Abstract
A fosmid library from genomic DNA of Streptomyces viridochromogenes DSM 40736 was constructed and screened for the presence of genes known to be involved in the biosynthesis of phosphinothricin tripeptide (PTT). Eight positives were identified, one of which was able to confer PTT biosynthetic capability upon Streptomyces lividans after integration of the fosmid into the chromosome of this heterologous host. Sequence analysis of the 40,241-bp fosmid insert revealed 29 complete open reading frames (ORFs). Deletion analysis demonstrated that a minimum set of 24 ORFs were required for PTT production in the heterologous host. Sequence analysis revealed that most of these PTT genes have been previously identified in either S. viridochromogenes or S. hygroscopicus (or both), although only 11 out of 24 of these ORFs have experimentally defined functions. Three previously unknown genes within the cluster were identified and are likely to have roles in the stepwise production of phosphonoformate from phosphonoacetaldehyde. This is the first report detailing the entire PTT gene cluster from any producing streptomycete.
The nonproteinogenic amino acid phosphinothricin is a structural glutamate analogue and potent inhibitor of glutamine synthetase, an enzyme central to nitrogen regulation in some plant cell types, making it an effective and widely used herbicide (4, 26). This compound is notable because it contains a reduced phosphorus center resulting from two direct carbon-to-phosphorus (C-P) bonds and is the only known naturally occurring compound in which the C-P-C bond motif is found. Phosphinothricin is produced by at least three species of actinomycete as a component of non-ribosomally synthesized peptides. These include phosalacine, produced by Kitasatosporia phosalacinea (35) and phosphinothricin tripeptide (PTT), also known as bialaphos, which is produced by Streptomyces hygroscopicus ATCC 21705 (43) and Streptomyces viridochromogenes DSM 40736 (4).
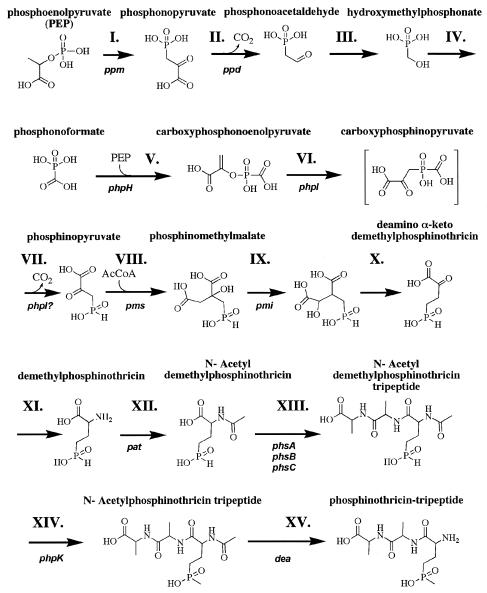
Research interest in PTT biosynthesis stems from both its herbicidal activity and the incorporation of the unique phosphinic acid moiety. Previous studies in either S. hygroscopicus or S. viridochromogenes have shown that the biosynthesis of PTT involves more than 13 discrete enzymatically catalyzed reactions, (Fig. 1) linked to a chromosomal gene cluster about 35 kb in length (50). Many of these biosynthetic steps have been accounted for, including those involved with nonribosomal peptide synthesis (Fig. 1, step XIII), the formation of both C-P bonds (steps I and XIV) and those with remarkable homology to portions of the tricarboxylic acid cycle (steps VIII-X). PTT biosynthesis has consequently become one of the only models for reduced-phosphorus antibiotic production. Important biosynthetic questions, however, remain regarding the nature of PTT biosynthesis despite the elucidation of most of the steps. For example, the genes and corresponding enzymes involved in the stepwise oxidation of phosphonoacetaldehyde, an early intermediate, to phosphonoformate (steps III and IV) are currently unaccounted for. Similarly, the mechanism of carboxyphosphonoenolpyruvate synthesis from phosphonoformate also remains largely uncharacterized (step V). Finally, the entire biosynthetic gene cluster has heretofore never been sequenced in its entirety from either producer.
FIG. 1.
Model of phosphinothricin tripeptide biosynthesis, adapted from Thompson and Seto (50). The steps referred to in the text are indicated by roman numerals. The genes sequenced here with equivalents to those with experimentally assigned function from previous work in S. viridochromogenes or S. hygroscopicus are listed below their corresponding steps.
To further our understanding of PTT biosynthesis and reduced-phosphorus biochemistry, we report here the intact cloning, sequencing, and analysis of the PTT biosynthetic gene cluster from S. viridochromogenes DSM 40736 and its expression in the heterologous host Streptomyces lividans 66.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions. The strains and plasmids used in this study are listed in Table 1. All streptomycete cultures were grown at 30°C unless otherwise indicated. S. viridochromogenes DSM 40736 and S. lividans 66 were propagated on ISP2 medium (Difco, Becton Dickinson Microbiology Systems, Sparks, Md.). For heterologous PTT production assays, S. lividans hosts were grown in 25 ml of S medium (34) supplemented with 10 mM alanine and 0.0001% CoCl2 incubated in 250-ml flasks equipped with glass beads (to provide mechanical disaggregation of mycelia) under vigorous agitation (250 rpm). All Escherichia coli strains were grown in Luria-Bertani (LB) liquid medium and on TYE solid medium with appropriate antibiotics (53). Chloramphenicol (12 μg/ml), streptomycin and spectinomycin in combination (35 μg/ml each), apramycin (50 μg/ml), and ampicillin (100 μg/ml) were used in solid and liquid media for the propagation of plasmids. Bioassays for PTT production were carried out with the sensitive strain Bacillus subtilis ATCC 6633 as previously done by Alijah et al. (1). B. subtilis spore suspensions were created by the nutrient exhaustion method (14).
TABLE 1.
Microorganisms and plasmids used in this study
| Strain or plasmid | Relevant characteristic | Source or referencea |
|---|---|---|
| Escherichia coli | ||
| DH10B | φ80dlacZΔM15/araD139 Δ(ara-leu)7697 ΔlacX74 galU galK rpsL deoR endA1 nupG recA1 mcrA Δ(mrr hsdRMS mcrBC) | 10 |
| DH5α | φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | 10 |
| DH5α/λpir | λpir/φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | 30 |
| GeneHogs(trfA) | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15/araD139 Δ(ara-leu)7697 ΔlacX74 galU galK rpsL deoR endA1 nupG recA1 trfA AmprfhuA::IS2 | Invitrogen |
| BW26678 | laclqrrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78/pKD46 | 6 |
| GM119 | dam-3 dcm-9 metB1 galK2 galT27 lacY1 tsx-78 supE44 thi-1 mel-1 tonA31 | 52 |
| WM3780 | dam-3 dcm-9 metB1 galK2 galT27 lacY1 tsx-78 supE44 thi-1 mel-1 tonA31 attHK::pJK202Δ(oriR6K-aadA)::Frt | This study |
| WM3321 | BL21(DE3)/pNR69 | 58 |
| Streptomyces viridochromogenes | Wild type (DSM 40736) | DSM |
| Streptomyces lividans | ||
| 66 | Wild type (NRRL B-16148) | ARS culture collection |
| WM 4366 | S. lividans with pJVD9 integrated into φC31 attB site | This study |
| WM 4367 | S. lividans with pJVD9/fosmid 5-9G integrated into φC31 attB site | This study |
| WM4368 | S. lividans with pJVD9/fosmid 5-9GΔ(orf416-orf571′)::kan integrated into φC31 attB site | This study |
| Bacillus subtilis | ||
| ATCC 6633 | PTT-sensitive bioassay tester strain | ATCC |
| WM 4445 | Spontaneous PTTr mutant | This study |
| Plasmids | ||
| pAM34 | Ampr Strr cloning vector, IPTG-dependent ColE1 oriR, lac1q | 8 |
| pAH144 | Strr, oriR6K, HKattP integrating cloning vector | 11 |
| RP4 | Broad-host-range plasmid | 36 |
| pJK190 | 53,981-bp BamHI-BglII fragment of RP4 cloned into BamHI-cut pAH144 | This study |
| pJK202 | Deletion of two adjacent BstBI fragments of pJK190 | This study |
| pMP45 | PCR template for oriR6K amplification in pJVD1 construction | 38 |
| pOJ436 | Aprr, oriT, φC31 int, φC31 attP, cosvector | 5 |
| pWM357 | oriF, dual cosvector, Cmr | 60 |
| pJVD1 | pWM357 unique ClaI site cut and ligated to ClaI site-flanked oriR6K and HKattB cassette PCR product cloned from pMP45 with primers oriR6K/HK attB FOR and oriR6K REV | This study |
| pJVD8 | pAH144 cut with NotI/NcoI to remove oriR6K, T4 DNA polymerase blunted, and ligated to T4 DNA polymerase-blunted BamHI pAM34 fragment containing replicon, lac1q, Ampr Strr | This study |
| pJVD9 | pJVD8 SmaI unique site cut and ligated to pOJ436 DraI fragment containing Aprr, oriT, φC31 int, φC31 attP | This study |
| fosmid 5-9G | S. viridochromogenes genomic DNA cloned into pJVD1 fosmid vector; contains PTT biosynthetic genes | This study |
| pJVD9/fosmid 5-9G | pJVD9 fosmid 5-9G cointegrant via HKattP/attB site-specific recombination | This study |
| pJVD9/fosmid 5-9GΔ(orf416-orf571′)::kan | pJVD9-fosmid 5-9G cointegrant via HKattP/attB site-specific recombination with orf416-orf571′ replaced by Kanr marker | This study |
| pWM303 | Pir-dependent cloning vector | 29 |
| pPB37 | oriR6K, bla-containing PCR product from pWM303 obtained with primers ori-BglII and bla-BamHI: circularized after cutting with BamHI and BglII | This study |
| pJK78 | Unique XbaI site in pPB37 destroyed by cutting with XbaI followed by treatment with T4 DNA polymerase and dNTPs | This study |
| pJK90 | AscI-cut PCR product obtained from GeneJumperOriV template and primer MuR-A/E/B (anneals to both sides of replicon) cloned into AscI site of pJK78 | This study |
| Supercos1 | Kmr cosmid vector | Stratagene |
| pJK91 | Replacement of BamHI fragment from pJK90 with BamHI-cut PCR product obtained from Supercos1 template and primers Supercos-aphF and Supercos-aphR | This study |
| pIJ702 | ThiorStreptomyces cloning vector | 23 |
| pJK92 | NotI/XbaI-cut PCR product obtained from pIJ702 template with primers Tsr-L and Tsr-R cloned into NotI/XbaI sites of pJK91 | This study |
| pJK93 | AscI-cut PCR product obtained from pJK93 template and primer MuR-A/M (anneals to both sides of replicon) cloned into AscI site of pJK90 | This study |
| pJK94 | Unique BglII site in pJK93 destroyed by cutting with BglII followed by treatment with T4 DNA polymerase and dNTPs | This study |
| pJK95 | AscI-cut PCR product obtained from pJK94 template and primer MuR-A/E/B (anneals to both sides of replicon) cloned into AscI site of pJK90: donor plasmid for mini-Mu-JK4790 | This study |
DSM, Deutsche Sammlung von Mikroorganismen, Braunschweig, Germany; ARS, U.S. Department of Agriculture Agricultural Resource Service, Peoria, Ill.; ATCC, American Type Culture Collection, Manassas, Va.
Construction of the Escherichia coli conjugative donor strain WM3780.
A plasmid-independent, DNA methylase-deficient conjugative donor, E. coli WM3780, was constructed to facilitate the introduction of DNA into Streptomyces strains. This strain was constructed by insertion of plasmid pJK202, which carries the tra functions from plasmid RP4, into the chromosome of the DNA methylase-deficient E. coli strain GM119 (52). Plasmid pJK202 was constructed in several steps as follows (see also Table 1). Initially, a 53,981-bp BamHI-BglII fragment comprising the majority of RP4 (36), except the Tn1-oriV region (nucleotides 6873 to 12991 based on the complete sequence of plasmid RP4, GenBank accession L27758), was cloned into the BamHI site of the integrating plasmid pAH144 (11) to create pJK190. Subsequently, two internal BstBI fragments comprising the IS21-aphA region of RP4 (RP4 nucleotides 36198 to 39431) were deleted, giving rise to pJK202. pJK202 was then inserted into the GM119 chromosome by site-specific recombination into the HK phage attB site as described (11). Finally, the region containing the aadA gene and oriR6K originating from pAH144 were removed from the integrated plasmid by the oligonucleotide-directed method of Datsenko and Wanner (6) with primers pAH144delP1 and pAH144delP2 (Table 2); the kanamycin resistance cassette used in this construction was removed by flp-mediated recombination as described (6). The complete genotype of WM3780 is shown in Table 1; the full sequence of pJK202 has been submitted to GenBank.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | DNA sequencea |
|---|---|
| PnPy F2 | CCBGGCRHRMMVGACGARCCCCAGCA |
| PnPy R1 | CCSCCGRYRSWKTCRTGSRCVCCGTTGTYGA |
| Pepmut F1 | GCCBVTBVTYKTBGAYGGHGACACSGGR |
| Pepmut R2 | CGGCVCGSAKTHKGTGRTTSGCSYARAT |
| phsA FOR | ACCCGTACATGGCGTTACTC |
| phsA REV | CTCCAGATCTCACCCACCTG |
| phsB 5′ end FOR | AGGGTCAGTACTCGGTGTGG |
| phsB 5′ end REV | CGAAGTGGATGGTGTAGGTG |
| pmi FOR | ACGTCCGGATGAACTACCTC |
| pmi REV | CTACGGAGGGGAATTCAGGT |
| pepmut FOR | GCTGAAGAACCTGCTGCAC |
| pepmut REV | TGTAGCGGTTCTCGTCCTG |
| pat FOR | CCGGGGACGACTTCTTCT |
| pat REV | GCCGTGCAGGTACAGCAG GGCGCGCCATCGATGGTGCACTTTAGGTG |
| oriR6K/HK attB FOR | AAAAAGGTTGAGTCGCTAACTGTCAGCCCGCC |
| oriR6K REV | GGCGCGCCATCGATAATTCACTGGG GGCAATTC |
| F5-9G, downstream | TACCGTCTGCTATGGTCCTCCGTCTCCT |
| KO F | GTGGGTGAGTGTAGGCTGGAGCTGCTTCG |
| F5-9G, downstream | GAGAATTCGCGGCCGCATAATACGACT |
| KO R | CACTATAGGTTCCGGGGATCCGTCGACCTG |
| RS1 check F | AACACTTAACGGCTGACATGG |
| RS1 check R | GAGTAGCCGCTTTCAAATGG |
| 8BT7F | GCGGTACTGTCATGGATGC |
| 8BT7R | CTAGTCCGGCCAGCAGTTC |
| 8BT3F | CCGTCGAGGTTGAAGGAGTA |
| 8BT3R | GATGAAGACCTACGCCAAGC |
| Mu-Seq L1 | CGATAAGCGCCTCTGTTCCT |
| Mu-Seq R1 | GGACTCTGGGGTTCGAAATG |
| Mu-Seq R5 | TCTATCGCCTTCTTGACGAG |
| pAH144 DEL P1 | GTAGGTCATTATTAGTCAAAATAAAATCATTTG TCGATTTCAATTTTGTCTGTGTAGGCTGGAGCTGC TTCG |
| pAH144 DEL P2 | TGAGCCTTTCGTTTTATTTGATGCCTGGCAGTTC CCTACTCTCGCATGGGATTCCGGGGATCCGTCG ACC |
| ori-BglII | CGCGCGAGATCTAATTCTGTCAGCCGTTAAG |
| bla-BamHI | CGCGCGGATCCTCTAGAGGCGCGCAGGTGG |
| MuR-A/E/B | GGCGCGCCGAATTCAGATCTGAAGCGGCGCACGAAAA ACG |
| MuR-A/M | GGCGCGCCACGCGTGAAGCGGCGCACGAAAAACG |
| Supercos-aphF | GGCGCGCCGGATCCTTGGCAGAACATATCCATCG |
| Supercos-aphR | GGCGCGCCGGATCCTCATTTCGAACCCCAGAGTC |
| Tsr-L | GGCGCGCCTCTAGACGGTGATCAAGGCGAATACT |
| Tsr-R | GGCGCGCCGCGGCCGCTCCGAGGAACAGAGGCGCTT |
Standard abbreviations are used: R = A or G, Y = C or T, M = A or C, K = G or T, S = C or G, W = A or T, H = A or C or T, B = C or G or T, V = A or C or G, and D = A or G or T.
Isolation of a PTT-resistant Bacillus subtilis mutant.
A 6-mm paper disk soaked with 9 μl (100 mg/ml) of commercially available PTT (Research Products Incorporated, Mt. Prospect, Ill.) was applied to a confluent lawn of Bacillus subtilis ATCC 6633 spores plated on minimal medium as done for the bioassays described above. Resistant mutants found growing in the zone of inhibition were streaked for purity and tested for the maintenance of the PTT-resistant phenotype.
DNA isolation and manipulation.
All cloning was performed by established methods (40). Endonucleases, T4 DNA polymerase, and T4 DNA ligase were purchased from Invitrogen (Carlsbad, Calif.) and New England Biolabs (Beverly, Mass.). Shrimp alkaline phosphatase was purchased from Roche Diagnostics GmbH (Mannheim, Germany). Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, Iowa). DNA fragments for cloning were isolated after gel purification with the Agarace enzyme (Promega, Madison, Wis.) according to the manufacturer's recommendations. Plasmids were isolated by the use of Qiagen (Valencia, Calif.) Miniprep or Maxiprep kits. Fosmids were isolated by CsCl gradient ultracentrifugation. For the isolation of high-molecular-weight chromosomal DNA, cultures of S. viridochromogenes were grown under the conditions published previously for protoplasting (48). These were incubated with vigorous agitation for 50 h prior to harvesting cells for lysis. Cells were lysed in TE25S buffer by combined proteinase K and sodium dodecyl sulfate treatment as outlined by Kieser et al. (24). Protein was removed from DNA suspensions by repeated extraction with phenol-chloroform before ethanol precipitation.
PCR amplifications involving Streptomyces DNA were performed with FailSafe PCR PreMix buffers (Epicentre, Madison, Wis.). Amplifications to screen exconjugants were performed via the colony PCR method established by Van Dessel et al. (51). Exconjugant screenings were routinely performed with Taq polymerase. The oligonucleotide PCR primers used in this study are listed in Table 2.
Construction and screening of an S. viridochromogenes genomic library.
S. viridochromogenes DNA was partially digested by Sau3AI, phosphatase treated, and ligated into the BamHI site of the fosmid vector pJVD1 (Table 1). This fosmid vector contains a low-copy-number origin of replication to ensure insert stability and can be modified, as described below, after cloning to allow introduction of new plasmid functions, e.g., the ability to be moved via conjugation and to integrate into the chromosome of various Streptomyces strains via the φC31 site-specific recombination system. Adding these functions to the plasmid vector after library construction maximizes the size of plasmid inserts obtainable with this fosmid vector, which is limited by the amount of DNA that can be contained in phage lambda particles.
Fosmid constructs were packaged with Gigapack III XL (Stratagene, La Jolla, Calif.), and the resulting phage were used to transduce E. coli DH10B to chloramphenicol resistance. Clones containing genes associated with PTT biosynthesis were isolated by PCR screening with the degenerate primer sets PnPy F2 and R1 and Pepmut F1 and R2 to detect the phosphonopyruvate decarboxylase and phosphoenolpyruvate phosphomutase genes, respectively. Positive clones were further characterized by PCR to detect DNA fragments internal to previously published S. viridochromogenes gene sequences associated with the PTT biosynthetic pathway, namely phsA, phsB, pmi, ppm, and pat with primers phsA FOR and phsA REV, phsB 5′ end FOR and phsB 5′ end REV, pmi FOR and pmi REV, pepmut FOR and pepmut REV, and pat FOR and pat REV, respectively (Table 2). Fosmids containing PTT biosynthetic genes were then sequenced through the cloning junction with the BigDye terminator kit v. 3.0 (ABI Prism, Foster City, Calif.), to screen for constructs where cloning did not disrupt known PTT biosynthetic genes.
DNA sequencing and analysis.
To provide priming sites for DNA sequencing reactions, fosmid 5-9G (Table 1) was mutagenized with transposon mini-Mu-JK4740 in in vitro reactions with Mu transposase as recommended (MJ Research, San Francisco, Calif.). Mini-Mu-JK4740 carries two antibiotic resistance markers that are functional in Streptomyces, the kanamycin resistance gene (aph) of Tn5 obtained from Supercos1 (Stratagene) and the thiostrepton resistance gene (tsr) of pIJ702 (23), as well as the conditional oriV replication origin to allow increasing the copy number of plasmids into which the transposon inserts (54). The transposon is carried on pJK95 and is flanked by BglII sites to allow its excision for use in in vitro transposition reactions. The construction of pJK95 is described in Table 1, and the sequence of the plasmid has been deposited in GenBank.
Sequencing reactions were performed at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois from primers (Mu-SEQ L1 and Mu-SEQ R1 or Mu-SEQ R5, Table 2) that read out of each side of the transposon. The sequence data from each insertion were compiled with Sequencher 4.0 (Gene Codes Co., Ann Arbor Mich.), and deduced open reading frames were analyzed with the BLAST (2), FASTA (37), and InterProScan (59) programs at NCBI and EMBL.
Construction of the pJVD9/5-9G and pJVD9/5-9GΔ(orf416-orf571)::kan integrating fosmids.
Fosmid clones derived from pJVD1 were modified to allow their insertion into the chromosome of appropriate Streptomyces species by retrofitting with plasmid pJVD9 (Table 1). Plasmid pJVD9 carries the Streptomyces phage φC31 integration and apramycin resistance determinants, oriT site from the conjugal plasmid RP4, and E. coli HK022 phage attachment site (attP), whereas pJVD1 carries the E. coli phage HK022 bacterial attachment site (attB). Site-specific recombination between HK022 attB and attP results in cointegration of the two plasmids and was performed in vitro as previously described for lambda phage recombination assays (33), except that HK022 integrase was substituted for the lambda equivalent. HK022 integrase was provided by cell extract from WM3321 (Table 1).
Extracts were obtained by inducing mid-log-phase cells of WM3321 with 1 mM isopropylthiogalactopyranoside (IPTG) for 4 h, resuspension of the cells in 50 mM Tris HCl-10% sucrose buffer at pH 7.4, and cell lysis with a French pressure cell. Lysates were cleared by centrifugation at 13,000 × g for 30 min and stored at −70°C. Recombination between pJVD9 and fosmid 5-9G gave rise to pJVD9/5-9G. The deletion of open reading frames 416 to 571′ in pJVD9/5-9G was carried out in E. coli with the PCR-mediated gene replacement technique of Datsenko and Wanner as described (6) with primers F5-9G and downstream KO Forward and Reverse (Table 2). Deletion of the desired region from pJVD9/5-9G gave rise to pJVD9/5-9GΔ(orf416-orf571′)::kan, the structure of which was confirmed by PCR with primers RS1Check F and RS1Check R.
Construction of S. lividans heterologous PTT-producing strains.
Plasmids pJVD9/5-9G and pJVD9/5-9GΔ(orf416-orf571′)::kan were transformed into the E. coli conjugal donor strain WM3780. Intergenic conjugation between E. coli donors and S. lividans germinating spores, after heat shock, was performed as previously described by Wohlleben and Pielsticker (57), except that 100 μl of an E. coli mid-logarithmic-phase culture was used as the donor and conjugation was allowed to proceed on nitrocellulose filter disks overnight at 37°C. S. lividans exconjugants were selected on TYE plates supplemented with apramycin and nalidixic acid (50 μg/ml each). Exconjugants were purified at least twice on selective medium with the same antibiotics before growing nonselectively for PTT production bioassays.
Detection of heterologous PTT production by S. lividans.
Broth cultures of S. lividans carrying PTT biosynthetic genes were assayed daily by disk diffusion bioassay as previously described (1). For subsequent analysis, cells were removed from bioactive cultures by centrifugation, and methanol was added to the supernatant to 70%. After chilling on ice, particulate matter was removed via centrifugation, and the supernatant was concentrated in vacuo. 31P nuclear magnetic resonance (NMR) analyses of concentrated supernatants were carried out in 20% D2O at the Varian Oxford Center for Excellence in NMR laboratory (University of Illinois at Urbana-Champaign) with a 5-mm Nalorac Quad probe equipped with a Varian Unity U500 spectrometer tuned for phosphorus at 202.28 MHz. The 31P NMR shift values reported have been externally referenced to an 85% phosphoric acid standard (0 ppm). The presence of PTT in bioactive supernatants was confirmed by the addition of genuine PTT and phosphinothricin (Research Products Incorporated, North Prospect, Ill.) to a final calculated concentration of 400 μg/ml each.
Nucleotide sequence accession number.
The sequence of the fosmid 5-9G insert containing the PTT biosynthetic gene cluster has been deposited in GenBank under accession number AY632461. The nucleotide sequences of pJK202 and pJK95 were compiled from known sequences and have been deposited in GenBank under accession numbers AY741093 and AY738638, respectively.
RESULTS
Cloning and identification of the PTT biosynthetic gene cluster.
A fosmid library of Streptomyces viridochromogenes DSM 40736 genomic DNA was constructed in E. coli, and 2,880 individual clones were screened for the presence of the ppm and ppd genes, which encode the PTT biosynthetic enzymes phosphoenolpyruvate phosphomutase and phosphonopyruvate decarboxylase, respectively. This effort resulted in the isolation of eight fosmids that were found to have overlapping BamHI digestion patterns, suggesting that each clone contained parts of the same chromosomal region. These fosmids were further characterized by PCR with primers designed to amplify internal fragments of other previously sequenced S. viridochromogenes PTT biosynthetic genes, including phsA, phsB, pmi, ppd, and pat. This specific set was chosen because previous restriction mapping and complementation analysis of mutants of both S. viridochromogenes and S. hygroscopicus indicated that these genes were probably distributed widely across the PTT biosynthetic gene cluster (15, 42, 55). Thus, if a single fosmid contained all of these genes, it would be a likely candidate to contain the entire gene cluster. Although most of the fosmids were found to contain all of the PCR-screened genes, subsequent sequencing of the cloning junction showed that many had disrupted known PTT biosynthetic genes. However, one clone, designated fosmid 5-9G, contained all known PTT genes that were screened for and none of them were disrupted, based on sequencing of the cloning junctions.
Heterologous expression of PTT biosynthetic genes in Streptomyces lividans.
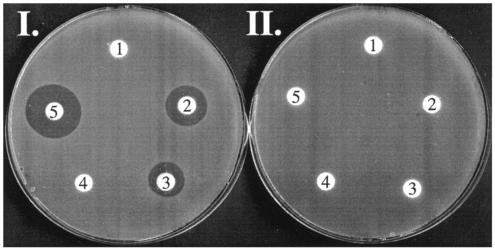
To find if fosmid 5-9G contained the intact PTT biosynthetic gene cluster, we inserted the plasmid into the Streptomyces lividans chromosome (after retrofitting with ΦC31 integration functions and an apramycin resistance determinant as described above) and assayed the recombinant strain for PTT biosynthesis. Bioassays showed that S. lividans with fosmid 5-9G integrated into the chromosome (WM4367) produced a bioactive compound that was not produced by S. lividans containing the vector alone (WM4366) (Fig. 2, plate I). To show that the bioactive compound was PTT and not another S. lividans natural product, the supernatant from this strain was tested against a PTT-resistant Bacillus subtilis 6633 mutant, which was not inhibited by either WM4367 supernatant or authentic PTT (Fig. 2, plate II). Authentic phosphinothricin (no alanyl residues) did not inhibit either strain.
FIG. 2.
Bioassay of WM4367 and WM4368 culture supernatants against PTT-sensitive (plate I) and PTT-resistant (plate II) Bacillus subtilis indicator strains. Disks designated 1 are soaked with WM4366 (pJVD9) supernatant; disks designated 2 are soaked with WM 4367 (pJVD9/5-9G) supernatant; disks designated 3 are soaked with WM 4368/pJVD9/5-9GΔ(orf416-orf571′)::kan supernatant; disks designated 4 are soaked with phosphinothricin (10 μg/ml); and disks designated 5 are soaked with PTT (10 μg/ml).
Further evidence for WM4367 PTT production was gathered by 31P NMR analysis. PTT production can be readily monitored by the strong chemical shift (ca. 42 ppm) in the 31P signal produced by the C-P-C bonding arrangement (45). 31P NMR analysis of bioactive WM4367 culture supernatants showed a distinct shift corresponding closely to the chemical shifts indicative of S. viridochromogenes and S. hygroscopicus PTT production (data not shown). In combination with the bioassay data, these data strongly suggest that fosmid 5-9G contains all the genes needed to confer PTT biosynthesis on S. lividans hosts.
DNA sequence analysis of fosmid 5-9G.
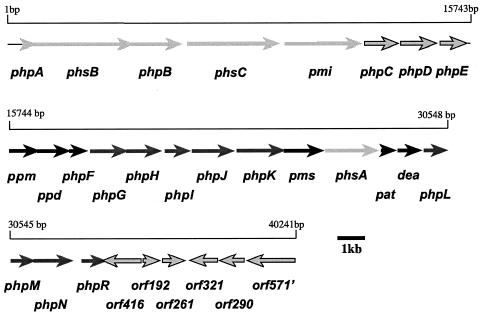
The 40,241-bp insert of fosmid 5-9G was found to contain 29 complete open reading frames (ORFs) and one partial ORF after double-strand DNA sequencing (Fig. 3). BLAST searches against GenBank revealed that most of these ORFs had been previously sequenced in either S. viridochromogenes or S. hygroscopicus or both. A portion of one ORF had been previously sequenced in S. hygroscopicus. Each gene was assigned a php locus name (for phosphinothricin tripeptide production), except that previously assigned names based upon experimental evidence were preserved. The ORFs found on fosmid 5-9G and their identity scores to Swiss-Prot homologs are presented in Table 3.
FIG. 3.
Open reading frame map of the fosmid 5-9G sequenced insert. ORFs with sequences previously published from S. viridochromogenes analysis alone are shown in light gray. ORFs with previously published sequences from S. hygroscopicus alone are shown in dark gray. ORFs with sequences previously published in both producers are shown in black. ORFs with sequences unique to this study are shown with dark outlines. The nonsequential base pair numbering at the end of phpL and the beginning of phpM indicates that the sequences of these genes overlap by 4 bp.
TABLE 3.
Summary of fosmid 5-9G open reading frames
| ORF | No. of amino acids | Protein homologya (Swiss-Prot accession no.) | % Amino acid identityb |
|---|---|---|---|
| phpA | 69 | S. viridochromogenes putative protein Orf1 (Q9KWY9) | 98.5 |
| Pseudomonas aeruginosa putative MbtH-like protein (Q9I169) | 51.5 | ||
| phsB | 1,189 | S. viridochromogenes peptide synthetase III (Q9KWY8) | 99.8 |
| phpB | 553 | S. viridochromogenes putative protein OrfM (Q8KLJ4) | 100 |
| Myxococcus xanthus putative membrane protein (Q9S433) | 35.9 | ||
| phsC | 1,086 | S. viridochromogenes peptide synthetase II (Q9K WY7) | 99.5 |
| pmi | 894 | S. viridochromogenes phosphinomethylmalate isomerase (AcnP) (Q9RIL3) | 99.8 |
| phpC | 395 | Amycolatopsis orientalis putative iron-dependent alcohol dehydrogenase (Q9XBE8) | 33.1 |
| phpD | 431 | No significant homology | N/A |
| phpE | 336 | Methanopyrus kandleri phosphoglycerate dehydrogenase homolog (Q8TYK0) | 39 |
| ppm | 313 | S. viridochromogenes phosphoenolpyruvate phosphomutase (O86937) | 96.8 |
| S. hygroscopicus phosphoenolpyruvate phosphomutase (BcpB) (P29247) | 83.3 | ||
| ppd | 397 | S. viridochromogenes phosphonopyruvate decarboxylase (O86938) | 100 |
| S. hygroscopicus phosphonopyruvate decarboxylase (BcpC) (O54271) | 84.3 | ||
| phpF | 184 | S. viridochromogenes putative protein OrfX (O86939) | 100 |
| S. hygroscopicus putative protein OrfX (Q9LCB1) | 91.1 | ||
| Pyrococcus furiosus nicotinamide-nucleotide adenylyltransferase (Q8U3K8) | 33.7 | ||
| phpG | 419 | S. hygroscopicus OrfZZ (364 aa) (Q9LCB3) | 84.2 |
| Aeropyrum pernix 2,3-BPGi phosphoglycerate mutase homolog (Q9YBI2) | 38.4 | ||
| phpH | 398 | S. hygroscopicus carboxyphosphonopyruvate synthase (BcpE, 333 aa) (Q54274) | 82.9 |
| Pseudomonas syringae pv. tomato enolase 1 homolog (Q886M3) | 38.9 | ||
| phpI | 296 | S. hygroscopicus carboxyphosphonopyruvate phosphonomutase (BcpA) (P11435) | 95.5 |
| phpJ | 466 | S. hygroscopicus unnamed PTT cluster protein (416 aa) (Q54272) | 90.1 |
| Sinorhizobium meliloti putative aldehyde dehydrogenase (Q92UV7) | 44.4 | ||
| phpK | 549 | S. hygroscopicus N-acetyldemethylphosphinothricin tripeptide P-methylase (BcpD) (Q54273) | 92.1 |
| pms | 440 | S. viridochromogenes phosphinomethylmalate synthase fragment (205 aa) (Q65169) | 100 |
| S. hygroscopicus phosphinomethymalate synthase (PmmS) (Q9LCB4) | 91.3 | ||
| phsA | 622 | S. viridochromogenes peptide synthetase 1 (Q56170) | 99.1 |
| pat | 183 | S. viridochromogenes demethylphosphinothricin N-acetyltransferase (Q57146) | 100 |
| S. hygroscopicus bialaphos resistance protein (Bar) (P16426) | 84.6 | ||
| dea | 299 | S. viridochromogenes N-acetylphosphinothricin tripeptide deacetylase (Q56171) | 99.3 |
| S. hygroscopicus bialaphos acetylhydrolase (Bah) (Q01109) | 87.8 | ||
| phpL | 253 | S. hygroscopicus orf1 gene product (Q03093) | 82.4 |
| Streptomyces coelicolor thioesterase II (Q9LAS9) | 37.6 | ||
| phpM | 260 | S. hygroscopicus orf2 gene product (Q03094) | 81 |
| Streptomyces avermitilis thioesterase (Q93H55) | 45.3 | ||
| phpN | 477 | S. hygroscopicus orf3 gene product (Q03095) | 74.6 |
| S. coelicolor putative transmembrane transport protein (Q9XAH4) | 60.4 | ||
| phpR | 261 | S. hygroscopicus bialaphos regulatory protein (BrpA) (Q01108) | 63.9 |
| orf416 | 416 | S. coelicolor putative transmembrane protein (Q9L223) | 51.7 |
| orf192 | 192 | Bacillus halodurans putative DNA-binding protein (Q9K8U3) | 33.7 |
| orf261 | 261 | Mycobacterium paratuberculosis putative formamidopyrimidine-DNA glycoslylase (AAS03645) | 54.2 |
| orf321 | 321 | S. coelicolor putative integral membrane protein (Q9RJN5) | 67.6 |
| orf290 | 290 | S. avermitilis putative acyl coenzyme A thioesterase (Q82MT5) | 94 |
| orf571′ | 571 | S. coelicolor putative helicase carboxyl terminus fragment (571 aa) (Q9KZJ3) | 94.7 |
aa, amino acids; 2,3-BPGi, bisphosphoglycerate independent.
Percent identity and closest homologs were based on FASTA searches conducted 21 May 2004. N/A, not available.
Deletion of ORFs downstream of phpR.
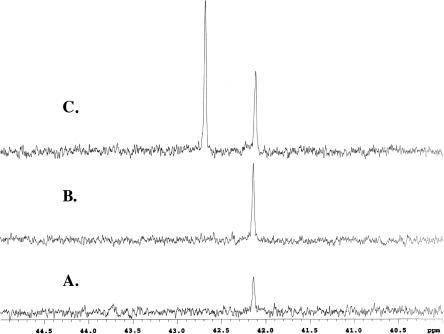
Based on the lack of homology to any known PTT biosynthetic genes and on the observation that no mutations causing PTT deficiency had been mapped to this region, we hypothesized that the ORFs downstream of the phpR gene would not be required for antibiotic production. To test this, the entire region downstream of phpR was deleted, and the shortened construct was moved into the chromosome of S. lividans as described above. The resulting strain (WM4368) was then assayed for PTT production by both bioassay and 31P NMR (Fig. 2, plates I and II, and Fig. 4, spectrum A, respectively). Addition of authentic PTT did not result in additional 31P signals but instead increased the intensity of the previously observed peak (Fig. 4, spectrum B). Addition of phosphinothricin produced a clearly distinguishable signal downfield of the PTT peak, indicating that the NMR signal in the bioactive supernatant was not from this structurally similar biosynthetic side product (Fig. 4, spectrum C). The results demonstrate that, like the full-length cosmid, the deleted construct also confers antibiotic production upon S. lividans. Therefore, a subset of the genes that we sequenced comprising the region from phpA through phpR represent the full PTT biosynthetic gene cluster.
FIG. 4.
31P NMR spectra of (A) concentrated WM4368 culture supernatant, (B) the same sample spiked with PTT with a concomitant gain in signal intensity at the same frequency, and (C) the sample shown in B spiked with phosphinothricin, showing the acquisition of a new phosphorus signal.
DISCUSSION
The data presented here define a ≈33.8-kb cluster of genes from S. viridochromogenes that confers the biosynthesis of PTT on a heterologous Streptomyces host. The results presented here indicate that most, if not all, genes required for PTT production are present on fosmid 5-9G. However, it is possible that other genes not present in our clone may have a role in PTT biosynthesis in the native host. If so, such genes would also have to be replaced by functionally equivalent genes in the heterologous host. Likewise, we cannot rule out the possibility that additional genes affect the level of PTT production. Indeed, a host carrying our plasmid with a deletion of the genes downstream of phpR appears to produce less PTT than a host with the full-length insert (Fig. 2). We are uncertain whether this is a function of the particular heterologous strain or whether a deleted ORF could have influenced PTT production in a nonessential manner. Many of the genes and enzymes of the proposed PTT biosynthetic pathway have been previously characterized and are readily identifiable in our sequence (Fig. 1 and 3). Other steps have not yet been solved, although possible candidates are suggested based on previous genetic studies and our analysis of as yet uncharacterized genes in our sequence. Together these data allow a plausible reconstruction of the PTT biosynthetic pathway.
The S. viridochromogenes genes and enzymes responsible for the initiation of phosphinothricin biosynthesis and production of phosphonoacetaldehyde (steps I and II), involving genes ppm and ppd, have been described previously (17, 32, 42) and were readily identified in our sequence; however, relatively little is known about the next two steps, the conversion of phosphonoacetaldehyde to hydroxymethylphosphonate (step III) and the subsequent oxidation of hydroxymethylphosphonate to phosphonoformate (step IV). S. hygroscopicus mutants blocked at both steps in the pathway could be complemented in trans by an unsequenced region of DNA closely linked to the ppm and ppd genes from either S. hygroscopicus (21, 31) or S. viridochromogenes (13). The arrangement of the complementing genes within the unsequenced region was refined by restriction to at least two separate ORFs upstream of ppm, with step III complementing DNA localizing directly upstream from the step IV complementing sequence (31).
Three open reading frames that localized to this region of the PTT cluster were identified by our analysis, phpC, phpD, and phpE. The location of phpE corresponds to the step IV complementing region and is a phosphoglycerate dehydrogenase homolog which could conceivably play a role in the oxidation of hydroxymethylphosphonate to phosphonoformate. Either or possibly both phpC and phpD could be assigned a function, based upon location, in the conversion of phosphonoacetaldehyde to hydroxymethylphosphonate (step III). phpC is an alcohol dehydrogenase homolog, whereas phpD is not homologous to any known proteins. Kuzuyama and Seto hypothesized that the unusual biochemistry involved in step III could be achieved by Baeyer-Villiger oxidation of phosphonoacetaldehyde (44), but homology to enzymes known to catalyze such reactions was not discovered here.
CPEP biosynthesis, hypothesized by Hidaka et al. (19) to involve the direct replacement of the phosphate group of PEP by phosphonoformate, was found to involve the bcpE gene product by DNA complementation of a S. hygroscopicus blocked mutant (27). The S. viridochromogenes homolog of this gene was found to be phpH.
Carboxyphosphoenolpyruvate (CPEP) is the substrate for CPEP phosphonomutase in a reaction that yields phosphinopyruvate as a product (combined steps VI and VII). It is unknown whether the decarboxylation of the presumed carboxyphosphinopyruvate intermediate (step VII) of this reaction is the result of enzyme catalysis or inherent product instability despite in vitro study of the enzyme (7); further enzymatic studies may help clarify the mechanism. CPEP phosphonomutase is encoded by bcpA in S. hygroscopicus (18), corresponding to phpI in S. viridochromogenes.
Phosphinopyruvate was found to be converted into phosphinomethylmalate by the addition of an acetate group (step VIII) from arising from acetyl coenzyme A by phosphinomethylmalate synthase (PmmS) in S. hygroscopicus (20, 46), a homolog of the S. viridochromogenes pms gene product (55). Phosphinomethylmalate isomerase, the pmi gene product, was previously shown to rearrange the structure of phosphinomethylmalate (15) (step IX) for subsequent oxidation and decarboxylation (step X) by an unknown enzyme into deamino-α-keto-demethylphosphinothricin (DAKDMPT). Comparison of the products of the genes found in the cluster based on homology to proteins of known function failed to identify a possible candidate for the enzyme responsible for the reaction predicted in step X. It was previously predicted that this reaction would take place by an enzyme similar to (or perhaps identical to) isocitrate dehydrogenase (15). Likewise, an aminotransferase homolog was also not found in the PTT biosynthetic gene cluster, which would likely be required for the conversion of DAKDMPT to demethylphosphinothricin (step XI). Unpublished results cited by Seto and Thompson (50) indicate that both of these steps could be catalyzed by microorganisms that do not produce PTT; thus, it is probable that these steps are catalyzed by ubiquitous, generic enzymes that can be found in most microorganisms.
It has previously been shown that the acetylation of demethylphosphinothricin (step XII) is catalyzed by demethylphosphinothricin N-acetyltransferase, corresponding to the pat (47) or the homologous S. hygroscopicus bar gene product (49). N-Acetyltransferase activity provides the substrate for the alanylation steps (collectively shown in Fig. 1 as step XIII, further discussed below) as well as a mechanism of detoxification against free phosphinothricin (25) (56) that may be produced within the cell. After N-acetylphosphinothricin tripeptide is nonribosomally synthesized, it has been shown that the phpK homolog from S. hygroscopicus, bcpD, encodes the P-methyltransferase that creates the second C-P bond (16, 22), yielding N-acetylphosphinothricin tripeptide (step XIV). The final step in PTT biosynthesis was found to be the deacetylation of N-acetylphosphinothricin tripeptide (step XV) by the dea gene product (39, 55) to produce the intact PTT molecule.
The addition of the two alanine residues to phosphinothricin, producing PTT, has been shown to occur by a nonribosomal peptide synthesis mechanism (9), and a large segment of the minimal gene cluster is dedicated to nonribosomal peptide synthesis activities (Fig. 1, step XIII) (for a review of nonribosomal peptide synthesis, see reference 28). The product of the first ORF found in the insert of fosmid 5-9G, phpA, formerly published in GenBank without analysis as orf1 by Schwartz et al. (accession number Y17268), is highly homologous to the product of an mbtH-like gene. MbtH homologs are typically found in association with nonribosomal peptide synthesis gene clusters, though the functions of these small peptides are not currently known. Thus, the presence of such a gene is expected in the PTT biosynthetic gene cluster, especially given its proximity to phsB and phsC. The proteins that correspond to these two genes, phosphinothricin tripeptide synthetase (PTTS) III and II, respectively, have been biochemically characterized and suggested by Grammel et al. (9) to be the alanine-activating enzymes required for nonribosomal peptide assembly with the phosphinothricin precursor N-acetyldemethylphosphinothricin.
InterProScan analysis of the peptide sequence of the nonribosomal peptide synthesis proteins revealed that PTTS III has phosphopantetheine-binding domains near the amino and carboxy termini that flank a condensation domain and adenylation/activation domain. PTTS II has an identical domain arrangement except that it lacks the amino-terminal phosphopantetheine-binding domain. The enzyme responsible for the activation of N-acetyldemethylphosphinothricin is PTTS I (9), encoded by phsA (41), a gene which is not in close proximity to phsB and phsC in the PTT biosynthetic gene cluster (Fig. 3).
The arrangement of the nonribosomal peptide synthesis genes on fosmid 5-9G confirms the predictions of Hara et al. (12) and Schwartz et al. (41), who noted the likely spatial separation of PTT nonribosomal peptide synthesis genes after complementation analysis of multiple independent nonribosomal peptide synthesis-deficient S. hygroscopicus and S. viridochromogenes mutants, respectively. PTTS I appears to function only as an activating module, because analysis of phsA revealed characteristic adenylation and phosphopantetheine domains (41) but no conserved condensation domain; thus, peptide bond synthesis most likely takes place in the condensation domains of PTTSs II and III. It is interesting that InterProScan domain searches of these proteins did not locate a strongly conserved thioesterase domain in any of the three peptide synthetases. This implies that thioesterase activity, required to release the mature tripeptide peptide from the terminal nonribosomal peptide synthesis module, is likely provided by another enzyme in trans; phpL and phpM are thioesterase gene homologs that could be involved in this step. This observation bolsters the prediction of Raibaud et al. (39) that the equivalents of these genes from S. hygroscopicus, orf1 and orf2, could encode nonribosomal peptide synthesis-related thioesterase activity.
A PTT-specific transport protein is predicted to be associated with the PTT gene cluster because Kyte-Doolittle hydropathy plotting of the protein encoded by dea, responsible for the deacetylation of inactive N-acetylphosphinothricin tripeptide to the active PTT molecule, indicates that this enzyme probably does not span the membrane extensively, if at all, and thus would not have transporter activity. This implies that PTT is released inside the cell in the bioactive form and then exported. Two open reading frames in the PTT biosynthetic gene cluster could have a role in PTT export. phpN has homology to the S. hygroscopicus orf3 gene, a transmembrane transporter homolog which had been previously suggested to be a PTT exporter, or involved in the uptake of biosynthetic substrates (39). phpB has homology to a putative membrane protein, and protein family predicted structure analysis lists it as a member of the OPT oligopeptide transporter protein family.
The production of PTT biosynthetic intermediates in S. hygroscopicus has previously been shown to be under the transcriptional control of a protein encoded by brpA, which has been analyzed by sequencing and mutagenesis (3, 39). A homolog of brpA found in S. viridochromogenes was cloned, sequenced, and found to have 62% identity at the protein level to the S. hygroscopicus gene according to Thompson and Seto (50) in a citation of unpublished data. The same source also indicates that the sequenced gene from S. viridochromogenes could also complement a brpA mutant. From our sequence analysis of fosmid 5-9G, phpR has homology to LuxR-type transcriptional regulators, and its translated product was found to have 63.9% protein identity to S. hygroscopicus brpA; phpR was therefore designated the PTT transcriptional regulatory gene in S. viridochromogenes. brpA was previously found to contain a TTA leucine codon at amino acid position 250, leading to the implication that brpA may be under translational regulation by bldA expression (39). We similarly located a TTA codon within phpR, although the position of that codon, at amino acid position 131, does not correspond with that found in brpA.
Sequence comparison of the S. viridochromogenes PTT biosynthetic cluster to published portions of the corresponding S. hygroscopicus cluster further substantiate previous mapping efforts that predicted the architecture of the two gene clusters were identical (13). The amino acid identity of S. viridochromogenes genes compared to published S. hygroscopicus homologs range from very highly conserved at 95.5% for the bcpA/phpI homolog pair from the core region of the PTT cluster to 63.9% for the phpR/brpA homologs located at the far downstream flank of the gene cluster. The significance of the apparent divergence of the downstream genes cannot be ascertained at this time.
Eleven of 24 ORFs in the gene cluster do not have experimental evidence to support the roles currently assigned. Some, for example the phpL and phpM thioesterase homologs, have proposed functions based on homology. Others cannot currently be assigned biosynthetic roles including phpJ, an aldehyde dehydrogenase homolog and phpG, a bisphosphoglycerate mutase homolog (Table 3). One of the most interesting discoveries resulting from previous studies on PTT biosynthesis was the analogy drawn between the reactions involved in the stepwise conversion of phosphinopyruvate to deamino-alphaketo-demethylphosphinothricin (steps VII through X, Fig. 1) to those corresponding closely to steps of the tricarboxylic acid cycle involved in the stepwise conversion of oxaloacetate to α-ketoglutarate (15). Evolutionary implications regarding the origin of these enzymes were discussed previously by Thompson and Seto (50), who noted that enzymes common to central metabolism may have been modified for specialized secondary metabolic reactions.
We noted two putative gene products in the PTT cluster with homologs involved in central metabolism, namely the glycolytic reactions leading to the production of PEP; PhpG is a homolog of bisphosphoglycerate mutase and PhpH a homolog of enolase. The respective predicted stop (TGA) and start (ATG) nucleotide sequences of the corresponding genes overlap, possibly indicating cotranscription. It is interesting that a mutant of the phpH homolog in S. hygroscopicus, bcpE, was found to be deficient in the production of carboxyphosphonoenolpyruvate (27), a phosphonoenolpyruvate homolog. Biosynthesis of the structural homologs phosphonoenolpyruvate and carboxyphosphonoenolpyruvate by enzymes also showing homology suggests that carboxyphosphonoenolpyruvate biosynthesis may be more complex than the direct phosphate replacement mechanism postulated by Hidaka et al. (19). Further investigation into carboxyphosphonoenolpyruvate biosynthesis is warranted to determine if the phpG product is involved.
Acknowledgments
This work was supported by National Institute of General Medical Sciences grant GM59334.
We thank M. J. Thomas (University of Wisconsin, Madison) for plasmid pOJ436, E. Yagil (Tel Aviv University, Israel) for strain WM3321, and V. Mainz (University of Illinois, Urbana-Champaign) for invaluable NMR instruction and advice.
REFERENCES
- 1.Alijah, R., J. Dorendorf, S. Talay, A. Pühler, and W. Wohlleben. 1991. Genetic analysis of the phosphinothricin-tripeptide biosynthetic pathway of Streptomyces viridochromogenes Tü494. Appl. Microbiol. Biotechnol. 34:749-755. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., G. W. Miller, E. W. Myers, and D. J. Lipmann. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anzai, H., T. Murakami, S. Imai, A. Satoh, K. Nagaoka, and C. J. Thompson. 1987. Transcriptional regulation of bialaphos biosynthesis in Streptomyces hygroscopicus. J. Bacteriol. 169:3482-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer, E., K. H. Gugel, K. Hagele, H. Hagenmaier, S. Jessipow, W. A. Konig, and H. Zahner. 1972. Phosphinothricin und phosphinothricyl-alanyl-alanin. Helv. Chim. Acta 55:224-239. [DOI] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman, S., S. Pollack, and J. R. Knowles. 1992. Synthesis of the unusual metabolite carboxyphosphonoenolpyruvate. Cloning and expression of carboxyphosphononenolpyurvate mutase. J. Am. Chem. Soc. 114:377-378. [Google Scholar]
- 8.Gil, D., and J. P. Bouché. 1991. ColE1-type vectors with fully repressible replication. Gene 105:17-22. [DOI] [PubMed] [Google Scholar]
- 9.Grammel, N., D. Schwartz, W. Wohlleben, and U. Keller. 1998. Phosphinothricin-tripeptide synthetases from Streptomyces viridochromogenes. Biochemistry 37:1596-1603. [DOI] [PubMed] [Google Scholar]
- 10.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara, O., H. Anzai, S. Imai, Y. Kumada, T. Murakami, R. Itoh, E. Takano, A. Satoh, and K. Nagaoka. 1988. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: cloning and analysis of the genes involved in the alanylation step. J. Antibiot. 41:538-547. [DOI] [PubMed] [Google Scholar]
- 13.Hara, O., T. Murakami, S. Imai, H. Anzai, R. Itoh, Y. Kumada, E. Takano, E. Satoh, A. Satoh, K. Nagaoka, and et al. 1991. The bialaphos biosynthetic genes of Streptomyces viridochromogenes: cloning, heterospecific expression, and comparison with the genes of Streptomyces hygroscopicus. J. Gen. Microbiol. 137:351-359. [DOI] [PubMed] [Google Scholar]
- 14.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, West Sussex, England.
- 15.Heinzelmann, E., G. Kienzlen, S. Kaspar, J. Recktenwald, W. Wohlleben, and D. Schwartz. 2001. The phosphinomethylmalate isomerase gene pmi, encoding an aconitase-like enzyme, is involved in the synthesis of phosphinothricin tripeptide in Streptomyces viridochromogenes. Appl. Environ. Microbiol. 67:3603-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidaka, T., M. Hidaka, T. Kuzuyama, and H. Seto. 1995. Sequence of a P-methyltransferase-encoding gene isolated from a bialaphos-producing Streptomyces hygroscopicus. Gene 158:149-150. [DOI] [PubMed] [Google Scholar]
- 17.Hidaka, T., M. Hidaka, and H. Seto. 1992. Studies on the biosynthesis of bialaphos (SF-1293). 14. Nucleotide sequence of phosphoenolpyruvate phosphomutase gene isolated from a bialaphos producing organism, Streptomyces hygroscopicus, and its expression in Streptomyces lividans. J. Antibiot. 45:1977-1980. [DOI] [PubMed] [Google Scholar]
- 18.Hidaka, T., M. Hidaka, T. Uozumi, and H. Seto. 1992. Nucleotide sequence of a carboxyphosphonoenolpyruvate phosphonomutase gene isolated from a bialaphos-producing organism, Streptomyces hygroscopicus, and its expression in Streptomyces lividans. Mol. Gen. Genet. 233:476-478. [DOI] [PubMed] [Google Scholar]
- 19.Hidaka, T., S. Imai, O. Hara, H. Anzai, T. Murakami, K. Nagaoka, and H. Seto. 1990. Carboxyphosphonopyruvate phosphonomutase, a novel enzyme catalyzing C-P bond formation. J. Bacteriol. 172:3066-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hidaka, T., K. W. Shimotohno, T. Morishita, and H. Seto. 1999. Studies on the biosynthesis of bialaphos (SF-1293). 18. 2-Phosphinomethylmalic acid synthase: a descendant of (R)-citrate synthase? J. Antibiot. 52:925-931. [DOI] [PubMed] [Google Scholar]
- 21.Imai, S., H. Seto, T. Sasaki, T. Tsuruoka, H. Ogawa, A. Satoh, S. Inouye, T. Niida, and N. Otake. 1984. Studies on the biosynthesis of bialaphos (SF-1293). 4. Production of phosphonic acid derivatives, 2-hydroxyethylphosphonic acid, hydroxymethylphosphonic acid and phosphonoformic acid by blocked mutants of Streptomyces hygroscopicus SF-1293 and their roles in the biosynthesis of bialaphos. J. Antibiot. 37:1505-1508. [DOI] [PubMed] [Google Scholar]
- 22.Kamigiri, K., T. Hidaka, S. Imai, T. Murakami, and H. Seto. 1992. Studies on the biosynthesis of bialaphos (SF-1293) 12. C-P bond formation mechanism of bialaphos: discovery of a P-methylation enzyme. J. Antibiot. 45:781-787. [DOI] [PubMed] [Google Scholar]
- 23.Katz, E., C. J. Thompson, and D. A. Hopwood. 1983. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans 66. J. Gen. Microbiol. 129:2703-2714. [DOI] [PubMed] [Google Scholar]
- 24.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Centre, Norwich, England.
- 25.Kumada, Y., H. Anzai, E. Takano, T. Murakami, O. Hara, R. Itoh, S. Imai, A. Satoh, and K. Nagaoka. 1988. The bialaphos resistance gene (bar) plays a role in both self-defense and bialaphos biosynthesis in Streptomyces hygroscopicus. J. Antibiot. 41:1838-1845. [DOI] [PubMed] [Google Scholar]
- 26.Lea, P. J., K. W. Joy, J. L. Ramos, and M. G. Guerrero. 1984. The action of 2-amino-4-(methylphosphonyl)-butanoic acid (phosphinothricin) and its 2-oxo-derivative on the metabolism of cyanobacteria and higher plants. Phytochemistry 23:1-6. [Google Scholar]
- 27.Lee, S. H., T. Hidaka, H. Nakashita, and H. Seto. 1995. The carboxyphosphonoenolpyruvate synthase-encoding gene from the bialaphos-producing organism Streptomyces hygroscopicus. Gene 153:143-144. [DOI] [PubMed] [Google Scholar]
- 28.Mariahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 29.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: Liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its usein construction of insertion mutants: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami, T., H. Anzai, S. Imai, A. Satoh, K. Nagaoka, and C. J. Thompson. 1986. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: Molecular cloning and characterization of the gene cluster. Mol. Gen. Genet. 205:42-50. [Google Scholar]
- 32.Nakashita, H., K. Kozuka, T. Hidaka, O. Hara, and H. Seto. 2000. Identification and expression of the gene encoding phosphonopyruvate decarboxylase of Streptomyces hygroscopicus. Biochim. Biophys. Acta 1490:159-162. [DOI] [PubMed] [Google Scholar]
- 33.Nash, H. A. 1983. Purification and properties of the bacteriophage lambda Int protein. Methods Enzymol. 100:210-216. [DOI] [PubMed] [Google Scholar]
- 34.Okanishi, M., K. Suzuki, and H. Umezawa. 1974. Formation and reversion of streptomycete protoplasts: cultural condition and morphological study. J. Gen. Microbiol. 80:389-400. [DOI] [PubMed] [Google Scholar]
- 35.Omura, S., M. Murata, H. Hanaki, K. Hinotozawa, R. Oiwa, and H. Tanaka. 1984. Phosalacine, a new herbicidal antibiotic containing phosphinothricin. Fermentation, isolation, biological activity and mechanism of action. J. Antibiot. 37:829-835. [DOI] [PubMed] [Google Scholar]
- 36.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FastP and FastA. Methods Enzymol. 183:63-68. [DOI] [PubMed] [Google Scholar]
- 38.Pritchett, M. A., J. K. Zhang, and W. W. Metcalf. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 70:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raibaud, A., M. Zalacain, T. G. Holt, R. Tizard, and C. J. Thompson. 1991. Nucleotide sequence analysis reveals linked N-acetyl hydrolase, thioesterase, transport, and regulatory genes encoded by the bialaphos biosynthetic gene cluster of Streptomyces hygroscopicus. J. Bacteriol. 173:4454-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Schwartz, D., R. Alijah, B. Nussbaumer, S. Pelzer, and W. Wohlleben. 1996. The peptide synthetase gene phsA from Streptomyces viridochromogenes is not juxtaposed with other genes involved in nonribosomal biosynthesis of peptides. Appl. Environ. Microbiol. 62:570-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz, D., J. Recktenwald, S. Pelzer, and W. Wohlleben. 1998. Isolation and characterization of the PEP-phosphomutase and the phosphonopyruvate decarboxylase genes from the phosphinothricin tripeptide producer Streptomyces viridochromogenes Tü494. FEMS Microbiol. Lett. 163:149-157. [DOI] [PubMed] [Google Scholar]
- 43.Seto, H., S. Imai, T. Tsuruoka, A. Satoh, M. Kojima, S. Inouye, T. Sasaki, and N. Otake. 1982. Studies on the biosynthesis of bialaphos (SF-1293). 1. Incorporation of 13C- and 2H-labeled precursors into bialaphos. J. Antibiot. 35:1719-1721. [DOI] [PubMed] [Google Scholar]
- 44.Seto, H., and T. Kuzuyama. 1999. Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat. Prod. Rep. 16:589-596. [DOI] [PubMed] [Google Scholar]
- 45.Seto, H., T. Sasaki, S. Imai, T. Tsuruoka, H. Ogawa, A. Satoh, S. Inouye, T. Niida, and N. Otake. 1983. Studies on the biosynthesis of bialaphos (SF-1293). 2. Isolation of the first natural products with a C-P-H bond and their involvement in the C-P-C bond formation. J. Antibiot. 36:96-98. [DOI] [PubMed] [Google Scholar]
- 46.Shimotohno, K., H. Seto, and N. Otake. 1998. Studies on the biosynthesis of bialaphos. Purification and characterization of 2-phosphinomethylmalic acid synthase from Streptomyces hygroscopicus SF-1293. J. Antibiot. 41:1057-1065. [DOI] [PubMed] [Google Scholar]
- 47.Strauch, E., W. Wohlleben, and A. Pühler. 1988. Cloning of a phosphinothricin N-acetyltransferase gene from Streptomyces viridochromogenes Tü494 and its expression in Streptomyces lividans and Escherichia coli. Gene 63:65-74. [DOI] [PubMed] [Google Scholar]
- 48.Strauch, E., W. Wohlleben, and A. Pühler. 1987. Development of a plasmid-cloning system for Streptomyces viridochromogenes Tü494. J. Basic Microbiol. 27:449-455. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, C. J., R. N. Movva, R. Tizard, R. Crameri, J. E. Davies, M. Lauwereys, and J. Botterman. 1987. Characterization of of the herbicide gene bar from Streptomyces hygroscopicus. EMBO J. 9:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, C. J., and H. Seto. 1995. Bialaphos, p. 197-222. In L. C. Vining and C. Stuttard (ed.), Genetics and biochemistry of antibiotic production. Butterworth-Heinemann, Newton, Mass.
- 51.Van Dessel, W., N. Van Mellaert, and J. A. Geukens. 2003. Improved PCR-based method for the direct screening of Streptomyces transformants. J. Microbiol. Methods 53:401-403. [DOI] [PubMed] [Google Scholar]
- 52.Wanner, B. L. 1987. Control of phoR-dependent bacterial alkaline phosphatase clonal variation by the phoM region. J. Bacteriol. 169:900-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wanner, B. L. 1994. Gene expression in bacteria using TnphoA and TnphoA′ elements to make and switch phoA, lacZ(op), and lacZ(pr) fusions, p. 291-310. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 3. Academic Press, Orlando, Fla. [Google Scholar]
- 54.Wild, J., and W. Szybalski. 2004. Copy-control pBAC/oriV vectors for genomic cloning. Methods Mol. Biol. 267:145-154. [DOI] [PubMed] [Google Scholar]
- 55.Wohlleben, W., R. Alijah, J. Dorendorf, D. Hillemann, B. Nussbaumer, and S. Pelzer. 1992. Identification and characterization of phosphinothricin-tripeptide biosynthetic genes in Streptomyces viridochromogenes. Gene 115:127-132. [DOI] [PubMed] [Google Scholar]
- 56.Wohlleben, W., W. Arnold, I. Broer, D. Hillemann, E. Strauch, and A. Pühler. 1988. Nucleotide sequence of the phosphinothricin N-acetyltransferase gene from Streptomyces viridochromogenes Tü494 and its expression in Nicotiana tabacum. Gene 70:25-37. [DOI] [PubMed] [Google Scholar]
- 57.Wohlleben, W., and A. Pielsticker. 1989. Presented at the DECHEMA Biotechnology Conferences 3.
- 58.Yagil, E., S. Dolev, J. Oberto, N. Kislev, N. Ramaiah, and R. A. Weisberg. 1989. Determinants of site-specific recombination in the lambdoid coliphage HK022. An evolutionary change in specificity. J. Mol. Biol. 207:695-717. [DOI] [PubMed] [Google Scholar]
- 59.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, J. K., A. K. White, H. C. Kuettner, P. Boccazzi, and W. W. Metcalf. 2002. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J. Bacteriol. 184:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]