Abstract
Combined overexpression of xylulokinase, pentose-phosphate-pathway enzymes and a heterologous xylose isomerase (XI) is required but insufficient for anaerobic growth of Saccharomyces cerevisiae on d-xylose. Single-step Cas9-assisted implementation of these modifications yielded a yeast strain expressing Piromyces XI that showed fast aerobic growth on d-xylose. However, anaerobic growth required a 12-day adaptation period. Xylose-adapted cultures carried mutations in PMR1, encoding a Golgi Ca2+/Mn2+ ATPase. Deleting PMR1 in the parental XI-expressing strain enabled instantaneous anaerobic growth on d-xylose. In pmr1 strains, intracellular Mn2+ concentrations were much higher than in the parental strain. XI activity assays in cell extracts and reconstitution experiments with purified XI apoenzyme showed superior enzyme kinetics with Mn2+ relative to other divalent metal ions. This study indicates engineering of metal homeostasis as a relevant approach for optimization of metabolic pathways involving metal-dependent enzymes. Specifically, it identifies metal interactions of heterologous XIs as an underexplored aspect of engineering xylose metabolism in yeast.
In conventional feedstocks for fermentative production of fuel ethanol, such as corn starch and cane sugar, carbohydrates predominantly occur as dimers or polymers of hexose sugars. These hexose sugars can be efficiently and rapidly fermented by Saccharomyces cerevisiae. Economically feasible ethanol production from non-food lignocellulosic feedstocks additionally requires efficient, anaerobic fermentation of d-xylose and l-arabinose1,2. Although wild-type S. cerevisiae strains cannot ferment these pentose sugars, they can slowly convert d-xylulose3. In yeast species that can grow on d-xylose, such as Scheffersomyces stipitis, its metabolism is initiated by a two-step conversion into d-xylulose by the combined activity of a xylose reductase (XR) and a xylitol dehydrogenase (XDH)4. The different redox cofactor preferences of XR and XDH represent a challenge in their use for constructing d-xylose-fermenting S. cerevisiae strains. This redox problem causes the production of substantial amounts of xylitol by anaerobic cultures of such engineered strains5,6,7. Elegant engineering strategies in which cofactor specificities of XR and/or XDH were altered, have not yet completely eliminated the formation of this by-product8,9.
In bacteria, d-xylose conversion is often initiated by its direct isomerization to d-xylulose, catalysed by xylose isomerase (XI) (EC 5.3.1.5). Until 2003, attempts to express heterologous XI genes in S. cerevisiae yielded no or very low XI activities under physiologically relevant conditions10,11,12,13. Then, multi-copy expression of a newly discovered xylose isomerase gene (xylA) from the anaerobic fungus Piromyces sp. E214 was shown to yield high XI activity in cell extracts of S. cerevisiae15,16. Piromyces xylA shows strong sequence similarity with Bacteroides XI genes, suggesting that the fungus acquired the gene by horizontal gene transfer. Indeed, expression of XI genes from Bacteroides species also yielded XI activity in S. cerevisiae17,18.
Consistent with the slow growth of wild-type S. cerevisiae strains on d-xylulose3, functional expression of xylA by itself only enabled very slow aerobic growth on xylose15,16. Kuyper et al. (2005a)19 reported that expression of xylA combined with constitutive overexpression of the genes encoding the native S. cerevisiae xylulokinase (XKS1, EC 2.7.1.17), ribulose 5-phosphate epimerase (RPE1, EC 5.3.1.1), ribulose 5-phosphate isomerase (RKI1, EC 5.3.1.6), transketolase (TKL1, EC 2.2.1.1) and transaldolase (TAL1, EC 2.2.1.2) was sufficient to enable anaerobic growth on d-xylose, at a specific growth rate of 0.07 h−1. Several subsequent studies confirmed that overexpression of a heterologous XI, combined with overexpression of xylulokinase and the enzymes of the non-oxidative pentose-phosphate pathway, is required for fast anaerobic fermentation of d-xylose20,21).
Laboratory evolution experiments designed to further improve the kinetics of xylose fermentation revealed expression of the heterologous XI as a key factor, as reflected by amplification of the XI gene via formation of extra-chromosomal circular DNA22 or increased numbers of XI genes on the yeast chromosomes21. Other studies demonstrated improved xylose fermentation in yeast strains in which XI expression was increased by random mutagenesis, codon optimization or by mutations influencing protein folding23,24,25. Additional mutations that improve pentose-fermentation kinetics, mainly identified by resequencing of laboratory-evolved strains, affected structural genes encoding native yeast hexose transporters26,27,28,29,30 and in the ‘secondary’ transaldolase and transketolase isoenzymes NQM1 and TKL220,31.
Over a decade of intensive research on d-xylose fermentation by XI-based, engineered S. cerevisiae strains yielded many important insights into their physiology. However, one important and industrially relevant aspect remains incompletely understood. While an initial study19 reported that combined overexpression of xylA, xylulokinase and non-oxidative pentose-phosphate pathway enzymes was sufficient to enable anaerobic growth of S. cerevisiae on d-xylose, subsequent reports indicated that anaerobic growth on xylose required additional, as yet unidentified mutations21,23.
The aim of the present study was to investigate the molecular basis for anaerobic growth of engineered xylA-expressing, d-xylose-metabolizing S. cerevisiae. To this end, we used CRISPR-Cas9 mediated genome editing for single-step construction of an S. cerevisiae strain that grew aerobically on d-xylose as sole carbon source. After adaptation to anaerobic growth in xylose-grown bioreactor batch cultures, we showed that mutations in a single gene enabled anaerobic growth on xylose. Via a combination of physiological and enzymological analyses, we investigated how these mutations affected intracellular metal homeostasis and d-xylose metabolism.
Results
One-step construction of a xylose-utilizing Saccharomyces cerevisiae strain
To construct a xylose-metabolizing S. cerevisiae strain, nine copies of an expression cassette containing Piromyces xylA, as well as single expression cassettes for constitutive overexpression of the native yeast genes for xylulokinase (XKS1) and for the enzymes of the non-oxidative branch of the pentose-phosphate pathway (RKI1, RPE1, TKL1, TKL2 and TAL1) were introduced in S. cerevisiae CEN.PK113-7D. Additionally, an expression cassette for NQM1, a paralog of TAL1 whose duplication has been shown to enhance pentose fermentation by engineered S. cerevisiae31, was introduced. Combination of in vivo assembly32 and CRISPR/Cas9-mediated chromosomal integration33 enabled a one-step introduction of all expression cassettes in the GRE3 locus, thereby inactivating GRE3, which encodes a non-specific aldose reductase that can reduce xylose to xylitol34. The nine copies of the xylA cassette were introduced as tandem repeats to facilitate adaptation of the xylA copy number by homologous recombination. Transformants obtained after plating on xylose synthetic medium (SMX) plates were restreaked thrice on the same medium. The genome of the resulting strain IMX696 (Table 1), in which correct integration of the cassettes was confirmed by diagnostic PCR using primers listed in Table S3, was sequenced to assess whether mutations had occurred during growth on SMX plates. No single-nucleotide polymorphisms (SNPs), insertion/deletions in coding regions or changes in chromosomal copy numbers were observed. However, read-depth analysis revealed the presence of 36 rather than 9 copies of the xylA cassette. This amplification of xylA is consistent with earlier reports that showed a positive impact of high xylA copy numbers on xylose metabolism by engineered S. cerevisiae strains21,22,25. In aerobic shake-flask cultures on SMX, strain IMX696 exhibited a specific growth rate of 0.21 h−1 (Fig. 1).
Table 1. Saccharomyces cerevisiae strains used in this study.
| Strain | Relevant genotype/description | Reference |
|---|---|---|
| CEN.PK 113-7D | MATa MAL2-8c SUC2 | 55 |
| IMX581 | MATa ura3-52 MAL2-8c SUC2 can1Δ::cas9-natNT2 | 33 |
| IMX696 | MATa ura3-52 MAL2-8c SUC2 CAN1::cas9-natNT2 gre3:: [pTDH3_RPE1- pPGK1_TKL1- pTEF1_TAL1- pPGI1_NQM - pTPI1_RKI1- pPYK1_TKL2- (pTPI1_xylA_tCYC1)*36 pTEF1_XKS1] pUDE335 | This study |
| IMS0488 | Single-cell line isolated after adaptation of IMX696 to anaerobic growth on xylose (reactor 1) | This study |
| IMS0489 | Single-cell line isolated after adaptation of IMX696 to anaerobic growth on xylose (reactor 2) | This study |
| IMX906 | MATa ura3-52 MAL2-8c SUC2 CAN1::cas9-natNT2 gre3:: [pTDH3_RPE1- pPGK1_TKL1- pTEF1_TAL1- pPGI1_ NQM1-pTPI1_RKI1- pPYK1_TKL2- (pTPI1_xylA_tCYC1)*36 pTEF1_XKS1] pmr1Δ::amdSYM pUDE335 | This study |
| IMX979 | IMX906 with PMR1 reintegrated at PMR1 locus | This study |
| IMK692 | MATa MAL2-8c SUC2 pmr1Δ::amdSYM | This study |
Native gene terminator sequences were used for expression of RPE1, TKL1, TAL1, NQM1, RKI1, TKL2 and XKS1.
Figure 1. Growth of S. cerevisiae strains with different PMR1 alleles in aerobic cultures on xylose.
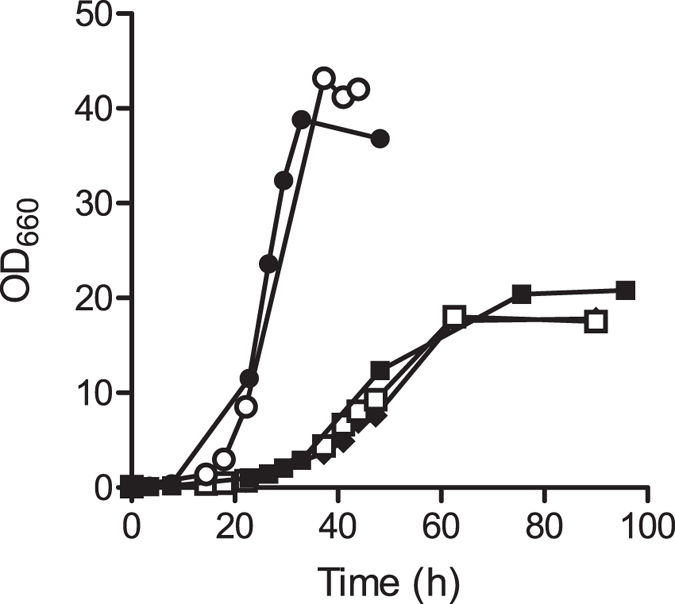
Aerobic growth curves in shake-flask cultures grown on synthetic medium with 20 g l−1 xylose. Symbols indicate the following S. cerevisiae strains: ●, IMX696 (xylA, PPP↑, XKS1↑), ■, IMX906 (xylA, PPP↑, XKS1↑, pmr1Δ), ○, IMX979 (xylA, PPP↑, XKS1↑, PMR1),◆, IMS0488 (isolate from IMX696 culture adapted to anaerobic growth on xylose carrying Pmr1G249V mutation) and □, IMS0489 (isolate from IMX696 culture adapted to anaerobic growth on xylose carrying Pmr1W387* mutation). Data shown are from a single flask experiment for each strain. For all strains, data obtained from independent duplicate experiments differed by less than 5%.
Anaerobic growth on xylose requires prolonged adaptation
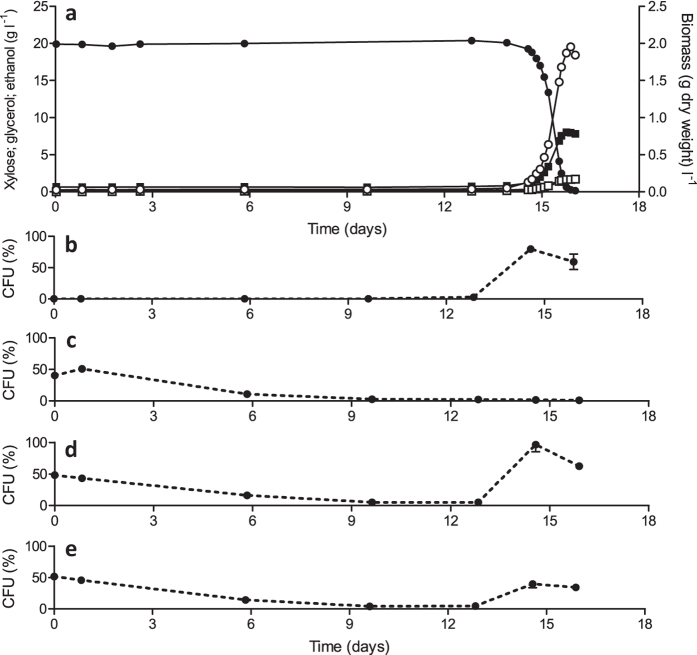
Anaerobic growth of the engineered xylose-fermenting strain IMX696 was investigated in nitrogen-sparged bioreactor cultures on SMX, supplemented with the anaerobic growth factors Tween-80 and ergosterol. In duplicate experiments, CO2 production, which was continuously monitored in the off-gas of the bioreactors, was only observed after 12 days of incubation (Supplementary Fig. S1). To investigate this slow adaptation to anaerobic growth on xylose in more detail, the experiment was repeated, with regular analysis of culture viability, metabolite concentrations and growth (Fig. 2). Again, no significant xylose consumption occurred during the first 12 days of the experiment. A subsequent increase in biomass concentration coincided with the conversion of xylose to ethanol and glycerol. The specific growth rate after the onset of anaerobic growth was estimated at 0.11 h−1 based on biomass dry weight measurements during the mid-exponential growth phase. Biomass and ethanol yields on xylose were 0.086 ± 0.01 g biomass (g xylose)−1 and 0.382 ± 0.01 g ethanol (g xylose)−1, respectively (Fig. 2a, Supplementary Fig. S2a). The dynamics of adaptation to anaerobic growth were further investigated by plating culture samples on synthetic medium with either glucose (SMD) or xylose (SMX). Colony counts on these plates were determined after aerobic and anaerobic incubation (Fig. 2b–e). On anaerobic SMX plates, colonies were first observed after 10 d, at which time they represented a fraction of only 1.8∙10−4 of the number of cells that were plated. Subsequently, consistent with the exponential growth observed by biomass dry weight measurements, the fraction of cells capable of anaerobic growth of xylose rapidly increased (Fig. 2b). When culture samples were plated on SMD, aerobic and anaerobic plates showed similar trends in colony counts (Fig. 2d,e). Conversely, plating on SMX revealed a strong trade-off between the ability to grow aerobically and anaerobically on xylose. On aerobically incubated SMX plates cell counts did not increase, not even when exponential growth on xylose took off during the final days of the bioreactor experiments and strongly increasing colony counts were observed on anaerobically incubated SMX plates (Fig. 2c).
Figure 2. Anaerobic growth of S. cerevisiae IMX696 (xylA, PPP↑, XKS1↑) on xylose requires a prolonged adaptation period.
(a) Growth, xylose consumption and product formation after inoculation of aerobically pregrown cells in anaerobic bioreactors containing synthetic medium with xylose (20 g l−1). Symbols: ●, xylose, ■, ethanol, ○, biomass, □, glycerol. (b) Colonyforming units (CFU) on anaerobically incubated xylose medium reflect adaptation to growth on xylose in the absence of oxygen. (c) CFU on aerobically incubated xylose medium reflect trade-off between aerobic and anaerobic growth on xylose. (d) and (e) CFU on anaerobically and aerobically incubated glucose medium, respectively, showing that oxygen sensitivity of cells adapted to anaerobic growth on xylose is not carbon-source dependent. Data shown in this figure are from one of two independent replicates, the replicate experiment is shown in Supplementary Fig. S2.
Adaptation to anaerobic growth on xylose coincides with mutations in PMR1
The dynamics of colony counts on SMX plates (Fig. 2b) suggested that adaptation to anaerobic growth might have involved one or more mutations. To test this hypothesis, the genomes of strains IMS0488 and IMS0489, which were isolated from the independent anaerobic adaptation experiments shown in Fig. 2 and Supplementary Fig. S2, were sequenced. Read-depth analysis of both strains revealed a decrease of the xylA copy number to 24 and 25, respectively, as compared to 36 in the parental strain IMX696. No other changes in chromosomal copy numbers were observed. Strikingly, both strains carried non-synonymous SNPs in the coding region of PMR1 (Table 2), which encodes a high-affinity Golgi Ca2+/Mn2+ P-type ATPase35. These mutations caused a single amino acid change (Pmr1G249V) in strain IMS0488 and introduced a premature stop codon (Pmr1W387*) in strain IMS0489.
Table 2. Single-nucleotide mutations in engineered S. cerevisiae strains adapted to anaerobic growth on xylose.
| Strain | Gene | Nucleotide change | Amino acid change | Change in codon |
|---|---|---|---|---|
| IMS0488 | PMR1 | G746T | G249V | gGt/gTt |
| IMS0489 | PMR1 | G1161A | W387* | tgG/tgA |
Strains IMS0488 and IMS0489 were isolated from independent anaerobic batch cultures of strain IMX696 (xylA, PPP↑, XKS1↑). The genome sequence of IMX696 was used as a reference. *Introduction of stop codon.
Mutations in PMR1 enable anaerobic growth on xylose
To investigate the role of the PMR1 mutations in the adaptation to anaerobic growth on xylose, the gene was deleted in the parental strain IMX696. In replicate anaerobic bioreactor cultures on xylose, the resulting strain IMX906 grew within 24 h and completely consumed all sugar within 70 h (Fig. 3). The specific growth rate of both cultures was 0.08 h−1, while biomass and ethanol yields on xylose were 0.086 g ± 0.01 biomass (g xylose)−1 and 0.40 g ± 0.01 ethanol (g xylose)−1, respectively. To further investigate the role of the PMR1 deletion in the instantaneous anaerobic growth of strain IMX906 on xylose, the wild-type PMR1 allele was reintegrated in this strain. The resulting strain IMX979 showed a lag phase of over 250 h in duplicate anaerobic bioreactor cultures on xylose (Supplementary Fig. S1), thereby confirming the key role of PMR1 inactivation in the ability of engineered, XylA-based S. cerevisiae to grow anaerobically on xylose.
Figure 3. Deletion of PMR1 enables anaerobic growth on xylose of engineered S. cerevisiae without prior adaptation phase.
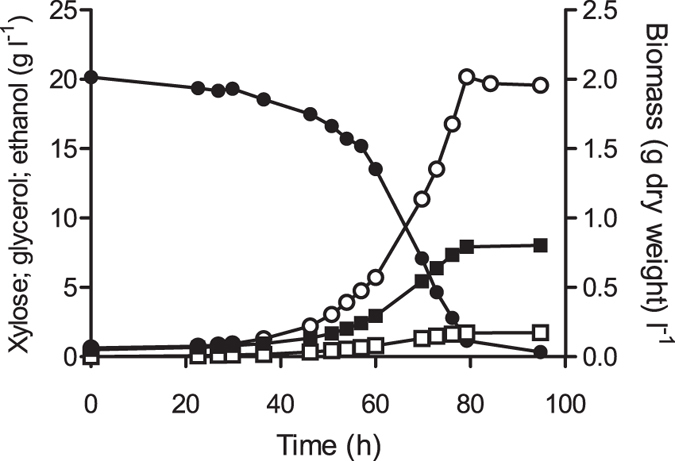
Growth and product formation of S. cerevisiae strain IMX906 (xylA, PPP↑, XKS1↑, pmr1Δ on xylose (20 g l−1) in anaerobic bioreactors. Symbols: ●, xylose, ■, ethanol, ○, biomass, □, glycerol. The data shown are from one of two independent replicates.
The plate count experiments during the anaerobic adaptation phase on xylose suggested a trade-off between aerobic and anaerobic growth on xylose (Fig. 2b,c). This possible trade-off was further explored by growth experiments in aerobic shake flasks on SMX. In these experiments, strains in which PMR1 was mutated or deleted consistently showed a lower specific growth rate than strains that carried a wild-type PMR1 allele (0.10 h−1 and 0.21 h−1, respectively; Fig. 1). Furthermore, aerobic xylose-grown shake-flask cultures of strains with mutated PMR1 alleles accumulated ethanol to 3–4-fold higher concentrations than corresponding cultures of strains with wild-type PMR1 alleles (Supplementary Fig. S3). Consistent with previous results36, aerobic shake flask cultures growing on glucose also revealed an approximately 50% reduced growth rate of the pmr1Δ strain IMK692 (Supplementary Fig. S4).
Mutations in PMR1 affect intracellular metal concentrations in xylose-metabolizing S. cerevisiae strains
Pmr1 is an ATP-dependent transporter that imports Ca2+ and Mn2+ into the Golgi complex37. Based on the observation that pmr1 null mutants accumulate Ca2+ and Mn2+ intracellularly, Pmr1 has also been implicated in secretion of divalent metal ions via the Golgi complex38. To explore a possible relation between metal homeostasis and anaerobic growth on xylose, we analysed intracellular concentrations of Ca2+, Mn2+, Mg2+ and Fe2+ in biomass samples from anaerobic mid-exponential phase bioreactor cultures using inductively coupled plasma mass spectrometry. Contents of Mg2+, Ca2+ and Fe2+ were similar in all analysed strains, with Mg2+ accounting for over 80% of the analysed divalent metal ions, followed by Ca2+, and with Fe2+ accounting for less than 1% of the measured metals. Conversely, large differences were observed for the Mn2+ content. While in strains with a wild-type PMR1 allele, Mn2+ represented less than 0.2% of the measured metal ions, 12- to 29-fold higher Mn2+ contents were observed in strains with mutated PMR1 alleles, irrespective of whether they were grown on xylose or glucose (Table 3). The observation that mutations in PMR1 affected cellular contents of Mn2+ but not those of Ca2+ is consistent with a previous study39.
Table 3. Impact of the deletion of PMR1 on intracellular metal ion concentrations.
| S. cerevisiae strain (relevant genotype) | Carbon source | μmol (g dry biomass)−1 | ||||
|---|---|---|---|---|---|---|
| Mg2+ | Ca2+ | Fe2+ | Mn2+ | Total | ||
| IMX696 (xylA, PPP↑, XKS1↑) | glucose | 60 ± 1 | 8.8 ± 1.6 | 0.56 ± 0.02 | 0.069 ± 0.001 | 69 ± 2 |
| IMX906 (xylA, PPP↑, XKS1↑, pmr1Δ) | glucose | 64 ± 1 | 12 ± 2 | 0.51 ± 0.02 | 1.2 ± 0.1 | 77 ± 2 |
| IMX906 (xylA, PPP↑, XKS1↑, pmr1Δ) | xylose | 50 ± 1 | 4.0 ± 0.7 | 0.36 ± 0.01 | 1.4 ± 0.1 | 56 ± 1 |
| CEN.PK113-7D (reference strain) | glucose | 61 ± 1 | 6.8 ± 1.3 | 0.44 ± 0.01 | 0.046 ± 0.001 | 68 ± 2 |
| IMK692 (pmr1Δ) | glucose | 72 ± 1 | 6.7 ± 1.2 | 0.46 ± 0.02 | 0.86 ± 0.06 | 80 ± 2 |
S. cerevisiae strains were grown in anaerobic bioreactors on xylose or glucose (20 g l−1). Data represent average and mean deviation calculated from analyses on independent duplicate cultures.
Activity and metal content of xylose isomerase expressed in S. cerevisiae strains
Laboratory evolution studies have identified XI activity as a key factor in rapid fermentation of xylose to ethanol16,21,22. XI enzymes are known to be metal dependent, with pronounced differences in metal binding and impact of metal identity on enzyme kinetics40. To examine the impact of Mn2+ on XylA activity, XI activities were assayed in cell extracts of strains IMX906 (pmr1Δ) and its parental strain IMX696 after aerobic and anaerobic growth on SMD in shake-flask cultures (Supplementary Fig. S5). Cell extracts from both strains exhibited similar activities in assays without added metal ions. These activities do not necessarily reflect in vivo metal binding as they may, for example, have been influenced by binding of metals released during preparation of cell extracts, e.g. by disruption of vacuoles. Addition of Mn2+ and, to a lesser extent, of Mg2+ to the XI assays yielded significantly higher XI activities than observed in the absence of added metals. Conversely, addition of Ca2+ led to lower activities.
For a further analysis of the effect of Mn2+ on XylA activity, we purified the enzyme from the controlled anaerobic bioreactor cultures that were also used to determine cellular metal contents (Supplementary Fig. S6). Concentrations of Mg2+, Ca2+, Fe2+ and Mn2+ were measured in purified protein samples and the amount of each metal per enzyme active site was calculated (Table 4). These analyses showed that the isolated enzymes contained fewer than two metal ions per subunit, indicating that their metal binding sites were not fully occupied. In independent replicate experiments, large and consistent differences were observed in the Mn2+ contents of XylA isolated from strains with wild-type and mutated PMR1 alleles (0.017 and 0.30 mol Mn (mol XylA subunit)−1, respectively, Table 4). The higher Mn2+ content of XylA isolated from xylose- or glucose-grown cells of the pmr1Δ strain coincided with a ca. 2-fold higher specific activity than measured with enzyme purified from the PMR1 strain (Table 4). Although metal binding may have changed during cell disruption and enzyme purification, this correlation does indicate that Mn2+-loaded XylA is a better catalyst than the Mg2+-loaded enzyme. Addition of 1 mM MgCl2 to purified enzyme preparations enhanced their XI activities, consistent with incomplete metal loading in the cell and/or metal loss during purification and activity assays.
Table 4. Impact of PMR1 deletion on metal content and activity of XylA.
| S. cerevisiae strain (relevant genotype) | Carbon source | mol metal/mol XylA monomer | Sp. activity (U/mg XylA protein) | ||||
|---|---|---|---|---|---|---|---|
| Mg2+ | Ca2+ | Fe2+ | Mn2+ | no metal added | Mg2+ added (1 mM) | ||
| IMX906 (xylA, PPP↑, XKS1↑, pmr1Δ) | xylose | 0.18 ± 0.01 | 0.54 ± 0.11 | 0.06 ± 0.01 | 0.38 ± 0.04 | 2.39 ± 0.61 | 2.68 ± 0.5 |
| IMX906 (xylA, PPP↑, XKS1↑, pmr1Δ) | glucose | 0.20 ± 0.06 | 0.66 ± 0.07 | 0.064 ± 0.02 | 0.30 ± 0.03 | 1.35 ± 0.14 | 2.10 ± 0.1 |
| IMX696 (xylA, PPP↑, XKS1↑) | glucose | 0.22 ± 0.06 | 0.93 ± 0.41 | 0.086 ± 0.04 | 0.017 ± 0.004 | 0.60 ± 0.24 | 1.68 ± 0.1 |
XylA protein was isolated from S. cerevisiae cultures grown on xylose or glucose (20 g l−1) in anaerobic bioreactors. Data represent average and mean deviation of analyses on XylA isolated from independent duplicate cultures.
Mn2+ binding results in superior catalytic efficiency of XylA
To accurately analyse the effect of different metals on catalytic properties of XylA, apoenzyme was prepared from XylA isolated from xylose-grown cultures of strain IMX906. Subsequently, XI activities were measured after reconstitution of apo-XylA with Mg2+, Ca2+ or Mn2+ (Table 5). The activities of Mn2+- and Mg2+ -reconstituted apo-XylA were higher than activities in non-metal-supplemented assays with XylA purified from yeast cultures (Table 4). The reconstituted enzyme showed the highest catalytic efficiency in the presence of Mn2+, with a kcat/KM ratio that was 4-fold and 1500-fold higher than with Mg2+ and Ca2+, respectively. Both the highest kcat and the lowest KM were observed with Mn2+ and contribute to the superior catalytic efficiency with this metal cofactor (Table 5). When XylA apoenzyme was reconstituted with mixtures of divalent metals that resembled those observed in intracellular metal content analyses (Table 3) of strains IMX696 (PMR1) and IMX906 (pmr1Δ), an 80–90% increase of XI activity was observed as the fraction of Mn2+ was increased from 0.002 to 0.01 (Supplementary Table S1).
Table 5. Kinetic parameters of XylA measured after reconstituting apo-XylA with different divalent metal ions.
| Metal | Kinetic parameters | ||
|---|---|---|---|
| kcat (s−1) | KM (mM) | kcat/KM (s−1 · M−1) | |
| Mg2+ | 2.8 ± 0.2 | 5.5 ± 0.4 | 500 |
| Mn2+ | 7.8 ± 0.1 | 3.9 ± 0.2 | 2000 |
| Ca2+ | 0.6 ± 0.06 | 420 ± 90 | 1.3 |
kcat and KM values represent average and mean deviation of independent duplicate experiments, calculated for each metal.
Discussion
One-step, Cas9-assisted integration of a heterologous XI (Piromyces XylA) and overexpression of native yeast genes encoding xylulokinase and enzymes of the non-oxidative pentose-phosphate pathway (PPP) enabled fast aerobic growth on xylose by S. cerevisiae, thus illustrating the efficiency of Cas9-based genome editing in this yeast33,41. Nine copies of the xylA cassette were incorporated in tandem to facilitate expansion or compression of the xylA copy number by homologous recombination. This approach was validated by the four-fold higher xylA copy number in transformants isolated on xylose medium and its decrease in independent replicate cultures after subsequent adaptation to anaerobic growth. The observed amplification of xylA was consistent with the previously reported positive impact of high xylA copy numbers on xylose metabolism20,21,22. In line with earlier studies21,23, this metabolic engineering strategy did not enable anaerobic growth on xylose. In principle, the engineered XI-based pathway should allow for efficient, redox-cofactor-balanced alcoholic fermentation on this sugar. However, anaerobic growth on xylose requires much higher fluxes through XI since the ATP yield of anaerobic, fermentative metabolism of this sugar is approximately eight-fold lower than that of its aerobic, respiratory dissimilation (assuming an in vivo P/O ratio of 1.0)42.
In independent replicate cultures, anaerobic growth on xylose required a two-week adaptation, which was shown to reflect the accumulation of spontaneous mutants with single-nucleotide mutations in PMR1. The observation that single, easily acquired point mutations enabled this adaptation may explain an earlier report that overexpression of XylA, xylulokinase and PPP enzymes sufficed to enable anaerobic growth of S. cerevisiae on xylose19. Here, we demonstrate that inactivation of PMR1 caused both a strongly elevated intracellular Mn2+ concentration and an increased loading of heterologously expressed XylA with Mn2+. Moreover, in vitro studies showed that loading of XylA apoenzyme with Mn2+ led to higher enzyme activities than binding of other divalent metal ions present in the yeast cytosol.
Consistent with the conclusion that intracellular Mn2+ homeostasis affects anaerobic xylose metabolism through its impact on in vivo XylA activity, none of the five S. cerevisiae enzymes that subsequently convert d-xylulose into glycolytic intermediates (xylulokinase, ribulose-5-phosphate isomerase, ribulose-5-phosphate 3- epimerase, transaldolase and transketolase) have been documented to be Mn2+ dependent (BRENDA database43). Our results do not exclude the possibility that altered Mn2+ levels influenced pentose metabolism by mechanisms other than influencing XylA activity. However, a key role of XylA is consistent with the observation that acquisition of mutations in PMR1 coincided with a decrease of the xylA copy number from 36 to 25. This decrease, which occurred during the course of a single batch culture, suggests that mutations in PMR1 may have affected a trade-off between the need for a high in vivo activity of XylA and the metabolic burden associated with its high-level synthesis. As demonstrated in a study on the energetic impacts of galactose-induced synthesis of the enzymes of the Leloir pathway44, such a metabolic burden is much more pronounced in anaerobic cultures than in aerobic, respiring cultures due to the lower ATP yield from fermentative sugar dissimilation.
The mutations in PMR1 that enabled anaerobic growth on xylose negatively affected aerobic growth. High intracellular Mn2+ concentrations have previously been implicated in impaired mitochondrial function36, which is consistent with the increased accumulation of ethanol in aerobic shake-flask cultures of pmr1 strains (Supplementary Fig. 3). Moreover, TORC1 signalling, which is involved in regulation of mitochondrial respiratory functions, is inhibited by Mn2+ and Pmr1 has been identified as a negative regulator of TOR1, which encodes a subunit of the TORC1 complex45. Additionally, Mn2+-induced apoptosis mediated by Ndi146,47, a mitochondrial NADH dehydrogenase, may have contributed to low colony counts observed when cultures adapted to anaerobic growth on xylose were plated under aerobic conditions (Fig. 1c). The reduced growth rate in aerobic cultures of strains carrying PMR1 mutations should be considered when their anaerobic industrial application is preceded by an aerobic biomass propagation phase.
Despite the pivotal role of the functional expression of a heterologous XI in S. cerevisiae18 and the well documented role of metal ions in the active sites of XIs from taxonomically diverse organisms48, the impact of metal loading on the performance of heterologously expressed XIs in S. cerevisiae has previously not been investigated. Similar to bacterial XIs40, apo-XylA isolated from yeast could be activated with different metals. The results of this study suggest that metal loading can have a large effect on the in vivo catalytic performance of the enzyme.
The pronounced influence of cellular metal content on XI activity was in agreement with its promiscuity towards metal cofactors found in the in vitro analyses. However, while the fraction of the XI-bound Mn2+ increased by more than 10-fold in strains that carried mutations in PMR1, cellular contents of Mn2+ were still at least 40 times lower than the combined Mg2+ and Ca2+ contents (Tables 3 and 4). This observation suggests that the affinity of XylA for Mn2+ is higher than for the other divalent metal ions. Functional expression of heterologous XIs in S. cerevisiae initially represented a formidable challenge in engineering S. cerevisiae for anaerobic xylose fermentation10,11,12,13. After Piromyces xyla14, XI genes from several eukaryotes and prokaryotes sources were shown to also be functionally expressed in S. cerevisiae, including those from Orpinomyces sp49, Arabidopsis thaliana50, Clostridium phytofermentans23, Bacteroides thetaiotaomicron18, Bacteroides stercoris, Prevotella ruminicula TC2-2451 and Sorangium cellosum52. In view of the key catalytic role of metal ions in all known xylose isomerases, we expect that in vivo activity of these and other XIs in yeast cells will also be affected by engineering of metal homeostasis.
Mutations in PMR1 have been identified in two previous studies on adaptive laboratory evolution of XylA-based, engineered S. cerevisiae strains. Klaassen et al.53 identified a mutation in PMR1 (Y38C) in a strain evolved for fermentation of l-arabinose and xylose to ethanol. Recently, Hou et al.25, reported a mutation in PMR1 (G698V) in a respiratory-deficient XylA based S. cerevisiae strain obtained by adaptive laboratory evolution on xylose medium. Our results strongly suggest that, in both studies, the mutations in PMR1 may have contributed to the selected phenotypes. Additionally, the superior catalytic efficiency of Mn2+-loaded XylA may explain a recent report that MnSO4 supplementation enhanced growth on xylose of acetate-stressed cultures of a XylA-based xylose-fermenting S. cerevisiae strain54.
Our study demonstrates the importance of metal homeostasis and enzyme loading in XI-based yeast metabolic engineering strategies for anaerobic conversion of xylose-containing lignocellulosic feedstocks into fuels and chemicals. Inactivation of PMR1, combined with overexpression of PPP enzymes, xylulokinase and xylA was shown to be sufficient to enable anaerobic growth of S. cerevisiae on xylose. Beyond xylose utilization, engineering of metal homeostasis has the potential to improve in vivo performance of other metal-dependent heterologous enzymes or pathways.
Methods
Strains and maintenance
All S. cerevisiae strains used in this study (Table 1) originate from the CEN.PK lineage55,56. Frozen stock cultures were stored at −80 °C in 30% (vol/vol) glycerol.
Plasmid and strain construction
Plasmids used in this study are presented in Supplementary Table S2. Expression cassettes required for xylose fermentation were introduced into the GRE3 locus of S. cerevisiae strain IMX581 by simultaneous in-vivo assembly and integration32. Expression cassettes for RPE1, RKI1, TAL1, NQM1, TKL1, TKL2 and XKS1 were obtained by fusing constitutive promoter sequences, ORFs and terminator sequences amplified from CEN.PK113-7D in a fusion-PCR57 using the primers specified in Supplementary Table S3. Plasmid pYM-N1858 was used as a template for the TEF1 promoter. The resulting fragments were cloned into pJET-1.2 blunt-end vectors. Correct assembly was verified by sequencing as described below. PCR amplification of expression cassettes and plasmids was performed using Phusion Hot Start II High Fidelity DNA Polymerase (Thermo Scientific, Waltham, MA), according to the manufacturer’s protocol. Integration in GRE3 locus was mediated by a chimeric CRISPR/Cas9 editing system33,41 with gRNA expressed from an episomal plasmid33. The plasmid backbone was PCR amplified from pMEL10 using primers 5792–5980 (Supplementary Table S3). A plasmid insert containing the 20 bp gRNA-targeting sequence was obtained by PCR amplification with primers 5978–5979 using pMEL10 as template. The resulting fragment was fused to the plasmid backbone with the Gibson Assembly Cloning kit (New England Biolabs, Ipswich, MA), yielding plasmid pUDE335. E. coli DH5a cells were transformed with 1 μL of the Gibson-assembly mix using a Gene PulserXcell Electroporation System (Biorad, Hercules, CA). Plasmid DNA was isolated from E. coli cultures using a Sigma GenElute Plasmid kit (Sigma-Aldrich, St. Louis, MO). The presence of the GRE3 cutting gRNA was confirmed by PCR-amplification using primer pair 2528–960 followed by digestion with FastDigest ClaI (Thermo Scientific).
The coding region of the Piromyces sp. E2 xylose isomerase gene [Genbank: CAB76571.1] was codon optimized according to the codon preference of highly expressed glycolytic genes in S. cerevisiae59. The codon-optimized sequence, flanked by the constitutive TPI1 promoter and CYC1 terminator, was synthesized by GeneArt GmbH (Regensburg, Germany). After subsequent transformation of the pMK-RQ (GeneArt) based vector pUDR350 into E. coli, nine different expression cassettes of xylA were made, flanked by 60 bp synthetic recombinant sequences (Supplementary Fig. S7). For XylA expression in E. coli, a codon-optimized synthetic xylA was cloned into pBAD/myc-His-derived plasmid.
Yeast transformation was performed using the lithium acetate protocol60. Strain IMX696 was obtained by adding 200 pmol of each of the 15 fragments combined with 500 ng of plasmid pUDE335. After one hour of incubation in synthetic medium with glucose (SMD) the cells were plated on SM plates with xylose as the carbon source (SMX). Correct assembly of all fragments in the GRE3 locus was confirmed by diagnostic PCR (Dreamtaq, Thermo Scientific) using primers listed in Table S3. Deletion of PMR1 in S. cerevisiae strains IMX696 and CEN.PK113-7D was done by integrating an amdSYM-based deletion cassette61, which was derived by PCR amplification from pUG-amdSYM using primers 8638/8639 as template. After transformation, cells were plated on glucose synthetic medium with acetamide as the nitrogen source (SMD-Ac). Gene deletion was confirmed by diagnostic PCR and the resulting strains were named IMX906 and IMK692, respectively. To reintegrate PMR1, the PMR1 ORF was PCR-amplified from CEN.PK113-7D and transformed into strain IMX906. After overnight incubation in SMD-Ac, cells were plated on SMD plates supplemented with 2.3 g l−1 fluoroacetamide (SMD-Fac). Correct integration of PMR1 in the resulting strain, IMX969, was confirmed by diagnostic PCR.
Cultivation and media
Shake-flask cultures were grown at 30 °C in an orbital shaker at 200 rpm, using 500-ml flasks containing 100 ml medium. Physiological characterization of aerobic growth was performed in shake flasks containing SMX or SMD with urea as sole nitrogen source to prevent acidification. Prior to filter sterilization, media were adjusted to pH 5.0 with 2 M KOH. For pre-cultures, SM adjusted to pH 6.0 was autoclaved at 12 °C for 20 min after which a 50 w/v % solution of sterile glucose or xylose was added to obtain a final sugar concentration of 20 g l−1, together with filter-sterilized vitamin solution62. Glucose and xylose solutions were autoclaved separately (20 min at 110 °C). For plates, 2% agar was added to media prior to autoclaving. Frozen stocks (1 ml aliquots in 30% glycerol) were inoculated directly into pre-culture shake flasks. In late exponential phase an aliquot was transferred to a second pre-culture to obtain an initial OD660 of 0.1. Flasks or anaerobic bioreactors used for characterization were inoculated from these cultures at an initial OD660 of between 0.1 and 0.2. Anaerobic batch cultures were conducted in 2-l bioreactors (Applikon, Delft, The Netherlands) with a working volume of 1 l. Biomass for metal content analysis was grown in 3-l bioreactors (Applikon) with a working volume of 2 l were used. Bioreactor cultures were grown at 30 °C, pH 5.0, and stirred at 800 rpm. To ensure anaerobic conditions, bioreactors were equipped with Viton O-rings and Norprene tubing. During cultivation, nitrogen gas (<10 ppm oxygen) was continuously sparged through the cultures at 0.5 l min−1. After autoclaving, synthetic medium used for anaerobic cultivation was supplemented with 0.2 g l−1 sterile antifoam C (Sigma-Aldrich), as well as Tween 80 (420 mg l−1) and ergosterol (10 mg l−1) dissolved in ethanol63.
Analytical methods
Cell dry weight (CDW) measurements were done using pre-weighed nitrocellulose filters (pore size, 0.45 μm; Gelman Laboratory, Ann Arbor, MI) to filter 10 ml of culture. Before weighing the sample, filters were washed with demineralised water and dried in a microwave oven (Bosch, Stuttgart, Germany) for 20 min at 360 W. Growth was monitored by optical density (OD) measurements at a wavelength of 660 nm using a Libra S11 spectrophotometer (Biochrom, Cambridge, United Kingdom). A correlation between OD measurements and CDW was used to estimate CDW in samples for which no direct CDW measurements were taken. This correlation was based on at least six points during the exponential phase.
CO2 and O2 concentrations in bioreactor exhaust gas were measured using an NGA 2000 analyzer (Rosemount Analytical, Orrville, OH) after the gas was cooled by a condenser (2 °C) and dried with a Permapure MD-110-48P-4 dryer (Permapure, Toms River, NJ). Metabolite levels in culture supernatants obtained by centrifugation were measured via high-performance liquid chromatography (HPLC) analysis on an Agilent 1260 HPLC (Agilent Technologies, Santa Clara, CA) fitted with a Bio-Rad HPX 87 H column (Bio-Rad, Hercules, CA). The column was eluted at 60 °C with 0.5 g l−1 H2SO4 at a flow rate of 0.6 ml min−1. Detection was by means of an Agilent refractive-index detector and an Agilent 1260 VWD detector. Correction for ethanol evaporation were done for all bioreactor experiments as described previously64.
Viability of strain IMX696 during anaerobic cultivation was assessed by plating culture samples. The number of cells per ml was measured using a Z2 Coulter Counter (Beckman Coulter, Woerden, The Netherlands) after which dilutions were plated in duplicate on SMX and SMG agar plates and incubated at 30 °C. To limit exposure to oxygen, cells that were used to determine anaerobic viability measurements were sampled directly into a container flushed with argon and immediately transferred into an anaerobic chamber (5% H2, 6% CO2, and 89% N2, Sheldon MFG Inc., Cornelius, OR) for plating and incubation. Colony-forming units (CFU) were counted after incubation at 30 °C for 4 days (aerobic growth) or 8 days (anaerobic growth).
DNA sequence analysis
Genomic DNA of strains IMX696, IMS0488 and IMS0489 was isolated using the QIAGEN Blood & Cell Culture DNA Kit with 100/G Genomics-tips (QIAGEN, Valencia, CA) according to the manufacturer’s protocol. From these DNA samples, 350-bp insert libraries were constructed using the Nextera XT DNA kit (Illumina, San Diego, CA). Paired-end sequencing (100-bp reads) of genomic or plasmid DNA was performed with an Illumina HiSeq 2500 sequencer (Baseclear BV, Leiden, The Netherlands). Data were mapped to the CEN.PK113-7D genome or to in silico-generated plasmid sequences using the Burrows-Wheeler alignment tool65 and processed with Pilon66. Identified single-nucleotide differences were inspected with the Integrated Genomics Viewer67 (IGV). The chromosomal copy number variance (CNV) was estimated using the Poisson mixture model based algorithm Magnolya68. The copy number of xylA was estimated by comparing the read depth to the average read depth of all chromosomes. Raw sequence data of strains IMX696, IMS0488 and IMS0489 are deposited at the NCBI Sequence Read archive (www.ncbi.nlm.nih.gov/sra) under BioProject ID PRJNA349142.
Purification of xylose isomerase
Cell pellets were resuspended in 10 mM MOPS, pH 7.0, containing protease inhibitors (cOmplete ULTRA tablets, Roche) and disrupted using a high pressure homogenizer (Constant Systems Ltd, Low March, United Kingdom). Samples were passed through the apparatus twice at 39 kpsi and cell debris was removed by centrifugation at 35,000 × g for 45 min at 4 °C. A single-step purification procedure based on anion-exchange chromatography was applied to minimize the loss of protein-bound metals. Cell-free extracts were loaded on a strong anion-exchange column (Resource Q, GE Healthcare, Chicago, IL) equilibrated with 10 mM MOPS, pH 7.0. A gradient elution was applied using 10 mM MOPS, pH 7.0, containing 0–200 mM KCl. XylA eluted at approximately 40 mM KCl. Protein concentrations were determined using the theoretical extinction coefficient at 280 nm (ε280, XI = 73,800 M−1 cm−1) calculated by the ProtParam tool (http://web.expasy.org/protparam/).
Metal content analysis
Metal concentrations were analysed with an inductively coupled plasma mass spectrometer (ICP-MS, Varian 820). All measurements were performed 5 times for each sample and yttrium was used as an internal standard. Purified protein samples were lyophilized and analysed for contents of magnesium, calcium, iron and manganese. Prior to measurement, samples were dissolved in 1% nitric acid solution. All analyses were performed on protein samples isolated from two replicate cultures. For intracellular metal analysis, cells were prepared with a protocol adopted from Eide et al.69. The harvested cells were washed three times each with 1 μM EDTA solution and subsequently with deionized water (Milli-Q) and suspended in 1 ml 30% (w/v) nitric acid and incubated at 6 °C for 4 h. Cell lysates were centrifuged at 16,000 × g and supernatants were collected. Pellets were washed with 1 ml deionized water and the supernatants were collected as before. The 2 ml of final sample solution containing approximately 15% (w/v) nitric acid were then subjected to the measurements. The metal content was determined with samples from two separate bioreactor batch cultures.
Preparation of cell extracts
Cell extracts were prepared following a previously published procedure with minor modifications16. To limit loss of metals during preparation, no EDTA was added prior to sonication. Cells were washed and suspended in 10 mM MOPS buffer pH 7.0 to avoid precipitation of MnCl2 and 10 mM DTT was added. After sonication (4 bursts of 30 s with 30 s intervals at 0 °C, amplitude 8 μm) using a Soniprep 150 sonicator (Beun de Ronde BV, Abcoude, The Netherlands), cell debris was removed by centrifugation (4 °C, 20 min at 48,000 g) and the clear supernatant was used for XylA assays.
Enzyme activity assays
Activity of XylA was measured with a coupled enzyme assay using d-sorbitol dehydrogenase70. d-sorbitol dehydrogenase (SDH) was obtained from Roche Diagnostics GmbH (Mannheim, Germany). Reactions were performed at 30 °C and pH 7.0 (20 mM MOPS buffer). The decrease in absorbance at 340 nm was monitored in either a spectrophotometer (Jasco, Easton, MD) or a Synergy Mx microtiter plate reader (BioTek Instruments, Winooski, VT). Reaction mixtures included 5, 200 or 500 mM xylose, 250 μM NADH and U ml−1 of SDH. Addition of 0.03 to 1 μM (depending on the substrate concentration and the metal added) XI or cell free extract into the mixture initiated the reaction. For measuring XylA activity in the presence of different metal cofactors, samples of apo-XylA were prepared by overnight incubation of the purified enzyme with 10 mM EDTA. Subsequently, EDTA was removed by buffer exchange to 20 mM MOPS, pH 7.0 and 1 mM of divalent metal solutions (MgCl2, MnCl2 or CaCl2) were added in the reaction. For kinetic analyses, d-xylose was added at concentrations ranging from 0.5 mM to 1.50 M.
Additional Information
How to cite this article: Verhoeven, M. D. et al. Mutations in PMR1 stimulate xylose isomerase activity and anaerobic growth on xylose of engineered Saccharomyces cerevisiae by influencing manganese homeostasis. Sci. Rep. 7, 46155; doi: 10.1038/srep46155 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was performed within the BE-Basic R&D Program (http://www.be-basic.org/), which is financially supported by an EOS Long Term grant from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I). The authors thank Hanna Dudek (RUG), Jasmine M. Bracher (TUD), Paul de Waal (DSM), Hans de Bruijn (DSM) and Paul Klaassen (DSM) for their input in this project.
Footnotes
The authors declare no competing financial interests.
Author Contributions M.D.V., J.M.D., A.J.A.v.M., M.L., D.B.J. and J.T.P. together designed this study; M.D.V., L.K. and M.L. designed and performed all wet-lab experiments; M.v.d.B. performed all analyses of genome sequencing data; M.D.V., M.L., D.B.J., A.J.A.v.M. and J.T.P. wrote the manuscript. All authors read and approved the submitted version of the manuscript.
References
- Alper H. & Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nature Rev Microbiol 7, 715–723 (2009). [DOI] [PubMed] [Google Scholar]
- van Maris A. J. et al. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Van Leeuwenhoek 90, 391–418 (2006). [DOI] [PubMed] [Google Scholar]
- Wang P. Y. & Schneider H. Growth of yeasts on D-xylulose. Can J Microbiol 26, 1165–1168 (1980). [DOI] [PubMed] [Google Scholar]
- Jeffries T. W. Utilization of xylose by bacteria, yeasts, and fungi. In Pentoses and Lignin(eds Fiechter A., Jeffries T. W.). (Springer, 1983). [DOI] [PubMed] [Google Scholar]
- Jeffries T. W. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 17, 320–326 (2006). [DOI] [PubMed] [Google Scholar]
- Hahn-Hägerdal B. et al. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. In Metabolic engineering(eds Nielsen J. et al.). (Springer, 2001). [DOI] [PubMed] [Google Scholar]
- Kötter P. & Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38, 776–783 (1993). [Google Scholar]
- Runquist D., Hahn-Hägerdal B. & Bettiga M. Increased ethanol productivity in xylose-utilizing Saccharomyces cerevisiae via a randomly mutagenized xylose reductase. Appl Environ Microbiol 76, 7796–7802 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S. et al. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology 153, 3044–3054 (2007). [DOI] [PubMed] [Google Scholar]
- Walfridsson M. et al. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol 62, 4648–4651 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy A. et al. Expression of the Escherichia coli xylose isomerase gene in Saccharomyces cerevisiae. Appl Environ Microbiol 53, 1996–2000 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes C. J., Pretorius I. S. & van Zyl W. H. Cloning and expression of the Clostridium thermosulfurogenes D-xylose isomerase gene (xylA) In Saccharomyces cerevisiae. Biotechnol Lett 18, 269–274 (1996). [Google Scholar]
- Gárdonyi M. & Hahn-Hägerdal B. The Streptomyces rubiginosus xylose isomerase is misfolded when expressed in Saccharomyces cerevisiae. Enzyme Microb Technol 32, 252–259 (2003). [Google Scholar]
- Harhangi H. R. et al. Xylose metabolism in the anaerobic fungus Piromyces sp. strain E2 follows the bacterial pathway. Arch Microbiol 180, 134–141 (2003). [DOI] [PubMed] [Google Scholar]
- Kuyper M., Winkler A. A., van Dijken J. P. & Pronk J. T. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res 4, 655–664 (2004). [DOI] [PubMed] [Google Scholar]
- Kuyper M. et al. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by. Saccharomyces cerevisia? FEMS Yeast Res. 4, 69–78 (2003). [DOI] [PubMed] [Google Scholar]
- Ha S.-J., Kim S. R., Choi J.-H., Park M. S. & Jin Y.-S. Xylitol does not inhibit xylose fermentation by engineered Saccharomyces cerevisiae expressing xylA as severely as it inhibits xylose isomerase reaction in vitro. Appl Microbiol Biotechnol 92, 77–84 (2011). [DOI] [PubMed] [Google Scholar]
- Van Maris A. J. et al. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. In Biofuels(ed. Olsson L.). (Springer, 2007). [DOI] [PubMed] [Google Scholar]
- Kuyper M. et al. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 5, 399–409 (2005). [DOI] [PubMed] [Google Scholar]
- Demeke M. M. et al. Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6, 89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Cheng J.-S., Wang B. L., Fink G. R. & Stephanopoulos G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng. 14, 611–622 (2012). [DOI] [PubMed] [Google Scholar]
- Demeke M. M., Foulquié-Moreno M. R., Dumortier F. & Thevelein J. M. Rapid evolution of recombinant Saccharomyces cerevisiae for xylose fermentation through formation of extra-chromosomal circular DNA. PLoS Genet 11, e1005010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat D., Boles E. & Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microb 75, 2304–2311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-M., Jellison T. & Alper H. S. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast. Saccharomyces cerevisiae. Appl Environ Microbiol. 78, 5708–5716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Jiao C., Peng B., Shen Y. & Bao X. Mutation of a regulator Ask10p improves xylose isomerase activity through up-regulation of molecular chaperones in. Saccharomyces cerevisiae. Metab Eng. 38, 241–250 (2016). [DOI] [PubMed] [Google Scholar]
- Farwick A., Bruder S., Schadeweg V., Oreb M. & Boles E. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. Proc Natl Acad Sci 111, 5159–5164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijland J. G. et al. Engineering of an endogenous hexose transporter into a specific D-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol Biofuels. 7, 168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. Y. et al. An engineered cryptic Hxt11 sugar transporter facilitates glucose–xylose co-consumption in. Saccharomyces cerevisiae. Biotechnol Biofuels 8, 176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel A. R., Ouellet M., Szmidt-Middleton H., Keasling J. D. & Mukhopadhyay A. Evolved hexose transporter enhances xylose uptake and glucose/xylose co-utilization in Saccharomyces cerevisiae. Sci Rep 6, 19512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper M. et al. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res 5, 925–934 (2005). [DOI] [PubMed] [Google Scholar]
- Wisselink H. W. et al. Metabolome, transcriptome and metabolic flux analysis of arabinose fermentation by engineered. Saccharomyces cerevisiae. Metab Eng. 12, 537–551 (2010). [DOI] [PubMed] [Google Scholar]
- Kuijpers N. G. et al. One-step assembly and targeted integration of multigene constructs assisted by the I-SceI meganuclease in Saccharomyces cerevisiae. FEMS Yeast Res 13, 769–781 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R. et al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications In. Saccharomyces cerevisiae. FEMS Yeast Res. 15, fov004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träff K., Cordero R. O., Van Zyl W. & Hahn-Hägerdal B. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Environ Microbiol 67, 5668–5674 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph H. K. et al. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ATPase family. Cell 58, 133–145 (1989). [DOI] [PubMed] [Google Scholar]
- Lapinskas P. J., Cunningham K. W., Liu X. F., Fink G. R. & Culotta V. C. Mutations in PMR1 suppress oxidative damage in yeast cells lacking superoxide dismutase. Mol Cell Biol 15, 1382–1388 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin A., Rosas G. & Rao R. P. M. R. 1. a Ca2+-ATPase in yeast Golgi, has properties distinct from sarco/endoplasmic reticulum and plasma membrane calcium pumps. J Biol Chem 272, 9895–9901 (1997). [DOI] [PubMed] [Google Scholar]
- Culotta V. C., Yang M. & Hall M. D. Manganese transport and trafficking: lessons learned from Saccharomyces cerevisiae. EC 4, 1159–1165 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D. et al. High-resolution genome-wide scan of genes, gene-networks and cellular systems impacting the yeast ionome. BMC genomics 13, 623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bastelaere P., Vangrysperre W. & Kersters-Hilderson H. Kinetic studies of Mg2+-, Co2+-and Mn2+-activated D-xylose isomerases. Biochem J 278, 285–292 (1991). [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41, 4336–4343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B. M. et al. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25, 15–37 (2001). [DOI] [PubMed] [Google Scholar]
- Chang A. et al. BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res 43, D439–D446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink J. et al. Energetic limits to metabolic flexibility: responses of Saccharomyces cerevisiae to glucose–galactose transitions. Microbiology 155, 1340–1350 (2009). [DOI] [PubMed] [Google Scholar]
- Devasahayam G., Ritz D., Helliwell S. B., Burke D. J. & Sturgill T. W. Pmr1, a Golgi Ca2+/Mn2+-ATPase, is a regulator of the target of rapamycin (TOR) signaling pathway in yeast. Proc Natl Acad Sci 103, 17840–17845 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Zhao S., Wu Z., Dai P. & Zhou B. Mitochondrial release of the NADH dehydrogenase Ndi1 induces apoptosis in yeast. Mol Biol Cell 23, 4373–4382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Mol Biol Cell 17, 1802–1811 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley B. S., Hanlon N., Jackson R. J. & Rangarajan M. Glucose isomerase: insights into protein engineering for increased thermostability. Biochim Biophys Acta, Protein Struct Mol Enzymol 1543, 294–335 (2000). [DOI] [PubMed] [Google Scholar]
- Madhavan A. et al. Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl Microbiol Biotechnol 82, 1067–1078 (2009). [DOI] [PubMed] [Google Scholar]
- Dun B., Wang Z., Ye K., Zhang B., Li G. & Lu M. Functional expression of Arabidopsis thaliana xylose isomerase in Saccharomyces cerevisiae. Xinjiang Agric Sci 49, 681–686 (2012). [Google Scholar]
- Hector R. E., Dien B. S., Cotta M. A. & Mertens J. A. Growth and fermentation of D-xylose by Saccharomyces cerevisiae expressing a novel D-xylose isomerase originating from the bacterium Prevotella ruminicola TC2-24. Biotechnol Biofuels 6, 84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J. et al. Characterization and evolution of xylose isomerase screened from the bovine rumen metagenome in Saccharomyces cerevisiae. J Biosci Bioeng 121, 160–165 (2016). [DOI] [PubMed] [Google Scholar]
- Klaassen P. et al. Yeast cell capable of converting sugars including arabinose and xylose Patent US 20140141473 A1 (2012).
- Ko J. K., Um Y. & Lee S.-M. Effect of manganese ions on ethanol fermentation by xylose isomerase expressing Saccharomyces cerevisiae under acetic acid stress. Bioresource Technology 222, 422–430 (2016). [DOI] [PubMed] [Google Scholar]
- Entian K.-D. & Kötter P. 25 Yeast genetic strain and plasmid collections. Method Microbiol 36, 629–666 (2007). [Google Scholar]
- Nijkamp J. F. et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN. PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 11, 36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchuk N. A. et al. Construction of long DNA molecules using long PCR‐based fusion of several fragments simultaneously. Nucleic Acids Res 32, e19–e19 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C. et al. A versatile toolbox for PCR‐based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004). [DOI] [PubMed] [Google Scholar]
- Wiedemann B. & Boles E. Codon-optimized bacterial genes improve L-arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl Environ Microbiol 74, 2043–2050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D. & Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350, 87–96 (2002). [DOI] [PubMed] [Google Scholar]
- Solis-Escalante D. et al. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res 13, 126–139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduyn C., Postma E., Scheffers W. A. & van Dijken J. P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous‐culture study on the regulation of respiration and alcoholic fermentation. Yeast 8, 501–517 (1992). [DOI] [PubMed] [Google Scholar]
- Verduyn C., Postma E., Scheffers W. A. & van Dijken J. P. Physiology of Saccharomyces cerevisiae in Anaerobic Glucose-Limited Chemostat Cultures. Microbiology 136, 395–403 (1990). [DOI] [PubMed] [Google Scholar]
- Guadalupe Medina V., Almering M. J., van Maris A. J. & Pronk J. T. Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microbiol 76, 190–195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS one 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T. & Mesirov J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijkamp J. F. et al. De novo detection of copy number variation by co-assembly. Bioinformatics 28, 3195–3202 (2012). [DOI] [PubMed] [Google Scholar]
- Eide D. J. et al. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol 6, R77 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersters-Hilderson H., Callens M., Van Opstal O., Vangrysperre W. & De Bruyne C. K. Kinetic characterization of d-xylose isomerases by enzymatic assays using d-sorbitol dehydrogenase. Enzyme Microb Technol 9, 145–148 (1987). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.