Abstract
NorA is a Staphylococcus aureus multidrug transporter that confers resistance to structurally distinct compounds. The MgrA global regulatory protein is reported to augment norA expression when mgrA is overexpressed from an undefined plasmid-based promoter. Further details about norA regulatory mechanisms are scant. A chromosomal norA::lacZ transcriptional fusion was constructed in different S. aureus strains, and allele replacement was used to define the relevance of promoter region sequences to norA expression. The effect of mgrA overexpression in wild-type and mutant backgrounds was also determined. Contrary to existing data, overexpression of mgrA repressed norA transcription in all parent and selected norA promoter mutant strains in a dose-dependent fashion. Disruption of a near-perfect inverted repeat or other putative regulatory protein binding sites did not affect norA transcription, but the repressive effect of mgrA overexpression was blunted in these mutants. This result, and the conservation of all of these motifs in S. aureus, suggests that their presence is required for the full effect of MgrA, or other regulatory proteins, on norA expression. Mutations at the +5 nucleotide of norA mRNA (flqB mutations) had a major impact; all resulted in markedly increased norA expression that was significantly reversed by mgrA overexpression. The flqB position of norA mRNA is part of a conserved imperfect inverted repeat; it is feasible that this motif could be a binding site for a norA regulatory protein.
Membrane-based efflux proteins, hereafter referred to as pumps, can contribute to antimicrobial agent resistance in bacteria (25, 43). Some of these drug pumps have a narrow substrate profile, whereas others are capable of removing many structurally unique compounds. These latter proteins are referred to as multidrug resistance (MDR) efflux pumps. MDR pumps have been shown to contribute to the intrinsic multidrug-resistant phenotype characteristic of Pseudomonas aeruginosa (29). In fact, significant effort has been invested in the search for inhibitors of P. aeruginosa MDR pumps, as combining them with pump substrates may result in the return of clinically relevant activity of those agents.
Gram-positive organisms also possess MDR efflux pumps, with Bmr of Bacillus subtilis and NorA of Staphylococcus aureus being the subjects of intensive research efforts. The sequences of Bmr and NorA are 44% identical and 67% similar and, thus, are relatively closely related in evolutionary terms. Expression of bmr is affected by BmrR, a protein encoded immediately downstream of bmr (1). When BmrR binds substrates of Bmr, it interacts with the bmr promoter and activates gene expression. A similar mechanism of gene activation occurs for another B. subtilis MDR transporter, Blt, which is homologous to Bmr (52% sequence identity; 39% identical and 62% similar to NorA) and has a similar substrate profile (2). The expression of blt is enhanced by the binding of BltR (encoded by bltR, found immediately upstream of blt) to the blt promoter region. This binding is thought to be affected by the interaction of substrates with BltR, although the specific activator substrates have not been identified. Rhodamine, which is a substrate for both Bmr and Blt, activates bmr but not blt expression. BmrR and BltR also differ with respect to their putative inducer binding domains. These data suggest that, despite having similar substrate profiles, Bmr and Blt probably have independent functions. This position is borne out by the fact that Blt has been shown to transport the natural polyamine spermidine, whereas Bmr does not have this function (44).
In addition to the specific regulators just described, the expression of bmr and blt is also affected by MtaN, a global regulator that interacts with their promoters, inducing transcription (5). MtaN consists of the N-terminal 109 residues of a larger protein, Mta (257 residues); the intact parent protein does not activate bmr or blt transcription. It is hypothesized that upon interacting with an inducer (as yet unidentified), the N- and C-terminal domains of Mta are functionally separated, allowing it to function as a transcriptional activator.
QacA and QacB are nearly identical S. aureus MDR efflux pumps encoded by plasmid-based genes that confer resistance to selected biocides (38). These pumps are related to NorA by being functionally dependent on the proton motive force and have some overlap in substrate profile with it. All qacA/B determinants are regulated by the divergently transcribed QacR repressor protein (12). Similar to BmrR and probably BltR, QacR interacts with substrates and in so doing dissociates from its operator site. This results in augmented expression of qacA/B.
The understanding of NorA regulation is less advanced than that of Bmr, Blt, and QacA/B. There is an open reading frame (SA0649 in the S. aureus N315 genome) on the opposite strand and immediately upstream of norA that encodes a putative protein that gives BLAST hits with a number of DNA-binding proteins. The putative promoter of SA0649 overlaps with that of norA, suggesting that the two genes may be coordinately regulated. No data have been published regarding the possible role of the SA0649 protein in norA regulation.
Recently, an 18-kDa protein that binds upstream of the −35 motif of the norA promoter has been identified and appears to augment norA expression in the presence on the chromosome of a transposon insertion into arlS (9). The arlR-arlS locus encodes a two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of S. aureus (10). The exact mechanism by which disruption of arlS modifies norA expression is not clear, nor is the site(s) at which the 18-kDa protein binds within the norA promoter clear. However, experimental data suggest that repeats consisting of the consensus sequence TTAATT may be involved. Four such hexamers are located within 60 bp upstream of the −35 motif (9) (Fig. 1B).
FIG. 1.
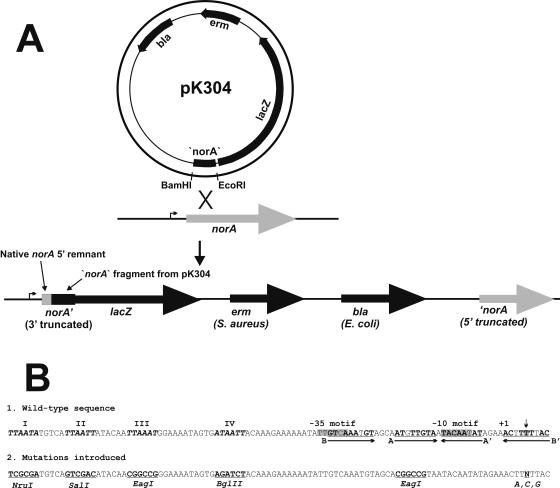
(A) Construction of the norA::lacZ fusion. In the presence of erythromycin, a single crossover at the site of homology between pK304 and the chromosome occurs, disrupting norA and transcriptionally fusing the norA promoter and about 400 bp of 5′ norA sequence with lacZ. erm expresses only in S. aureus, and bla expresses in E. coli. Truncated (3′ and 5′) refers to norA lacking 3′ and 5′ sequences, respectively. (B, 1) Wild-type norA promoter region and upstream sequence. Consensus TTAATT sequences are bold, italicized, and identified by Roman numerals, the −35 and −10 motifs and the +5 position of norA mRNA (site of flqB mutations) are highlighted (the +5 position is also noted by the downward arrow), the +1 position of norA mRNA is indicated, and the inverted repeats (A and B) are underlined and indicated by arrows. Perfectly matching bases in each repeat are in bold. (2) Mutations introduced into the norA promoter region by allele replacement.
The identity of the 18-kDa protein, initially named NorR, was recently established (42). NorR is a 147-residue protein which has modest homology with other regulatory proteins such as MarR and SarR (3, 31). One group has shown that the binding of NorR to the norA promoter is modified in an arlS-negative strain such that increased norA expression is observed. The same group has also shown that overexpression of norR from an uncharacterized plasmid-based promoter in an arlS+ background also appears to result in an increase in the level of norA mRNA and increased MICs of compounds that are NorA substrates (9, 42). In fact, these investigators found that only when norR was overexpressed was an effect on norA transcript level observed in a wild-type genetic background. These data suggest that a mutation in arlS appears to augment, but is not required for, a NorR effect on the norA promoter and that wild-type levels of NorR have little effect on norA expression. Highly fluoroquinolone-resistant strains of S. aureus have been described in which norA expression is enhanced in the absence of any modification of the arlR-arlS loci or change in norR expression, indicating that factors other than arlR-arlS and norR must be involved in the regulation of norA expression (10, 35, 41, 42). Work performed in our laboratory has also demonstrated that substrate exposure can augment norA expression, but the mediator(s) of this effect are not currently known (20, 23).
Subsequent work has revealed that NorR is not a specific regulator of norA expression but rather is a global regulator that, in addition to altering norA transcription when overexpressed, also affects the transcription of several known autolytic regulators including ArlR-ArlS (17, 30). This protein, which independently has been named Rat (regulator of autolytic activity) and MgrA (multiple gene regulator), is transcribed optimally from two promoters, positively regulates its own expression, and acts at the transcriptional level to enhance the expression of numerous genes, the products of which negatively impact the expression of murein hydrolases. All investigators involved in the study of this regulator (A. L. Cheung, D. C. Hooper, and C. Y Lee) have agreed on the designation of its gene and protein as mgrA and MgrA, respectively, and we will hereafter use these names.
Several years ago we observed the emergence of resistance to ciprofloxacin in a rabbit with experimental S. aureus endocarditis being treated with ciprofloxacin (18). The involved strain (SA-1199B) had a T→A transversion 11 bp downstream of the −10 motif of the norA promoter that subsequently was shown to correlate with an apparent increase in norA mRNA (flqB mutation) (19) (Fig. 1B and 2). The association of this mutation with increased transcription was supported by its introduction into S. aureus RN4220 by plasmid integration followed by quantitative analysis of norA mRNA (21). Others showed that a T→G transversion and a T→C transition at this position resulted in the same phenomenon (35, 36). This mutation lies in the 5′ untranslated region (5′ UTR) of norA mRNA and could alter its secondary structure and, as a result of this, its half-life. We have found that in SA-1199B (T→A flqB mutation) the half-life of the norA message is short (40 s) and not different from that of its parent strain (SA-1199) (unpublished data). However, utilizing a reverse transcription-PCR approach, others have found that the T→G flqB mutation in a different strain results in a prolonged half-life for the norA message (7 min [parent] versus 34 min [mutant]) (11). These authors proposed that norA mRNA is more stable in the presence of an flqB mutation and that this was the basis for the prolonged half-life observed. Why such vastly different half-lives are found for T→A and T→G flqB mutants versus their respective parent strains is not known.
FIG. 2.
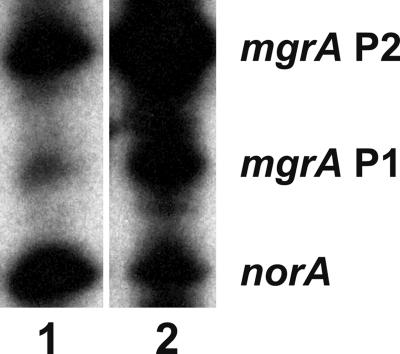
norA primer extension analysis. SH1000, parent strain; SA-K2124, SH1000 norA::lacZ; SA-K2161, SH1000 norA::lacZ flqB (T→A); SA-K2173, inverted repeat mutant of SA-K2124; SA-K2242, TTAATT consensus sequence mutant of SA-K2124.
The present study was undertaken in an effort to identify new factors important for the regulation of norA expression, and establish with greater certainty those previously identified, by utilizing a genetic system that creates desired changes in a single copy on the chromosome. Such a system is more representative of what may occur naturally and improves on previous work done with multicopy plasmids. We used information that is known or presumed about the importance of selected individual nucleotides or sequences in the norA promoter region and examined the effect(s) of mutagenizing those positions by allele replacement. Because MgrA appears to affect norA expression only when its gene is overexpressed, the expression of mgrA from plasmids by using defined promoters in mutant strains and the appropriate control strains also was evaluated.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, reagents, and growth conditions.
The strains and plasmids employed in this study are listed in Table 1. Unless otherwise noted, all reagents were the highest grade available and were obtained from Sigma Chemical Co., St. Louis, Mo. Growth media (brain heart infusion broth, tryptic soy broth, and Mueller-Hinton II broth) were obtained from BD Biosciences, Inc., Sparks, Md. pK404 was constructed by cloning an EcoRI-HindIII fragment of pTL2989, which contains mgrA and both of its promoters (P1 and P2), into pCU1. pK410 was constructed by amplifying mgrA, its Shine-Dalgarno sequence, and all of its putative transcription terminator sequences from S. aureus NCTC 8325-4 and then cloning the product into pALC2073 (6, 42). pALC2073 is an S. aureus-Escherichia coli shuttle vector possessing a tetracycline-inducible promoter controlling the expression of cloned genes. pCU1, pK404, and pK410 were electroporated into RN4220 and then transferred to other strains by transduction with phage 85 (8). All experiments with strains possessing any of these plasmids were done in the presence of chloramphenicol (10 μg/ml) to ensure plasmid maintenance.
TABLE 1.
Study strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference(s) |
|---|---|---|
| S. aureus strains | ||
| NCTC 8325-4 | Wild-type strain cured of known prophages, rsbU mutant | 27, 37 |
| RN4220 | NCTC 8325-4 r-; capable of stably maintaining recombinant plasmids | 26 |
| SA-K2069 | RN4220 norA::lacZ | This study |
| SH1000 | rsbU+ derivative of NCTC 8325-4 | 15 |
| SA-K2124 | SH1000 norA::lacZ | This study |
| SA-K2173 | SA-K2124 IR mutant | This study |
| SA-K2161, -K2291, -K2293 | SA-K2124 flqB (T→A, C, or G, respectively) | This study |
| SA-K2242 | SA-K2124 TTAATT mutant | This study |
| ISP794 | NCTC 8325 pig-131; rsbU mutant | 40 |
| SA-K2382 | ISP794 norA::lacZ | Q.-C. Truong-Boldoc and D. C. Hooper |
| SA-K2383 | SA-K2382 flqB (T→A) | Q.-C. Truong-Boldoc and D. C. Hooper |
| Newman | Easily transducible clinical strain; rsbU+ | 7 |
| SA-K2083 | Newman norA::lacZ | This study |
| SA-K2159 | SA-K2083 flqB (T→A) | This study |
| Plasmids | ||
| pAZ106 | Suicide vector for construction of lacZ fusions, Emr | 24 |
| pK304 | pAZ106 containing 5′ internal norA fragment | This study |
| pTS2tetK | Temperature-sensitive vector used for allele replacement, Tcr | J. Higgins and T. Foster |
| pCU1 | S. aureus-E. coli shuttle vector, Cmr | 4 |
| pTL2989 | pCL52.2 containing mgrA (P1+P2) cloned between EcoRI and HindIII sites | C. Y. Lee |
| pK404 | pCU1 containing EcoRI-HindIII fragment of pTL2989 (mgrA [P1+P2]) | This study |
| pALC2073 | S. aureus-E. coli shuttle vector containing a tetracycline-inducible promoter controlling expression of cloned genes, Cmr | 6 |
| pK410 | pALC2073 containing mgrA under control of its tetracycline-inducible promoter | This study |
TTAATT, putative consensus sequences to which a norA regulatory protein(s) binds; Emr, Tcr, Cmr, erythromycin, tetracycline, and chloramphenicol resistance selection; P1+P2, both promoters of mgrA present.
Genome data.
S. aureus genome data were obtained from online sources (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/genomes/MICROBES/complete.html], The Institute for Genomic Research [http://www.tigr.org], the Sanger Institute [http://www.sanger.ac.uk], and the University of Oklahoma [http://www.genome.ou.edu/staph.html]). Data from these sources will be referred to collectively as genome data.
Determination of antimicrobial susceptibilities.
Antimicrobial susceptibilities were determined by using Mueller-Hinton II broth and microdilution techniques according to NCCLS guidelines (34).
Nucleotide sequencing and Southern blots.
Nucleotide sequences were determined by using the Applied Biosystems 377 automated capillary-based system (Perkin-Elmer Applied Biosystems, Inc., Foster City, Calif.) at the Center for Molecular Medicine and Genetics Macromolecular Core Facility, Wayne State University, Detroit, Mich. Southern blots were performed by using standard techniques (39). The norA probe used was labeled with [α-32P]ATP (800 Ci/mmol; NEN Life Science Products, Inc., Boston, Mass.) by using the RadPrime DNA labeling system and the procedures recommended by the manufacturer (Invitrogen, Carlsbad, Calif.).
Primer extension.
Relative amounts of norA or mgrA transcripts were determined by primer extension. Strains were grown under the same conditions employed for β-galactosidase assays (see below), and cells were collected at an optical density at 600 nm (OD600) of 1. RNA for this and other experiments for which it was required was isolated by using the RNeasy midi kit (QIAGEN Inc., Valencia, Calif.). mRNAs were labeled by using the primer extension system-avian myeloblastosis virus reverse transcriptase kit (Promega Corp., Madison, Wis.). The oligonucleotides used for primer extension were end-labeled with [γ-32P]dATP (3,000 Ci/mmol; NEN) according to procedures recommended by the manufacturer of the primer extension system. Product band intensities were digitally quantitated by using a phosphorimaging system (Storm 860; Molecular Dynamics, Sunnyvale, Calif.) and Phoretix one-dimensional advanced software (version 5.20; Nonlinear Dynamics Ltd., Newcastle upon Tyne, United Kingdom).
Determination of norA mRNA half-life.
The half-life of norA mRNA was determined by quantifying primer extension products produced and analyzed as described above. Briefly, cells were grown to an OD550 of 1.0, and at T = 0, a 1-ml aliquot was removed, 0.5 ml NaN3 was added, and the mixture was snap-frozen at −70°C. Rifampin (150 μg/ml) was added to the culture, and additional aliquots were removed at frequent intervals and treated as described above. Samples were thawed on ice, cells were pelleted and lysed, and RNA was isolated by using the RNeasy midi kit. The mRNA half-life was determined by calculating the rate of reduction of band intensities of primer extension products following rifampin addition.
Construction of a norA::lacZ fusion.
The chromosomal norA::lacZ fusion was constructed by amplifying 400 bp internal to the near 5′ region of norA with primers incorporating BamHI and EcoRI sites. Using these enzymes, the product was cloned into pAZ106, producing pK304 (Table 1). pAZ106 is a suicide vector in S. aureus that has a multiple cloning site 5′ to a promoterless lacZ gene (24). Since the plasmid cannot replicate in S. aureus, introduction of pK304 into S. aureus RN4220 in the presence of erythromycin forces integration into the chromosome within the norA gene (Fig. 1A). The integration disrupts norA (resulting in a functional knockout mutation) and creates, in a single copy on the chromosome, a transcriptional fusion between norA and lacZ (producing SA-K2069) (Table 1). DNA sequencing and Southern analysis verified the proper construct, and phage 85 was used to transduce the fusion into other strains. Existing data reveal that norA transcripts are the appropriate size for norA alone and that it is not cotranscribed with any other open reading frame. Thus, the disruption of norA by fusing it with lacZ is unlikely to have a polar effect on the transcription of downstream genes (19, 20, 35).
β-Galactosidase assay.
The β-galactosidase assay employed was performed as described previously (23, 33). Briefly, strains grown overnight in tryptic soy broth were washed with phosphate-buffered saline (pH 7.0) and then diluted 1:200 in 100 ml of prewarmed brain heart infusion broth and grown at 37°C with agitation. Culture aliquots (0.5 ml) were obtained at intervals for measurement of OD600. The cells in a second aliquot were harvested by centrifugation, and the pellets were snap-frozen at −70°C. At the time of the assay, cells were thawed, resuspended in 0.5 ml of ABT (100 mM NaCl, 60 mM K2HPO4, 40 mM KH2PO4, 0.1% Triton X-100), and incubated at 37°C for 15 min, and then 50 μl of 4-methylumbelliferyl-β-d-galactopyranoside (MUG) (4-mg/ml stock) was added followed by an additional 1 h of incubation at room temperature. The reaction was stopped by the addition of 0.5 ml of 0.4 M Na2CO3. Samples were serially diluted in a 1:1 (vol/vol) mixture of ABT and Na2CO3 in 96-well white polystyrene microtiter plates (Corning, Inc., Corning, N.Y.). A range of concentrations of 4-methylumbelliferone was used to generate a standard curve, and β-galactosidase activity (expressed in MUG units; 1 unit = 1 pmol of MUG cleaved per min per OD600) was determined by fluorescence with a Bio-Tek FLx800 plate reader (Bio-Tek Instruments, Inc., Winooski, Vt.). Cumulative norA expression over the course of the experiment (10 h) was determined by integrating the area beneath expression curves with SigmaPlot, version 8.0 (Systat Software, Inc., Point Richmond, Calif.). For the sake of simplicity, β-galactosidase assays will be referred to hereafter as MUG assays.
Mutagenesis of norA promoter region.
There is a nearly perfect inverted repeat (IR) that includes the −10 motif of the norA promoter (repeat A) (Fig. 1B). The role played by this repeat in the regulation of norA expression was examined by allele replacement. PCR-based overlap extension was employed to create a product consisting of 350 bp upstream and 178 bp downstream of the norA start codon, with the 5′ portion of the repeat altered by substituting an EagI restriction site for the wild-type sequence (14) (Fig. 1B). The PCR product was cloned into pTS2tetK (Table 1), and this construct was introduced into SA-K2069 (RN4220 norA::lacZ) by electroporation. Growth of the recipient at 42°C in the presence of tetracycline results in integration of the plasmid into the chromosome at the site of shared homology. Subsequent cycles of growth at 30°C and then at 42°C in the absence of tetracycline results in excision of the plasmid and its ultimate loss, with a small proportion of excisants retaining the mutagenized norA promoter introduced with the plasmid. After the last temperature shift cycle, the norA promoter of tetracycline-susceptible strains was amplified by PCR, the product was digested with EagI, and a mutant that had an EagI site in its PCR product was identified. The IR mutation was transferred to S. aureus SH1000 by transduction, which also transferred norA::lacZ and produced strain SA-K2173 (Table 1).
In a similar manner, all possible flqB mutations were introduced into the chromosome of SA-K2069 (Fig. 1B). In generating the mutagenized PCR product, a unique EcoRI restriction site was created 138 bp upstream of the −35 motif to allow detection of strains having undergone allelic exchange. The silent nature of the introduced EcoRI site with respect to norA expression was verified by creation of a strain that possessed this mutation only (see below). Appropriate mutants were identified by digestion of PCR products encompassing the region in question with EcoRI. Transduction was employed to move the mutations and the associated norA::lacZ fusion into SH1000, producing SA-K2161, SA-K2291, and SA-K2293 (flqB = A, C, or G, respectively) (Table 1). The T→A flqB mutation and the norA::lacZ fusion were also transduced into ISP794 and Newman for mgrA overexpression studies (producing strains SA-K2383 and SA-K2159, respectively).
PCR-based overlap extension was employed to create a derivative of the norA promoter in which all four of the consensus TTAATT sequences upstream of the −35 motif were altered to unique restriction endonuclease sites (NruI, SalI, EagI, and BglII, respectively, for hexamers I, II, III, and IV) (Fig. 1B). The mutations were introduced into the chromosome of SA-K2069 by allelic exchange, and their presence was verified by the acquisition of the new restriction sites in a PCR product that included the region in question. The mutations were transduced along with norA::lacZ into S. aureus SH1000 (producing SA-K2242).
All mutants constructed as described above were verified by DNA sequencing and Southern blot analysis. The expression of norA in the mutant and appropriate control strains was determined by MUG assay. MUG assays were repeated three times, and results were expressed as means ± standard deviations in MUG units and were compared by employing the t test.
Overexpression of mgrA.
In the absence of an arlS mutation, overexpression of mgrA has been reported to augment norA expression (42). We wished to determine whether this effect was changed in any of our mutant strains. Plasmid pCU1 or pK404, as appropriate, was transduced into SA-K2124 (SH1000 norA::lacZ) and its inverted repeat (SA-K2173), flqB (SA-K2161; T→A), and TTAATT consensus sequence (SA-K2242) mutants. These plasmids also were transduced into ISP794 and Newman norA::lacZ (SA-K2382 and SA-K2083, respectively) and the flqB (T→A) mutants of these strains (SA-K2383 and SA-K2159, respectively). To eliminate possible confounding effects of positive autoregulation of plasmid-based mgrA via its native promoters on, and to determine if there is a dose dependency of, the effect of MgrA on norA expression, pK410, which contains mgrA under the control of a heterologous tetracycline-inducible promoter, was transduced into SA-K2124, SA-K2161, SA-K2382, and SA-K2083. norA expression was determined for all of these strains by MUG assay; for strains containing pK410, these assays were done by using inducing concentrations of tetracycline of 0, 25, 50, 75, and 100 ng/ml. pCU1 and pK404 were also transduced into wild-type SH1000, ISP794, and Newman, and MICs were determined for these strains and the corresponding norA::lacZ derivatives containing the same plasmids. The relationship of mgrA expression to that of norA was confirmed for strains with and without the norA::lacZ fusion expressing either pCU1 or pK404 by primer extension as described previously.
RESULTS AND DISCUSSION
MIC determinations.
Compared to SH1000, twofold reductions in MICs for a variety of NorA substrates were observed for SA-K2124 (SH1000 norA::lacZ). Identical results were observed for these same strains containing pCU1, which will not affect the MICs of the tested compounds (Table 2). This is consistent with what has been observed for other norA-disrupted strains (16, 22).
TABLE 2.
Effect of mgrA overexpression on susceptibility to NorA substrates
| Drug | MIC (μg/ml) for strain and plasmidb:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SH1000
|
SA-K2124a
|
ISP794
|
Newman
|
|||||
| pCU1 | pK404 | pCU1 | pK404 | pCU1 | pK404 | pCU1 | pK404 | |
| Acriflavine | 12.5 | 6.25 (2) | 6.25 | 6.25 | 3.13 | 3.13 | 6.25 | 6.25 |
| BACc | 1.25 | 1.25 | 0.63 | 0.63 | 0.63 | 0.63 | 1.25 | 1.25 |
| Cetrimide | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.16 (2) | 0.63 | 0.16 (4) |
| Ethidium bromide | 6.25 | 6.25 | 3.13 | 3.13 | 1.56 | 1.56 | 6.25 | 3.13 (2) |
| Norfloxacin | 1.25 | 0.63 (2) | 0.63 | 0.63 | 0.63 | 0.31 (2) | 0.31 | 0.16 (2) |
| Rhodamine | 0.63 | 0.63 | 0.31 | 0.31 | 0.31 | 0.31 | 0.63 | 0.63 |
| TPPd | 25 | 25 | 12.5 | 12.5 | 6.25 | 3.13 (2) | 12.5 | 12.5 |
SH1000 norA::lacZ.
Numbers in parentheses are reductions (n-fold) in MICs.
BAC, benzalkonium chloride.
TPP, tetraphenylphosphonium bromide.
MICs were determined for wild-type SH1000, ISP794, and Newman containing either pCU1 or pK404, and the presence of pK404 resulted in reproducible twofold reductions in MICs for some, but not all, NorA substrates in each strain background (Table 2). The effect was not consistent between strains, with the exception of that for norfloxacin. The reason(s) for the variable effect of mgrA overexpression on MICs between strains is not readily apparent, but it may be related to differential expression of other pumps unaffected by MgrA that have overlapping substrate profiles with NorA.
The expression of pK404 in SA-K2124 did not affect any MICs, with the same result observed in norA::lacZ fusion mutants of ISP794 and Newman (data not shown). These strains are norA knockout mutants, and the lack of an MgrA effect in this genetic background is consistent with the absence of the MDR pump that it regulates. These data also suggest that MgrA does not affect the transcription of other pumps having the tested compounds as substrates to any significant degree.
Our data are in conflict with the previously reported effect of mgrA overexpression, which revealed that MICs of norfloxacin, cetrimide, and ethidium bromide increased fourfold when mgrA was expressed from the temperature-sensitive plasmid pSK950 in the ISP794 background (42). Significant technical and methodological differences exist between this earlier work and ours. The mgrA clone we employed included its native promoters (P1 and P2), the expression vector was pCU1, and the incubation temperature was 37°C. Testing with strains bearing pSK950-based constructs was done at 30°C (permissive temperature), and the mgrA construct used lacked both native promoters, in which case expression of the gene had to occur by read-through from an undefined plasmid-based promoter. In subsequent experiments (described below), we found that strain-related issues, incubation temperature, and the presence of the native mgrA promoters do not contribute to this apparent conflict. The one remaining possibility is the different vectors employed. It is possible that expression of mgrA from pSK950 results in concentrations of MgrA much greater than that achieved by use of pCU1 (or pALC2073; see below). It is conceivable that there are binding sites within the norA promoter region that have high or low affinity for MgrA, and the effect on norA expression observed is dependent on which site(s) is occupied at any given time. High-affinity repressive binding sites may be occupied at low to moderate concentrations of MgrA, and low-affinity stimulatory sites may be occupied at very high MgrA concentrations.
mRNA quantitation.
The quantity of norA mRNA was not altered by fusion with lacZ (compare lanes 1 and 2 in Fig. 2, which represent SH1000 and SH1000 norA::lacZ [SA-K2124], respectively) or by disruption of the IR (SA-K2173, lane 4) or the TTAATT consensus sequences (SA-K2242, lane 5). The flqB mutation resulted in a quantitative increase in the norA message (SA-K2161, lane 3), with this effect seen in all flqB mutants constructed (data not shown). A similar increase in norA mRNA has been observed in all flqB mutants described to date (19, 35, 36).
A profound increase in mgrA transcripts was observed for strains containing pK404 compared to the same strains containing pCU1, and increased expression of mgrA correlated with a diminished norA transcript level for all tested strains. This effect is easiest to appreciate in flqB mutants, owing to the fact that these strains produce significantly more norA mRNA. For SA-K2161, mgrA overexpression resulted in a 2.3-fold decrease in norA transcripts (Fig. 3). The same effect was observed for strains without an flqB mutation and for SH1000, ISP794, and Newman without norA::lacZ fusions (data not shown). The variable reductions in NorA substrate MICs that we observed in the presence of mgrA overexpression are consistent with reduced norA transcription (Table 2).
FIG. 3.
Primer extension analysis of mgrA and norA expression of SA-K2161 (SH1000 norA::lacZ flqB [T→A]). Lane 1, plus pCU1; lane 2, plus pK404. Transcripts for both mgrA promoters are clearly evident.
mRNA half-life determinations.
Whether the apparent increase in norA mRNA observed for flqB mutants is the result of a true quantitative increase, a prolongation of mRNA half-life, or a combination of both is a controversial issue. We found that SA-K2124 (SH1000 norA::lacZ) and its T→A or G flqB mutants had similarly short half-lives (68, 43, and 53 s, respectively), indicating that augmented transcription and not increased mRNA stability was responsible for the increase in norA message observed in these flqB strains. The typical half-life for bacterial mRNA is about 2 min, and our data are reasonably consistent with this (28).
These results contrast with previous work that employed a strain having a T→G flqB mutation, which showed a fivefold increase in norA mRNA half-life compared to its parent (34 versus 7 min, respectively) (11). These half-lives are exceedingly long for bacterial mRNA, and the reason(s) for this remarkable observation escapes easy explanation. We did find that SA-K2291 (flqB T→C) had a modestly prolonged norA half-life compared to that of SA-K2124 (151 versus 68 s, respectively) that could have contributed to its increased norA message and its apparent increase in norA transcription as determined in MUG assays (see below).
Using the M-fold algorithm available at http://www.bioinfo.rpi.edu/applications/mfold, no flqB mutant had a predicted improvement in 5′ UTR stability versus the wild type (32, 45). In fact, all mutants had less favorable free energy of folding (ΔG) values. The ΔG for the 5′ UTR of norA mRNA for SA-K2124 was −11.8 kcal/mol, whereas those of its T→A, C, and G flqB mutants were −9.2, −10.8, and −8.9 kcal/mol, respectively. From these data, it is clear that nucleic acid folding algorithms are useful guides but their predictions may not correlate with experimentally generated data.
In all flqB mutants of SA-K2124, MUG assays indicated that norA expression was increased at least eightfold (see below), increased norA signal was evident in norA primer extension products (Fig. 2), and mRNA half-life was not prolonged in T→A or T→G flqB mutants. These data support the conclusion that in most cases the flqB mutation results only in a quantitative increase in norA message. In selected mutants, a modest prolongation of norA message half-life may be an additive factor.
MUG assays and mgrA overexpression.
The creation of an EcoRI site in a presumed silent location upstream of the norA promoter resulted in no change in norA expression, which for all strains peaked in the mid- to late logarithmic growth phase (data not shown) (23). Cumulative norA expression, as measured by MUG assays over a 10-h period for SA-K2124 and its norA promoter region mutants, and the effect of mgrA overexpression in the same strains are shown in Fig. 4.
FIG. 4.
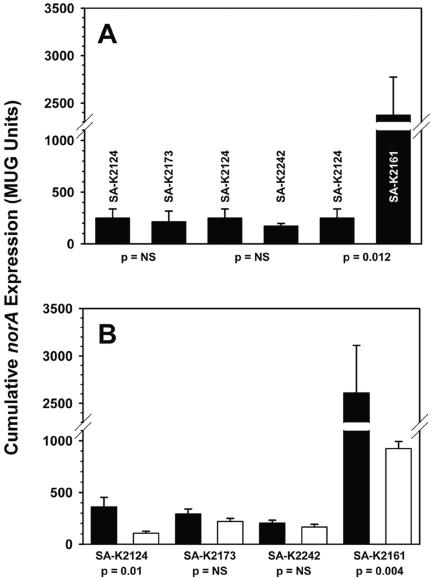
norA expression by β-galactosidase assay. (A) SA-K2124, wild-type norA promoter; SA-K2173, inverted repeat mutant; SA-K2242, TTAATT consensus sequence mutant; SA-K2161, flqB (T→A) mutant. (B) Filled and open bars represent the indicated strain with pCU1 or pK404 (pCU1-mgrA), respectively. Data represent the means of the results from three experiments ± standard deviations. NS, not significant.
In the presence of a wild-type norA promoter (SA-K2124), mgrA overexpression resulted in a significant reduction in norA expression (71%; P = 0.01). Similar results were observed in the Newman background (70%; P = 0.03) and to a lesser extent in the ISP794 background (38%; P = 0.06) (data not shown). To address the possibility that incubation temperature has a role in the apparent inconsistency between ours and the earlier data with respect to the MgrA effect on norA expression, a MUG assay was performed at 30°C by using SA-K2124 and SA-K2161 (flqB T→A) with either pCU1 or pK404. Reduced norA expression in the presence of pK404 still was observed (≥70%), ruling out a temperature effect (data not shown).
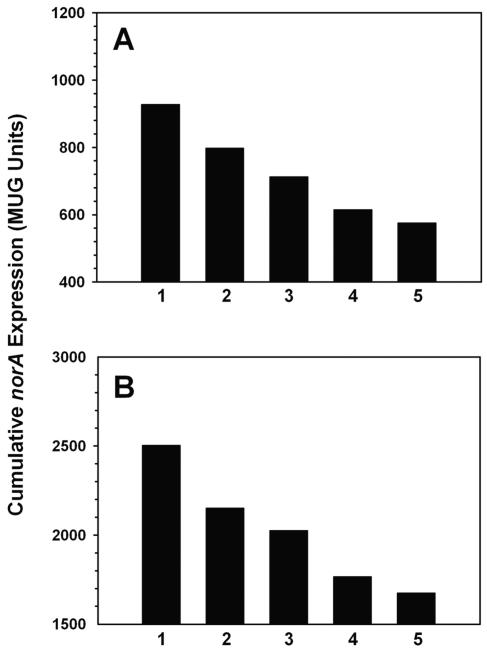
The possible role played by the presence of the native mgrA promoters in this apparent discrepancy was addressed by using strains containing pK410 and induced with variable concentrations of tetracycline. We found that mgrA expression had a dose-related repressive effect on norA transcription in SA-K2124 and SA-K2161 (Fig. 5). A similar effect was observed in the wild-type norA promoter norA::lacZ derivatives of ISP794 and Newman, but as was found with pK404, the effect was blunted for ISP794 (data not shown). The blunted effect of mgrA overexpression in ISP794 is most likely the result of uncharacterized genetic differences between it and SH1000 or Newman. However, it is possible that the SigB deficiency of ISP794 also contributes.
FIG. 5.
Effect of expression of mgrA from pK410 (pALC2073-mgrA) on norA expression by β-galactosidase assay. (A) SA-K2124 (SH1000 norA::lacZ); (B) SA-K2161 (SH1000 norA::lacZ flqB [T→A]). Bars: 1, no tetracycline; 2, 25 ng of tetracycline/ml; 3, 50 ng of tetracycline/ml; 4, 75 ng of tetracycline/ml; 5, 100 ng of tetracycline/ml. Data represent the means of the results from duplicate experiments.
Destruction of the near-perfect IR, which encompasses the −10 promoter motif (Fig. 1) and is conserved in all sequenced S. aureus strains, resulted in a nonsignificant reduction in norA expression (15%) (Fig. 4A [SA-K2173]). These data allow us to conclude that the IR probably has no role, or only a minor one, in the regulation of norA expression. mgrA overexpression in the IR mutant background resulted in a 25% reduction in norA expression (Fig. 4B); this effect was quite blunted compared to that observed in the presence of a wild-type promoter and did not reach statistical significance. It is possible that the IR mutation may have a minor effect on the interaction of MgrA with the norA promoter.
The TTAATT consensus sequences upstream of the norA promoter (Fig. 1) are completely conserved among sequenced S. aureus strains. These hexamers have been proposed as potential recognition sites for binding of a regulatory protein such as MgrA. Upon simultaneous disruption of all four of these motifs, a modest (32%, SA-K2242) but nonsignificant reduction in norA expression was observed. Overexpression of mgrA in the TTAATT mutant resulted in a minor reduction in norA expression (18%), less than that observed in the wild-type and IR mutant backgrounds. These data indicate that the TTAATT consensus sequences are important for maximal norA expression. Their elimination removes much of the repressive effect of mgrA overexpression and suggests that MgrA is likely to interact with one or more of these, or closely linked, sites within the norA promoter region.
Consistent with previous data indicating that the presence of an flqB mutation leads to augmented norA expression, we found that in the SH1000 background all flqB mutants demonstrated a highly significant ≥8-fold increase in norA expression (data for the T→A flqB mutant are shown in Fig. 4A). The same was also observed for SA-K2159 and SA-K2383, the T→A flqB mutants of Newman norA::lacZ and ISP794 norA::lacZ, respectively (data not shown). As noted previously, for all except the T→C flqB mutation, this effect appears to be related purely to augmented transcription. Overexpression of mgrA resulted in a significant (65%) reduction in norA expression in the presence of the flqB mutation in the SH1000 background.
Clearly, the regulation of norA expression is complex. The perfect IR that encompasses the −10 motif plays no great role. However, its conservation among sequenced S. aureus strains and the blunted MgrA effect observed in an IR-disrupted mutant suggests that it may be required for the full effect of MgrA on norA expression. As for the perfect IR, disruption of the TTAATT consensus sequences also resulted in a blunted MgrA effect on norA expression. These repeats may serve as recognition sites for the binding of MgrA or other norA-regulatory proteins.
We have established that the flqB mutation has a great effect on norA expression. In its presence, norA transcription is increased significantly. A modest prolongation in half-life was found for one flqB mutant (∼2-fold; SA-K2291), but it is probable and indeed likely based on data with other flqB mutants that increased transcription also occurs in this strain. The involvement of the flqB position in the regulation of norA expression is intriguing; it is possible that this position is part of a recognition site for the binding of either MgrA or an as yet unidentified regulatory protein. From Fig. 1, it can be seen that this position is part of an imperfect IR (repeat B) that is conserved in all sequenced S. aureus strains. Perhaps this single base change reduces the affinity of regulatory protein binding. There are numerous examples of regulatory proteins binding to operator regions that include 5′ sequences of mRNA. One of particular relevance is the S. aureus QacR repressor, which binds to an IR region positioned between bases −14 and +14 of the qacA gene encoding the QacA multidrug transporter (13).
The determination of the MgrA footprint in the norA promoter region will address many of the issues we have raised. Such experiments done by using variable MgrA concentrations will reveal if high- and low-affinity binding sites exist, as we have proposed. The blunted effect of mgrA overexpression that we observed in the perfect IR and TTAATT consensus sequence mutants may be related to diminished MgrA binding, which can be verified or refuted by footprinting experiments with strains with or without these mutations. Finally, it should be determined whether MgrA binds to the imperfect IR region and, if so, if that binding is affected by flqB mutations. The imperfect IR should also be employed as a target in the search for novel regulatory proteins that target the norA promoter.
Employing two different methods, including primer extension and reporter gene product activity, we have shown that mgrA overexpression results in a reduction in norA expression. It may be argued that our use of mgrA+ strains as hosts for mgrA-containing plasmids may have influenced the results we observed. This is exceedingly unlikely because, if anything, chromosomal mgrA expression would have increased as a result of positive autoregulation by MgrA. The quantity of MgrA contributed by this process would be small in comparison to that originating from the plasmid-based gene and would simply add to the overall quantity of MgrA present. This would not have affected our data in any significant way.
Efflux-related resistance may be clinically relevant by itself, but in the case of NorA, it may also favor the emergence of target-based mutations and high-level resistance by diminishing intracellular drug concentrations. It is important to understand the regulation of expression of efflux pumps such as NorA because that understanding may lead to the development of means to interfere with their activity.
Acknowledgments
This study was supported by VA Research Funds.
We thank J. Higgins and T. Foster, C. Y. Lee, and Ambrose Cheung for providing pTS2tetK, pTL2989, and pALC2073, respectively. We also thank Q.-C. Truong-Bolduc and D. C. Hooper for constructing strains SA-K2382 and SA-K2383.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Ahmed, M., L. Lyass, P. N. Markham, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1995. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J. Bacteriol. 177:3904-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Augustin, J., R. Rosenstein, B. Weiland, U. Schneider, N. Schnell, G. Engelke, K. Entian, and F. Götz. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149-1154. [DOI] [PubMed] [Google Scholar]
- 5.Baranova, N. N., A. Danchin, and A. A. Neyfakh. 1999. Mta, a global MerR-type regulator of the Bacillus subtilis multidrug efflux transporters. Mol. Micribiol. 31:1549-1559. [DOI] [PubMed] [Google Scholar]
- 6.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 8.Foster, T. J. 1998. Molecular genetic analysis of staphylococcal virulence. Methods Microbiol. 27:433-454. [Google Scholar]
- 9.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier, B., Q.-C. Truong-Bolduc, X. Zhang, and D. C. Hooper. 2001. A mutation in the 5′ untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J. Bacteriol. 183:2367-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grkovic, S., M. H. Brown, M. A. Schumacher, R. G. Brennan, and R. A. Skurray. 2001. The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol. 183:7102-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 15.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh, P.-C., S. A. Siegel, B. Rogers, D. Davis, and K. Lewis. 1998. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc. Natl. Acad. Sci. USA 95:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingavale, S. S., W. Van Wamel, and A. L. Cheung. 2003. Characterization of Rat, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 18.Kaatz, G. W., S. L. Barriere, D. R. Schaberg, and R. Fekety. 1987. The emergence of resistance to ciprofloxacin during treatment of experimental Staphylococcus aureus endocarditis. J. Antimicrob. Chemother. 20:753-758. [DOI] [PubMed] [Google Scholar]
- 19.Kaatz, G. W., S. M. Seo, and C. A. Ruble. 1993. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1086-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaatz, G. W., S. M. Seo, and T. J. Foster. 1999. Introduction of a norA promoter region mutation into the chromosome of a fluoroquinolone-susceptible strain of Staphylococcus aureus using plasmid integration. Antimicrob. Agents Chemother. 43:2222-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaatz, G. W., S. M. Seo, L. O'Brien, M. Wahiduzzaman, and T. J. Foster. 2000. Evidence for the existence of a multidrug efflux transporter distinct from NorA in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1404-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaatz, G. W., and S. M. Seo. 2004. Effect of substrate exposure and other growth condition manipulations on norA expression. J. Antimicrob. Chemother. 54:364-369. [DOI] [PubMed] [Google Scholar]
- 24.Kemp, E. H., R. L. Sammons, A. Moir, D. Sun, and P. Setlow. 1991. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J. Bacteriol. 173:4646-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, R. B. 2002. Transporters and xenobiotic disposition. Toxicology 181-182:291-297. [DOI] [PubMed] [Google Scholar]
- 26.Kreiswirth, B. N., M. S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 27.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewin, B. 1995. Messenger RNA is the template, p. 253-276. In B. Lewin (ed.), Genes V. Oxford University Press Inc., New York, N.Y.
- 29.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multi-drug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination chemotherapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 33.McAleese, F. M., E. J. Walsh, M. Sieprawska, J. Potempa, and T. J. Foster. 2001. Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 278:29969-29978. [DOI] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. 1999. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 35.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1994. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob. Agents Chemother. 38:1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noguchi, N., M. Tamura, K. Narui, K. Wakasugi, and M. Sasatu. 2002. Frequency and genetic characterization of multidrug-resistant mutants of Staphylococcus aureus after selection with individual antiseptics and fluoroquinolones. Biol. Pharm. Bull. 25:1129-1132. [DOI] [PubMed] [Google Scholar]
- 37.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen, I. T., M. H. Brown, T. G. Littlejohn, et al. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. USA 93:3630-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 40.Stahl, M. L., and P. A. Pattee. 1983. Confirmation of protoplast fusion-derived linkages of Staphylococcus aureus by transformation with protoplast DNA. J. Bacteriol. 154:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trucksis, M., J. S. Wolfson, and D. C. Hooper. 1991. A novel locus conferring fluoroquinolone resistance in Staphylococcus aureus. J. Bacteriol. 173:5854-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Truong-Boldoc, Q.-C., X. Zhang, and D. C. Hooper. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 185:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Bambeke, F., E. Balzi, and P. M. Tulkens. 2000. Antibiotic efflux pumps. Biochem. Pharmacol. 60:457-470. [DOI] [PubMed] [Google Scholar]
- 44.Woolridge, D. P., N. Vazquez-Laslop, P. N. Markham, M. S. Chevalier, E. W. Garner, and A. Neyfakh. 1997. Efflux of the natural polyamine spermidine facilitated by the Bacillus subtilis multidrug transporter Blt. J. Biol. Chem. 272:8864-8866. [DOI] [PubMed] [Google Scholar]
- 45.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark, (ed.), NATO ASI series. Kluwer Academic Publishers, Dordrecht, The Netherlands.