Abstract
Parkin is associated with various inflammatory diseases, including Parkinson's disease (PD) and rheumatoid arthritis (RA). However, the precise role of Parkin in RA is unclear. The present study addressed this issue by comparing the development of RA between non-transgenic (non-Tg) mice and PARK2 knockout (KO) mice. We found that cyclooxygenase-2 and inducible nitric oxide synthase expression and nuclear factor-κB activity were reduced but p53 activation was increased in PARK2 KO as compared to non-Tg mice. These effects were associated with reduced p53 degradation. Parkin was found to interact with p53; however, this was abolished in Parkin KO mice, which prevented p53 degradation. Treatment of PARK2 KO mice with p53 inhibitor increased Parkin expression as well as inflammation and RA development while decreasing nuclear p53 translocation, demonstrating that PARK2 deficiency inhibits inflammation in RA via suppression of p53 degradation. These results suggest that RA development may be reduced in PD patients.
Keywords: Parkin, Arthritis, p53, Ubiquitination
1. Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disease characterized by persistent inflammation of the joint synovium, which can lead to cartilage damage, bone erosion, and joint destruction [1]. The pathogenesis of RA is not fully understood, but involves an interaction between genetic and environmental factors [2]. Parkinson's disease (PD) is a neurodegenerative disorder that was previously often misdiagnosed as RA [3]. Patients with autoimmune diseases such as RA have chronically elevated levels of inflammatory mediators over long periods of time, and may be at increased risk for stroke and PD [4], [5], although the molecular link between PD and RA development remains unclear.
The p53 tumor suppressor gene inhibits the proliferation of abnormal cells such as cancer cells by triggering apoptosis [6]. It is activated by a variety of cellular stressors, including DNA damage, hypoxia, and oncogenes [7], but is also associated with inflammation. Lipopolysaccharide (LPS)-induced acute lung injury was exacerbated and interleukin (IL)−6 levels were found to be reduced in p53−/− as compared to p53+/+ mice [8], [9]. It was also reported that p53 suppresses inflammatory responses during RA development [10]. However, mutation of p53 stimulates the proliferation of the synovial cells and may contribute to the pathogenesis of chronic diseases [11]. The joints of p53−/− mice with collagen antibody-induced arthritis (CAIA) showed greater severity of arthritis than those of wild-type mice [12]. More severe arthritis—as evidenced by increased bovine type II collagen-stimulated T cell proliferation and interferon (IFN)-γ production—was also observed in p53−/− as compared to p53+/+ mice [13]. Conversely, p53 overexpression induced apoptosis and reduced leukocyte infiltration without affecting cartilage metabolism in cultured synovial cells and tissue in a rabbit model of arthritis [14]. Mutation of p53 has been reported to enhance the activation of nuclear factor (NF)-κB [15], a major regulator of inflammation and associated diseases [16]. Thus, p53 has important roles in the regulation of cell proliferation in the synovium.
Parkin is encoded by the PARK2 gene, which is expressed in multiple tissues and functions as a RING-between-RING E3 ligase [17]. Protein ubiquitination involves the concerted action of the E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin-protein ligases. The latter binds substrate proteins and E2s with high specificity to mediate the transfer of ubiquitin between these two molecules [18]. It also targets substrates for proteasomal destruction. It was previously supposed that the loss of Parkin function leads to accumulation of toxic substrates that damage dopaminergic neurons, resulting in Parkinsonism [19]. Parkin was shown to inhibit p53 expression and activity in TSM1 neuronal cells [20], and a recent study demonstrated that cytosolic p53 binds to the RING0 domain of Parkin [21]. Thus, it is possible that Parkin binds to and targets p53 for degradation, thereby providing a mechanistic basis for the development of RA.
To investigate this possibility, we examined the role of Parkin in the development of inflammatory arthritis using transgenic (Tg) PARK2 knockout (KO) mice.
2. Results
2.1. LPS-induced iNOS and COX-2 expression is decreased by Parkin knockdown and increased by p53 knockdown
To investigate how Parkin and p53 modulate inflammatory responses, we stimulated RAW 264.7 cells, a murine macrophage like cell line, and human fibroblast-like synoviocytes (FLS) derived from RA patients with LPS or TNF-α. iNOS and COX-2 were upregulated by both stimuli in a dose dependent manner whereas Parkin and p53 were downregulated by the treatment in both cell lines (Supplementary Fig. 1). To further assess the relationship between Parkin or p53 and the inflammatory response, RAW 264.7 cells and human FLS were transfected with siRNAs against Parkin or p53 for 24 h and then treated with LPS for 24 h. Parkin knockdown decreased LPS-induced expression of iNOS and COX-2 but increased that of p53 (Supplementary Fig. 2A, B). Conversely, p53 knockdown enhanced LPS-induced expression of iNOS and COX-2and increased that of Parkin (Supplementary Fig. 2C, D).
2.2. PARK2 deficiency reduces inflammatory arthritis
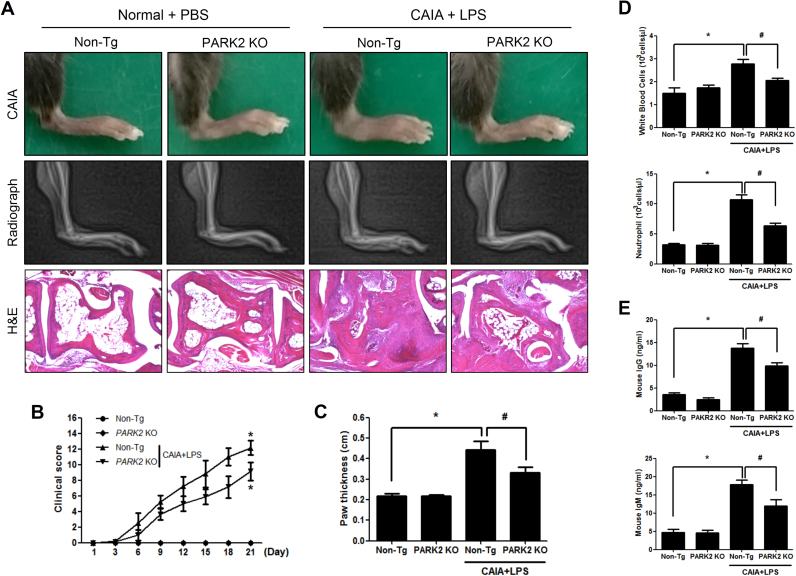
To investigate whether PARK2 deficiency leads to the development of arthritis, we compared the degree of arthritis in non-Tg and PARK2 KO mice. Following CAIA, hind paw edema was increased in non-Tg mice, but this effect was reversed in the mutants (Fig. 1A), accompanied by a decrease in the clinical score (Fig. 1B, C). A histopathological evaluation in the hind paws also showed reduced synoviocyte hyperplasia, bone erosion, and cartilage destruction in the joints of PARK2 KO as compared to non-Tg mice (Fig. 1A). The number of white blood cells and neutrophils in the blood was lower in CAIA and LPS-treated non-Tg as compared to PARK2 KO mice (Fig. 1D), which was associated with reduced IgG and IgM levels (Fig. 1E).
Fig. 1.
Inhibition of inflammatory arthritis in PARK2 KO mice. To generate the CAIA model, mice were injected with an antibody against type II collagen on day 0, followed by injection of LPS on day 3. Development of CAIA was monitored for 21 days by (A) radiography, (B) paw thickness measurements, and (C) clinical scores. (D–G) The number of white blood cells and neutrophils (D, E) and IgG (F) and IgM (G) levels in blood were determined in CAIA with LPS-induced non-Tg and PARK2 KO mice. (H) Histopathological analysis of joint of hind paws and bone destruction in PARK2 KO mice with CAIA and in non-Tg mice with or without CAIA. Values represent the mean ±SD of 10 mice. #P<0.05 vs. non-Tg mice with CAIA and LPS; *P<0.05 vs. non-Tg mice without CAIA.
2.3. P53 interacts with Parkin and is targeted for degradation in vitro and in vivo
To investigate the relationship between Parkin and p53, we performed a Gene Ontology (GO) analysis using GeneMania (http:www.genemania.org) to generate networks of interaction between Parkin and p53. This strategy uncovered tight networking of the these genes with 21 associated genes including NFкB, which is well-known inflammation regulatory factor (Supplementary Fig. 3). Since p53 suppresses the inflammatory response, we speculated that Parkin binds and targets p53 for degradation, resulting in loss of p53 function. Parkin was found to interact with p53 in RAW 264.7 cells, as determined by immunoprecipitation followed by immunoblot analysis (Fig. 2A) as well as by the Octet system (Supplementary Fig. 4). Molecular docking revealed that the p53 binding pocket of Parkin comprised Ser19, Ala46, Lys48, Leu50, Lys51, Asp53, Trp54, Thr55, Gln57, Asn58, Asp60, Leu61, Tyr391, Arg392, Asp394, Glu395, Arg396, and Ala397 (Fig. 2B).
Fig. 2.
Interaction between p53 and Parkin and p53 degradation in vitro and in vivo. (A) RAW264.7 cell lysates were immunoprecipitated with anti-p53 antibody and analyzed by immunoblotting with anti-Parkin antibody. (B) Molecular surface representation in the docking model of p53 with Parkin. (C) In vitro analysis of Parkin-mediated p53 ubiquitination. In vitro ubiquitination of p53 in the presence of Parkin. Reaction mixtures were analyzed by immunoblotting using the indicated antibodies. (D) In vivo ubiquitination assay in HEK293 cells transfected with plasmids expressing Flag-tagged p53, Myc-tagged Parkin, and HA-tagged ubiquitin. Cell lysates were immunoprecipitated with anti-p53 antibody and analyzed by immunoblotting with indicated antibodies. (E) In vivo ubiquitination assay of RAW 264.7 cells treated LPS (1 μg/ml). Cell lysates were immunoprecipitated with anti-ubiquitin antibody and analyzed by immunoblotting with indicated antibodies. (F) P53 expression in cytosolic and nuclear fractions. (G) P53 localization, as determined by immunofluorescence analysis and confocal microscope. Each band in the blots is representative of three experiments.
We investigated whether p53 is a target of Parkin-mediated ubiquitination by in vitro and in vivo assays. P53 was ubiquitinated in the presence of Parkin, but was not degraded upon Parkin knockdown (Fig. 2C). To determine whether Parkin mediates protein ubiquitination in vivo, HEK293 cells were transfected with plasmids expressing Myc-tagged Parkin and HA-tagged ubiquitin, and cell extracts were analyzed by immunoblotting with antibodies against Myc and HA. Parkin was expressed in transfected cells and caused an increase in protein ubiquitination, which was not observed in the absence of HA-tagged ubiquitin (Fig. 2D).
Parkin expression was downregulated by LPS stimulation in RAW 264.7 cells and human FLS; this corresponded to an increase in p53 degradation in RAW 264.7 cells (Fig. 2E). The decrease in inflammation observed in PARK2 KO relative to non-Tg mice was associated with p53 accumulation resulting from Parkin deficiency. CAIA and LPS-treated PARK2 KO mice showed only slightly higher levels of p53 in the cytoplasm as compared to non-Tg mice; however, nuclear expression of p53 was increased (Fig. 2F). To determine whether this was due to a reduction in p53 degradation, we examined p53 expression in ankle joint tissues by immunofluorescence analysis and confocal microscopy. Expression of p53 was increased in CAIA and LPS-treated PARK2 KO mice relative to their non-Tg counterparts (Fig. 2G).
2.4. PARK2 deficiency alters cytokine, COX-2, and iNOS levels in the spleen and paw joints
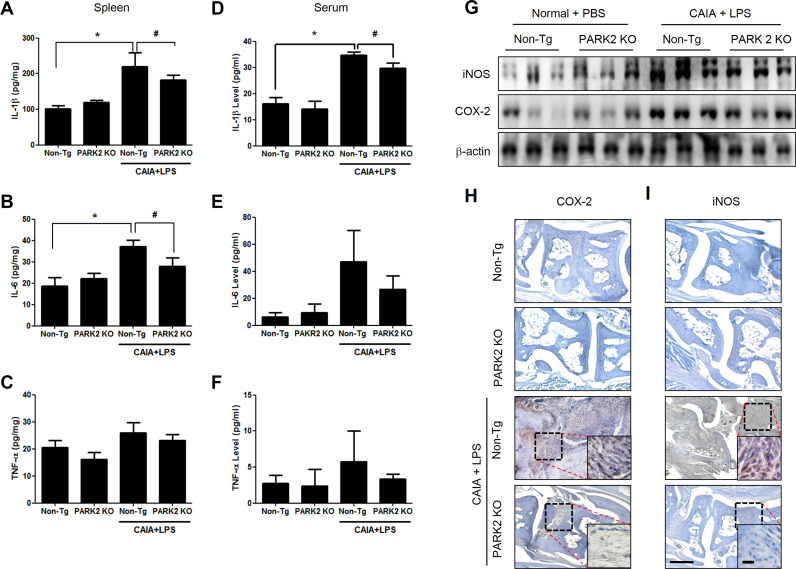
We next investigated whether loss of PARK2 alters cytokine levels in the immune organ (spleen) by ELISA, and examined whether this leads to the suppression of arthritis development in the paw joint. The levels of pro-inflammatory cytokines including IL-1β, IL-6, and TNF-α were increased in the spleen as well as in the serum of CAIA and LPS-treated non-Tg mice. However, in CAIA and LPS-treated PARK2 KO mice, IL-1β (Fig. 3A, D) and IL-6 (Fig. 3B, E) levels in the spleen and paw joint were reduced whereas that of TNF-α (Fig. 3C, F) was unaltered. In addition, iNOS and COX-2 were upregulated in CAIA and LPS-treated non-Tg mice, as determined by western blotting (Fig. 3G) and immunohistochemistry (Fig. 3H, I).
Fig. 3.
PARK2 deficiency alters cytokine, COX-2, and iNOS levels in the spleen and paw joint tissue. (A–F) IL-1β, IL-6, and TNF-α levels in the spleen (A–C) and serum (D–F) were measured by ELISA. Values represent mean±SD of 10 mice. #P<0.05 vs. non-Tg mice with CAIA and LPS; *P<0.05 vs. non-Tg mice without CAIA and LPS. (G) iNOS and COX-2 expression, a detected by western blotting. β-Actin served as a loading control. (H, I) Sections of joint tissue from mice were analyzed for iNOS and COX-2 expression by immunohistochemistry. Each band in the blots is representative of three experiments.
2.5. PARK2 deficiency inhibits NF-κB activation in the paw joint
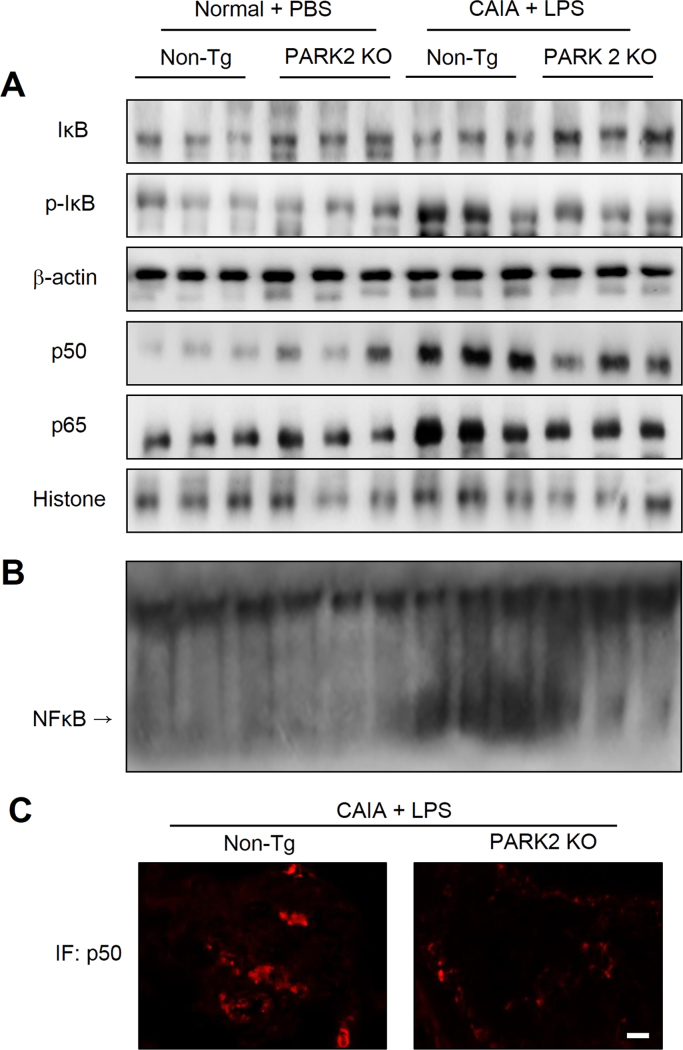
To determine whether loss of PARK2 inhibits NF-κB activation in paw joints, we examined the nuclear localization of p65 and p50 and IκB degradation by western blotting. The levels of p65 and p50 in the nucleus and p-IκB in the cytoplasm were increased in the paw joints of CAIA and LPS-treated non-Tg mice, but were decreased in PARK2 KO animals (Fig. 4A, C). In addition, the DNA binding activity of NF-κB was higher in the paw joints of non-Tg as compared to PARK2 KO CAIA mice (Fig. 4B).
Fig. 4.
Effect of PARK2 deficiency on NF-κB activity in the paw joint. (A) IκBα, p-IκBα, p50, and p65 levels were detected by western blotting. (B) DNA binding activity of NF-κB was measured by electrophoretic mobility shift assay (EMSA). β-Actin and histone protein were used as loading controls. (C) P50 localization was analyzed by immunofluorescence analysis and confocal microscopy. Each band in the blots is representative of three experiments.
2.6. P53 inhibition increases RA severity and p53 degradation in the absence of PARK2
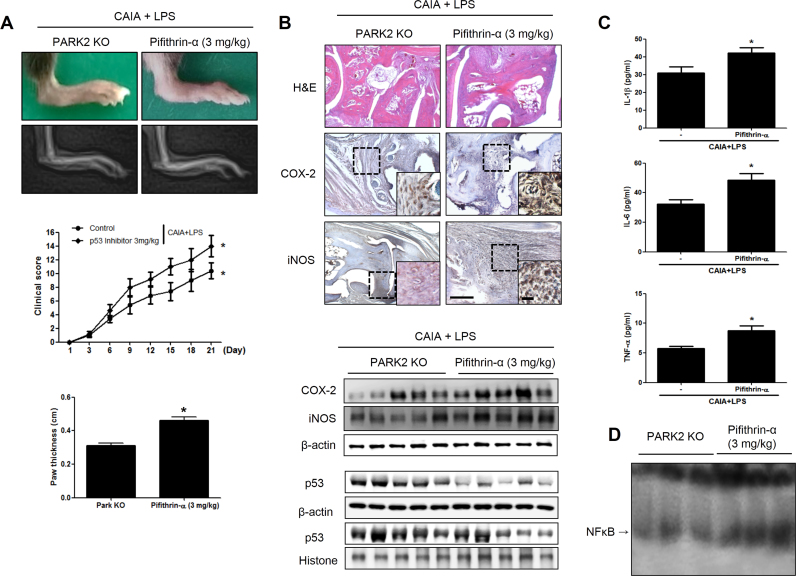
To further clarify the involvement of p53 in the development of RA, PARK2 KO mice with CAIA were treated with LPS along with the p53 inhibitor pifithrin-α. Interestingly, after 3 days of pifithrin-α administration, arthritis was exacerbated (Fig. 5A); synoviocyte hyperplasia, bone erosion, and cartilage destruction were increased, while iNOS and COX-2 expression in the joints was upregulated, as determined by immunohistochemistry (Fig. 5B). The decrease in p53 expression resulting from inhibitor treatment was associated with higher levels of IL-1β, IL-6, and TNF-α (Fig. 5C). CAIA and LPS-treated PARK2 KO mice also showed nuclear accumulation of p53 and reduced NF-κB activation.
Fig. 5.
Increased RA severity by p53 inhibition in PARK2-deficient mice. Mice were injected with anti-type II collagen antibody on day 0, followed by LPS and p53 inhibitor treatment on day 3. Control mice were not treated with p53 inhibitor. (A) Development of CAIA was monitored for 21 days as described in the legend for Fig. 1. (B, C) iNOS and COX-2 expression, as determined by immunohistochemistry. (D) P53 expression in joint tissue from mice with CAIA. β-Actin and histone served as loading controls. (E) Serum IL-1β, IL-6, and TNF-α levels, as measured by ELISA. Values represent mean±SD from five mice. *P<0.05 vs. PARK2 KO mice with CAIA and LPS. (F) DNA binding activity of NF-κB, as determined by electrophoretic mobility shift assay. Each band in the blots is representative of three experiments.
NF-κB activation has been shown to be suppressed by p53 [22]. To determine whether p53 regulates NF-κB activity in RA, NF-κB expression was examined in mice treated with pifithrin-α. NF-κB level was increased in CAIA and LPS-treated PARK2 KO mice upon pifithrin-α administration (Fig. 5D). The nuclear translocation of p65 and p50 and IκB degradation were also increased in the paw joint as a result of p53 inhibition (Supplementary Fig. 5).
3. Discussion
PD is characterized by the degeneration of dopaminergic neurons [23] and is related to inflammation and oxidative stress [24]. The relationship between the development of PD and RA has been controversial [25]. To address this issue, the presents study investigated the role of Parkin in the development of RA. We found that Parkin deficiency decreased LPS-induced COX-2 and iNOS expression in RAW 264.7 cells and human FLS. The opposite was observed upon p53 knockdown; moreover, Parkin was upregulated in the absence of p53, consistent with the reciprocal regulation between p53 and Parkin. It was previously demonstrated that Parkin depletion enhanced p53 expression in fibroblasts and mouse brain [20], [26], while Parkin overexpression had the opposite effect [20], [26]. P53 protein level was found to be increased by 17-fold in the brains of PD patients relative to controls [26].
We observed that nuclear translocation of p53 was increased in CAIA and LPS-treated PARK2 KO as compared to non-Tg mice, whereas Parkin knockdown increased LPS-induced p53 expression in RAW 264.7 cells and human FLS. P53 acts as a tumor suppressor; however, it has also been implicated in inflammatory diseases, including RA. In this study, we found that CAIA and LPS-treated PARK2 KO mice were less likely to develop arthritis as compared to non-Tg mice. These data indicate that loss of Parkin can reduce inflammatory responses and thereby prevent the development of arthritis.
A previous study reported that the joints of p53−/− CAIA mice showed more severe arthritis [12]. P53 was shown to inhibit innate immune responses and the development of autoimmune diabetes in p53 KO mice [27]. We demonstrated that pharmacological inhibition of p53 enhanced CAIA-induced arthritis in PARK2 KO mice, suggesting that p53 is responsible for the reduction in inflammatory arthritis in PARK2 KO mice.
P53 is a target of various E3 ubiquitin ligases, including Mouse double minute 2 homolog [28]. Pirh2 was shown to interact with the RING-H2 domain of p53 and promote p53 ubiquitination and degradation [29]. Parkin also interacts with the RING1 domain of p53 [20], and immunoprecipitation studies demonstrated that the RING0 domain of Parkin was essential for this interaction [21]. On the other hand, p53 inhibited Parkin-mediated autophagic degradation of damaged mitochondria, leading to impaired insulin secretion in pancreatic β cells of diabetic mice [30]. Based on these findings and the results of the in vitro and in vivo ubiquitination assays presented here, we propose that Parkin negatively regulates p53 via direct interaction and ubiquitination [29]; the consequent loss of p53 function may contribute to the development of arthritis.
P53 accumulation in the absence of Parkin altered NF-κB activation. NF-κB is a component of a pro-inflammatory signaling pathway and modulates the expression of cytokines, chemokines, and adhesion molecules [16]. As such, inactivation of NF-κB may be an effective strategy for the treatment of inflammatory diseases. P53 is a known inhibitor of NF-κB-mediated transcription; loss of p53 therefore resulted in an increase in inflammatory responses [31]. LPS increased the NF-κB-induced expression of IL-1, IL-6, IL-12, TNF-α, chemokine receptor (CCR)2, CCR5, monokine induced by IFN-γ, and IFN-γ-induced protein 10 in the thymus of p53 KO as compared to wild-type mice [9]. Another study showed that NF-κB activity was reduced in human metastatic melanoma cells with wild-type p53, but was enhanced in cells expressing mutant p53 [32]. In this study, NF-κB activation and p50 and p65 expression were decreased whereas nuclear p53 level was increased in CAIA PARK2 KO relative to non-Tg mice. P53 inhibition in CAIA and LPS-treated PARK2 KO mice increased p50 and p65 nuclear localization and NF-κB activation. Thus, Parkin deficiency leads to the development of arthritis due to the suppression of p53 degradation and consequent inactivation of NF-κB. Our results suggest that the development of PD and RA is closely related and that therapeutic strategies that target Parkin could reduce risk of RA development.
4. Materials and methods
4.1. Cell culture
RAW 264.7 mouse macrophage-like cells and HEK293 human embryonic kidney cells were obtained from the American Type Culture Collection (Manassas, VA, USA). The Clinical Research Ethics Committee of the College of Medicine, Soonchunhyang University Medical Center (Chungnam, Korea) approved the study protocol and the use of human tissue samples. Informed consent was obtained from all patients. Human fibroblast-like synoviocytes (FLS) and RAW 264.7 cells were cultured as previously described [33].
4.2. Animals and ethics statement
Male PARK2 knockout (PARK2 KO) and non-Tg (C57BL6/J) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Mice were housed and bred under specific pathogen–free conditions at the Laboratory Animal Research Center of Chungbuk National University, Korea, and maintained 10 per cage in a room at constant temperature of 22 °C±18 °C and relative humidity of 55%±10% on a 12:12-h light/dark cycle. The animals were provided drinking tap water and a standard rodent chow diet ad libitum (RodFeed, DBL, Chungbuk, Korea) throughout the experiment. All the animal experiments were conducted in accordance with the principles and procedures outlined in the Korean National Institute of Health Guide for the Care and Use of Laboratory Animals. The protocol for the animal procedures was approved by the Chungbuk National University Institutional Animal Care and Use Committee and compiled with the Korean National Institute of Health Guide for the Care and Use of Laboratory Animals (CBNUA-754-14-01).
4.3. CAIC induction
Mice were subjected to CAIA-induced arthritis as previously described [33]. Briefly, 5 mg of an anti-collagen II monoclonal antibody (Chondrex, Redmond, WA, USA) was administered by intraperitoneal injection into 12-week-old male PARK2 KO mice on day 0, followed by 50 μg LPS on day 3. The severity of arthritis was graded on a scale of 0–4 for each paw in a blinded fashion: 0, no evidence of erythema or swelling; 1, erythema and mild swelling confined to the midfoot (tarsals) or ankle joint; 2, erythema and mild swelling extending from the ankle to the midfoot; 3, erythema and moderate swelling extending from the ankle to the metatarsal joints; and 4, erythema and severe swelling encompassing the ankle, foot and digits. The scores for each of the four paws were added to obtain a final score, with a maximal severity score of 16, presented as the mean±standard error of the mean (SEM).
4.4. Histological analysis
Histological analysis was carried out as previously described [33].
4.5. Radiological evaluation
Radiological evaluation was performed as previously described [33].
4.6. Western blotting
Western blot analysis was performed as previously described [33]. Membranes were incubated with antibodies against the following proteins: inducible nitric oxide synthase (iNOS) and cyclooxygenase (COX)-2 (both 1:1000; Novus Biologicals, Littleton, CO, USA); and Parkin, p53, inhibitor of (I)κB, phosphorylated (p-)IκB, p50, p65, β-actin, and histone (all 1:1000; Santa Cruz Biotechnology, CA, USA). Immunoreactivity was visualized by enhanced chemiluminescence.
4.7. Short interfering (si)RNA transfection
RAW 264.7 cells and human FLS (5×104/well) were seeded in 96-well plates and transiently transfected with siRNA against Parkin or p53 using Lipofectamine 3000 reagent in Opti-MEM (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Transfected cells were treated with 1 μg/ml LPS for 1 or 24 h before cell viability and protein expression were analyzed.
4.8. Molecular docking
The docking of Parkin with p53 was examined using the rigid-body docking program ZDOCK 3.0.2 on the ZDOCK server (http://zdock.umassmed.edu). Parkin (PDB ID: 5C1Z monomer) and p53 (PDB ID: 1TUP monomer) without DNA were used for docking. Docking experiments were carried out without selecting or blocking residues.
4.9. Immunoprecipitation
Immunoprecipitation was carried out as previously described [34]. Briefly, cells were lysed in EBC buffer. The precleared soluble supernatant was mixed with a polyclonal anti-p53 antibody and incubated overnight at 4 °C. Protein A/G beads were then added to the reaction mixture. After washing the immune complexes, bound proteins were resuspended in sodium dodecyl sulfate (SDS) sample buffer, resolved by SDS–polyacrylamide gel electrophoresis, and analyzed by western blotting using a monoclonal antibody against Parkin.
4.10. DNA-binding activity assay by electromobility shift assay (EMSA)
The DNA-binding activities of STAT3 and NF-κB in the cultured cells and ankle joint tissues of CAIA mice were determined using an electromobility shift assay (EMSA) as described previously [35]. In brief, cells were treated with MMPP or DMSO. After incubation for 24 h, the cells were washed thrice with ice-cold PBS and the nuclear extracts were prepared for EMSA. For the measurement in mouse tissue, the ankle joint tissues were isolated from normal and CAIA mice treated with MMPP (5 mg/kg), indomethacin (5 mg/kg), or vehicle, and parts of the tissues were rinsed with ice-cold PBS, homogenised in ice-cold buffer A (10 mM potassium chloride [KCl], 0.2 mM ethylenediaminetetraacetic acid [EDTA], 1.5 mM magnesium chloride [MgCl2], 0.5 ml dithiothreitol [DTT], and 0.2 mM phenylmethanesulfonyl fluoride [PMSF]) and centrifuged for 5 min at 14,000×g. The residual pellet was resuspended in 100 μL buffer C (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 420 mM sodium chloride (NaCl), 1.5 mM MgCl2, 20% glycerol, 0.2 mM EDTA, 0.5 mM DTT and 0.2 mM PMSF). After incubation at 4 °C for 20 min, the lysate was centrifuged for 6 min at 14,000×g and the supernatants (nuclear extract) were collected, and EMSA was carried out as described above.
4.11. Octet analysis
Binding interactions were analyzed using the Octet system (Fortebio, Menlo Park, CA, USA) by measuring the wavelength shift in nanometers. Kinetic rate constants were determined with the HIS1K-Anti-penta-His sensor in the advanced kinetic mode. Recombinant His-tagged Parkin protein was immobilized on the sensor at 2 μg/ml in 10× kinetic buffer (Fortebio) and exposed for 1200 s to various concentrations of recombinant p53 protein covering the predicted dissociation constant (KD) followed by a 4000-s dissociation step in 10× kinetic buffer with shaking at 1000 rpm. Interferometry data were globally fitted to a 1:1 binding ratio to calculate kinetic parameters using Octet ForteBio software (Fortebio).
4.12. Measurement of IL-1β, IL-6, and tumor necrosis factor (TNF)-α levels
IL-1β, IL-6, and TNF-α levels in spleen and serum lysates were determined with enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, Minneapolis, MN, USA) according to the manufacturer's protocol. The optical density at 450 nm (OD450) was measured with an automated microplate reader.
4.13. In vitro and in vivo ubiquitination assays
The in vitro ubiquitination assay was carried out using a kit (Boston Biochem, Cambridge, MA, USA) according to the manufacturer's instruction. HEK293 cells were transfected with HA-ubiquitin (2 μg), Flag-p53 (1 μg), and Myc-Parkin (1 μg) expression plasmids using Lipofectamine 3000; 48 h later, cells were harvested and used for immunoblotting and for the ubiquitination assay. Eluted proteins were analyzed by immunoblotting with a monoclonal antibody against p53.
4.14. Immunoglobulin (Ig)G and IgM quantification
Serum levels of IgG and IgM were evaluated with ELISA kits (Komabiotech, Seoul, Korea) according to the manufacturer's protocol. OD450 was measured with an automated microplate reader.
4.15. Immunofluorescence analysis
Cells were permeabilized by treatment with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 2 min, then blocked with 5% bovine serum albumin in PBS at room temperature for 2 h. The cells were then incubated overnight at 4 °C with a rabbit polyclonal antibody against activated p53 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA). After washes with ice-cold PBS, cells were incubated with an Alexa Fluor 488-conjugated anti-rabbit secondary antibody (1:100; Molecular Probes, Eugene, OR, USA) for 2 h at room temperature. Images were acquired on a confocal laser scanning microscope (TCS SP2; Leica Microsystems, Wetzlar, Germany) at 200× magnification.
4.16. Immunohistochemistry
Immunohistochemistry was carried out by the avidin–biotin–peroxidase method [33]. Sections were stained with hematoxylin and eosin (Sigma-Aldrich) and labeled with antibodies against COX-2 and iNOS.
4.17. Statistical analysis
Data were analyzed using GraphPad Prism v.4.03 software (Graph Pad Inc., La Jolla, CA, USA). Data are presented as mean±SD. Differences between groups were assessed by two-way ANOVA analysis of variance; when the P value was statistically significant, the differences were evaluated with Bonferroni-adjusted t-test. P≤0.05 was considered to be statistically significant.
Author contributions
All authors were involved in drafting or critically revising the manuscript for intellectual content, and all authors approved the final version for publication. Y.Y. Jung and D.J. Son conducted most of the experiments, performed data analysis, and were the primary writer of the manuscript. H.L. Lee performed in vitro assays and Western blotting assays. D.H. Kim assisted in the in vivo functional assays and immunehistological studies. Y.W. Ham performed computational docking analysis. M.J. Song, Y. Kim, and S.B. Han provided advice throughout the project. M.H. Park and J.T. Hong supervised the entire project and had major roles in experimental design, data interpretation, and writing the manuscript. Drs. S.B. Han, Y. Kim, and J.T. Hong had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the analyses.
Competing interests
The authors declare no competing financial interests.
Acknowledgements
This work was supported by a National Research Foundation of Korea grant funded by the Korean Government (MRC, 2008-0062275).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.04.007.
Contributor Information
Mi Hee Park, Email: pmh5205@hanmail.net.
Jin Tae Hong, Email: jinthong@chungbuk.ac.kr.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Scott D.L., Pugner K., Kaarela K., Doyle D.V., Woolf A., Holmes J. The links between joint damage and disability in rheumatoid arthritis. Rheumatology. 2000;39:122–132. doi: 10.1093/rheumatology/39.2.122. [DOI] [PubMed] [Google Scholar]
- 2.McInnes I.B., Schett G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 3.Hrycaj P., Korczowska I., Lacki J.K. Severe Parkinson's disease in rheumatoid arthritis patient treated with infliximab. Rheumatology. 2003;42:702–703. doi: 10.1093/rheumatology/keg160. [DOI] [PubMed] [Google Scholar]
- 4.Rugbjerg K., Friis S., Ritz B., Schernhammer E.S., Korbo L., Olsen J.H. Autoimmune disease and risk for Parkinson disease: a population-based case-control study. Neurology. 2009;73:1462–1468. doi: 10.1212/WNL.0b013e3181c06635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liou T.H., Huang S.W., Lin J.W., Chang Y.S., Wu C.W., Lin H.W. Risk of stroke in patients with rheumatism: a nationwide longitudinal population-based study. Sci. Rep. 2014;4:5110. doi: 10.1038/srep05110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., Chumakov P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe S.W. Activation of p53 by oncogenes. Endocr.-Relat. Cancer. 1999;6:45–48. doi: 10.1677/erc.0.0060045. [DOI] [PubMed] [Google Scholar]
- 8.Liu G., Park Y.J., Tsuruta Y., Lorne E., Abraham E. p53 Attenuates lipopolysaccharide-induced NF-kappaB activation and acute lung injury. J. Immunol. 2009;182:5063–5071. doi: 10.4049/jimmunol.0803526. [DOI] [PubMed] [Google Scholar]
- 9.Komarova E.A., Krivokrysenko V., Wang K., Neznanov N., Chernov M.V., Komarov P.G. p53 is a suppressor of inflammatory response in mice. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2005;19:1030–1032. doi: 10.1096/fj.04-3213fje. [DOI] [PubMed] [Google Scholar]
- 10.Munoz-Fontela C., Mandinova A., Aaronson S.A., Lee S.W. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat. Rev. Immunol. 2016;16:741–750. doi: 10.1038/nri.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inazuka M., Tahira T., Horiuchi T., Harashima S., Sawabe T., Kondo M. Analysis of p53 tumour suppressor gene somatic mutations in rheumatoid arthritis synovium. Rheumatology. 2000;39:262–266. doi: 10.1093/rheumatology/39.3.262. [DOI] [PubMed] [Google Scholar]
- 12.Yamanishi Y., Boyle D.L., Pinkoski M.J., Mahboubi A., Lin T., Han Z. Regulation of joint destruction and inflammation by p53 in collagen-induced arthritis. Am. J. Pathol. 2002;160:123–130. doi: 10.1016/S0002-9440(10)64356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simelyte E., Rosengren S., Boyle D.L., Corr M., Green D.R., Firestein G.S. Regulation of arthritis by p53: critical role of adaptive immunity. Arthritis Rheum. 2005;52:1876–1884. doi: 10.1002/art.21099. [DOI] [PubMed] [Google Scholar]
- 14.Yao Q., Wang S., Glorioso J.C., Evans C.H., Robbins P.D., Ghivizzani S.C. Gene transfer of p53 to arthritic joints stimulates synovial apoptosis and inhibits inflammation. Mol. Ther.: J. Am. Soc. Gene Ther. 2001;3:901–910. doi: 10.1006/mthe.2001.0343. [DOI] [PubMed] [Google Scholar]
- 15.Scian M.J., Stagliano K.E., Anderson M.A., Hassan S., Bowman M., Miles M.F. Tumor-derived p53 mutants induce NF-kappaB2 gene expression. Mol. Cell. Biol. 2005;25:10097–10110. doi: 10.1128/MCB.25.22.10097-10110.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 18.Scheffner M., Nuber U., Huibregtse J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 19.Dawson T.M., Dawson V.L. The role of parkin in familial and sporadic Parkinson's disease. Mov. Disord.: Off. J. Mov. Disord. Soc. 2010;25(Suppl 1):S32–S39. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Costa C.A., Sunyach C., Giaime E., West A., Corti O., Brice A. Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson's disease. Nat. Cell Biol. 2009;11:1370–1375. doi: 10.1038/ncb1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshino A., Mita Y., Okawa Y., Ariyoshi M., Iwai-Kanai E., Ueyama T. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat. Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 22.Murphy S.H., Suzuki K., Downes M., Welch G.L., De Jesus P., Miraglia L.J. Tumor suppressor protein (p)53, is a regulator of NF-kappaB repression by the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA. 2011;108:17117–17122. doi: 10.1073/pnas.1114420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davie C.A. A review of Parkinson's disease. Br. Med. Bull. 2008;86:109–127. doi: 10.1093/bmb/ldn013. [DOI] [PubMed] [Google Scholar]
- 24.Hald A., Lotharius J. Oxidative stress and inflammation in Parkinson's disease: is there a causal link? Exp. Neurol. 2005;193:279–290. doi: 10.1016/j.expneurol.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Tufekci K.U., Meuwissen R., Genc S., Genc K. Inflammation in Parkinson's disease. Adv. Protein Chem. Struct. Biol. 2012;88:69–132. doi: 10.1016/B978-0-12-398314-5.00004-0. [DOI] [PubMed] [Google Scholar]
- 26.Sunico C.R., Nakamura T., Rockenstein E., Mante M., Adame A., Chan S.F. S-Nitrosylation of parkin as a novel regulator of p53-mediated neuronal cell death in sporadic Parkinson's disease. Mol. Neurodegener. 2013;8:29. doi: 10.1186/1750-1326-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng S.J., Lamhamedi-Cherradi S.E., Wang P., Xu L., Chen Y.H. Tumor suppressor p53 inhibits autoimmune inflammation and macrophage function. Diabetes. 2005;54:1423–1428. doi: 10.2337/diabetes.54.5.1423. [DOI] [PubMed] [Google Scholar]
- 28.Moll U.M., Petrenko O. The MDM2-p53 interaction. Mol. Cancer Res.: MCR. 2003;1:1001–1008. [PubMed] [Google Scholar]
- 29.Leng R.P., Lin Y., Ma W., Wu H., Lemmers B., Chung S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 30.Hoshino A., Ariyoshi M., Okawa Y., Kaimoto S., Uchihashi M., Fukai K. Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic beta-cell function in diabetes. Proc. Natl. Acad. Sci. USA. 2014;111:3116–3121. doi: 10.1073/pnas.1318951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudkov A.V., Gurova K.V., Komarova E.A. Inflammation and p53: a tale of two stresses. Genes Cancer. 2011;2:503–516. doi: 10.1177/1947601911409747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gulati A.P., Yang Y.M., Harter D., Mukhopadhyay A., Aggarwal B.B., Benzil D.L. Mutant human tumor suppressor p53 modulates the activation of mitogen-activated protein kinase and nuclear factor-kappaB, but not c-Jun N-terminal kinase and activated protein-1. Mol. Carcinog. 2006;45:26–37. doi: 10.1002/mc.20149. [DOI] [PubMed] [Google Scholar]
- 33.Kim D.H., Lee D.H., Jo M.R., Son D.J., Park M.H., Hwang C.J. Exacerbation of Collagen antibody-induced arthritis in transgenic mice overexpressing peroxiredoxin 6. Arthritis Rheumatol. 2015;67:3058–3069. doi: 10.1002/art.39284. [DOI] [PubMed] [Google Scholar]
- 34.Park M.H., Choi D.Y., Jin H.W., Yoo H.S., Han J.Y., Oh K.W. Mutant presenilin 2 increases beta-secretase activity through reactive oxygen species-dependent activation of extracellular signal-regulated kinase. J. Neuropathol. Exp. Neurol. 2012;71:130–139. doi: 10.1097/NEN.0b013e3182432967. [DOI] [PubMed] [Google Scholar]
- 35.Ban J.O., Kim D.H., Lee H.P., Hwang C.J., Shim J.H., Kim D.J. Anti-arthritis effects of (E)-2,4-bis(p-hydroxyphenyl)-2-butenal are mediated by inhibition of the STAT3 pathway. Br. J. Pharmacol. 2014;171:2900–2912. doi: 10.1111/bph.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material