Abstract
A pyrosequencing method for detection and quantification of macrolide resistance mutations at positions 2058 and 2059 (Escherichia coli numbering) of the 23S rRNA gene is described. The method was developed and tested for Streptococcus pneumoniae, Streptococcus pyogenes, Mycobacterium avium, Campylobacter jejuni, and Haemophilus influenzae.
Macrolides inhibit bacterial protein synthesis by binding to the peptidyl transferase region of the bacterial ribosome. Mutations mainly at position 2058 or 2059 of the 23S rRNA mediate macrolide resistance. Adenine-to-cytosine or adenine-to-guanine mutations are the most prevalent ones at these positions (13, 14). These mutations were first found in clinical isolates of species harboring one or two rRNA gene (rDNA) alleles but later also in bacteria harboring up to six rDNA operons (2, 9). Several techniques, such as real-time PCR (3), restriction fragment length polymorphism (12), and denaturing high-performance liquid chromatography (1), have been used to detect macrolide resistance mutations in 23S rDNA. However, for determination of the number of mutated alleles by sequencing, allele-specific amplification has been necessary (2, 11).
In pyrosequencing, a sequencing primer is annealed to a single-stranded PCR product, and nucleotides are given to the reaction. Incorporation of the nucleotide by DNA polymerase leads to the release of pyrophosphate, which is further processed by sulfurylase and luciferase, producing light in proportion to the amount of pyrophosphate. The light is detected by the equipment and presented as a pyrogram, in which the peak heights are proportional to the number of nucleotides incorporated. Excess nucleotides are enzymatically degraded before the following nucleotide is added (10).
Here we describe a pyrosequencing method for detection of mutations at positions 2058 and 2059 of the 23S rDNA. In this method, a universal PCR product is pyrosequenced using various primers targeting to different bacteria. Wild-type and mutant strains are readily distinguished, and the proportion or number of mutated alleles can be estimated by interpreting the pyrogram or more exactly determined by analyzing the numerical peak heights. The procedure is fast and is easily performed during a single working day; the most time-consuming steps are PCR and agarose gel electrophoresis. An actual pyrosequencing run lasts 15 min, and the preceding template preparation lasts approximately 30 to 45 min, depending on the number of samples processed. Other benefits of pyrosequencing are its low cost and the ease of both performing the sequencing without gels or capillars and analyzing the data.
The bacterial strains were grown on suitable media, and colonies were dissolved into water. The suspension was inactivated at 95°C for 10 min and was used as a sample in PCR containing 0.3 μM universal primers (Table 1), 0.015 U of Ampli TaqGold DNA polymerase/μl, 1× Geneamp PCR Gold buffer, 1.5 mM MgCl2 (Applied Biosystems, Foster City, Calif.), and 0.2 mM deoxynucleoside triphosphates (Amersham Biosciences, Piscataway, N.J.). The reaction volume was 50 to 100 μl, and the amount of sample was 1/10 of the reaction volume. The reactions were performed in a PTC-200 thermal cycler (MJ Research, Waltham, Mass.), using the following cycling: 95°C for 10 min followed by 35 cycles of 15 s at 95°C, 15 s at 61°C, and 30 s at 72°C. Pyrosequencing was performed by using streptavidin-coated Sepharose beads (Amersham Biosciences), an SQA reagent kit, a vacuum prep workstation, and a PSQ 96MA instrument (Biotage AB, Uppsala, Sweden) according to the instructions of the manufacturer. In one reaction, 20 μl of PCR product and 15 pmol of suitable sequencing primer (Table 1) were used. Alternatively, 12 pmol of all the sequencing primers could be added to one reaction. The standard dispensation order was TCGACGAGACATG. The dispensation order TACGAGACATG was designed for confirming the amount of mutated alleles at position 2058. Peak height of the first T dispensation was used as a background in numerical peak height analysis. The numerical peak heights could not be compared between runs due to the signal intensity differences. Therefore, signals of variable positions were compared to the signals of constant positions, and the results were expressed as numbers of incorporated nucleotides. The peak height data of Table 2 were averaged from three runs.
TABLE 1.
Primers used for PCR and pyrosequencing
| Primer | Positiona | Use | Sequenceb | Target species |
|---|---|---|---|---|
| 23SV_univF_1926 | 1926-1948 | PCR | TAAGGTAGCGAAATTCCTTGTCG | Universal primer; all species |
| Bio_23SV_univR_2259 | 2241-2259 | PCR | Bio-CGACCGCCCCAGTCAAACTc | Universal primer; all species |
| 23SV_gpos_seq | 2038-2057 | Pyrosequencing | GGTTACCCGCGACAGGACGG | Gram-positive bacteria |
| 23SV_gpos_seq | 2044-2057 | Pyrosequencing | CCGCGGCAAGACGG | Gram-negative bacteria |
| 23SV_Hinf_seq | 2044-2057 | Pyrosequencing | CCGCGGCTAGACGG | H. influenzae |
| 23SV_myco_seq | 2041-2057 | Pyrosequencing | TACGYGCGGCGGACGA | Mycobacteria |
Escherichia coli numbering.
Y = T/C.
Bio, biotin.
TABLE 2.
Theoretical and detected numbers of incorporated nucleotides of the bacterial strains analyzed in this study, using the standard dispensation order
| Strain | ERYf MIC (μg/ml) | Mutation | Mutated alleles | Reference | Theoretical and detected no. of added NTPsa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | G | A | C | G | A | G | A | C | Ab | Tb | |||||
| S. pneumoniae ATCC 49819 | 0.125 | 0/4 | 8 | 0 | 0 | 12 | 0 | 4 | 4 | 0 | 0 | 16 | 4 | 4 | |
| 0 | 0 | 11 | 0 | 4 | 4 | 0 | 0 | 15 | 4 | 4 | |||||
| S. pyogenes ATCC 700294 | 0.063 | 0/6 | 2 | 0 | 0 | 18 | 0 | 6 | 6 | 0 | 0 | 24 | 6 | 6 | |
| 0 | 0 | 20 | 0 | 6 | 7 | 0 | 0 | 23 | 6 | 6 | |||||
| S. pyogenes NI 4277 | >256 | A2058G | 5/6 | 2 | 0 | 5 | 13 | 0 | 6 | 6 | 0 | 0 | 24 | 6 | 6 |
| 0 | 5 | 15 | 0 | 6 | 7 | 0 | 0 | 24 | 6 | 6 | |||||
| S. pneumoniae r581 | 128 | A2059C | 1/4c | 0 | 0 | 10 | 1 | 3 | 4 | 1 | 1 | 16 | 4 | 4 | |
| 0 | 0 | 10 | 1 | 3 | 4 | 1 | 1 | 17 | 4 | 4 | |||||
| S. pneumoniae r506 | 256 | A2059C | 2/4 | 8 | 0 | 0 | 8 | 2 | 2 | 4 | 2 | 2 | 16 | 4 | 4 |
| 0 | 0 | 8 | 2 | 2 | 4 | 2 | 2 | 17 | 4 | 4 | |||||
| S. pneumoniae r771 | 256 | A2059G | 2/4 | 8 | 0 | 0 | 8 | 0 | 4 | 4 | 2 | 2 | 16 | 4 | 4 |
| 0 | 0 | 8 | 0 | 4 | 4 | 3 | 3 | 16 | 4 | 4 | |||||
| S. pneumoniae r1317 | >512 | A2050C | 3/4 | 8 | 0 | 0 | 6 | 3 | 1 | 4 | 3 | 3 | 16 | 4 | 4 |
| 0 | 0 | 6 | 3 | 1 | 4 | 3 | 3 | 16 | 4 | 4 | |||||
| S. pneumoniae r733 | >512 | A2059C | 4/4 | 8 | 0 | 0 | 4 | 4 | 0 | 4 | 4 | 4 | 16 | 4 | 4 |
| 0 | 0 | 4 | 4 | 0 | 4 | 4 | 4 | 17 | 4 | 4 | |||||
| S. pneumoniae 01-41 | 256 | A2058G | 2/4 | 0 | 2 | 10 | 0 | 4 | 4 | 0 | 0 | 16 | 4 | 4 | |
| 0 | 3 | 11 | 0 | 4 | 4 | 0 | 0 | 16 | 4 | 4 | |||||
| C. jejuni 62 | >256 | A2059G | 3/3 | 0 | 0 | 3 | 0 | 3d | 3d | 3 | 3 | 12 | 0 | 0 | |
| 0 | 0 | 4 | 0 | 3 | 3 | 3 | 3 | 12 | 0 | 0 | |||||
| H. influenzae 286 | >64e | A2058G | 6/6 | 0 | 6 | 12 | 0 | 6d | 6d | 0 | 0 | 24 | 0 | 0 | |
| 0 | 5 | 12 | 0 | 6 | 6 | 0 | 0 | 25 | 0 | 0 | |||||
| M. avium H0851/98 | >256e | A2058G | 1/1 | 0 | 1 | 2 | 0 | 1d | 1d | 0 | 0 | 4 | 0 | 0 | |
| 0 | 1 | 2 | 0 | 1 | 1 | 0 | 0 | 4 | 0 | 0 | |||||
| M. avium H0812/96 | ≤8e | 0/1 | 0 | 0 | 3 | 0 | 1d | 1d | 0 | 0 | 4 | 0 | 0 | ||
| 0 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 4 | 0 | 0 | |||||
Theoretical numbers of incorporated nucleotides (top row for each strain) are calculated as a sum of all alleles into which one or more dispensed nucleotides are incorporated. The detected numbers (bottom row for each strain) are mean values of three pyrosequencing runs.
Peaks used as a reference to calculate the numbers of other incorporated nucleotides in streptococci. The T peak is used for calculating the detected numbers of C, G, and T incorporations, and the A peak is used for the A nucleotide only. The heights of these peaks are equal to the copy number of the 23S rRNA gene in the strain, as one nucleotide is incorporated to each copy of the gene.
The genotype of this strain has been determined by using the ABI Prism BigDye Terminator kit (Applied Biosystems), using primers described by Tait-Kamradt et al. (8, 11).
These G and A peaks were used as a reference for C. jejuni, H. influenzae, and M. avium because these species do not possess the A and T nucleotides used as a reference for streptococci. These peaks can only be used as a reference in A2058 mutations, since in the case of an A2059 mutation the number of nucleotides incorporated to these positions does not remain constant.
Azithromycin MIC.
ERY, erythromycin.
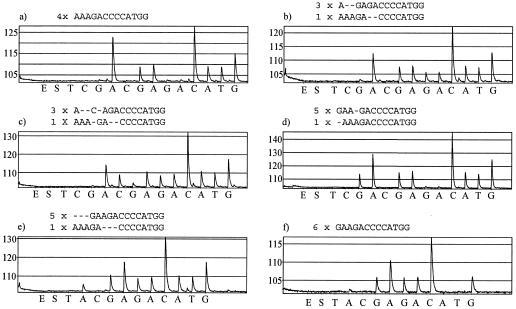
The pyrograms of wild-type strains began with a high A peak deriving from three adenine nucleotides incorporated at positions 2058 to 2060 of all alleles (Fig. 1a). After the A peak, lower G and A peaks due to single-nucleotide incorporations were obtained. Thereafter, a high C peak reflected the incorporation of four C nucleotides to all alleles. Mutation at position 2059 generated a different peak pattern (Fig. 1b and c), and the proportion of mutant alleles could be estimated from the pyrogram, since some peaks derived from all alleles, some from mutant alleles, and some from wild-type alleles. For example, in the pyrogram of Streptococcus pneumoniae r771 (Fig. 1b), the first G and A peaks (from the second G and A dispensations) derived from all alleles, and the following ones derived from mutant alleles. Since the heights of the latter peaks are approximately half of those of the former ones, half of the 23S rDNA alleles contained the A2059G mutation. In case of an A2058G mutation (Fig. 1d), the first peak obtained with the standard dispensation order was G, the height of which reflected the proportion of mutant alleles. For Streptococcus pyogenes NI 4277, the first G peak was slightly lower than the second G peak, suggesting fewer than six mutant alleles. The numerical data indicated the presence of one wild-type allele (Table 2), and by using the other dispensation order, a clear A peak was detected before the G peak, verifying the presence of one wild-type allele (Fig. 1e).
FIG. 1.
Pyrograms obtained with bacterial strains harboring different numbers of mutant and wild-type 23S rDNA alleles. Above each pyrogram, the nucleotide sequences and numbers of wild-type and mutant alleles of the given strain are presented. The nucleotides are aligned so that the nucleotides giving rise to a signal with the same nucleotide dispensation are one on top of the other. Pyrograms a to d were obtained by using the standard dispensation order, and pyrograms e to f were obtained by using the dispensation order designed for quantification of mutations at position 2058. (a) S. pneumoniae ATCC 49619; (b) S. pneumoniae r771; (c) S. pneumoniae r1317; (d-e) S. pyogenes NI 4277; (f) H. influenzae 286.
There are small discrepancies in the peak height data (Table 2). However, they are not severe, since the detected numbers of incorporations fit best with the correct number of mutant alleles. For example, the analysis of Haemophilus influenzae 286 results in five A2058G incorporations, whereas the number of consequent A nucleotides coincides with the hypothesis of six mutant alleles. Also, the pyrogram obtained with the dispensation order designed for the A2058 mutation verifies the absence of wild-type allele (Fig. 1f).
The first T peak was used as a background signal. Naturally, if the analyzed strain had an A2058T mutation, this peak could not be used as a background. A2058T mutations have been described in human mycobacterial (4, 6) and Staphylococcus aureus (9) isolates. However, the presence of the mutation in a mycobacterial strain would be evident from the height of the first T peak. Instead, if only a minority of 23S rDNA alleles had the mutation, the A2058T signal might be left unnoticed. Of course, the dispensation order can also be designed to more efficiently detect a suspected A2058T mutation. Macrolide resistance mutations also have been described at position 2057 in a few bacterial species (13, 14). These mutations cannot be identified using the sequencing primers of this study, but they can be indirectly detected, since no sequence is obtained due to the mismatch at the 3′ end of the sequencing primer.
Detection and quantification of these macrolide resistance mutations in Helicobacter pylori could be one important application of this technique, and the mutations could be even determined simultaneously with identification and subtyping of H. pylori (5). The quantification of mutations in general also has other important applications, such as detection of drug resistance mutations in human immunodeficiency virus for the choice of efficient treatment (7).
Acknowledgments
We thank the following persons for providing bacterial strains for this study: Merja Marjamäki (Mycobacterium avium), Antti Hakanen (Campylobacter jejuni), Pia Littauer (University of Tromsø, Tromsø, Norway; S. pneumoniae 01-41), and Laura Lindholm and Pauliina Kärpänoja (Päijat-Häme Central Hospital, Lahti, Finland; H. influenzae).
REFERENCES
- 1.Canu, A., A. Abbas, B. Malbruny, F. Sichel, and R. Leclercq. 2004. Denaturing high-performance liquid chromatography detection of ribosomal mutations conferring macrolide resistance in gram-positive cocci. Antimicrob. Agents Chemother. 48:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jalava, J., M. Vaara, and P. Huovinen. 2004. Mutation at the position 2058 of the 23S rRNA as a cause of macrolide resistance in Streptococcus pyogenes. Ann. Clin. Microbiol. Antimicrob. 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lascols, C., D. Lamarque, J.-M. Costa, C. Copie-Bergman, J.-M. Le Glaunec, L. Deforges, C.-J. Soussy, J.-C. Petit, J.-C. Delchier, and J. Tankovic. 2003. Fast and accurate quantitative detection of Helicobacter pylori and identification of clarithromycin resistance mutations in H. pylori isolates from gastric biopsy specimens by real-time PCR. J. Clin. Microbiol. 41:4573-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meier, A., P. Kirschner, B. Springer, V. A. Steingrube, B. A. Brown, R. J. Wallace, Jr., and E. C. Bottger. 1994. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob. Agents Chemother. 38:381-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monstein, H., S. Nikpour-Badr, and J. Jonasson. 2001. Rapid molecular identification and subtyping of Helicobacter pylori by pyrosequencing of the 16S rDNA variable V1 and V3 regions. FEMS Microbiol. Lett. 199:103-107. [DOI] [PubMed] [Google Scholar]
- 6.Nash, K. A., and C. B. Inderlied. 1995. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob. Agents Chemother. 39:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Meara, D., K. Wilbe, T. Leitner, B. Hejdeman, J. Albert, and J. Lundeberg. 2001. Monitoring resistance to human immunodeficiency virus type 1 protease inhibitors by pyrosequencing. J. Clin. Microbiol. 39:464-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pihlajamäki, M., J. Kataja, H. Seppalä, J. Elliot, M. Leinonen, P. Huovinen, and J. Jalava. 2002. Ribosomal mutations in Streptococcus pneumoniae clinical isolates. Antimicrob. Agents Chemother. 46:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prunier, A.-L., B. Malbruny, D. Tandé, B. Picard, and R. Leclercq. 2002. Clinical isolates of Staphylococcus aureus with ribosomal mutations conferring resistance to macrolides. Antimicrob. Agents Chemother. 46:3054-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronaghi, M., M. Uhlén, and P. Nyrén. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363, 365. [DOI] [PubMed] [Google Scholar]
- 11.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vacher, S., A. Ménard, E. Bernard, and F. Mégraud. 2003. PCR-restriction fragment length polymorphism analysis for detection of point mutations associated with macrolide resistance in Campylobacter spp. Antimicrob. Agents Chemother. 47:1125-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]