acrEF promoter but formed a new hybrid promoter, bringing together the −35 hexamer located in its terminal IRL and the acrEF original −10 hexamer (Fig. 1) (47). Finally, mutant M3 showed a particular situation where transposition of IS1 resulted in the deletion of the majority of acrS. IS1-mediated deletion has been described previously (48), in a study whose authors suggested that the deletion was formed by a mechanism similar to the one described by Campbell for the excision of integrated episomes (5). After IS1 has integrated into the target DNA, its excision can form two types of deletions. In type I deletion, the IS1 element is removed together with chromosomal sequences either to the right or to the left end of IS1. In type II deletion, only DNA sequences adjacent to the right or to the left end of IS1 are deleted, but IS1 itself is not deleted. Both types of deletions are supposed to be formed by recombination between two sites, one created by the integration of IS1 and the other being one of several sites located to the right or to the left end of IS1 (48). The case of mutant 102SA00 acrB::kan M3 consists of a type II deletion since (i) IS1 remains integrated and (ii) the deletion terminates exactly at the site of location of IS1. In this mutant, the acrEF promoter was still intact but a cryptic −10 sequence upstream of acrEF could be used with the −35 hexamer located in the terminal IRL of IS1 to provide an alternative promoter for acrEF (Fig. 1).
New promoter sequences created after IS integration were stronger than the original promoter for acrEF, as shown by β-galactosidase experiments. However, different situations were observed, with hybrid promoters created after IS1 insertion giving higher β-galactosidase activity than did the promoter provided by IS10. It is interesting that for revertant M1 carrying an IS10 element, the β-galactosidase activities were about the same whether the IS10 promoter alone or both the IS10 and the original acrEF promoters were tested. However, this significant difference in promoter strength did not correlate with the level of acrF expression measured by RT-PCR.
Gene activation or inactivation following integration of IS elements is a well-known phenomenon, and the creation of hybrid promoters by IS1 or the supply of an entire promoter sequence by IS10 has been reported for E. coli (8, 47, 51). However, it was not reported for serovar Typhimurium. IS1 or IS10 sequences are not present in the chromosome of the serovar Typhmimurium reference strain LT2, whose genome has completely been sequenced. Together with the fact that insertion of such elements upstream of acrEF were found only in FQ-resistant mutants of the DT204 phage type, this led us to investigate the occurrence of IS1 and IS10 in the chromosome of the serovar Typhimurium DT104 and DT204 strains used in this study. We thus detected by Southern blot hybridization 5 and 15 chromosomal copies of IS1 and IS10, respectively, in serovar Typhimurium DT204 strains. They were not detected in either serovar Typhimurium DT104 strains or, as expected, in the reference strain LT2. Carrying IS1 and/or IS10 in their chromosome could thus be a selective advantage for serovar Typhimurium DT204 strains as opposed to DT104 strains, for which no high-level FQ-resistance or insertional mutations were found. These results underline the importance of transposable elements in the acquisition of efflux-mediated FQ resistance, considering that integration of IS1 or IS10 upstream of acrE was quite a common event in the resistant phenotype of the strains selected in this study. Taken together, these results also underline the role of efflux pumps other than AcrAB, in this study AcrEF, in efflux-mediated FQ resistance of serovar Typhimurium.
Acknowledgments
We thank H. Imberechts and S. Rankin for providing the Salmonella field strains used in this study and S. Baucheron for supplying their acrB::kan mutants. We are grateful to R. W. Simons for his kind gift of plasmid pRS415. We also thank C. Mouline for his expert technical assistance and C. Schouler, P. Germon, I. Payant, and S. Payot-Lacroix for helpful suggestions.
This study was funded by the Direction Générale de l'Alimentation (Ministère de l'Agriculture), projet AQS 2000/S2, by INRA, projet Transversalité, and by a C.I.F.R.E. grant from Vetoquinol.
REFERENCES
- 1.Baucheron, S., S. Tyler, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serotype Typhimurium DT104. Antimicrob. Agents Chemother. 48:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baucheron, S., E. Chaslus-Dancla, and A. Cloeckaert. 2004. Role of TolC and parC mutation in high-level fluoroquinolone resistance in Salmonella enterica serotype Typhimurium DT204. J. Antimicrob. Chemother. 53:657-659. [DOI] [PubMed] [Google Scholar]
- 3.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, A. 1969. Episomes. Harper & Row, New York, N.Y.
- 6.Casin, I., J. Breuil, J. Darchis, C. Guelpa, and E. Collatz. 2003. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica Typhimurium isolates in humans. Emerg. Infect. Dis. 9:1455-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu, C. H., T. L. Wu, L. H. Su, C. Chu, J. H. Chia, A. J. Kuo, M. S. Chien, and T. Y. Lin. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis. N. Engl. J. Med. 346:413-419. [DOI] [PubMed] [Google Scholar]
- 8.Ciampi, M. S., M. B. Schmid, and J. R. Roth. 1982. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc. Natl. Acad. Sci. USA 79:5016-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloeckaert, A., and E. Chaslus-Dancla. 2001. Mechanisms of quinolone resistance in Salmonella. Vet. Res. 32:291-300. [DOI] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., and S. Schwarz. 2001. Molecular characterization, spread and evolution of multidrug resistance in Salmonella enterica Typhimurium DT104. Vet. Res. 32:301-310. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galas, D. J., and M. Chandler. 1989. Bacterial insertion sequences, p. 109-162. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 13.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerra, B., B. Malorny, A. Schroeter, and R. Helmuth. 2003. Multiple resistance mechanisms in fluoroquinolone-resistant Salmonella isolates from Germany. Antimicrob. Agents Chemother. 47:2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen, H., W. Rabsch, and A. P. Heisig. 2001. Effect of gemifloxacin on in vitro selected mutants and fields isolates of Salmonella spp. with mutations in gyrA, gyrB, parC and marR. 7th Int. Symp. New Quinolones, abstr. P80.
- 16.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic. Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heisig, P. 1993. High-level fluoroquinolone resistance in a Salmonella typhimurium isolate due to alterations in both gyrA and gyrB genes. J. Antimicrob. Chemother. 32:367-377. [DOI] [PubMed] [Google Scholar]
- 18.Heisig, P., B. Kratz, E. Halle, Y. Graser, M. Altwegg, W. Rabsch, and J. P. Faber. 1995. Identification of DNA gyrase A mutations in ciprofloxacin-resistant isolates of Salmonella Typhimurium from men and cattle in Germany. Microb. Drug. Resist. 1:211-218. [DOI] [PubMed] [Google Scholar]
- 19.Hsueh, P.-R., L.-J. Teng, S.-P. Tseng, C.-F. Chang, J.-H. Wan, J.-J. Yan, C.-M. Lee, Y.-C. Chuang, W.-K. Huang, D. Yang, J.-M. Shyr, K.-W. Yu, L.-S. Wang, J.-J. Lu, W.-C. Ko, J.-J. Wu, F.-Y. Chang, Y.-C. Yang, Y.-J. Lau, Y.-C. Liu, C.-Y. Liu, S.-W. Ho, and K.-T. Luh. 2004. Ciprofloxacin-resistant Salmonella enterica Typhimurium and Choleraesuis from pigs to humans. Taiwan. Emerg. Infect. Dis. 10:60-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imberechts, H., I. D'Hooghe, H. Bouchet, C. Godard, and P. Pohl. 2000. Apparent loss of enrofloxacin resistance in bovine Salmonella typhimurium strains isolated in Belgium, 1991 to 1998. Vet. Rec. 147:76-77. [DOI] [PubMed] [Google Scholar]
- 21.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnsrud, L. 1979. DNA sequence of the transposable element IS1. Mol. Gen. Genet. 169:213-218. [DOI] [PubMed] [Google Scholar]
- 23.Kawamura-Sato, K., K. Shibayama, T. Horii, Y. Iimuma, Y. Arakawa, and M. Ohta. 1999. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol. Lett. 179:345-352. [DOI] [PubMed] [Google Scholar]
- 24.Klein, J. R., B. Henrich, and R. Plapp. 1991. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol. Gen. Genet. 230:230-240. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, K., N. Tsukagoshi, and R. Aono. 2001. Suppression of hypersensitivity of Escherichia coli acrB mutant to organic solvents by integrational activation of the acrEF operon with the IS1 or IS2 element. J. Bacteriol. 183:2646-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling, J. M., E. W. Chan, A. W. Lam, and A. F. Cheng. 2003. Mutations in topoisomerase genes of fluoroquinolone-resistant salmonellae in Hong Kong. Antimicrob. Agents Chemother. 47:3567-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomovskaya, O., and K. Lewis. 1992. emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomovskaya, O., K. Lewis, and A. Matin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lontie, M., J. Verhaegen, M. L. Chasseur-Libotte, and L. Verbist. 1994. Salmonella typhimurium serovar Copenhagen highly resistant to fluoroquinolones. J. Antimicrob. Chemother. 34:845-846. [DOI] [PubMed] [Google Scholar]
- 31.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, D., D. N. Cook, J. E. Hearst, and H. Nikaido. 1994. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 33.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Garcia, E., J. M. Navarro-Llorens, and A. Tormo. 2003. Identification of an unknown promoter, OUTIIp, within the IS10R element. J. Bacteriol. 185:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazzariol, A., Y. Tokue, T. M. Kanegawa, G. Cornaglia, and H. Nikaido. 2000. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 44:3441-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 37.Members of the SFM Antibiogram Committee. 2003. Comité de l'antibiogramme de la société française de microbiologie report 2003. Int. J. Antimicrob. Agents 21:364-391. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Nakaya, H., A. Yasuhara, K. Yoshimura, Y. Oshihoi, H. Izumiya, and H. Watanabe. 2003. Life-threatening infantile diarrhea from fluoroquinolone-resistant Salmonella enterica Typhimurium with mutations in both gyrA and parC. Emerg. Infect. Dis. 9:255-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohtsubo, H., and E. Ohtsubo. 1978. Nucleotide sequence of an insertion element, IS1. Proc. Natl. Acad. Sci. USA 75:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen, S. J., E. E. DeBess, T. E. McGivern, N. Marano, T. Eby, S. Mauvais, V. K. Balan, G. Zirnstein, P. R. Cieslak, and F. J. Angulo. 2001. A nosocomial outbreak of fluoroquinolone-resistant salmonella infection. N. Engl. J. Med. 344:1572-1579. [DOI] [PubMed] [Google Scholar]
- 43.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769-775. [DOI] [PubMed] [Google Scholar]
- 44.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 45.Piddock, L. J. 2002. Fluoroquinolone resistance in Salmonella serovars isolated from humans and food animals. FEMS Microbiol. Rev. 26:3-16. [DOI] [PubMed] [Google Scholar]
- 46.Piddock, L. J., D. G. White, K. Gensberg, L. Pumbwe, and D. J. Griggs. 2000. Evidence for an efflux pump mediating multiple antibiotic resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:3118-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prentki, P., B. Teter, M. Chandler, and D. J. Galas. 1986. Functional promoters created by the insertion of transposable element IS1. J. Mol. Biol. 191:383-393. [DOI] [PubMed] [Google Scholar]
- 48.Reif, H. J., and H. Saedler. 1975. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol. Gen. Genet. 137:17-28. [DOI] [PubMed] [Google Scholar]
- 49.Renau, T. E., R. Leger, E. M. Flamme, J. Sangalang, M. W. She, R. Yen, C. L. Gannon, D. Griffith, S. Chamberland, O. Lomovskaya, S. J. Hecker, V. J. Lee, T. Ohta, and K. Nakayama. 1999. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 42:4928-4931. [DOI] [PubMed] [Google Scholar]
- 50.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 51.Simons, R. W., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1983. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell 34:673-682. [DOI] [PubMed] [Google Scholar]
- 52.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 53.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]