Abstract
Previously, we identified three porcine circovirus type 2 (PCV2) strains in buffalo meat samples from southern China. In this study, we confirmed the reappearance of those buffalo-origin-like PCV2 strains in swine herds in this region, which supported the possible cross-species infection of PCV2 between buffalos and pigs under field conditions.
Keywords: Buffalo-origin, cross-species infection, porcine circovirus type 2, southern China, swine-origin
Porcine circovirus type 2 (PCV2) is an important virus for the worldwide swine industry, because it can cause a variety of clinical diseases including postweaning multisystemic wasting syndrome, porcine dermatitis and nephropathy syndrome, porcine respiratory disease complex, proliferative and necrotizing pneumonia and reproductive disorders [1]. However, in recent years, it was confirmed that PCV2 widely existed in non-porcine animals including cattle, rodents, insects and even humans [1]. Although PCV2 was reported in some diseased animals [2], [3], [4], the relationship between PCV2 and the disease in non-porcine animals was unclear. However, one animal experimental study demonstrated that calves were susceptible to PCV2 and displayed clinical signs including lymph node swelling, reddening of oral and ocular mucosa and diarrhoea [5]. Moreover, another study suggested that PCV2 might be closely related to the death of minks clinically [6], indicating that PCV2 is also an important pathogen in minks, not just in pigs.
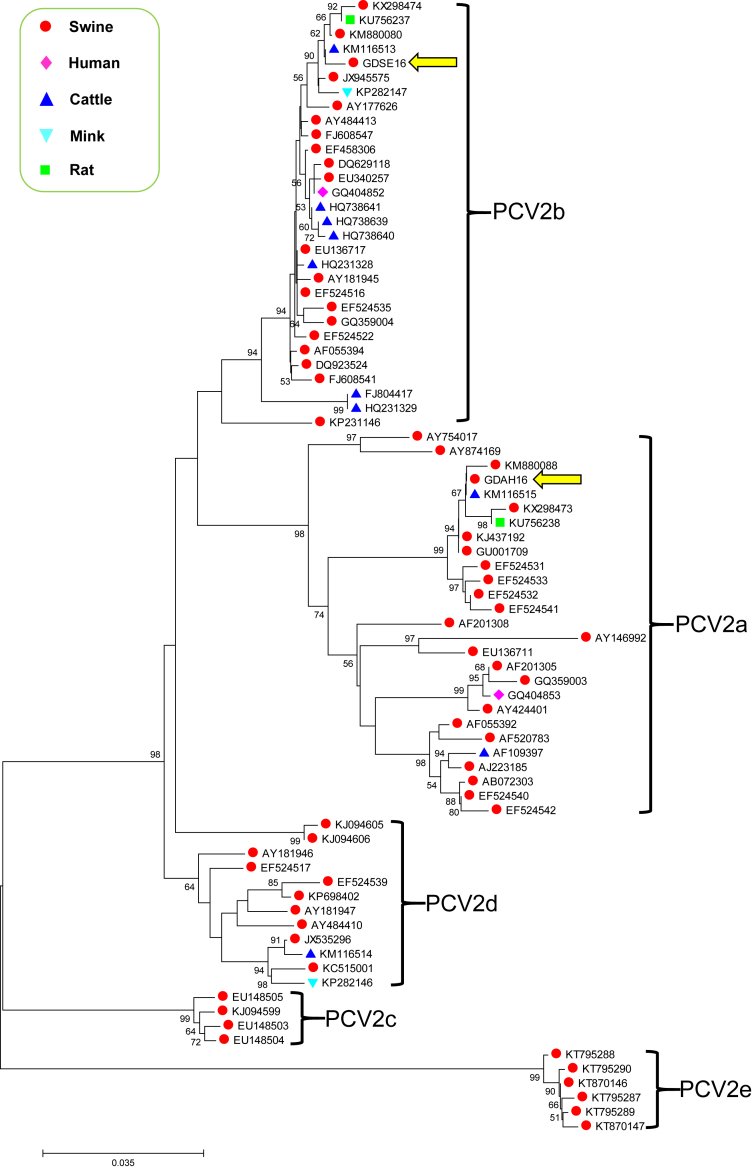
In our previous study, we identified three different PCV2 strains (Buffalo1, Buffalo2 and Buffalo3) in buffalo meat samples bought from one fresh meat market of southern China. Genetic analysis showed that they belonged to three different PCV2 genotypes [7]. During the epidemiological survey of 2016, we obtained two significantly different PCV2 sequences (designated GDAH16 and GDSE16) from lung samples of dead pigs with respiratory disease according to a previously described method [7], [8]. GDAH16 was 1768 nucleotides in length and had two major open reading fragments (ORFs). ORF1 and ORF2 were targeted at position 51→995 and 1735→1034, and encoded the predicted replicase and capsid proteins, respectively. Online BLAST alignment analysis showed that GDAH16 shared 99.72% nucleotide similarity (only five nucleotides different) with Buffalo3 (GenBank Accession no. KM116515). However, GDSE16 was 1767 nucleotides long. Its ORF1-targeted position was the same as GDAH16, but its ORF2-targeted position (1734→1033) was different. Similarly, online BLAST alignment analysis revealed 99.55% nucleotide identity (only eight nucleotides different) between GDSE16 and Buffalo1 (GenBank Accession no. KM116513). Further alignment analysis suggested that GDAH16 and GDSE16 only had 95.6% nucleotide similarity (about 78 nucleotides different). In addition, phylogenetic analysis indicated that they were clustered in different branches (PCV2a and PCV2b) and had close genetic relationships with Buffalo-origin PCV2 isolates (Buffalo3 and Buffalo1) (GenBank Accession nos. KM116515 and KM116513), respectively (Fig. 1).
Fig. 1.
Phylogenetic analysis of porcine circovirus type 2 (PCV2). The phylogenetic tree based on open reading frame 2 (ORF2) gene sequences of PCV2 was constructed by the neighbour-joining method using MEGA 5.1 software. One thousand bootstrap replications and 0.035 distance scale length were used. Values <50% were hidden in the phylogenetic tree. The strains obtained in this study, GDAH16 and GDSE16, are indicated by the left arrows. Different origins of PCV2 are labelled by different symbols.
Possible cross-species transmission of PCV2 is reported in several previous studies [6], [8], [9], [10], [11], [12], [13], [14], which included pig–rodent transmission, pig–mink transmission, pig–human transmission and pig–insect transmission. In this study, we confirmed those buffalo-origin-like PCV2 strains in swine herds in southern China, which further supported the possible cross-species infection of PCV2 between buffalo and pig in this region.
Accession number(s)
The genome sequences of GDAH16 and GDSE16 have been deposited in GenBank under the Accession numbers KY347898, KY347899.
Conflict of interest
There is no conflict of interest.
Acknowledgements
This work was supported by Ministry of Science and Technology of the People's Republic of China (Grant no. 2015GA780010), Guangdong Provincial Department of Science and Technology (Grant nos. 2015A020208008, 2016A040403083 and 2016B020234006), and Guangzhou Science Technology and Innovation Commission (Grant no. 201508020055).
Contributor Information
S.-L. Zhai, Email: zhaishaolun@163.com.
D.-H. Lv, Email: lvdianhong@gdaas.cn.
References
- 1.Zhai S.L., Chen S.N., Xu Z.H., Tang M.H., Wang F.G., Li X.J. Porcine circovirus type 2 in China: an update on and insights to its prevalence and control. Virol J. 2014;11:88. doi: 10.1186/1743-422X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nayar G.P., Hamel A.L., Lin L., Sachvie C., Grudeski E., Spearman G. Evidence for circovirus in cattle with respiratory disease and from aborted bovine fetuses. Can Vet J. 1999;40:277–278. [PMC free article] [PubMed] [Google Scholar]
- 3.Li L., Kapoor A., Slikas B., Bamidele O., Wang C., Shaukat S. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol. 2010;84:1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kappe E.C., Halami M.Y., Schade B., Alex M., Hoffmann D., Gangl A. Bone marrow depletion with haemorrhagic diathesis in calves in Germany: characterization of the disease and preliminary investigations on its aetiology. Berl Munch Tierarztl Wochenschr. 2010;123:31–41. [PubMed] [Google Scholar]
- 5.Halami M.Y., Freick M., Shehata A.A., Müller H., Vahlenkamp T.W. Susceptibility of calves to porcine circovirus-2 (PCV2) Vet Microbiol. 2014;173:125–131. doi: 10.1016/j.vetmic.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Wang G.S., Sun N., Tian F.L., Wen Y.J., Xu C., Li J. Genetic analysis of porcine circovirus type 2 from dead minks. J Gen Virol. 2016;97:2316–2322. doi: 10.1099/jgv.0.000529. [DOI] [PubMed] [Google Scholar]
- 7.Zhai S.L., He D.S., Qi W.B., Chen S.N., Deng S.F., Hu J. Complete genome characterization and phylogenetic analysis of three distinct buffalo-origin PCV2 isolates from China. Infect Genet Evol. 2014;28:278–282. doi: 10.1016/j.meegid.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Zhai S.L., Chen S.N., Liu W., Li X.P., Deng S.F., Wen X.H. Molecular detection and genome characterization of porcine circovirus type 2 in rats captured on commercial swine farms. Arch Virol. 2016;161:3237–3244. doi: 10.1007/s00705-016-3004-7. [DOI] [PubMed] [Google Scholar]
- 9.Li L., Shan T., Soji O.B., Alam M.M., Kunz T.H., Zaidi S.Z. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J Gen Virol. 2011;92:768–772. doi: 10.1099/vir.0.028704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorincz M., Cságola A., Biksi I., Szeredi L., Dán A., Tuboly T. Detection of porcine circovirus in rodents—short communication. Acta Vet Hung. 2010;58:265–268. doi: 10.1556/AVet.58.2010.2.12. [DOI] [PubMed] [Google Scholar]
- 11.Blunt R., McOrist S., McKillen J., McNair I., Jiang T., Mellits K. House fly vector for porcine circovirus 2b on commercial pig farms. Vet Microbiol. 2011;149:452–455. doi: 10.1016/j.vetmic.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Yang X., Hou L., Ye J., He Q., Cao S. Detection of porcine circovirus type 2 (PCV2) in mosquitoes from pig farms by PCR. Pak Vet J. 2012;32:134–135. [Google Scholar]
- 13.Pinheiro A.L., Bulos L.H., Onofre T.S., de Paula Gabardo M., de Carvalho O.V., Fausto M.C. Verification of natural infection of peridomestic rodents by PCV2 on commercial swine farms. Res Vet Sci. 2013;94:764–768. doi: 10.1016/j.rvsc.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Truong Q.L., Seo T.W., Yoon B.I., Kim H.C., Han J.H., Hahn T.W. Prevalence of swine viral and bacterial pathogens in rodents and stray cats captured around pig farms in Korea. J Vet Med Sci. 2013;75:1647–1650. doi: 10.1292/jvms.12-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]