Abstract
Ketolides represent the latest group of macrolide antibiotics. Tight binding of ketolides to the ribosome appears to correlate with the presence of an extended alkyl-aryl side chain. Recently developed 6,11-bridged bicyclic ketolides extend the spectrum of platforms used to generate new potent macrolides with extended alkyl-aryl side chains. The purpose of the present study was to characterize the site of binding and the action of bridged macrolides in the ribosomes of Escherichia coli. All the bridged macrolides investigated efficiently protected A2058 and A2059 in domain V of 23S rRNA from modification by dimethyl sulfate and U2609 from modification by carbodiimide. In addition, bridged macrolides that carry extended alkyl-aryl side chains protruding from the 6,11 bridge protected A752 in helix 35 of domain II of 23S rRNA from modification by dimethyl sulfate. Bridged macrolides efficiently displaced erythromycin from the ribosome in a competition binding assay. The A2058G mutation in 23S rRNA conferred resistance to the bridged macrolides. The U2609C mutation, which renders E. coli resistant to the previously studied ketolides telithromycin and cethromycin, barely affected cell susceptibility to the bridged macrolides used in this study. The results of the biochemical and genetic studies indicate that in the E. coli ribosome, bridged macrolides bind in the nascent peptide exit tunnel at the site previously described for other macrolide antibiotics. The presence of the side chain promotes the formation of specific interactions with the helix 35 of 23S rRNA.
Macrolides inhibit protein synthesis in sensitive bacteria by binding to the large ribosomal subunit and blocking progression of the nascent peptides through the ribosome exit tunnel (33). These drugs had been clinically very successful until their utility was curbed by the spread of resistant strains. Many of these strains express Erm-type methyltransferases that modify rRNA within the macrolide binding site, thereby preventing drug binding (26, 35). Newer macrolides which exhibit more favorable pharmacological properties have been developed. The most prominent of these newer macrolides are ketolides, which exhibit stronger binding to ribosomes, including those from resistant strains (1, 8, 37).
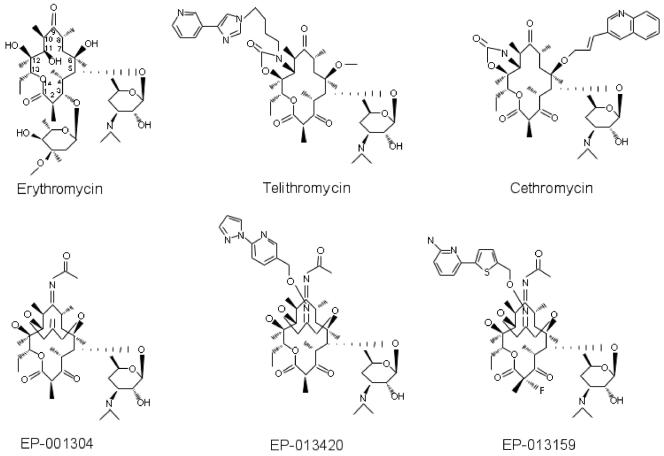
Therapeutically active macrolides are built of a lactone ring (14, 15, or 16 atoms long) ornamented with various side chains. (Fig. 1). A 14-member-ring erythromycin and a number of its derivatives contain a cladinose residue at the C-3 carbon atom of the lactone ring and a desosamine at position C-5. In ketolides, the C-3 cladinose sugar was replaced with the keto group. The ketolides telithromycin (formerly HMR 3647) and cethromycin (formerly ABT 773) carry a carbamate group formed in part from positions 11 and 12 of the lactone. In addition, a heteroaromatic side chain is linked via a flexible alkyl linker either to the nitrogen atom of the carbamate cycle (in telithromycin) or to O-6 on the other side of the lactone ring (in cethromycin) (1, 8, 14, 37).
FIG. 1.
Chemical structures of the macrolide antibiotics used in this study.
Macrolides bind to the large ribosomal subunit in the narrow part of the nascent peptide exit tunnel (5, 6, 16, 28, 29). The drug binding site is composed of several segments of 23S rRNA. Van der Waals interactions between the lactone ring and rRNA are important for drug binding; however, more than half of the binding free energy comes from interactions involving macrolide side chains (16, 29). One of the most important contacts between the 14-member-ring macrolides and the ribosome involves the C-5 desosamine sugar approaching adenines at positions 2057, 2058, and 2059 of 23S rRNA (5, 6, 16, 28, 29). Dimethylation of A2058 by Erm-type methyltransferases produces a steric clash and renders cells resistant to most macrolides (16, 29, 35). Mutations involving positions 2057, 2058, and 2059 also confer macrolide resistance (34). In contrast to desosamine, the C-3 cladinose sugar of erythromycin and its derivatives does not make any significant contacts with the ribosome; it is not surprising, therefore, that its replacement with a keto group in ketolides does not prevent drug binding. However, binding of ketolides appears to be significantly enhanced due to ketolide-specific contacts with 23S rRNA. One such contact involves U2609. The U2609C mutation confers resistance to telithromycin and cethromycin, while it slightly increases the susceptibility of Escherichia coli to erythromycin (13). Crystallographic studies of cethromycin complexed with the large ribosomal subunit of Deinococcus radiodurans showed a possible interaction of the carbamate nitrogen atom with O-4 of U2609 (28). However, such an interaction was not observed in the telithromycin complex with the D. radiodurans large ribosomal subunit (5). The most prominent ketolide-specific interaction involves the heteroaromatic side chain of ketolides, which in telithromycin and cethromycin is attached to different sides of the lactone ring (see Fig. 1). Mutational and footprinting studies indicated a possible interaction of this side chain with the loop of helix 35 that forms the wall of the exit tunnel opposite to A2058 (9, 12, 13, 17, 36). Protection of A752 in E. coli 23S rRNA from modification with dimethyl sulfate (DMS), which depended on the presence of the side chain, correlated with the increased affinity of ketolides to ribosomes (12, 36). Direct contact of the heteroaromatic side chain with the loop of helix 35 was not seen in crystallographic structures of ketolides complexed to the D. radiodurans ribosome; however, the side chain did approach the corresponding region of domain II and was seen in close proximity to a position equivalent to U790 in E. coli 23S rRNA (5, 28). Although macrolides are presumed to act at a single ribosomal site, binding of a 15-member-ring macrolide, azithromycin, to a second site in the exit tunnel of the D. radiodurans large ribosomal subunit, farther away from the peptidyltransferase center, was reported (28). Binding of a clinically active macrolide to the second ribosomal site could have very important medical implications, since it might help to overcome resistance mechanisms, like Erm-mediated methylation, that are targeted against the primary binding site. It remains unclear, however, whether the second macrolide site exists only in the ribosomes of D. radiodurans or is a common feature of all bacterial ribosomes.
The success of ketolides for the treatment of macrolide-resistant streptococcal and staphylococcal infections prompted a search for newer drug versions. One of the new platforms is represented by 6,11-O-bridged oxime macrolides developed by ENANTA Pharmaceuticals, Watertown, Mass. (D. Niu, G. Wang, Y. Qiu, N. H. Vo, Y. Wang, M. Busuyek, Y. Hou, Y. Peng, K. Amsler, A. Polemeropoulos, B. Scorneaux, L. T., Phan, and Y. S. Or, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1194, 2003; G. Wang, Y. Qiu, D. Niu, N. H. Vo, T. Beach, A. Polemeropoulos, B. Scorneaux, A. Arya, F. Schlünzen, J. M. Harms, R. Albrecht, A. Yonath, Y. Korkhin, L. T. Phan, and Y. S. Or, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1193, 2003). These compounds show good activities against both macrolide-sensitive pathogens and a number of macrolide-resistant pathogens. The most active bridged ketolides carry heteroaromatic side chains attached to the three-carbon-atom bridge.
The main purpose of this study was to characterize the binding of 6,11-O-bridged macrolides to the ribosome. We wanted to find out whether bridged macrolides bind to the primary and/or the secondary site in the E. coli ribosome, as previously observed in the cocrystal structure of azithromycin with the 50S ribosomal subunit of D. radiodurans. It was also important to understand whether the alkyl-aryl side chains of 6,11-bridged macrolides occupy a position in the ribosome similar to that taken by the side chains of the prototype ketolides, telithromycin and cethromycin, and whether interactions of the side chain with the ribosome improve the affinity of the bridged macrolides.
MATERIALS AND METHODS
Antibiotics and ribosomes.
All the bridged macrolides used in this study (EP-013420, EP-013159, and EP-001304) were provided by ENANTA Pharmaceuticals. Telithromycin was from Aventis Pharma (Romainville, France), cethromycin was from Abbott Laboratories (Abbott Park, Ill.), and erythromycin was purchased from Sigma (St. Louis, Mo.). Antibiotics were dissolved in dimethyl sulfoxide (DMSO) at 2.5 mM. For the MIC determinations, the antibiotics were diluted to higher concentrations in ethanol.
Salt-washed ribosomes were prepared from E. coli strain MRE 600 by standard protocols (23, 31).
RNA probing.
RNA probing was carried out by the basic procedure developed in Noller's laboratory (20, 22), with minor modifications.
For DMS modification, ribosomes were initially activated by incubation for 5 min at 37°C in buffer A (80 mM potassium cacodylate [pH 7.2], 20 mM MgCl2, 100 mM NH4Cl, 1.5 mM dithiothreitol). 70S ribosomes (final concentration, 200 nM) were combined with antibiotic (100 μM) in 50 μl of buffer A (control samples without antibiotics were supplemented with the corresponding amount of DMSO). The reaction mixtures were incubated for 10 min at 37°C, followed by 10 min at 20°C. Two microliters of DMS diluted 1:5 in ethanol was added, and after incubation for 10 min at 37°C, the reactions were quenched by the addition of 50 μl of 0.6 M sodium acetate (NaAc; pH 5.5), 1 M mercaptoethanol, and 300 μl of cold ethanol.
For carbodiimide modification, ribosomes were preincubated for 5 min at 37°C in buffer B (50 mM HEPES-KOH [pH 7.2], 20 mM MgCl2, 100 mM NH4Cl, 1.5 mM dithiothreitol). 70S ribosomes (400 nM) and antibiotic (200 μM) were combined in 25 μl of buffer B. The reaction mixtures were incubated for 10 min at 37°C, followed by 10 min at 20°C. RNA modification was initiated by the addition of 25 μl of 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT) dissolved to a concentration of 63 mg/ml in buffer B. The reaction mixtures were incubated for 10 min at 37°C, and the reactions were stopped by the addition of 100 μl of 0.6 M NaAc (pH 5.5) and 600 μl of cold ethanol. Ribosomes were precipitated by centrifugation at 20,000 × g for 10 min at 2°C.
Ribosome pellets were dissolved in 200 μl of 0.3 M NaAc (pH 5.5)-5 mM EDTA-0.5% sodium dodecyl sulfate. rRNA was isolated by extraction with equal volumes of phenol, phenol-chloroform, and chloroform, followed by ethanol precipitation.
The distributions of the modified nucleotides in 23S rRNA were assessed by primer extensions with a set of primers that allowed scanning of the entire 23S rRNA sequence (20).
Cell-free transcription-translation assay.
The transcription-translation assay was optimized for high-throughput format in 96-well plates (catalog no. 3600; Costar) in a final volume of 25 μl. The assay was performed in a luminescent format with the E. coli S30 Extract System for Circular DNA (catalog no. L1020; Promega). The reaction mixture was composed of 2.5 μl of an amino acids mixture, 10 μl of Premix reagent, 7.5 μl of S30 extract, 2.5 μl of pBESTLuc circular DNA at a 1:50 dilution, and 2.5 μl of a macrolide compound diluted to 0.1 to 50 μM in 10% DMSO. After a 1-h incubation at 37°C, 100 μl of luciferase detection reagent (catalog no. E1501; Promega) was added to the reaction mixture, immediately followed by luminescence readout on a Packard Fusion α-FP HT plate reader. The data were analyzed with the GraphPad Prizm (version 4.0) software package.
Competition binding studies.
Binding of erythromycin to the ribosome and competition with the bridged macrolides were assayed by size-exclusion chromatography in a spin-column format (19). Spin columns were prepared by equilibrating 10 g of Bio-Gel P30 (catalog no. 150-4154; Bio-Rad) with 95 ml of buffer C (20 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 150 mM NH4Cl, 6 mM 2-mercaptoethanol) overnight at room temperature. A total of 800 μl of gel slurry was placed into Micro Bio-Spin columns (catalog no. 732-6204; Bio-Rad) and stored at 4°C. The columns were spun in a swinging-bucket microcentrifuge for 1 min at 1,000 × g immediately prior to use.
In the erythromycin titration experiments, ribosomes at a concentration of 1 μM were combined with various concentrations of [14C]erythromycin (48.8 mCi/mmol; NEN) in 80 μl of buffer C. After incubation for 15 min at 37°C and 10 min at 20°C, the reaction mixtures were applied to the spin columns and immediately centrifuged at 1,000 × g for 1 min in a swinging-bucket microcentrifuge at room temperature. Ten microliters of the flowthrough solution was used to read the optical density (260 nm), and the amount of radioactivity in 30 μl of the solution was determined in a scintillation counter. The data were used to calculate the amount of ribosome-bound erythromycin.
In competition experiments, 80 μl of buffer C containing 1 μM ribosomes and 1 μM [14C]erythromycin (48.8 mCi/mmol; NEN) was preincubated for 15 min at 37°C and 10 min at 20°C. Competing antibiotics were added in 2 μl of DMSO, and the incubations were continued for 100 min at 20°C. Solutions were applied to the spin columns, and the amount of ribosome-bound erythromycin was determined as described above.
Effects of rRNA mutations on cell resistance.
Wild-type or mutant 23S rRNA was expressed from plasmids containing a complete copy of the rRNA operon under the control of the P1 and P2 rrn promoters and an ampicillin resistance marker (7). The A2058G mutation (30) was expressed from plasmid pSTL102 (which also carried a C1192U mutation in the 16S rRNA gene that did not affect susceptibility to macrolides) (32). The U2609 mutation (13) and the wild-type rRNA were expressed from the mutant or wild-type pKK3535 plasmids, respectively (7). All plasmids carrying rrn were introduced into TA531 cells that lacked chromosomal rrn alleles but that contained plasmid pHK-rrnC+ (Kanr) as a source of wild-type rRNA genes (3). The cells were cured of plasmid pHK-rrnC+ by previously described protocols (2, 3); and Ampr Kans cells, in which newly introduced plasmids served as the sole source of rRNA genes, were used in subsequent experiments.
MIC determinations.
For MIC determinations, cells were grown overnight in 1 ml of Luria-Bertani medium containing 100 μg of ampicillin per ml (in a round-bottom 15-ml culture tube). The cultures were diluted 100-fold into fresh Luria-Bertani medium containing 100 μg of ampicillin per ml and were grown for 2 h. Exponential-phase cultures were then diluted to an A600 of 0.002 and placed into 96-well plates at 100 μl/well. After the addition of antibiotics, the plates were incubated in a moist atmosphere for 15 h at 37°C without shaking and then for 3 h with shaking. The MIC was recorded as the lowest concentration of drug in the well with no obvious turbidity over that of the background.
RESULTS
Three types of 6,11-bridged macrolides were used in this study. Two of the drugs, EP-013420 and EP-013159, contained different heteroaromatic side chains attached through a linker to the 6,11 three-carbon-atom bridge (Fig. 1). The third compound, EP-001304, lacked the heteroaromatic side chain. Controls consisted of erythromycin, a representative cladinose-containing macrolide; telithromycin, a ketolide with the side chain attached to the 11,12-carbamate cycle; and (in some experiments) cethromycin, which contains the O-6-linked side chain.
RNA probing was used to gain insights into the interaction of bridged macrolides with the ribosome. Two types of chemical reagents were used in RNA footprinting experiments: DMS, which modifies N-1 of adenine and N-3 of cytosine, and CMCT, which can modify N-3 of uracil and N-1 of guanine (20). Since we were interested in examining the possibility that bridged macrolides might interact with more than one site in the large ribosomal subunit, an excess of antibiotic (100 μM) was used in the footprinting experiments.
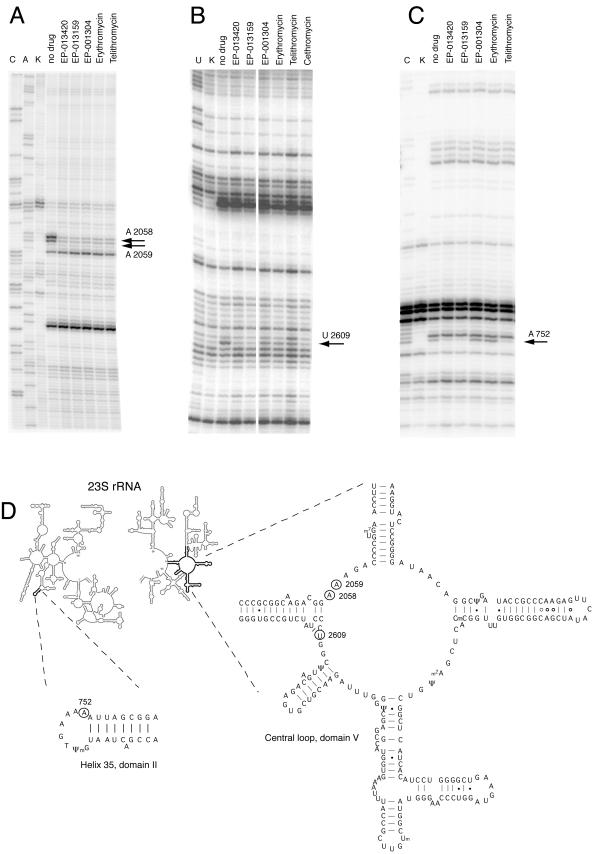
The most prominent effect observed in the DMS probing experiments was a strong protection of A2058 and A2059 in domain V of 23S rRNA by all the macrolides tested (Fig. 2A). In addition, ketolides with extended side chains (EP-013420, EP-013159, and telithromycin), but not EP-001304 or erythromycin, slightly enhanced the accessibility of A2062 to DMS. Conversely, in domain II, the extended-chain ketolides strongly protected A752 in the loop of helix 35, while binding of either EP-001304 or erythromycin slightly enhanced the accessibility of this base (Fig. 2C). No other major effects of macrolide binding on the accessibilities of RNA bases to DMS modification were observed in the rest of the 23S rRNA.
FIG. 2.
Chemical probing of the ribosome-macrolide complexes. (A and C) DMS probing; (B) CMCT probing. K, unmodified control; no drug, ribosomes modified in the absence of antibiotic; C, A, and U, sequencing lanes. All other samples contained the indicated macrolide antibiotic at 100 μM. The nucleotides whose accessibility to chemical modification is affected by the drugs are indicated by arrows, and their positions in the 23S rRNA secondary structure (4, 15) are shown in panel D.
CMCT probing was limited to the central loop of domain V (the primary macrolide site) and the region from positions 600 to 800 in domain II of 23S rRNA that might contribute to macrolide binding in the putative second site. All the macrolides tested protected U2609 (Fig. 2B). However, bridged macrolides with the extended side chain as well as cethromycin afforded much stronger protection than telithromycin and drugs that lack alkyl-aryl side chains.
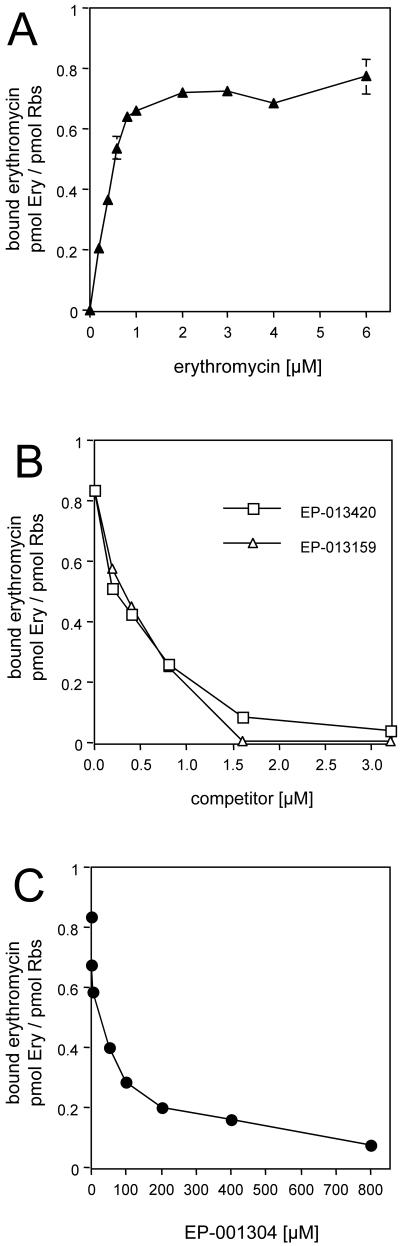
RNA probing indicated that bridged macrolides and erythromycin bind to the same or overlapping sites in the ribosome. This conclusion was further affirmed in the competition binding experiments. Erythromycin is known to bind to a single site in the E. coli ribosome with a dissociation constant of ca. 10−8 M (11, 24). In agreement with these data, titration of 1 μM ribosomes with [14C]erythromycin reached a plateau when the molar concentration of the drug roughly equaled that of the ribosomes (Fig. 3A), indicating binding of one molecule of erythromycin per ribosome. In competition experiments, ribosomes (1 μM) were first allowed to bind to [14C]erythromycin (1 μM), which was then competed out by using increased concentrations of another macrolide. As the curves in Fig. 3B show, all the antibiotics that contain heteroaromatic side chains effectively displaced erythromycin from its binding site in the E. coli ribosome. This thus confirms the binding of the bridged macrolides to the primary macrolide site. In the case of EP-001304, an approximately 100-fold higher concentration of the drug compared to those of the ketolides with extended side chains, EP-013420, EP-013159, or telithromycin, was required to displace erythromycin from the ribosome.
FIG. 3.
Competition of ketolides with erythromycin (Ery) for binding to the ribosome. Ribosomes (1 μM) were preincubated with 1 μM [14C]erythromycin, followed by incubation with increasing concentrations of the competing drugs (see Material and Methods for details).
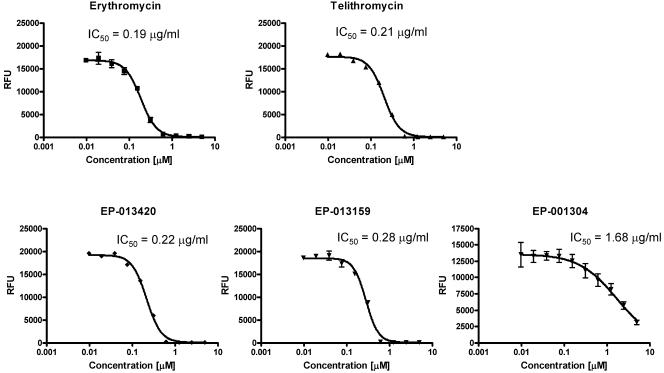
The importance of the bicyclic side chain for the activity of bridged macrolides was further established in cell-free transcription-translation experiments. The inhibitory activitiesof the bridged macrolides devoid of the extension arm (EP-001304) were an order of magnitude lower that those of the bridged macrolides decorated with a bicyclic arm or those of erythromycin and telithromycin (Fig. 4). Interestingly, despite the higher in vivo potencies of the extended-arm ketolides compared to that of erythromycin (Table 1), the 50% inhibitory concentrations (IC50s) of all compounds tested in the cell-free transcription-translation system were very similar (with the exception of EP-001304).
FIG. 4.
Effects of various macrolides on protein expression in E. coli transcription-translation cell-free system. The activity of the reporter protein (luciferase) is shown as raw fluorescence units (RFU), and the IC50s of the compounds calculated from the experimental data are shown above the inhibition curves.
TABLE 1.
Effects of rRNA mutations on susceptibility of E. coli to antibiotics
| Mutationa | MIC (μg/ml)b
|
|||||
|---|---|---|---|---|---|---|
| ERY | EP-001304 | EP-013420 | EP-013159 | TEL | CET | |
| Wild type | 128 | >512 | 32 | 16 | 16 | 8 |
| A2058G | >512 | >512 | >128 | >128 | >128 | >128 |
| U2609C | 128c | >512 | 64 | 16 | 64 | 64 |
Wild-type or mutant RNAs were expressed in the TA531 strain of E. coli (3) from plasmid-borne genes that were the single source of rRNA in the cell. Wild-type cells contained plasmid pKK3535 (7), the A2058G mutant contained plasmid pSTL102 (32), and the U2609C mutant contained plasmid pKK3535(2609C) (13).
ERY, erythromycin; TEL, telithromycin; CET, cethromycin.
The optical density of the U2609C cell culture grown in the presence of 128 μg erythromycin per ml was less than one-third of the density of the wild-type culture grown in the presence of the drug at this concentration.
In order to characterize the interactions of bridged macrolides with the ribosome in the cell, we investigated the effects of resistance mutations in the 23S rRNA on cell susceptibilities to the compounds. Two previously isolated macrolide resistance mutations, A2058G and U2609C (13, 30), were expressed in an E. coli strain lacking chromosomal rRNA alleles (2, 3). All the ribosomes in these cells carry rRNA that is encoded in plasmid-borne wild-type or mutant rRNA genes. As shown in Table 1, the A2058 mutation conferred resistance to all of the macrolides tested, whereas the U2609C mutation conferred relatively high levels of resistance to cethromycin and telithromycin, low-level resistance (1 dilution) to EP-013420, and no resistance to EP-013159. In accordance with the previous observation that the U2609C mutation renders cells more sensitive to erythromycin (13), the density of the U2609C cell culture grown in the presence of 128 μg of erythromycin per ml was approximately threefold lower than the density of the wild-type culture grown in the presence of the drug at this concentration. The effect of the mutations on cell susceptibility to EP-001304 could not be evaluated because of the low level of activity of the drug against E. coli. EP-001304 did not inhibit the growth of either wild-type or mutant E. coli cells, even at 512 μg/ml, the highest concentration of the drug tested.
DISCUSSION
The purpose of the present investigation was to characterize the interactions of a new type of macrolide antibiotics, 6,11-bridged macrolides, with the ribosome. The site of action of bridged macrolides in the ribosome and the mode of their binding were studied by using a combination of biochemical and genetic techniques. The experimental data revealed binding of this new class of ketolide antibiotics to the main macrolide site in the ribosome, in the vicinity of the peptidyltransferase center.
All the bridged macrolides used in this study strongly protected A2058 and A2059 from modification by DMS (Fig. 2A). Protection of these two adenines is a signature footprint of all macrolides studied to date (12, 13, 17, 21, 27, 36). The desosamine sugar of 14- and 15-member-ring macrolides bound in the primary macrolide site contacts A2058 and A2059 and causes direct protection of these bases from modification by DMS (16, 29). Putative drug binding exclusively to a more distant second site seen in complexes of azithromycin with large ribosomal subunits of D. radiodurans would place the macrolide molecule too far away from A2058 and A2059 (minimal distance, 9.5 Å) to affect their accessibility to DMS (28). Thus, protection of A2058 and A2059 by the bridged macrolides shows the binding of the drugs in the primary macrolide site in the vicinity of the peptidyltransferase center and rules out the possibility that the bridged macrolides bind exclusively to the secondary site.
This conclusion is further corroborated by the results of competition experiments, in which all bridged macrolides tested were able to compete with erythromycin for binding to the ribosome. Erythromycin is known to occupy a single site in the E. coli ribosome that corresponds to the primary site of macrolide action (11, 24). Therefore, competition of bridged macrolides with erythromycin is compatible with their binding to the primary site.
RNA probing and drug competition experiments demonstrated interaction of the bridged macrolides with the primary macrolide binding site in the isolated ribosome. Genetic evidence confirmed that it is also the primary site of action of the bridged macrolides in the living cell. The A2058G mutation, which is known to render cells resistant to erythromycin, telithromycin, and cethromycin by reducing the affinities of the drugs for the primary macrolide site, also rendered E. coli resistant to the bridged macrolides with heteroaromatic side chains (Table 1). Dimethylation of A2058 by Erm-type methyltransferase was reported to increase the rates of resistance of Streptococcus pneumoniae to EP-001304 more than 100-fold (35). Thus, in both gram-positive and gram-negative bacteria, A2058 appears to be the component of the site of action of bridged macrolides, which leads us to believe that bridged macrolides act upon the same site in the ribosome targeted by other macrolides.
In footprinting experiments, in addition to shielding of A2058 and A2059 from DMS modification, all the compounds protected U2609 from modification by CMCT. Similar to A2058 and A2059, U2609 belongs to domain V of 23S rRNA and contributes to the formation of the primary macrolide site (13, 25). While all the drugs afforded almost complete protection of A2058 and A2059, the level of protection of U2609 critically depended on the drug structure. Binding of cethromycin and bridged macrolides with heteroaromatic side chains (EP-013420 and EP-013159) strongly protected U2609 from modification by CMCT, whereas telithromycin and compounds that lacked alkyl-aryl side chains (erythromycin and EP-001304) afforded only partial protection (Fig. 2B). The special role of U2609 in the structure-specific binding of macrolides is further supported by mutational studies in which the U2609C mutation conferred the highest level of resistance to cethromycin, a somewhat lower level of resistance to telithromycin, an even lower level of resistance to EP-013420, no resistance to EP-013159, and increased susceptibility to erythromycin (Table 1) (13). Given the resolution limits of the X-ray structures of ketolide-ribosome complexes presently available, it is difficult to conclude which side of the drug contacts U2609: the C-3-linked functional groups (keto group or cladinose sugar) and C-11- and C-12-linked groups appear to be equally good candidates for interaction partners with the nitrogen base of U2609 (5, 28). This uncertainty is further exacerbated by the lack of a clear correlation between the level of resistance to a specific drug conferred by the U2609C mutation and the extent of U2609 protection (Table 1 and Fig. 2B). This seeming discrepancy can readily be resolved if one assumes that the orientation of the drug complexed to the ribosome in the test tube may not precisely represent its positioning in the translating ribosome in the living cell. Evidence for such differences exists for other ribosome-targeting antibiotics and might easily apply to macrolides as well (10).
In addition to the interactions with the domain V rRNA residues, bridged macrolides appear to form intimate contacts with nucleotides in domain II of 23S rRNA. Both bridged macrolides that contain alkyl-aryl side chains strongly protected A752 in the loop of helix 35 of E. coli 23S rRNA from DMS modification; in contrast, EP-001304, which lacks the side chain, slightly enhanced the accessibility of A752 to modification with DMS (Fig. 2C). This result agreed with footprinting data obtained with other types of ketolides and macrolides: the ketolides telithromycin and cethromycin, which carry alkyl-aryl side chains, strongly protected A752, whereas binding of erythromycin and its derivatives, which lack an alkyl-aryl side chain, enhanced the accessibility of A752 to DMS (12, 13, 17, 36). Interestingly, crystallographic structures of cethromycin and telithromycin with the D. radiodurans large ribosomal subunit did not reveal direct contacts of the side chain with the loop of helix 35 (5, 28). Instead, the side chains of both ketolides studied closely approached the ribose 2′-OH of the nucleotide corresponding to U790 in E. coli 23S rRNA. Although U790 was accessible for modification with CMCT in the E. coli ribosome, no strong drug-related effects were observed at this position (data not shown). The question remaining to be clarified is whether the lack of a direct contact between the macrolides' alkyl-aryl side chain and the loop of helix 35 is a phenomenon specific to D. radiodurans ribosomes or, conversely, whether the changes in accessibility of this loop to DMS modification are caused by repositioning of helix 35 upon drug binding. The effects of mutations and posttranscriptional modification in the loop of helix 35 on cell susceptibility to antibiotics seem to support a direct-interaction model (9, 18, 36).
Interactions of the alkyl-aryl side chain with hairpin 35 in domain II were previously proposed to account for the high affinities of ketolides to the ribosome and the high therapeutic potencies of ketolides (12, 17, 36). Our data obtained for the bridged macrolides are in good agreement with this conclusion. Both bridged macrolides with the extended heteroaromatic side chain strongly protect A752 from DMS modification, which, in our view, suggests the interaction of the side chain with this rRNA element. The same ketolides efficiently competed erythromycin out of the ribosome and readily inhibited E. coli growth. In contrast, EP-001304, which lacks the side chain and which failed to protect A752, did not inhibit the growth of E. coli strain TA531 used in this study. Furthermore, EP-001304 was a much weaker erythromycin competitor than the bridged macrolides with the alkyl-aryl side chains. In the cell-free transcription-translation assay, the IC50 of EP-001304 was six to eight times higher than the IC50s of ketolides with an extended side chain or erythromycin (Fig. 4). Thus, the extended alkyl-aryl side chains attached in the middle of a 6,11 three-carbon bridge in the EP compounds appear to play the same stimulatory role on drug binding and potency as the carbamate-linked side chain of telithromycin or the O-6-linked chain in cethromycin.
The footprinting, competition, and mutational data presented in this study all argue in favor of binding of the bridged macrolides at the primary site of macrolide action. When they are bound there, the bridged macrolides interact with elements of domain V, including nucleotides 2058, 2059, and 2609. The extended alkyl-aryl side chain appears to reach toward the loop of helix 35, where its interactions with rRNA increase the affinity of the drug to the ribosome, resulting in a high potency of the bridged macrolides. Although footprinting experiments did not reveal any indication of the binding of bridged macrolides to the second site suggested by crystallographic studies, it should be noted that our data cannot entirely rule out such binding (28; Wang et al., Abstr. 43rd ICAAC). The drug bound in the second site may interact with nucleotides inaccessible to DMS or may contact the atomic positions in adenines or cytosines that are not critical for modification. It is also possible that the second macrolide site exists in ribosomes of D. radiodurans but not in E. coli. To clarify the existence of the second macrolide site, it would be of interest to study binding and probe the interactions of macrolides with the D. radiodurans ribosome in solution and to look for resistance mutations in the putative second macrolide site by using experimental organisms that have a single rRNA operon.
Acknowledgments
We thank Ada Yonath (Weizman Institute of Science, Rehovot, Israel) for helpful discussions, John Chosay (Pfizer) for advice with the antibiotic binding assay, and Shannon Foley (UIC) for help with making the text less irritating for native English speakers.
This work was supported in part by NIH grant U19 AI056575 (to A.S.M.).
REFERENCES
- 1.Alvarez-Elcoro, S., and M. J. Enzler. 1999. The macrolides: erythromycin, clarithromycin, and azithromycin. Mayo Clin. Proc. 74:613-634. [DOI] [PubMed] [Google Scholar]
- 2.Asai, T., C. Condon, J. Voulgaris, D. Zaporojets, B. H. Shen, M. Al-Omar, C. Squires, and C. L. Squires. 1999. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181:3803-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai, T., D. Zaporojets, C. Squires, and C. L. Squires. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. USA 96:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 5.Berisio, R., J. Harms, F. Schluenzen, R. Zarivach, H. A. Hansen, P. Fucini, and A. Yonath. 2003. Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 185:4276-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berisio, R., F. Schluenzen, J. Harms, A. Bashan, T. Auerbach, D. Baram, and A. Yonath. 2003. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 10:366-370. [DOI] [PubMed] [Google Scholar]
- 7.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 8.Bryskier, A. 2001. Telithromycin—an innovative ketolide antimicrobial. Jpn. J. Antibiot. 54(Suppl. A):64-69. [PubMed] [Google Scholar]
- 9.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colca, J. R., W. G. McDonald, D. J. Waldon, L. M. Thomasco, R. C. Gadwood, E. T. Lund, G. S. Cavey, W. R. Mathews, L. D. Adams, E. T. Cecil, J. D. Pearson, J. H. Bock, J. E. Mott, D. L. Shinabarger, L. Xiong, and A. S. Mankin. 2003. Crosslinking in the living cell locates the site of action of oxazolidinone antibiotics. J. Biol. Chem. 278:21972-21979. [DOI] [PubMed] [Google Scholar]
- 11.Douthwaite, S., and C. Aagaard. 1993. Erythromycin binding is reduced in ribosomes with conformational alterations in the 23S rRNA peptidyl transferase loop. J. Mol. Biol. 232:725-731. [DOI] [PubMed] [Google Scholar]
- 12.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183-193. [DOI] [PubMed] [Google Scholar]
- 13.Garza-Ramos, G., L. Xiong, P. Zhong, and A. Mankin. 2002. Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA. J. Bacteriol. 183:6898-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaynor, M., and A. S. Mankin. 2003. Macrolide antibiotics: binding site, mechanism of action, resistance. Curr. Top. Med. Chem. 3:949-961. [DOI] [PubMed] [Google Scholar]
- 15.Gutell, R. R., B. Weiser, C. R. Woese, and H. F. Noller. 1985. Comparative anatomy of 16S-like ribosomal RNA. Prog. Nucleic Acids Res. Mol. Biol. 32:155-216. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, J., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, L. H., P. Mauvais, and S. Douthwaite. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31:623-632. [DOI] [PubMed] [Google Scholar]
- 18.Liu, M., and S. Douthwaite. 2002. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc. Natl. Acad. Sci. USA 99:14658-14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNicholas, P. M., D. J. Najarian, P. A. Mann, D. Hesk, R. S. Hare, K. J. Shaw, and T. A. Black. 2000. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 44:1121-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merryman, C., and H. F. Noller. 1998. Footprinting and modification-interference analysis of binding sites on RNA, p. 237-253. In C. W. J. Smith (ed.), RNA:protein interactions, a practical approach. Oxford University Press, Oxford, United Kingdom.
- 21.Moazed, D., and H. F. Noller. 1987. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie 69:879-884. [DOI] [PubMed] [Google Scholar]
- 22.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 23.Moazed, D., and H. F. Noller. 1989. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 57:585-597. [DOI] [PubMed] [Google Scholar]
- 24.Pestka, S., and R. A. Lemahieu. 1974. Effect of erythromycin analogues on binding of [14C]erythromycin to Escherichia coli ribosomes. Antimicrob. Agents Chemother. 6:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulsen, S. M., C. Kofoed, and B. Vester. 2000. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304:471-481. [DOI] [PubMed] [Google Scholar]
- 26.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Fonseca, C., R. Amils, and R. A. Garrett. 1995. Fine structure of the peptidyl transferase centre on 23S-like rRNAs deduced from chemical probing of antibiotic-ribosome complexes. J. Mol. Biol. 247:224-235. [DOI] [PubMed] [Google Scholar]
- 28.Schlunzen, F., J. M. Harms, F. Franceschi, H. A. Hansen, H. Bartels, R. Zarivach, and A. Yonath. 2003. Structural basis for the antibiotic activity of ketolides and azalides. Structure (Cambridge) 11:329-338. [DOI] [PubMed] [Google Scholar]
- 29.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 30.Sigmund, C. D., and E. A. Morgan. 1982. Erythromycin resistance due to mutation in a ribosomal RNA operon of Escherichia coli. Proc. Natl. Acad. Sci. USA 79:5602-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spedding, G. 1990. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles, p. 1-29. In G. Spedding (ed.), Ribosomes and protein synthesis, a practical approach. Oxford University Press, Oxford, United Kingdom.
- 32.Triman, K., E. Becker, C. Dammel, J. Katz, H. Mori, S. Douthwaite, C. Yapijakis, S. Yoast, and H. F. Noller. 1989. Isolation of temperature sensitive mutants of 16S rRNA in Escherichia coli. J. Mol. Biol. 209:645-653. [DOI] [PubMed] [Google Scholar]
- 33.Vazquez, D. 1975. The macrolide antibiotics, p. 459-479. In J. W. Corcoran and F. E. Hahn (ed.), Antibiotics. III. Mechanism of action of antimicrobial and antitumor agents. Springer-Verlag, New York, N.Y.
- 34.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S ribosomal RNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong, L., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633-639. [DOI] [PubMed] [Google Scholar]
- 37.Zhong, P., and V. Shortridge. 2001. The emerging new generation of antibiotic: ketolides. Curr. Drug Targets Infect. Disord. 1:125-131. [DOI] [PubMed] [Google Scholar]