Abstract
We have previously shown that the Enterococcus faecalis lsa gene, encoding the putative ABC protein Lsa, influences resistance to quinupristin-dalfopristin (Q-D) and clindamycin (CLI). We have now found that, while cloned lsa from E. faecalis strain V583 (lsaV) fully restored resistance to Q-D, CLI, and dalfopristin (DAL) lost by the OG1 lsa disruption mutant TX5332 and also caused increased MICs for Lactococcus lactis LM2301, cloned lsa from OG1 (lsaOG) did not cause any increase in MICs for either species. Sequencing of ca. 2 kb of these two lsa alleles found differences between lsaOG and lsaV in the upstream region as well as in the 5′ and 3′ halves of the lsa gene. To investigate the reason for the phenotypic differences expressed by the two cloned loci, 5′ half plus 3′ half hybrid constructs were created. When introduced into both TX5332 and L. lactis, cloned lsaV5′OG3′ conferred increases in MICs of Q-D, CLI, and DAL similar to those of cloned lsaV while cloned lsaOG5′V3′ showed a moderate increase in MICs relative to those of lsaOG, indicating that both halves of the locus can influence resistance expression. After site-directed mutagenesis of the cloned lsa alleles at positions −131 and −133 (relative to the putative Lsa start codon ATG), which converted two A's of lsaV to the G and T of lsaOG and vice versa, MIC testing showed that mutagenized lsaOG (lsaOG-M) was strongly influenced by these changes in terms of conferring increased MICs of Q-D, CLI, and DAL relative to lsaOG while the phenotype of mutagenized lsaV (lsaV-M) was less influenced, with moderately decreased MICs, primarily to CLI, relative to lsaV. In conclusion, this study found that changes in different regions of the E. faecalis lsa locus influence the ability of cloned lsa to confer resistance to Q-D, CLI, and DAL.
Quinupristin-dalfopristin (Q-D) is a mixture of streptogramin B and streptogramin A, respectively, and studies have shown that the species Enterococcus faecalis has intrinsic resistance to this compound (2, 8, 19, 25). Our previous study of Lsa (which stands for lincosamide and streptogramin A resistance), a predicted ABC protein homologue, implicated it as being the cause of the intrinsic resistance of E. faecalis to clindamycin (CLI), dalfopristin (DAL), and Q-D (25). Specifically, TX5332, a mutant of OG1RF (henceforth referred to as OG1) with a disruption in lsa (formerly abc23) (6) showed a marked increase in susceptibility to Q-D, CLI, and DAL, and complementation of TX5332 with an intact lsa gene (lsaV) from the sequenced E. faecalis strain V583 (17) restored resistance to Q-D, CLI, and DAL (25). Consistent with this finding, a subsequent study by Dina et al. (7) found that the lsa alleles of two clinical isolates of E. faecalis susceptible to lincosamides and dalfopristin contained mutations within lsa that produced premature termination codons, further supporting the role of lsa in Q-D resistance.
In the present study, we attempted complementation of the mutant TX5332 with a recombinant plasmid carrying an intact lsa allele from wild-type E. faecalis strain OG1 (lsaOG) and found that this strain's gene did not restore resistance, unlike the equivalent fragment from strain V583 in our previous study (25). We compared the lsa sequences derived from both E. faecalis V583 and OG1, constructed hybrid fragments by combining different regions of lsa from each strain, generated site-directed mutagenized DNA fragments of lsa, and then tested the impact of these cloned elements on the susceptibility of TX5332 to Q-D, CLI, quinupristin, and DAL. We also tested these constructs in another gram-positive host, Lactococcus lactis LM2301 (27), and PCR amplified, sequenced, and studied lsa and upstream sequences from six Q-D- and CLI-susceptible E. faecalis isolates of different origins.
MATERIALS AND METHODS
Bacterial strains.
The strains and plasmids used in the study are listed in Table 1. These include E. faecalis strains OG1RF (14) (referred to here as OG1) and V583 (17, 22), L. lactis LM2301 (27), four Q-D- and CLI-susceptible E. faecalis isolates TX4107 to TX4110 from animal feed (provided courtesy of S. Simjee, U.S. Food and Drug Administration, Bethesda, Md.), and two Q-D-susceptible clinical E. faecalis isolates, TX0263 and TX0271, isolated from the Houston area. Mutant strains of E. faecalis used as controls to determine the effect of kanamycin (KAN; used for selection of the OG1 lsa disruption mutant) on MICs of test compounds include TX5076 (30), TX5243 (18), and TX5248 (18), which were constructed by using cloned intragenic gene fragments in the suicide vector pTEX4577 containing aph(3′)-llla (24), the method used for the construction of the OG1 lsa disruption mutant, TX5332 (6, 25).
TABLE 1.
Bacterial strains and plasmids used in the study
| Strains or plasmid | Purpose and relevant characteristic(s) | Reference(s) or source |
|---|---|---|
| E. faecalis strain(s) | ||
| OG1RF | Used for lsa insertional mutagenesis and to amplify lsa for complementation experiments; Rifr, Fusr | 14 |
| V583 | TIGR-sequenced strain; used to amplify lsa for complementation experiments | 17, 22 |
| TX4107-TX4110 | Q-D- and CLI-susceptible isolates from animal feed | This study |
| TX0263, TX0271 | Q-D- and CLI-susceptible clinical isolates | This study |
| TX5076 | Antigen gene disruption mutant generated with pTEX4577 containing aph(3′)-llla; Kanr | 30 |
| TX5243 | sprE gene disruption mutant [OG1RF sprE::pTEX4577 containing aph(3′)-llla]; Kanr | 18 |
| TX5248 | orf1 gene disruption mutant [OG1RF orf1::pTEX4577 containing aph(3′)-llla]; Kanr | 18 |
| TX5332 | lsa gene disruption mutant [OG1RF lsa::pTEX4577 containing aph(3′)-llla]; Kanr | 25 |
| TX5332(pTEX5333.04) (contains lsaV); Kanr, Chlr | 25 | |
| TX5332(pTEX5333.03) (contains lsaOG); Kanr, Chlr | This study | |
| TX5332(pTEX5333.08) (contains lsaOG5′V3′); Kanr, Chlr | This study | |
| TX5332(pTEX5333.09) (contains lsaV5′OG3′); Kanr, Chlr | This study | |
| TX5332(pTEX5333.12) (contains a mutagenized gene, lsaV-M); Kanr, Chlr | This study | |
| TX5332(pTEX5333.13) (contains a mutagenized gene, lsaOG-M); Kanr, Chlr | This study | |
| L. lactis strains | ||
| LM2301 | Used as a gram-positive host for E. faecalis lsa | 27 |
| LM2301(pTEX5333.04) (contains lsaV5); Chlr | This study | |
| LM2301(pTEX5333.03) (contains lsaOG); Chlr | This study | |
| LM2301(pTEX5333.08) (contains lsaOG5′V3′); Chlr | This study | |
| LM2301(pTEX5333.09) (contains lsaV5′OG3′); Chlr | This study | |
| LM2301(pTEX5333.12) (contains lsaV-M); Chlr | This study | |
| LM2301(pTEX5333.13) (contains lsaOG-M); Chlr | This study | |
| TX5392 | LM2301(pWM401); Chlr | This study |
| Plasmids | ||
| pTEX4577 | pBluescript SK(−) with aph(3′)-IIIa inserted into the ScaI site; Kanr, used for insertional mutagenesis | 24 |
| pWM401 | Shuttle vector; Chlr, Tetr | 29 |
| pCR2.1 vector | PCR product cloning vector | Invitrogen |
| pTEX5333.03 | pWM401::lsaOG, contains a ca. 2-kb lsa fragment from OG1RF, used for complementation of TX5332 and transformation of L. lactis LM2301; Chlr | This study |
| pTEX5333.04 | pWM401::lsav, contains a ca. 2-kb lsa fragment from V583, used for complementation of TX5332 and transformation of L. lactis LM2301; Chlr | 25 |
| pTEX5333.08 | pWM401::lsaOG5′V3′, contains a hybrid gene with the 5′ half of lsaOG and 3′ half of lsav, used for complementation of TX5332 and transformation of L. lactis LM2301; Chlr | This study |
| pTEX5333.09 | pWM401::lsaV5′OG3′, contains a hybrid gene with the 5′ half of lsaV and 3′ half of lsaOG, used for complementation of TX5332 and transformation of L. lactis LM2301; Chlr | This study |
| pTEX5333.12 | pWM401::lsaV-M, contains ca. 2-kb lsaV fragment mutagenized in its upstream region at positions −131 and −133 with A's replaced with a G and a T, used for complementation of TX5332 and transformation of L. lactis LM2301; Chlr | This study |
| pTEX5333.13 | pWM401::lsaOG-M, contains ca. 2-kb lsaOG fragment mutagenized in its upstream region at positions −131 and −133 with G and T replaced by A's, four additional nucleotide changes were found at −109, +1248 (silent), +1572 (downstream), and +1517 (downstream), used for complementation of TX5332 and transformation of L. lactis LM2301; Chlr | This study |
MIC studies.
MICs were determined by the broth microdilution method (15, 16) or by E-test (PDM Epsilometer test; AB BIODISK North America, Inc., Piscataway, N.J.) by using Mueller-Hinton II broth (MHB) or Mueller-Hinton II agar (MHA) (Becton Dickinson Company, Sparks, Md.) as test media following the manufacturer's instruction. KAN, CLI, and chloramphenicol (CHL) were purchased from Sigma Chemical Co., St. Louis, Mo., and quinupristin, DAL, and Q-D were provided by Aventis Pharma S.A., Vitry-sur-Seine Cedex, France. For the lsa disruption mutant TX5332 and for TX5332 derivatives containing the shuttle vector pWM401 (29), KAN (2,000 μg/ml) and KAN (2,000 μg/ml) plus CHL (8 μg/ml), respectively, were added to the MIC test media; for LM2301 derivatives with pWM401, CHL (8 μg/ml) was added to the test media. For wild-type strains, only test compounds were added to the media.
DNA extraction, PCR, sequencing, and cloning.
DNA extraction was done by using previously published methods (12, 28). PCRs were performed by using the optimized buffer B kit (Invitrogen, San Diego, Calif.). PCR was generally performed in volumes of 50 μl, with an initial denaturation at 94°C for 2 min followed by 25 to 30 cycles consisting of 1 min of denaturation at 94°C, 2 min of annealing at 55°C, 3 min of extension at 72°C, and a 10-min final extension at 72°C. Primers used for amplification and sequencing are listed in Table 2 and were initially designed by using the V583 genome database (www.tigr.org). PCR products were analyzed by automated DNA sequencing at the Microbiology and Molecular Genetics core facility, University of Texas Medical School, Houston. Sequence analysis was done by using the BLAST network service of the National Center for Biotechnology Information, and DNASTAR software (Madison, Wis.) was used to compare similarities among other sequences. Cloning was done by using the pCR2.1 vector of the TA cloning kit following the manufacturer's instructions or with standard methods (23) into the shuttle vector pWM401 (29).
TABLE 2.
Primers used in the study
| Primer | Positiona | Sequence (5′-3′)b |
|---|---|---|
| abc2Fc | −309, −288 | +GGCAATCGCTTGTGTTTTAGCG |
| abc2R | +1735, +1713 | −GTGAATCCCATGATGTTGATACC |
| abc3Fd | +108, +129 | +GATTGGCCGCAATGGCCGTGGG |
| abc3R | +1123, +1101 | −GGTGAGCCAAAGTGGCTTCGCC |
| abc3F2 | +1349, +1372 | +GATGAACCCCTTAATTACTTGG |
| abc7F | −203, −182 | +GACGAAAAGAGGGATTCGTGCG |
| abc7R | −11, −32 | −CCATAAAGCAAAATTGATGCAG |
| abc2Fe | −309, −288 | +GGCAATCGCTTGTGTTTTAGCG |
| Isa mut | −105, −85 | −GAATTCTTTAAGCTAATTTC |
| Mega primer F (OG1) (double stranded)f | −309, −85 | GGCAATCGCTTGTGTTTTAGCGAATAATTTAAATTTTTATCAGAATGAAAGCAAGGGGCCTTCGTTGACAAATCAGTGAAGAGATGCTAAATTCTAGATAATAAAAGACGAAAAGAGGGATTCGTGCGTTGAAAATTTAAGTTAATTTTTTAGAGAAACTGTATAACTTTGCCTGATTGTAACGGGTGGAGTGTGCGGATTTTCGGAAATTAGCTTAAAGAATTC |
| Mega primer F (V583) (double stranded)g | −309, −85 | GGCAATCGCTTGTGTTTTAGCGAATAATTTAAATTTTTATCAGAATGAAAGCAAGGGGCCTTCGTTGACAAATCAGTGAAGAGATGCTAAATTCTAGATAATAAAAGACGAAAAGAGGGATTCGTGCGTTGAAAATTTAAGTTAATTTTTTAGAGAAACTGTATAACTTTGCCTGAATATAACGGGTGGAGTGTGCGGATTTTCGGAAATTAGCTTAAAGAATTC |
Bases relative to ATG.
+, sense primer; −, antisense primer.
Primers abc2F and abc2R were used to amplify intact lsa on a 2-kb fragment.
Primers abc3F to abc7R are intragenic primers used to confirm the lsa sequence.
Primers abc2F and lsa mut were used for site-directed mutagenesis (to generate mega primer).
Mega primer F(OG1) and abc2R were used to generate lsaV-M. Boldfaced T and G were targeted in site-directed mutagenesis.
Mega primer F(V583) and abc2R were used to generate lsaOG-M. Boldfaced A and A were targeted in site-directed mutagenesis.
Generation of OG1 lsa and hybrid lsa fragments.
A ca. 2-kb OG1 fragment consisting of intact lsa (lsaOG) equivalent to the previously published ca. 2-kb V583 lsa fragment (lsaV) (25) was PCR amplified with primers abc2F and abc2R from wild-type E. faecalis strain OG1, cloned into the pCR2.1 vector, and designated pTEX5333.01. The lsaOG fragment was then excised from pTEX5333.01 by digestions with XbaI and BamHI and was recloned into the shuttle vector pWM401, resulting in pTEX5333.03. The ca. 2-kb V583 lsa fragment (lsaV) previously cloned into pCR2.1 was designated pTEX5333.02 and, in pWM401, pTEX5333.04 (Table 1).
Using the single KpnI site located in pCR2.1 and a SmaI site located in approximately the middle of the lsa gene of both OG1 and V583, we exchanged the two halves (including the upstream region with the 5′ coding half). The resulting plasmid constructs in pCR2.1 were designated pTEX5333.06, containing the upstream plus 5′ half (nucleotides −309 to +1102 relative to the predicted start codon) of OG1 lsa plus the 3′ half (nucleotides +1103 to +2044) of lsa of V583 (lsaOG5′V3′), and pTEX5333.07, containing the upstream plus 5′ half (nucleotides −309 to +1102) of V583 lsa plus the 3′ half (nucleotides +1103 to +2044) of lsa of OG1 (lsaV5′OG3′). The cloned fragments were excised from the pCR2.1 vector by digestion with BamHI and EcoRV and recloned into the shuttle vector pWM401. The recombinant shuttle plasmids containing hybrid lsa genes were designated pTEX5333.08 (containing lsaOG5′V3′) and pTEX5333.09 (containing lsaV5′OG3′) (Table 1). In cloning experiments, we noticed many deletions by restriction enzyme digestions; therefore, all of the cloned lsa derivatives in the shuttle vector were resequenced in both directions before use for complementation experiments.
Site-directed mutagenesis of lsa.
Two nucleotides in the regions immediately upstream of V583 and of OG1 lsa were targeted for mutagenesis by using the previously published mega primer method (5). The primers used for mutagenesis experiments are listed in Table 2. The A's at positions −131 and −133 (bases relative to the predicted ATG start codon) in the upstream region of the ca. 2-kb lsaV fragment were replaced with a G and a T, respectively. Similarly, the same nucleotide positions were targeted in the ca. 2-kb lsaOG fragment for changing the G and T at −131 and −133, respectively, to A's. Mutagenized lsa fragments were generated by PCR. Both mutagenized lsa fragments were cloned into pCR2.1 and then into pWM401. The resulting recombinant shuttle plasmids were designated pTEX5333.12 (lsaV-M) and pTEX5333.13 (lsaOG-M), respectively (Table 1), and their sequences were reconfirmed by sequencing.
Complementation of the OG1 lsa disruption mutant, TX5332, and transformation of L. lactis LM2301.
Electrocompetent cells of TX5332 and LM2301 were prepared by following the previously published method (9), and the electroporation conditions used were the same as those previously published (10). Recombinant plasmids pTEX5333.03 (lsaOG), pTEX5333.04 (lsaV), pTEX5333.08 (lsaOG5′V3′), pTEX5333.09 (lsaV5′OG3′), pTEX5333.12 (lsaV-M), and pTEX5333.13 (lsaOG-M) were electroporated into competent cells of TX5332 and LM2301. Complemented colonies were recovered on Todd-Hewitt agar (Difco Laboratories, Sparks, Md.) plus KAN (2,000 μg/ml) plus CHL (8 μg/ml) and Todd-Hewitt agar plus CHL (8 μg/ml) for TX5332 and LM2301, respectively. The presence of lsa and the identity of the host were confirmed by pulsed-field gel electrophoresis (PFGE) (13) and hybridization (14) to an intragenic lsa DNA probe. The resulting complemented colonies in TX5332 and LM2301 were designated as described in Table 1.
Q-D- and CLI-susceptible E. faecalis isolates.
For Q-D- and CLI-susceptible isolates (TX4107 to TX4110, TX0263, and TX0271), PCR-amplified lsa-containing fragments were sequenced in both directions, and the sequences were aligned with each other as well as with lsa from OG1 and V583 (The Institute for Genomic Research [TIGR] genome database [www.tigr.org] and our own results). The relatedness of the isolates was examined by PFGE (14, 26).
RESULTS
Sequencing of lsaOG and lsaV and use for complementation of TX5332 and LM2301.
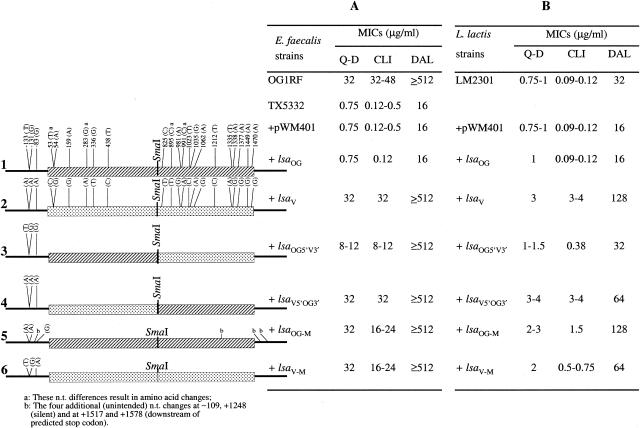
Our previously published study showed that the intact lsaV gene on a ca. 2-kb DNA fragment cloned into a shuttle vector (pTEX5333.04) was able to fully restore the MIC decreases seen with TX5332 (Fig. 1A) for Q-D (from 0.75 to 32 μg/ml), CLI (from 0.12 to 0.5 μg/ml to 32 to 48 μg/ml) and DAL (from 16 to 512 μg/ml) (25) (Fig. 1A, row 2). In the present study, cloned lsaOG on the equivalent ca. 2-kb fragment (pTEX5333.03) did not increase the resistance of TX5332 to Q-D, CLI, or DAL (Fig. 1A, row 1). We also determined the effect of lsaOG and lsaV on MICs of Q-D, CLI, and DAL in another gram-positive host LM2301 (Fig. 1B, rows 1 and 2), and again, no increase in MICs was conferred by lsaOG versus wild-type LM2301 or LM2301(pWM401) while lsaV conferred an increase in MICs of Q-D (from 0.75 to 1 μg/ml to 3 μg/ml), CLI (from 0.094 to 0.125 μg/ml to 3 to 4 μg/ml) and of DAL (from 32 to 128 μg/ml), consistent with the results with the E. faecalis lsa disruption mutant, TX5332. MICs of quinupristin were the same for LM2301 and LM2301 containing the recombinant plasmids, as was also the case with TX5332 (data not shown).
FIG. 1.
Susceptibility of E. faecalis and L. lactis derivatives to Q-D, DAL, and CLI. Rows 1 to 6 show the lsa gene constructs and nucleotide differences of lsaOG (ca. 2-kb lsa from OG1RF), lsaV (ca. 2-kb lsa from V583), lsaOG5′V3′ (hybrid lsa with 5′ half of OG1RF and 3′ half of V583), lsaV5′OG3′ (hybrid lsa with 5′ half of V583 and 3′ half of OG1RF), lsaOG-M (mutagenized lsaOG at positions −131 and −133 where G and T were replaced with A's [and four unintended changes were also found]), and lsaV-M (mutagenized lsaV at positions −131 and −133 where A's were replaced with G and T, respectively) cloned into pWM401 and their influence on susceptibility of TX5332 (E. faecalis OG1 lsa disruption mutant) (A) and L. lactis LM2301 (B) to Q-D, DAL, and CLI. n.t., nucleotide.
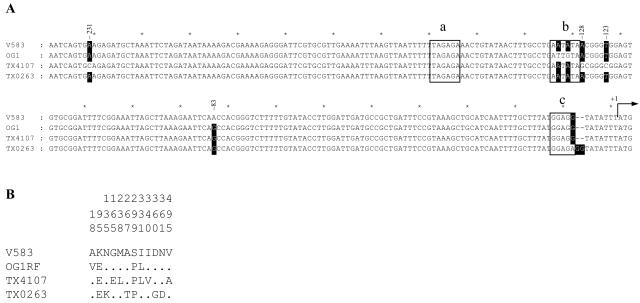
In an effort to understand these differences, we determined the nucleotide sequence of lsaOG and lsaV, which showed that the ca. 2-kb lsaOG (accession no. AY587982) fragment differed by 22 nucleotides from the lsaV sequence (Fig. 1, rows 1 and 2, and 2A). In the upstream region, we found differences at positions −131 and −133 (relative to the predicted ATG start codon), a possible strong promoter region for lsaV, where the A's present in lsaV were replaced with a G and a T in lsaOG (Fig. 2A), and at −83 where A in lsaV was replaced with a G in lsaOG. The peptide sequence of LsaOG also showed 4 amino acid changes versus LsaV; two of those are in the N-terminal half (V18 and E95 in OG1 versus A18 and K95 in V583) while the other two amino acid differences are in the C-terminal half (P299 and L331 in OG1 versus S299 and I331 in V583) (Fig. 2B).
FIG. 2.
Sequence differences among four strains of E. faecalis. (A) Region upstream (from −239 to +1) of lsa (including putative start codon) derived from E. faecalis strains V583, OG1RF, TX4107 (representative of feed clone), and TX0263 (a human clinical isolate). The nucleotide differences are highlighted (black with white letters). The nucleotide sequence from −309 to −240 is not shown, as it was identical among all strains. Predicted (from V583) −35 (a), −10 (b), and ribosomal binding sites (c) of V583 are marked by boxes. (B) Amino acid differences encoded by the lsa alleles. The amino acids that are shared by all of the strains are not shown. The amino acids present in E. faecalis V583 at each of the variable sites are shown (B). The position of each variable amino acid within the sequenced fragment is shown in the numbers above the amino acids, read vertically. The sequence of lsa from TX4107 is identical to that of TX4108, TX4109, and TX4110, and the sequences of TX0263 and TX0271 are also identical.
In an effort to determine the influence of the difference in these three regions of lsaOG versus lsaV, we first exchanged the two halves of lsa. The hybrid lsa fragment of pTEX5333.09 (containing lsaV5′OG3′) (Fig. 1A, row 4) fully restored the MICs of Q-D (from 0.75 to ≥32 μg/ml), CLI (from 0.125 to 0.5 μg/ml to 32 μg/ml), and DAL (from 16 μg/ml to 256 to >512 μg/ml) to levels equivalent to those of lsaV (Fig. 1, row 2). Similarly, this plasmid in LM2301 also conferred an increase in MICs of Q-D similar to the MIC levels of these drugs against LM2301 containing cloned lsaV (Fig. 1B, row 4), with slightly less increase for DAL (64 versus 128 μg/ml for lsaV). This suggests that the presence of A's at the −131, −133, and −83 positions of lsaV and the two amino acid changes present in the 5′ half are sufficient to fully complement the lsa disruption in TX5332 as well as cause the increased MICs of Q-D and CLI in LM2301 (Fig. 1, row 4).
TX5332 complemented with a second hybrid fragment (pTEX5333.08 containing lsaOG5′V3′) showed a moderate increase in MICs of Q-D and CLI (from 0.75 and 0.125 μg/ml, respectively, to 8 to 12 μg/ml for both drugs) (Fig. 1A, row 3) and higher MICs of DAL (from 16 to >512 μg/ml). This indicates that the amino acid changes in the 3′ half of lsa can also contribute to resistance. LM2301 containing cloned lsaOG5′V3′ showed little to modest increases in MICs relative to lsaOG of Q-D (1 versus 1 to 1.5 μg/ml), DAL (16 versus 32 μg/ml), and CLI (0.09 to 0.12 versus 0.38 μg/ml) (Fig. 1B, row 3), implying that the host background can also influence the effect of the lsa gene.
To explore the independent contribution of the nucleotide changes at the −131 and −133 positions (which could generate a strong promoter in V583) versus the changes in the coding region, we mutagenized the lsaOG and lsaV fragments with primers directed toward this region. The lsa nucleotide sequences of two mutagenized fragments, pTEX5333.12 containing lsaV-M, and pTEX5333.13, containing lsaOG-M, were reconfirmed in both directions. No nucleotide changes were found in lsaV-M except the ones targeted at nucleotide positions −131 and −133 (Fig. 1, row 6). However, in lsaOG-M (Fig. 1, row 5), four unintended nucleotide changes were found (one upstream of lsa at nucleotide position −109, one at +1248 [silent] in the lsa coding region, and two downstream of the putative stop codon at positions +1517 and +1572). We did not evaluate the specific effect of each unintended nucleotide change.
The lsa disruption mutant, TX5332, complemented separately with lsaV-M and lsaOG-M, showed full restoration of MICs of Q-D (from 0.75 to ≥32 μg/ml) and DAL (from 16 to ≥512 μg/ml) (Fig. 1A), similar to MICs of Q-D and DAL for wild-type E. faecalis OG1 and the lsa disruption mutant complemented with lsaV. Both lsaV-M and lsaOG-M also conferred an increase in MICs of CLI (from 0.12 to 0.5 μg/ml to 16 to 24 μg/ml); however, this increase was less than that seen with the cloned lsaV (from 0.12 to 0.5 μg/ml to 32 μg/ml) (Fig. 1A). These results show that the changes in the coding region, as well as those in the upstream region, can independently influence resistance to Q-D, DAL, and/or CLI.
In L. lactis, both lsaOG-M and lsaV-M increased the MICs of Q-D (from 0.75 to 1 μg/ml to 2 to 3 μg/ml), DAL (from 32 μg/ml to 64 to 128 μg/ml), and CLI (from 0.09 to 0.12 μg/ml to 0.5 to 1.5 μg/ml) (Fig. 1B); the latter are somewhat lower than those conferred by lsaV or lsaV5′OG3′, consistent with the effect of these plasmids on CLI MICs when present in TX5332. Preliminary reverse transcription (RT)-PCR results (unpublished data) implied much lower expression of lsa in lsaV-M than lsaOG-M, consistent with the increased levels of drug resistance of lsaOG-M in L. lactis. However it is also possible that some of the unintended nucleotide changes may influence RNA stability.
Even though MIC comparisons were done to derivatives also grown in KAN with and without CHL, we also tested other E. faecalis OG1-derived strains, TX5076 (30), TX5243 (18), and TX5248 (18), which have genes inactivated with the help of suicide vector pTEX4577 containing aph(3′)-llla (conferring KAN resistance), the same vector used in construction of TX5332 (6, 25). All three mutants showed MICs of Q-D and CLI on MHA plus KAN (2,000 μg/ml) plates similar to those determined with wild-type E. faecalis OG1 on MHA, indicating that the presence of KAN in the test media did not decrease the Q-D and CLI MICs (data not shown) for KAN-resistant mutants. Similarly, both LM2301(pWM401) in MHB plus CHL (8 μg/ml) and LM2301 in MHB showed equivalent MICs of test drugs (Fig. 1B), suggesting that the presence of the shuttle vector or CHL (8 μg/ml) in test media did not affect the MICs of all of the drugs tested, except for a slight decrease in MICs of DAL with pWM401.
Q-D- and CLI-susceptible E. faecalis isolates.
To investigate the Q-D and CLI susceptibility noted among several other E. faecalis isolates, their lsa sequences were analyzed. Nucleotide sequences of lsa from a group of animal feed isolates TX4107 (accession no. AY737525) to TX4110 (MICs: Q-D, 0.5 μg/ml; CLI, 0.38 to 0.5 μg/ml; DAL, 8 μg/ml; quinupristin, 8 to 16 μg/ml) were found to be identical with each other; by PFGE, TX4107 and TX4108 appeared clearly related (a three-band difference) while TX4109 and TX4110 are possibly related to TX4107 and TX4108 (four- to five-band differences). The two clinical isolates TX0263 (accession no. AY737526) and TX0271 (MICs for both: Q-D, 1 μg/ml; CLI, 2 to 4 μg/ml; DAL, 128 to 256 μg/ml; quinupristin, 8 to 16 μg/ml) had identical nucleotide sequences with each other; however, they were different (a >7-band difference) from each other and from the animal feed isolate by PFGE. In the lsa allele of the feed isolates (e.g., TX4107) (Fig. 2A), four and five nucleotide changes were found versus the upstream sequence of the lsa alleles of V583 and OG1, respectively. Like lsaV, the feed allele has −131A and −133A and, like lsaOG, −83G; at the other upstream positions, where the feed lsa allele differs, lsaV and lsaOG are the same (Fig. 2A). In the coding region of the feed isolates, Lsa differs from LsaV by 7 amino acids (three in the N-terminal half and four in the C-terminal half) (Fig. 2B); three of these amino acids (at positions 95, 299, and 331) are also found in LsaOG. There were 23 silent nucleotide changes in lsaTX4107 versus lsaV.
The lsa sequence of the susceptible human isolate (TX0263), in comparison to lsaV, had −83G (like lsaOG) and one nucleotide difference in the putative ribosomal binding site, followed immediately by two additional nucleotides (Fig. 2A). In the Lsa coding region, there were five amino acid differences versus LsaV (two in the N-terminal half and three in the C-terminal half) with K95E (like LsaOG and the feed isolate), S299P (like LsaOG and the feed isolate), D360G, and N361D (Fig. 2B). In lsaTX0263, there were 15 silent nucleotide changes versus lsaV. We did not find any frameshift mutations in lsa of TX4107 or TX0263, as has been reported previously as the cause of susceptibility of E. faecalis isolates to lincosamide and streptogramin A compounds (7). While we note the consistent changes in all susceptible strains at −83 and residues 95 and 299, these changes have not been further evaluated.
DISCUSSION
We previously identified 34 putative transporter system components in the E. faecalis V583 sequence at TIGR and made mutants of 31 of these in strain OG1, some of which showed increased susceptibility to various compounds including novobiocin, pentamidine, daunorubicin, norfloxacin, Q-D, and CLI (6). One of these mutants, abc23, with reduced susceptibility to Q-D, DAL, and CLI, was renamed lsa when we showed, with an E. faecalis OG1 disruption mutant as well as with complementation, that this gene influenced resistance to lincosamide (e.g., CLI) and streptogramin A compounds (e.g., DAL) (25). We also described the presence of conserved elements of ABC proteins, including the Walker A and B motifs, which are involved in the binding and hydrolysis of ATP, as well as an ABC signature sequence thought to be involved in energy transduction (11, 25). Lsa does not have any obvious transmembrane domains (7, 25), and it appears that the predicted two ATP-binding regions of Lsa represent a fused single protein, as has been previously described for Msr(A) (21). In the literature, both Msr(A) and Lsa, along with Vga(B), Vga(A), Orf5, and MsrC, have been classified into the class 2 type of ABC proteins which have duplicated fused ABC domains and lack identifiable transmembrane domains, and it has been suggested that these systems may be involved in cellular processes other than transport (4, 20).
In the present study, we found that, in contrast to cloned lsa from V583, the cloned lsa allele of OG1 did not increase resistance to Q-D, CLI, and DAL when introduced into TX5332 or L. lactis. This is despite the fact that disruption of lsa eliminated resistance to these antibiotics in OG1. Sequencing found several differences between the OG1 and V583 lsa loci, including the N-terminal half of Lsa, where two amino acid changes were identified, the first of which (V18A) is located before the first conserved Walker A motif (amino acid positions 40 to 48) and the second of which (E95K) is located between the Walker A and Walker B motifs (amino acid positions 141 to 144) of the first ABC domain. The C-terminal Lsa half also contains two amino acid changes (P299S and L331I), both of which are located in the interdomain sequences before the Walker A (amino acid positions 347 to 351) motif of the second ABC domain. In considering other ABC homologues, we note that in a study on P-glycoprotein, the replacement of a small number of residues near the Walker B motif of nucleotide binding domain 1 by the corresponding nucleotide binding domain 2 residues caused profound alterations of the drug resistance profile both in mammalian and yeast cells, showing the importance of amino acids and their positions for proper functioning of nucleotide binding domain 1 (3). Also, in another published study (20), the functionality and importance of the N- and C-terminal domains of Msr(A), both of which are essential, were demonstrated when independently cloned domains did not function when introduced into S. aureus strain RN4220 separately and did not complement if expressed from separate plasmids, suggesting the essential structural or functional role of the Q-linker region that joins the two ATP-binding domains (1, 20). These observations suggested that the changes we found may indeed influence the function of Lsa.
While we did not interconvert specific residues, our experiments show that, in the face of what would be a poor −10 region (ATTGTA in OG1), the amino acid sequence encoded by lsa from OG1 did not cause an increase in MICs to the agents tested when cloned and inserted into either TX5332 or L. lactis LM2301. Preliminary RT-PCR results (unpublished data) also indicate considerably less lsa transcript from an OG1-like promoter region than a V583-like promoter region. However, even this predicted a poor −10 region which, together with the amino acids changes found in LsaV, can substantially restore resistance (lsaV-M) (Fig. 1, row 6). The higher MICs with lsaOG5′V3′ versus lsaOG indicate that the 3′ amino acids can exert an effect independent of the 5′ half of the locus, although the effect is less than that when both the 5′ and 3′ coding regions of V583 occur together. Conversely, when present together with the strong promoter-like region found in V583, the amino acids found in LsaOG restored MICs (lsaOG-M) (Fig. 1, row 5) almost to the levels of wild-type lsaV, and the presence of the V583 upstream sequence plus the V583 5′ half were sufficient to fully increase the MIC to those levels conferred by the entire lsaV locus. A contribution of the N-terminal amino acid differences, independent of the different upstream nucleotides, is suggested by the higher MICs of lsaV-M (Fig. 1, row 6) versus lsaOG5′V3′ (Fig. 1, row 3), which differ at residues 18 and 95, although these alleles also differ at the −83 position.
Although the cloned lsa locus from OG1 does not confer resistance, we know that disruption of lsa in OG1 causes loss of resistance. Since it appears very unlikely from sequence analysis that there is a polar effect, this suggests that another promoter or an upstream cis-acting element is functional in the natural setting in OG1, causing its resistance to Q-D, CLI, and DAL. Additional sequence analysis of lsa and preliminary mRNA and RT-PCR experiments also indicate that there is another potential promoter further upstream of lsa, which may drive expression of the native gene. Future studies, including mapping of the transcriptional start site(s), will assess these potential promoter regions and transcriptional differences in the wild-type strains versus recombinant genes.
We also analyzed other isolates of Q-D-susceptible E. faecalis. In a study by Dina et al. (7), it has been found previously with Q-D- and CLI-susceptible E. faecalis that frameshift mutations in lsa resulting in a premature termination codon were the cause of susceptibility in these isolates. In the present study of six Q-D- and CLI-susceptible E. faecalis isolates, among which we found two clonal types, we did not detect frameshift mutations or premature termination codons in our isolates. However, in addition to unique changes not shared with LsaOG or each other, there were common amino acids (95E and 299P) found among LsaOG and the representative types, LsaTX4107 and LsaTX0263, which were different than that of LsaV and, additionally, 331L shared by LsaOG and LsaTX4107. These two lsa loci also share the −83G with lsaOG, a position we did not study here. Whether these or other changes contribute to the lowered MICs for Q-D and CLI resistance in these isolates is also an area of further interest.
In conclusion, we demonstrated that changes in different regions of lsa (the upstream region of lsa which can create a strong promoter, the rest of the lsa 5′ half, and the lsa 3′ half) can influence the MICs for Q-D, CLI, and DAL. We have also shown that lsa is functional in the L. lactis LM2301 strain where changes in all three regions influenced the MICs of Q-D, CLI, and DAL to various degrees. Further studies of mRNA expression, start site mapping of lsaOG and lsaV, and efflux experiments to determine whether active transport occurs versus ribosomal protection, as suggested may occur with class 2 ABC systems (20), should help clarify the mechanisms by which these phenotypic differences occur.
Acknowledgments
This work was supported in part by NIH grant R37 AI 47923 from the Division of Microbiology and Infectious Diseases, NIAID, to B.E.M.
We thank Fang Teng for helpful discussions, Sreedhar R. Nallapareddy for help with preparation of computer graphics, and S. Simjee (U.S. Food and Drug Administration) for providing some of the Q-D- and CLI-susceptible isolates.
REFERENCES
- 1.Allignet, J., and N. El Solh. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202:133-138. [DOI] [PubMed] [Google Scholar]
- 2.Allington, D. R., and M. P. Rivey. 2001. Quinupristin/dalfopristin: a therapeutic review. Clin. Ther. 23:24-44. [DOI] [PubMed] [Google Scholar]
- 3.Beaudet, L., and P. Gros. 1995. Functional dissection of P-glycoprotein nucleotide-binding domains in chimeric and mutant proteins. Modulation of drug resistance profiles. J. Biol. Chem. 270:17159-17170. [DOI] [PubMed] [Google Scholar]
- 4.Dassa, E., and P. Bouige. 2001. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152:211-229. [DOI] [PubMed] [Google Scholar]
- 5.Datta, A. K. 1995. Efficient amplification using ‘megaprimer’ by asymmetric polymerase chain reaction. Nucleic Acids Res. 23:4530-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, D. R., J. B. McAlpine, C. J. Pazoles, M. K. Talbot, E. A. Alder, C. White, B. M. Jonas, B. E. Murray, G. M. Weinstock, and B. L. Rogers. 2001. Enterococcus faecalis multi-drug resistance transporters: application for antibiotic discovery. J. Mol. Microbiol. Biotechnol. 3:179-184. [PubMed] [Google Scholar]
- 7.Dina, J., B. Malbruny, and R. Leclercq. 2003. Nonsense mutations in the lsa-like gene in Enterococcus faecalis isolates susceptible to lincosamides and streptogramins A. Antimicrob. Agents Chemother. 47:2307-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duh, R. W., K. V. Singh, K. Malathum, and B. E. Murray. 2001. In vitro activity of 19 antimicrobial agents against enterococci from healthy subjects and hospitalized patients and use of an ace gene probe from Enterococcus faecalis for species identification. Microb. Drug Resist. 7:39-46. [DOI] [PubMed] [Google Scholar]
- 9.Friesenegger, A., S. Fiedler, L. A. Devriese, and R. Wirth. 1991. Genetic transformation of various species of Enterococcus by electroporation. FEMS Microbiol. Lett. 63:323-327. [DOI] [PubMed] [Google Scholar]
- 10.Li, X., G. M. Weinstock, and B. E. Murray. 1995. Generation of auxotrophic mutants of Enterococcus faecalis. J. Bacteriol. 177:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 12.Malathum, K., K. V. Singh, G. M. Weinstock, and B. E. Murray. 1998. Repetitive sequence-based PCR versus pulsed-field gel electrophoresis for typing of Enterococcus faecalis at the subspecies level. J. Clin. Microbiol. 36:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grows aerobically; approved standard, 6th ed. NCCLS document M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 16.National Committee for Clinical Laboratory Standards. 2003. MIC testing—supplemental tables. NCCLS document M100-S13 (M7). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 18.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rende-Fournier, R., R. Leclercq, M. Galimand, J. Duval, and P. Courvalin. 1993. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob. Agents Chemother. 37:2119-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds, E., J. I. Ross, and J. H. Cove. 2003. Msr(A) and related macrolide/streptogramin resistance determinants: incomplete transporters? Int. J. Antimicrob. Agents 22:228-236. [DOI] [PubMed] [Google Scholar]
- 21.Ross, J. I., E. A. Eady, J. H. Cove, and S. Baumberg. 1995. Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene 153:93-98. [DOI] [PubMed] [Google Scholar]
- 22.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 25.Singh, K. V., G. M. Weinstock, and B. E. Murray. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 46:1845-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh, P. M., and L. L. McKay. 1982. Restriction endonuclease analysis of the lactose plasmid in Streptococcus lactis ML3 and two recombinant lactose plasmids. Appl. Environ. Microbiol. 43:1006-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. M. David, J. G. Scidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Green Publishing Associates, Brooklyn, N.Y. [DOI] [PubMed]
- 29.Wirth, R., F. An, and D. B. Clewell. 1987. Highly efficient cloning system for Streptococcus faecalis: protoplast transformation, shuttle vectors, and applications, p. 25-27. In J. J. Ferretti and R. Curtiss III (ed.), Streptococcal genetics. American Society for Microbiology, Washington, D.C.
- 30.Xu, Y., L. Jiang, B. E. Murray, and G. M. Weinstock. 1997. Enterococcus faecalis antigens in human infections. Infect. Immun. 65:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]