Abstract
A 1.8-million-member d-octapeptide combinatorial library was constructed in which each member comprised a diversity-containing N-terminal pentapeptide and a C-terminal amidated triarginine motif. The C-terminal motif concentrated the library members at the fungal cell surface. A primary screen for inhibitors of Saccharomyces cerevisiae and Candida albicans growth, together with an in vitro secondary screen with the S. cerevisiae plasma membrane ATPase (Pma1p) as a target, identified the antifungal d-octapeptide BM0 (d-NH2-RFWWFRRR-CONH2). Optimization of BM0 led to the construction of BM2 (d-NH2-RRRFWWFRRR-CONH2), which had broad-spectrum fungicidal activity against S. cerevisiae, Candida species, and Cryptococcus neoformans; bound strongly to the surfaces of fungal cells; inhibited the physiological activity of Pma1p; and appeared to target Pma1p, with 50% inhibitory concentrations in the range of 0.5 to 2.5 μM. At sub-MICs (<5 μM), BM2 chemosensitized to fluconazole (FLC) S. cerevisiae strains functionally hyperexpressing fungal lanosterol 14α-demethylase and resistance-conferring transporters of azole drugs. BM2 chemosensitized to FLC some FLC-resistant clinical isolates of C. albicans and C. dubliniensis and chemosensitized to itraconazole clinical isolates of C. krusei that are intrinsically resistant to FLC. The growth-inhibitory concentrations of BM2 did not cause fungal cell permeabilization, significant hemolysis of red blood cells, or the death of cultured HEp-2 epithelial cells. BM2 represents a novel class of broad-spectrum, surface-active, Pma1p-targeting fungicides which increases the potencies of azole drugs and circumvents azole resistance.
Cationic peptides produced by multicellular organisms are an evolutionarily ancient and rapidly mobilized primary defense against infections caused by a broad range of microbes (8). The cationic antimicrobial peptides are ribosomally synthesized, proteolytically processed species of 12 to ∼50 amino acids that comprise about 50% hydrophobic residues and that have a net excess of positive charge (9). They are usually found on epithelial cell surfaces and in phagocytic cells at sites of microbial invasion, and only a few instances of constitutive or induced resistance to cationic peptides have been detected (for a review, see reference 44). Although the cationic peptides are subdivided into several structural classes (10), they are, in general, amphipathic molecules that preferentially bind to acidic phospholipids, acidic polysaccharides, and lipopolysaccharides on the exterior of the lipid bilayer of invading microbes rather than to the cholesterol-rich and neutral plasma membrane surfaces of mammalian host cells. The bound cationic peptides are then thought to kill target microbes, including fungi (4), by forming assemblies that alter the lipid bilayer structure and disrupt the functional properties of the microbial membrane. A few cationic peptides may affect intracellular targets, including mitochondria and DNA and RNA metabolism; but apart from the binding of the salivary histatin 5 to a cell surface receptor in Candida albicans (16), there is no evidence of direct effects on fungal cell surface proteins. We hypothesized that the incorporation of a cationic peptide-like motif into an antifungal would enhance its potency by concentrating the compound at fungal cell surfaces. This idea has been validated in the present study by obtaining a membrane-impermeant and surface-active cationic peptide inhibitor of the fungal plasma membrane proton-pumping ATPase (Pma1p), an essential enzyme involved in fungal energy transduction (36).
Pma1p is an ∼100-kDa electrogenic, polytopic integral membrane protein of the P-type ATPase class which contributes 10 to 20% of the yeast plasma membrane protein. It generates the plasma membrane electrochemical gradient that is required for the maintenance of intracellular pH, cellular ion balance, and the uptake of numerous nutrients (36). The amount of functional Pma1p is tightly regulated (5), and yeast growth requires at least 25% of normal Pma1p activity (35). Pma1p was postulated to be a target for surface-mediated, broad-spectrum antifungal intervention because of the structural similarity between cell surface loops in Pma1ps from fungal cells and their dissimilarity to the comparable loops in P-type ATPases from other organisms, as well as the specificity achieved with therapies targeting mammalian P-type ATPases (28). Pma1p was validated as an antifungal target by demonstrating that acid-activated omeprazole is a fungicidal Pma1p inhibitor that acts from outside the cell (25, 37). This paper describes a drug discovery strategy that targets Pma1p. Screening of a compact 324-pool d-octapeptide library, which comprises 1.8 million combinatorial pentapeptides linked to a C-terminal amidated triarginine motif, has identified a potent, broad-spectrum, surface-active fungicidal Pma1p inhibitor that circumvents three clinically important (43) mechanisms of energy-dependent azole resistance. Such inhibitors may provide a novel and timely avenue for antifungal drug discovery.
MATERIALS AND METHODS
Strains and culture conditions.
The strains of Saccharomyces cerevisiae, Candida species, and Cryptococcus used in this study are listed in Table 1. The S. cerevisiae, C. albicans and Cryptococcus neoformans strains were stored at −80°C in YPD medium (1% [wt/vol] yeast extract [Difco, Becton Dickinson, Sparks, Md.], 2% [wt/vol] Bacto Peptone [Difco], 2% [wt/vol] glucose) containing 15% glycerol. S. cerevisiae strains were routinely maintained on complete synthetic medium (CSM) without uracil (CSM − ura) containing 0.67% (wt/vol) Yeast Nitrogen Base (Difco), 0.077% (wt/vol) CSM−URA (Bio 101, Vista, Calif.), 2% (wt/vol) glucose, and 2% (wt/vol) agar (pH 7.0). Candida species and the C. neoformans strain were maintained on YPD agar containing 2% (wt/vol) agar (pH 5.5). Growth inhibition and chemosensitization assays used CSM − ura liquid medium buffered to pH 7.0 with 10 mM morpholineethanesulfonic acid (MES) and 20 mM HEPES. Cells were grown at 30°C with shaking (150 rpm).
TABLE 1.
Yeast strains used in this study
| Species and strain | Genotype or source and comments |
|---|---|
| S. cerevisiae | |
| AD/PDR5− | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG, pdr5Δ ::hisG; designated strain AD1234567 in a previous report (3) |
| AD/PDR5+ | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG PDR5; designated strain AD124567 in a previous report (3) |
| AD1-8u− | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ ::hisG pdr5Δ::hisG (23) |
| AD/PDR5 | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ ::hisG pdr5Δ::PDR5-URA3 (30) |
| AD/CaCDR1 | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ ::hisG pdr5Δ::CDR1-URA3 (23) |
| AD/CaCDR2 | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ ::hisG pdr5Δ::CDR2-URA3; E. Lamping, University of Otago (23) |
| AD/CgCDR1-1B | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ ::hisG pdr5Δ::CgCDR1-URA3 (41) |
| AD/CgCDR2-4 | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ ::hisG pdr5Δ::CgCDR2-URA3 (41) |
| AD/MDR1 | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ ::hisG pdr5Δ::MDR1-URA3; designated strain AD/BEN in a previous report (23) |
| AD/ERG11 | Matα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ ::hisG pdr5Δ::CaERG11-URA3 (23) |
| T48 | HO/HO Mata/Matα ade6-1/ade6-1 trp5-1/trp5-1 leu2-1/leu2-1 lys1-1/lys1-1 ura3-1/ura3-1 PMA1-URA3/PMA-URA3 (19) |
| ΔN | HO/HO Mata/Matα ade6-1/ade6-1 trp5-1/trp5-1 leu2-1/leu2-1 lys1-1/lys1-1 ura3-1/ura3-1 pma1Δ1-27-URA3/pma1Δ1-27-URA3 (19) |
| ΔC | HO/HO Mata/Matαade6-1/ade6-1 trp5-1/trp5-1 leu2-1/leu2-1 lys1-1/lys1-1 ura3-1/ura3-1 pma1 Δ901-918-URA3/pma1Δ901-918-URA3 (19) |
| ΔNΔC | HO/HO Mata/Matα ade6-1/ade6-1 trp5-1/trp5-1 leu2-1/leu2-1 lys1-1/lys1-1 ura3-1/ura3-1 pma1Δ1-27, Δ901-918-URA3/pma1Δ1-27, Δ901-918-URA3 (19) |
| CTM1+2 | HO/HO Mata/Matα ade6-1/ade6-1 trp5-1/trp5-1 leu2-1/leu2-1 lys1-1/lys1-1 ura3-1/ura3-1 pma1-URA3/pma1-URA3 (20) |
| CTM3+4 | HO/HO Mata/Matα ade6-1/ade6-1 trp5-1/trp5-1 leu2-1/leu2-1 lys1-1/lys1-1 ura3-1/ura3-1 pma1-URA3/pma1-URA3 (20) |
| CTM1+2+3+4 | HO/HO Mata/Matα ade6-1/ade6-1 trp5-1/trp5-1 leu2-1/leu2-1 lys1-1/lys1-1 ura3-1/ura3-1 pma1-URA3/pma1-URA3 (20) |
| C. albicans | |
| ATCC 10261 | American Type Culture Collection, Manassas, Va. |
| FR2 | Derived from SGY-243 (1) |
| KB | Pfizer, Sandwich, United Kingdom |
| C. dubliniensis | |
| CD36 | Clinical isolate; D. C. Coleman, School of Dental Science, University of Dublin, Dublin, Republic of Ireland |
| CD43 | Clinical isolate; D. C. Coleman, School of Dental Science, University of Dublin, Dublin, Republic of Ireland |
| C. glabrata | |
| CBS138 | Schimmelecultures, Baarn, The Netherlands |
| 850821 | ESR: Health, NZCDCa |
| C. krusei | |
| 850920 | ESR: Health, NZCDC, Wellington, NZ |
| B2399 | ESR: Health, NZCDC, Wellington, NZ |
| IFO 0011 | Institute for Fermentation, Osaka, Japan |
| 89.102 | ESR: Health, NZCDC |
| C. parapsilosis | |
| 90.493 | ESR: Health, NZCDC |
| 425 | School of Dentistry, Kagoshima University, Japan |
| C. tropicalis IFO 0618 | Institute for Fermentation, Osaka, Japan |
| C. neoformans ATCC 90112 | New Zealand Reference Culture Collection, ESR, Wellington, NZ |
ESR, Institute of Environmental Sciences and Research Limited; NZCDC, New Zealand Centre for Disease Control, Wellington, New Zealand.
Chemicals, antifungals, and reagents.
The following chemicals and antifungals used in this study were obtained from the indicated sources: fluconazole (FLC; Diflucan), Pfizer Laboratories Limited, Auckland, New Zealand; itraconazole (ITC), Janssen Research Foundation, Beerse, Belgium; oligomycin, aurovertin B, sodium metavanadate, sodium azide, ATP, HEPES, tetramethylrhodamine isothiocyanate (TRITC), and dog kidney Na+,K+-ATPase, Sigma, St. Louis, Mo.; phenylmethylsulfonyl fluoride, Roche Diagnostics NZ Limited, Auckland, New Zealand; and l-ascorbic acid, ammonium molybdate, and dimethyl sulfoxide (DMSO), BDH, Poole, United Kingdom. ITC, oligomycin, and the peptide combinatorial library pools and subpools were prepared as stock solutions dissolved in DMSO. Agarose, minimum essential medium with Earle's salts, fetal bovine serum, and l-glutamine for tissue culture were from Gibco (Invitrogen Corporation, Auckland, New Zealand). The Live/Dead Viability/Cytotoxicity kit was from Molecular Probes (Eugene, Oreg.).
Peptide and combinatorial library synthesis.
The d-octapeptide combinatorial library and peptides were synthesized manually by using solid-phase 9-fluorenylmethoxy carbonyl chemistry (31, 33). The peptides identified in stage 4 and those used in lead optimization studies were manually synthesized and purified by high-pressure liquid chromatography (HPLC) with a Shimadzu LC-6 system equipped with a 15-μm, 300-Å Phenomenex guard column and a separation column (250 by 21.20 mm). Acetonitrile gradients were used to separate the naked peptide from the mono-substituted 4-methoxy-2,3,6-trimethylbenzensulfonyl (Mtr) peptides and any other contaminants. The following buffers were used: buffer A, 2% acetonitrile and 0.1% trifluoroacetic acid (TFA); buffer B, 90% acetonitrile and 0.1% TFA. The peptides were analyzed with a Waters Alliance HT 2790 HPLC instrument coupled to a Micromass ZMD 4000 electrospray mass spectrometer.
Yeast whole-cell assays. (i) Peptide MICs and MFCs.
The MICs of peptide pools, subpools, or peptides were determined by a microdilution method in 96-well microplates, as described by Niimi et al. (31). The minimum fungicidal concentration (MFC) was determined by plating 5-μl samples from wells with no visible growth on nonselective YPD medium. The MFC was the minimum concentration that gave no CFU. For the time course experiments, the percent cell death was determined by calculating the number of CFU obtained from treated and untreated samples at suitable time intervals. The initial cell density was an optical density at 600 nm (OD600) of 0.01, or ∼105 CFU per ml, in CSM at pH 7.0. Fungicidal activity was defined as the activity that resulted in >98% cell death.
(ii) Glucose-dependent proton pumping.
A 100-ml culture of T48 cells grown in YPD medium to mid-log phase (OD600 = 3) was washed twice in 150 ml of ice-cold deionized distilled water and stored overnight in the same volume of ice-cold water. The cells were washed twice in deionized distilled water and resuspended at an OD600 of 16 in 2 mM HEPES-NaOH (pH 7.0). Samples of 1 ml were incubated in the presence or absence of peptide for 30 min at room temperature and washed twice with 1 ml of ice-cold deionized distilled water adjusted to pH 5.0 with 2 mM HCl. Three duplicate 50-μl portions from each sample of cells were placed in the wells of a 96-well microplate containing 35 μg of bromophenol blue/ml (from a 140-μg/ml stock in 0.04 M NaOH adjusted to pH 5.0 with 0.2 M HCl) and 50 mM KCl at pH 5.0. The KCl abolished the membrane potential of the plasma membrane and increased the net rate of glucose-dependent proton pumping by about threefold. To make a final volume of 200 μl, 50 μl of 8% d-glucose (pH 5.0) was added to all wells except a pair of control wells, to which deionized distilled water at pH 5.0 was added. The cell density baseline wells also contained 0.5 mM HCl to give a pH of ∼3.5. The bromophenol blue indicator was monitored at 590 nm at 20-s intervals in an EL340 microplate reader, with the plates shaken at low speed for 5 s immediately prior to each measurement. The mean maximum rate of glucose-dependent proton pumping was determined for the duplicates of each cell sample by using Delta Soft software (Biometalics Inc, Princeton, N.J.).
(iii) Drug chemosensitization assay.
Checkerboard drug chemosensitization assays were performed to evaluate the effects on the cells of simultaneous exposure to a peptide and an azole (FLC or ITC), as described previously (31).
(iv) Agarose diffusion chemosensitization assay.
CSM − ura (20 ml) containing FLC (120 μg/ml) was solidified with 0.6% agarose in a culture plate (Nunc, Roskilde, Denmark). AD/PDR5+ cells (1 × 105 to 2 × 105) in 5 ml of melted CSM − ura top medium containing 0.4% agarose and FLC (at the same concentration used in the base medium) were overlaid on the agar. An appropriate volume of peptide (<10 μl) was applied to a sterile 3MM blotting paper disk (diameter, 5 mm; Whatman), dried at 37°C for 1 to 2 h, and placed onto the solidified medium surface. The cells were incubated at 30°C for 48 h. FLC-free medium was also used in the assay to assess the direct effects of the test peptide on the growth of strains AD/PDR5+ and AD/PDR5−.
(v) Preparation of TRITC-labeled peptides and confocal microscopy of labeled cells.
The peptide d-NH2-ASARRR-CONH2 was labeled with TRITC, as described previously (31).
TRITC-labeled BM2 was prepared by reacting resin-bound BM2 (120 mg, ∼34 nmol) with TRITC (5 mg dissolved in 2 ml of N-methylpyrrolidone [NMP]), followed by the addition of diisopropylethylamine (23 μmol). After 24 h of shaking in the dark, the reddish resin was washed with NMP and then CH2Cl2-CH3OH (50:50) and dried under vacuum overnight. TRITC-BM2 was cleaved from the resin by treatment with TFA-trimethylsilylbromide-thioanisole-ethanedithiol-p-cresol (69:12.5:11:5.5:2) at 0°C for 45 min. The free peptide was diluted with H2O (50 ml), and the aqueous layer washed with diethyl ether (twice, with 60 ml each time), followed by ethyl acetate (twice, with 60 ml each time). After neutralization with (NH4)2CO3, the red aqueous layer was freeze-dried. The crude peptide was purified by preparative HPLC with a C18 reverse-phase 15-μm column (Synchroprep 15R503), a flow rate of 2 ml/min, and a linear gradient from 5% buffer B at 0 min to 23% buffer B at 5 min and 60% buffer B at 45 min (buffer A-H2O-CH3CN-TFA [98:2:0.1]; buffer B-H2O-CH3CN-TFA [10:90:0.1]). Fractions containing TRITC-BM2 were detected at 555 nm, combined, and freeze-dried.
Yeast cells (OD600 = 1.0) were incubated in 50 μl of CSM − ura (pH 7.0) containing 10 μM TRITC-peptide at room temperature for 30 min. Some samples were also harvested by centrifugation (5,600 × g) and washed with 100 μl of CSM − ura. The cells in 10-μl samples were visualized by confocal microscopy with a Zeiss LSM510 confocal laser scanning microscope with a Zeiss Axiovert 200 M inverted microscope configuration (Zeiss, Jena, Germany).
(vi) Rh6G uptake and efflux.
Rhodamine 6G (Rh6G) uptake in the absence of glucose and Rh6G efflux in the presence and the absence of glucose were measured for AD/PDR5+ and AD/PDR5− cells, as described previously (31).
In vitro assays. (i) Isolation of plasma membranes.
Plasma membranes from S. cerevisiae strain T48 were used in the Pma1p ATPase activity screens, while the Candida and C. neoformans strains used for Pma1p ATPase assays are listed in Table 1. Plasma membranes obtained from strains AD/PDR5+ and AD/PDR5− were used for Pdr5p ATPase assays (31). Cells were grown in YPD liquid medium at 30°C with shaking at 200 rpm and were harvested when they were in the diauxic phase of growth (OD600 ≈ 7). Plasma membranes were separately prepared for the Pma1p ATPase and the Pdr5p ATPase assays by previously described methods (31). The protein concentration was determined by a micro-Bradford assay (Bio-Rad Laboratories, Hercules, Calif.) with bovine gamma globulin as the standard. Protein profiles were examined in sodium dodecyl sulfate-polyacrylamide (8%) gels stained with Coomassie blue.
(ii) Pma1p ATPase inhibition assay.
Pma1p ATPase assays were carried out at pH 7.0 on plasma membrane preparations with 2 to 5 μg of protein per assay and incubation at 30°C for 20 min, as described previously (24), with the inclusion of oligomycin (10 μM) and aurovertin B (10 μg/ml). The peptides were preincubated with membranes in the assay cocktail for 5 min, and the reaction was initiated by the addition of 6 mM ATP together with excess (7 mM) MgCl2. In ATP protection experiments, 6 mM ATP was added 5 min prior to peptide addition, and the reaction was initiated by the addition of 7 mM MgSO4 after a further 5 min of incubation.
(iii) Pdr5p ATPase inhibition assay.
The Pdr5p ATPase inhibition assays were carried out as described previously (31). The oligomycin-sensitive ATPase-specific activity of hyperexpressed Pdr5p was corrected for the activities of other ATPases by subtraction of the approximately fivefold lower background oligomycin-sensitive specific activity detected in plasma membranes from strain AD/PDR5− (31).
(iv) Dog kidney Na+,K+-ATPase inhibition assay.
Dog kidney Na+,K+-ATPase inhibition assays were carried out in a 120-μl reaction mixture containing 20 μg of dog kidney Na+,K+-ATPase, 2.4 mM ATP, 3.6 mM MgCl2, 150 mM NaCl, and 15 mM KCl in 59 mM MES-Tris buffer (pH 7.0) for 30 min at 37°C. Peptides were preincubated for 5 min with the dog kidney enzyme, and the reaction was started by the addition of 25 μl of Mg-ATP. The Pi released was quantitated as described previously (24).
Toxicity for human cells. (i) Hemolysis.
The lytic effects of the peptides on human erythrocytes were assessed at 405 nm by the method of Helmerhorst et al. (11).
(ii) Toxicities of peptides for cultured human epithelial cells.
Assays for determination of the toxicities of peptides for cultured human epithelial cells were conducted as described previously (31).
RESULTS
Preparation and properties of the d-octapeptide combinatorial library.
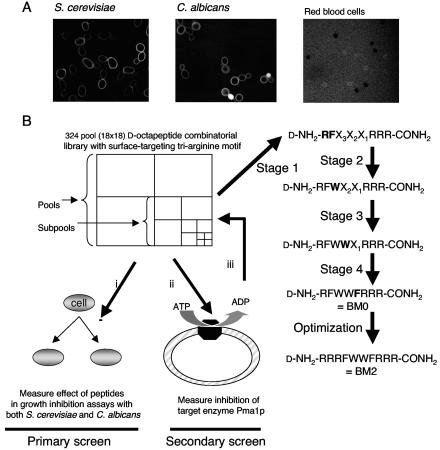
A soluble, 1.8-million-member, 324-pool, combinatorial peptide library of the form d-NH2-A-B-X3-X2-X1-RRR-CONH2 (in which A and B are known) was prepared by using manual solid-phase 9-fluorenylmethoxy carbonyl chemistry by adapting a divide, synthesize, and mix strategy to a resin displaying the d-triarginine motif (31, 33). Peptides contained d-form rather than l-form amino acids (with cysteine and glycine excluded) to provide maximum biological stability during extended incubation with fungal cells. The C-terminal amidated triarginine motif was designed to mimic the patches of positive charge found in many cationic antimicrobial peptides. Arginine was chosen instead of histidine or lysine to maintain the positive charges over a wide, physiologically relevant pH range. The triarginine motif was expected to selectively target microbial cell surfaces and also to confer membrane impermeability because recent studies suggest that the passage of d-peptides across plasma membranes requires at least six consecutive arginine residues in the peptide (21). Experiments with model TRITC-labeled d-hexapeptides showed that constructs with an amidated C-terminal triarginine motif were excluded from intact red blood cells and fungal cells, such as those of S. cerevisiae and Candida species (C. albicans, C. glabrata, C. krusei, and C. tropicalis), and were concentrated in the fungal cell wall (Fig. 1A) due to an ionic interaction with the negatively charged phosphomannan that characterizes all known fungal cell walls. Binding of the TRITC-labeled model peptide d-ASARRR-CONH2 was observed with the isolated cell walls of S. cerevisiae but not with protoplasts or cell walls that had been treated to degrade endogenous phosphomannan (data not shown).
FIG. 1.
Screening of a d-octapeptide combinatorial library containing a C-terminal triarginine motif. (A) The TRITC-labeled synthetic peptide d-NH2-ASARRR-CONH2 (5 μM), prepared as described in Materials and Methods, was incubated in 5 mM HEPES buffer (pH 7.0) with S. cerevisiae T48 or C. albicans ATCC 10261 or in phosphate-buffered saline with washed human red blood cells for 30 min; and the mixture was viewed by confocal microscopy. (B) In stage 1, a 1.8-million-member d-octapeptide combinatorial library consisting of 324 pools that each contained approximately equal quantities of 5,832 peptide sequences of the form d-NH2-ABX3X2X1RRR-CONH2 was screened (i) for inhibitors of the growth of both S. cerevisiae T48 and C. albicans ATCC 10261 by using peptide concentrations of 25 and 400 μg/ml (by weight), respectively. In a secondary screen (ii), the pools that showed the most potent inhibition of the growth of both S. cerevisiae and C. albicans strains were assayed for inhibition of the ATP-protected ATPase activity of S. cerevisiae Pma1p by using permeable plasma membrane vesicles. This identified the peptide pool d-NH2-RF X3X2X1RRR-CONH2 as the best candidate for deconvolution (iii). Three cycles of resynthesis, guided by the primary and secondary screens (stages 2 to 4), identified d-NH2-RFWWFRRR-CONH2 as a possible lead peptide sequence. This was confirmed by showing that HPLC-purified d-NH2-RFWWFRRR-CONH2 was a potent antifungal inhibitor of Pma1p. The amino acid(s) selected at each stage is given in boldface.
d-Octapeptide combinatorial library deconvolution. (i) Screens.
Because the essential and surface-exposed Pma1p enzyme is the major plasma membrane protein, we expected that inhibition of Pma1p by surface-active d-octapeptide combinatorial library pools would be detected by simple cell growth assays and in vitro assays of Pma1p activity (Fig. 1B). The primary screen (i) identified the most active peptide library pools (stage 1) by measuring growth inhibition with both S. cerevisiae T48 and C. albicans ATCC 10261. A plating test was used to measure the viabilities of cells in wells in which growth was eliminated by exposure to peptide pools. A secondary screen (ii) measured the inhibition of the ATP-protected, vanadate-sensitive ATPase activity of a permeable plasma membrane fraction from S. cerevisiae T48 (26). The most suitable pool (iii) identified in stage 1 was then deconvoluted by cycles of resynthesis and bioassay that sequentially determined the optimal amino acids for positions X3 (stage 2), X2 (stage 3), and X1 (stage 4). Informative supplementary assays included measurement of glucose-dependent proton pumping in S. cerevisiae cells and germ tube formation in C. albicans (27).
(ii) Screening of combinatorial library pools and subpools.
Primary screens in CSM − ura buffered to pH 7.0 with 10 mM MES-HEPES determined the potency of each pool after 48 h of growth of S. cerevisiae T48 (25 μg of peptide pool/ml) and after 24 h of growth of C. albicans ATCC 10261 (400 μg of peptide pool/ml). The growth of the S. cerevisiae strain was >16-fold more sensitive to active combinatorial library pools than the C. albicans strain. For both test strains only 3% of the pools gave >90% growth inhibition, and pools with an N-terminal alanine (position A) and pools with tryptophan at position B were growth inhibitory at the greatest frequencies. The growth inhibition profiles showed that the primary sequences of the A and B amino acids in each d-octapeptide were important. For example, the AR pool but not the RA pool inhibited growth at the concentrations of peptide tested. Pools AR, AM, AF, AW, RF, and LW, which showed >90% inhibition of S. cerevisiae and C. albicans growth at 25 and 400 μg/ml, respectively, were tested for inhibition of the ATP-protected, vanadate-sensitive plasma membrane ATPase activity of strain T48. This secondary screen was designed to identify pools that targeted Pma1p without affecting its cytoplasmic ATP binding site. Under the assay conditions at pH 7.0, the model peptide d-NH2-ASARRR-CONH2 had no effect on ATP-protected ATPase activity, indicating that the amidated triarginine motif was not an inhibitor. In contrast, the AR, AM, AF, AW, RF, and LW d-octapeptide pools inhibited Pma1p ATPase activity with 50% inhibitory concentrations (IC50s) <25 μg/ml, while pools like AN, which were not growth inhibitory in the stage 1 screen, had IC50s >100 μg/ml. Three pools (pools AW, AM, and RF), which best represented the most active growth-inhibitory and ATPase-inhibitory pools, were selected for deconvolution. These pools were fungicidal in microdilution assays, inhibited glucose-dependent proton pumping by S. cerevisiae, and blocked germ tube formation in C. albicans (data not shown). The solubilities of the peptides from the AM and AW pools caused difficulties during bioassays, which made their deconvolution too problematic. Deconvolution of the RF (d-NH2-RFX3X2X1RRR-CONH2) pool (Fig. 1B, stages 2 to 4) selected the antifungal Pma1p inhibitor d-NH2-RFWWFRRR-CONH2 (BM0).
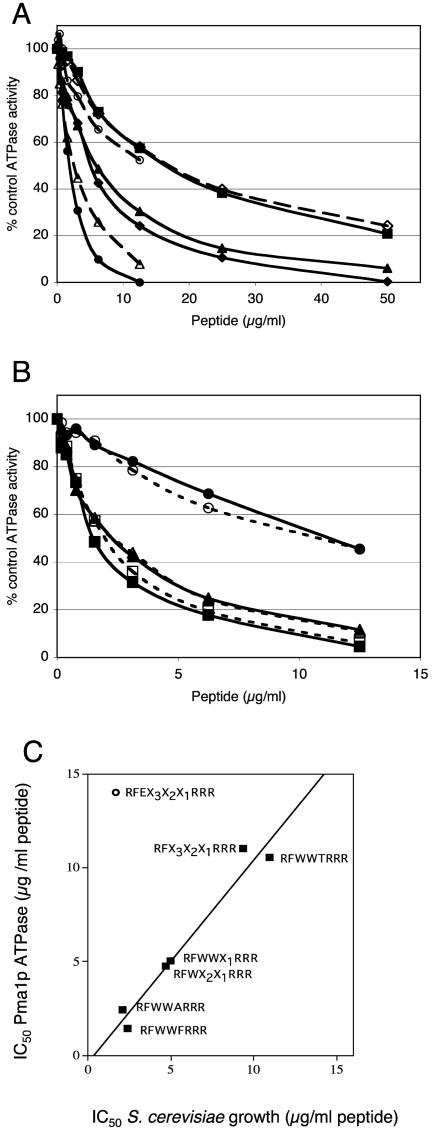
The most active peptide pools and subpools gave steep cell growth inhibition profiles (generally an eightfold difference in peptide concentration between normal growth and no visible growth), consistent with a single target. The inhibition of Pma1p ATPase activity was also consistent with a 1-to-1 interaction, with complete inhibition generally occurring within a 10-fold range of the peptide concentration (Fig. 2A). Inhibition of Pma1p ATPase activity and inhibition of S. cerevisiae growth by individual peptides in the d-NH2-RFWWX1RRR-CONH2 subpool showed a similar pattern. The peptides fell into three main groups. d-NH2-RFWWFRRR-CONH2 (IC50 = 1.4 μg/ml) was the strongest ATPase inhibitor, d-NH2-RFWWARRR-CONH2 (IC50 = 2.4 to 5 μg/ml) and most other peptides had intermediate inhibitory activities, and d-NH2-RFWWTRRR-CONH2 (IC50 > 11 μg/ml) was the weakest inhibitor. The strong correlation between the IC50 for inhibition of ATPase activity (ATP protected; see below) and the IC50 for inhibition of S. cerevisiae growth by members of the d-NH2-RFX3X2X1RRR-CONH2 pool (Fig. 2C) identified Pma1p as the most likely growth-inhibitory target of the peptides selected from this pool. A discarded growth-inhibitory subpool, d-NH2-RFEX2X1RRR-CONH2, did not show this correlation, indicating that its primary target was not Pma1p. d-NH2-RFWWFRRR-CONH2, d-NH2-RFWWARRR-CONH2, and d-NH2-RFWWTRRR-CONH2 all inhibited Pma1p with apparent affinities that were not affected by the presence of protective 6 mM ATP (Fig. 2B). This result suggested noncompetitive enzyme inhibition at a site other than the active site.
FIG. 2.
Stage 2 to 4 secondary screens of the d-NH2-RFX3X2X1RRR-CONH2 pool, and the relationship between inhibition of growth and Pma1p ATPase activity. (A) The Pma1p ATPase inhibition profiles for plasma membranes from S. cerevisiae strain T48 were measured under conditions of ATP protection, as described in Materials and Methods for selected pools and subpools. Symbols: ▪, d-NH2-RFX3X2X1RRR-CONH2; ▴, d-NH2-RFWX2X1RRR-CONH2; ♦, d-NH2-RFWWX1RRR-CONH2; ⋄, d-NH2-RFEX2X1RRR-CONH2; ○, d-NH2-RFWWTRRR-CONH2; ▵, d-NH2-RFWWARRR-CONH2; •, d-NH2-RFWWFRRR-CONH2. (B) The Pma1p inhibition profiles for the plasma membrane from S. cerevisiae strain T48 were measured in the presence and the absence of ATP protection, as described in Materials and Methods for selected subpools from stage 4 of the deconvolution. Symbols: ▴ and ▵, d-NH2-RFWWARRR-CONH2; ▪ and □, d-NH2-RFWWFRRR-CONH2; • and ○, d-NH2-RFWWTRRR-CONH2; filled lines and symbols, presence of ATP; dashed lines and empty symbols, absence of ATP. (C) The relationships between the IC50 for S. cerevisiae strain T48 and the IC50 for the ATP-protected Pma1p activity for T48 plasma membranes were obtained from the MIC profiles and the data in panels A and B.
Comparison with related peptides.
The peptide d-NH2-RFWWFRRR-CONH2 (denoted BM0) was synthesized, purified and quantitated spectrophotometrically by assuming a molar extinction coefficient of 5,600 cm−1 for each tryptophan at 280 nm. BM0 inhibited the growth of several S. cerevisiae mutants and C. albicans (Tables 2 and 3). Purified BM0 at pH 7.0 showed comparable growth-inhibitory activity (MIC = 0.9 to 1.8 μM) with control strain T48, mutants of Pma1p with significant N-terminal and C-terminal deletions, a control strain (AD/PDR5−) from which multiple membrane transporters were deleted, and an isogenic derivative of strain AD/PDR5− that overexpressed the Pdr5p multidrug efflux pump (strain AD/PDR5+). These data indicated that purified peptide BM0 is an active principal compound in the library that inhibited growth independently of the glucose-dependent signaling pathways that modify Pma1p activity via its C-terminal negative regulatory domain. Furthermore, hyperexpression of a functional ABC transporter in strain AD/PDR5+, which confers high-level resistance to the antifungal FLC (600 μg/ml), did not significantly alter the growth-inhibitory activity of this peptide. A derivative of BM0 (denoted BM20), which contains an arginine side chain-blocking Mtr group, showed antifungal activity that was about twofold weaker than that of BM0. Although BM20 was a major component of the tryptophan-containing library pools (up to two-thirds of the product in the d-NH2-RFWWFRRR-CONH2 subpool), it was more potent than BM0 against dog kidney Na+,K+-ATPase and was therefore discarded as a drug candidate.
TABLE 2.
Effects of target modification on BM0 activity against S. cerevisiae
| Strain | MIC (μM) at pH 7.0 |
|---|---|
| T48 (wild type) | 0.91 |
| ΔN | 0.91 |
| ΔC | 0.91 |
| ΔNΔC | 0.91 |
| ATCC10261 | 15 |
| CTM1+2 | 0.46 |
| CTM3+4 | 0.91 |
| CTM1+2+3+4 | 0.91 |
| AD/PDR5+ | 1.82 |
| AD/PDR5− | 1.82 |
TABLE 3.
Effects of stereochemistry and structure on BM0 antifungal activity
| Peptide | MIC90 (μM)a
|
IC50 (μM) for S. cerevisiae Pma1p (7.0) | |||
|---|---|---|---|---|---|
| S. cerevisiae (7.3)b | C. albicans (7.5) | C. glabrata (7.5) | C. tropicalis (7.5) | ||
| d-NH2-RFWWFRRR-CONH2 (BMO) | 0.46 | 7.3 | 7.3 | 7.3 | 2.6 |
| d-NH2-RRRFWWFR-CONH2 | 1.6 | 6.3 | 3.1 | 6.3 | 1.8 |
| l-NH2-RFWWFRRR-CONH2 | 1.9 | 31 | 15 | 7.7 | 3.8 |
| l-NH2-RRRFWWFR-CONH2 | 2.0 | 16 | 8.0 | 4.0 | 2.2 |
| d-NH2-RFWWF-CONH2 | 11 | 88 | 44 | 44 | 17 |
| d-CH3CO-RFWWFRRR-CONH2 | 2.0 | 8.0 | 8.0 | 8.0 | 5.8 |
MIC90, minimum concentration at which there is >90% growth inhibition compared to no-peptide control.
Values in parentheses are pHs.
The ∼20-fold greater sensitivity of S. cerevisiae compared with that of C. albicans to growth inhibition by individual d-octapeptide pools and subpools during library deconvolution was reflected in studies with purified peptides (Tables 3 and 4). The purified peptides also showed the position-specific effects of amino acids expected from the deconvolution. Amino acids A (BM47) and F (BM0) at position X1 gave comparable MICs for S. cerevisiae (Table 4), but the compound with A gave a fivefold lower potency than that with F against C. albicans. The lower potency of BM0 against C. albicans (Table 3) was independent of specific features of transmembrane loops TM1-TM2 and TM3-TM4 in the C. albicans ATPase, either singly or in combination, when it was tested at pH 7.0 (Table 2). This was determined by using fully functional S. cerevisiae strains (strains CTM1+2, CTM3+4, and CTM1+2+3+4) containing chimeric Pma1p engineered with C. albicans Pma1p sequences that replace the homologous transmembrane loops in S. cerevisiae Pma1p (20).
TABLE 4.
Effects of BM0 sequence modifications on antifungal activity against S. cerevisiae
| Name of peptide | Sequencea | MIC/MFCb (μM) at pH 7.3 |
|---|---|---|
| BM0 | d-NH2-RFWWFRRR-CONH2 | 0.625s/2.5c |
| BM1 | d-NH2-RRFWWFRRR-CONH2 | 0.625s/1.25c |
| BM2 | d-NH2-RRRFWWFRRR-CONH2 | 0.625c |
| BM3 | d-NH2-RWWWFRRR-CONH2 | 0.625s/2.5c |
| BM4 | d-NH2-RFFWFRRR-CONH2 | 0.625s/2.5c |
| BM5 | d-NH2-RFLWFRRR-CONH2 | 0.625c/2.5c |
| BM6 | d-NH2-RFWFFRRR-CONH2 | 1.25c |
| BM7 | d-NH2-RFWNFRRR-CONH2 | 1.25c |
| BM8 | d-NH2-RFWKFRRR-CONH2 | 0.625s/5c |
| BM9 | d-NH2-RFWRFRRR-CONH2 | 1.25c |
| BM47 | d-NH2-RFWWARRR-CONH2 | 0.625s/2.5c |
Amino acid residues that differ from those in BM0 are given in boldface.
s, fungistatic; c, fungicidal.
The effects of BM0 and congeners of this peptide on S. cerevisiae and pathogenic Candida species were studied (Table 3). Removal of the C-terminal amidated triarginine motif diminished the potency of BM0 as an inhibitor of S. cerevisiae Pma1p and reduced the activities against all species tested. The triarginine motif was therefore critical for activity. BM0 modified at its N terminus with a carboxylmethyl group was two- to fourfold less potent against both S. cerevisiae cells and S. cerevisiae Pma1p, but this modification had no effect on the pathogenic fungi tested. BM0 modified as an isothiohydantoin with TRITC failed to inhibit either S. cerevisiae growth or S. cerevisiae Pma1p ATPase activity (data not shown). The effects of carboxymethylation and conjugation with TRITC on the activity of BM0 against S. cerevisiae indicated a preference for an N-terminal positive charge or an incompatibility with bulky residues. The l-amino acid version of the BM0 and both the d- and l-amino acid versions of the reverse peptide (NH2-RRRFWWFR-CONH2) were as potent as BM0 as inhibitors of S. cerevisiae Pma1p but were about fourfold less active against S. cerevisiae cells. l-NH2-RFWWFRRR-CONH2 was approximately two- to fourfold less active against both C. albicans and C. glabrata, while l-NH2-RRRFWWFR-CONH2 was approximately twofold less active against C. albicans. These results implied that proteolysis and/or peptide stereochemistry modestly affected the interaction between BM0 and its target. The activity of d-NH2-RRRFWWFR-CONH2 indicated that N-terminal extension of BM0 with one or more arginine residues might improve the potency of the peptide against the fungal pathogens. This possibility was tested with d-NH2-RRFWWFRRR-CONH2 (BM1) and d-NH2-RRRFWWFRRR-CONH2 (BM2). The addition of each arginine to the N terminus of BM0 increased the fungicidal activity of BM0 against S. cerevisiae by twofold (Table 4). Replacement of the amino acids within the hydrophobic core of BM0 with residues that either were second choices in S. cerevisiae deconvolution or were likely to increase potency against C. albicans but not S. cerevisiae did not give a significant (twofold or greater) change in the MIC or the MFC relative to that of the parent peptide. The cycles of resynthesis and bioassay used to identify BM0 therefore appear to involve a relatively robust process that is not deleteriously affected by the 18-fold changes of diversity at each step in the deconvolution. Once the primary pool had been identified, a more efficient positional scanning deconvolution strategy (34) could therefore have been applied.
Effects of lead peptides on the growth and Pma1p ATPase activities of S. cerevisiae and pathogenic fungi.
The MICs of BM0 and BM2 for S. cerevisiae strains at pH 7.0 and 7.3 and for pathogenic non-C. albicans Candida strains and C. neoformans at pH 7.0 and 7.5 are shown in Table 5. In general, BM2 was about twice as effective as BM0 at inhibiting the growth of these organisms, including an FLC-resistant strain of S. cerevisiae which overexpressed the Pdr5p multidrug efflux pump. BM2 was also severalfold more potent than BM0 as a fungicide against S. cerevisiae and each fungal pathogen tested by both microdilution and time course assays. In addition, BM2 was 5- to 10-fold more effective than BM0 as an inhibitor of the vanadate-sensitive Pma1p ATPase activities of S. cerevisiae and the fungal pathogens. The antifungal activities of both BM0 and BM2 were strongly pH dependent, but their inhibition of the isolated enzyme was not (data not shown). Since the peptides should retain an essentially constant overall positive charge in the pH range of 7.0 to 7.5 (arginine pKa, >12; α-amino acid pKa, >9), the pH dependence of their antifungal activities suggests that the accessible negative charge of the target cell, but not the target membrane, increases between pH 7.0 and 7.5. The most likely explanation is that the cell wall phosphomannan attracted a pH-dependent reservoir of positively charged BM0 and BM2 peptides to the vicinity of the ATPase.
TABLE 5.
Effects of BM0 and BM2 on growth, viability, and Pma1p activity for S. cerevisiae and fungal pathogens
| Species and strain | MIC90/MFCa (μM)
|
IC50 (μM) for Pma1p ATPase
|
Concn (μM)/time (h) to >98% cell deathb
|
|||||
|---|---|---|---|---|---|---|---|---|
| BM0
|
BM2
|
BM0 (pH 7.0) | BM2 (pH 7.0) | BM0 (pH 7.0) | BM2 (pH 7.0) | |||
| pH 7.0 | pH 7.3 (7.5)c | pH 7.0 | pH 7.3 (7.5)c | |||||
| S. cerevisiae | ||||||||
| T48 | 1.3s | 0.63s/2.5c | 5s | 0.63s/2.5c | 2.5 | 0.5 | 100/17.5 | 20/17.5 |
| AD/PDR5+ | 10c | 2.5c | 5c | 1.25c | ND | ND | ND | ND |
| AD/PDR5− | 10c | 1.3s/2.5c | 2.5s/10c | 0.63s/1.25c | ND | ND | ND | ND |
| Fungal pathogen | ||||||||
| C. albicans ATCC 10261 | >20 | 20s | >10 | 5s/10c | 20 | 2 | 100/2.0 | 40/2.0 |
| C. glabrata CBS138 | >20 | >10 | 10s | 5s/10c | 10 | 2 | 100/17.5 | 20/17.5 |
| C. krusei B2399 | >20 | 20s | >10 | 10s | 6 | 1 | 100/17.5 | 40/1.0 |
| C. tropicalis IF0168 | 20c | 10s/20c | 5s | 2.5s/5c | 5 | 0.6 | 25/0.5 | 5/0.5 |
| C. neoformans ATCC 90121 | 10c | 5s/10c | 5c | 2.5s/10c | 3 | 1.5 | 25/0.5 | 10/2.0 |
MIC90, minimum concentration at which there is >90% growth inhibition compared to no-peptide control (s, fungistatic); MFC, peptide concentration at which no CFU was recovered (c, fungicidal).
Concentration of peptide that gives >98% cell death/time required to achieve this effect. ND, not determined.
The pHs in parentheses are those used in tests with fungal pathogens other than S. cerevisiae. A single pH value indicates that the same pH was used for both S. cerevisiae and the other fungal pathogens.
The IC50s obtained from the inhibition profiles, with or without preincubation with 6 mM ATP, indicated that BM2 inhibition of S. cerevisiae, C. albicans, C. glabrata, C. krusei, C. tropicalis, and C. neoformans Pma1p was noncompetitive with ATP binding (Table 6). This was confirmed in an independent experiment that determined the Km of ATP for each of these ATPases after preincubation in the presence or the absence of BM2 at concentrations that inhibit enzyme activity by ∼50% (Table 6). The Km values of ATP changed less than twofold under these conditions, indicating the absence of strong competition between the inhibitor and the enzyme substrate. Thus, the site of inhibition of BM2 on the Pma1p of S. cerevisiae and the fungal pathogens tested may be remote from the cytoplasmic ATP binding site. This was supported by experiments with BM2 derivatized with TRITC via its N-terminal amino group. The MIC and the MFC of TRITC-BM2 for strain T48 were about 2.5 and 5 μM, respectively, at pH 7.3. The ATP-protected IC50 of TRITC-BM2 for S. cerevisiae Pma1p was 1.4 μM at pH 7.0. Apart from a small proportion of T48 cells (<20%) that appeared to be dead because they showed high concentrations of intracellular fluorescence after exposure to 5 μM TRITC-BM2, the labeled peptide bound almost exclusively and avidly to the surfaces of live cells. Unlike the fluorescently labeled model peptide TRITC-d-ASARRR-CONH2, which bound to the cell wall but which did not inhibit Pma1p (Fig. 1A), TRITC-BM2 was not removed by simple washing.
TABLE 6.
Effects of BM0 and BM2 on Pma1p and Na+,K+-ATPase kinetic propertiesa
| Strain | Vmax (μmol/min/mg of protein) | Km (mM) for ATP | IC50 (μM)
|
|
|---|---|---|---|---|
| Without ATP | With ATP | |||
| S. cerevisiae T48 | 1.2-1.3 | 1.4-2.0 | ||
| With BM0 (3 μM) | 0.62 | 1.9 | 2.5 | 2.5 |
| With BM2 (0.8 μM) | 0.73 | 3.3 | 0.5 | 0.5 |
| C. albicans ATCC 10261 | 0.91-1.1 | 3.5-3.8 | ||
| With BM0 (20 μM) | 0.74 | 6.8 | 22 | 22 |
| With BM2 (4 μM) | 0.49 | 7.5 | 2.3 | 2.3 |
| C. glabrata CBS138 | 0.22 | 1.8 | ||
| With BM2 (3 μM) | 0.10 | 1.7 | 1.3 | 1.8 |
| C. krusei B2399 | 2.6 | 3.5 | ||
| With BM2 (0.8 μM) | 1.8 | 4.0 | 0.6 | 1.0 |
| C. tropicalis IFO 0618 | 3.8 | 3.0 | ||
| With BM2 (0.5 μM) | 2.4 | 4.0 | 0.4 | 0.5 |
| C. neoformans ATCC 90112 | 0.20 | 0.66 | ||
| With BM2 (1.5 μM) | 0.09 | 0.65 | 1.0 | 1.3 |
| Dog Na+, K+-ATPase | ||||
| With BM0 | ND | ND | ND | 62 |
| With BM2 | ND | ND | ND | 25 |
All assays were conducted at pH 7.0. ND, not determined.
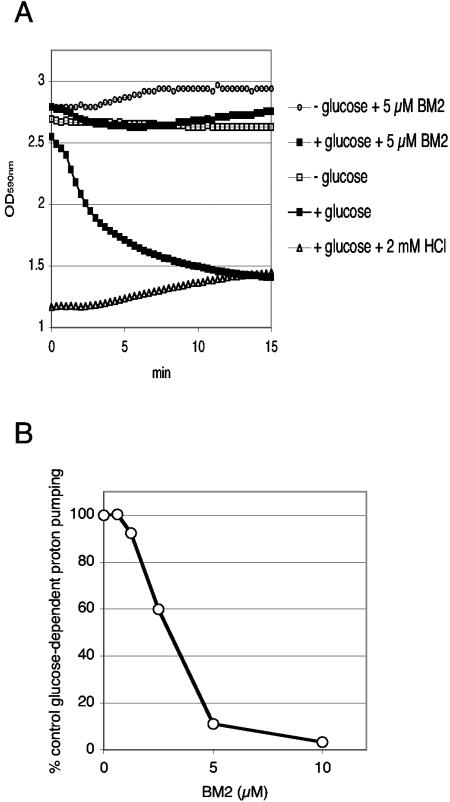
The stability of the BM2-yeast cell interaction was used to show that BM2 inhibited glucose-dependent proton efflux, which in S. cerevisiae is mediated by Pma1p (Fig. 3). Glucose-starved yeast cells resuspended in 2 mM HEPES (pH 7.0) were incubated at room temperature for 30 min in the presence or the absence 5 μM BM2. Samples of washed cells were then assayed for glucose-dependent proton pumping at pH 5.0 by a microplate assay with bromophenol blue as a sensitive pH reporter. The glucose-dependent proton efflux obtained in the presence of 50 mM KCl (mean maximum rate, 0.314 OD590/min) was almost completely abolished by the pretreatment with 5 μM BM2 (mean maximum rate, 0.063 OD590/min). The BM2 pretreatment gave a barely detectable rate of net proton uptake in the absence of glucose (mean maximal rate of leakage, −0.025 OD590/min, compared with 0.004 OD590/min for the control). Pretreatment with 10 μM BM2 abolished pumping and increased the net proton uptake rate by a further 40%. Other studies showed that T48 cells treated with 10 μM amphotericin B (9.2 μg/ml) rapidly buffered the bromophenol blue medium to a pH higher than 5. The inhibition of glucose-dependent proton pumping by preincubation with BM2 at pH 7.0 was dose dependent for both strain T48 and strain AD/PDR5+, with BM2 IC50s of about 3 μM.
FIG. 3.
Inhibition of glucose-dependent proton pumping in S. cerevisiae by BM2. (A) T48 cells were incubated at pH 7.0 in the presence or absence of 5 μM BM2 at pH 7.0 and were prepared for proton pumping, as described in Materials and Methods. Proton pumping was initiated at time zero by adding 2% glucose where indicated, and the OD590 between 3 and 1 was monitored as an indicator of pH in the range of 5.0 to 3.5. Typical results from a representative experiment are shown. (B) T48 cells preincubated with BM2 at pH 7.0 were assayed for glucose-dependent proton pumping at pH 5, as described for panel A.
Effects of lead peptides on host targets.
A commercially available dog kidney Na+,K+-ATPase preparation provided a convenient surrogate host P-type ATPase. Although BM0 and BM2 inhibited dog kidney Na+,K+-ATPase with IC50s of 62 and 25 μM, respectively, they were much stronger inhibitors of fungal Pma1ps at neutral pH (IC50 range, 0.5 to 2.5 μM) (Table 6). The ratios of IC50 for dog kidney Na+,K+-ATPase/IC50 for Pma1p obtained with BM0 and BM2 under ATP-protected conditions with the mammalian enzyme and the enzymes from S. cerevisiae and the range of fungal pathogens showed that the extension of the N terminus of BM0 with arginine residues increased the specificity of the interaction with all the fungal Pma1ps compared with that with the Na+,K+-ATPase. This was primarily achieved through an increased affinity for the fungal enzymes rather than by a decreased affinity for the dog kidney Na+,K+-ATPase.
BM2 was weakly hemolytic (Fig. 4A). Less than 3% of human red blood cells were hemolyzed by 50 μM BM2. Under identical conditions, 1.5 μM amphotericin B (1.4 μg/ml) caused >50% hemolysis of red blood cells. The effect of BM2 on host cells was also evaluated with the HEp-2 cultured human epithelial cell line as an experimental model. HEp-2 cells were exposed to 13.5, 27, or 54 μM BM2 for either 3 or 24 h before assay by confocal microscopy with the Molecular Probes live/dead reagent. Figure 4B shows that 27 μM BM2 was not toxic for the cultured cells, even after 24 h, but at 54 μM about 15% of the cells were no longer viable due to the effects of the peptide. The toxicity caused by 54 μM BM2 was rapid, with a similar percentage of the HEp-2 cells compromised after both 3 and 24 h of exposure.
FIG. 4.
Effects of BM2 on host cells. (A) Hemolysis of red blood cells by BM2 and amphotericin B was measured as described in Materials and Methods. (B) The effect of BM2 on the viability of cultured HEp-2 cells was measured after 3 and 24 h, as described in Materials and Methods.
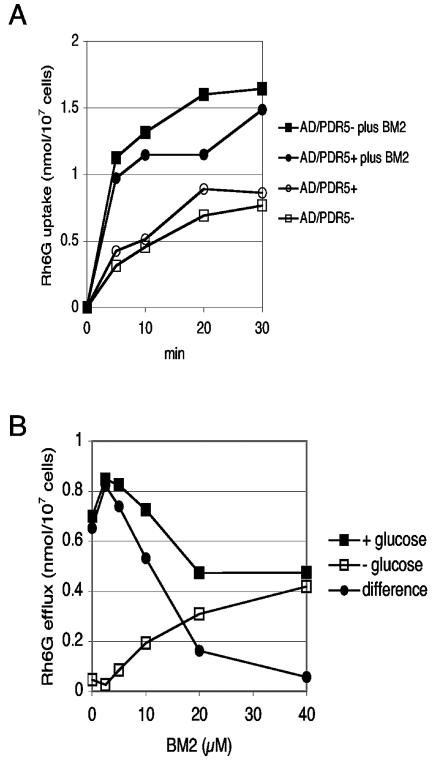
Effect of BM2 on Rh6G uptake and efflux.
Rh6G uptake by starved and deenergized strains AD/PDR5+ and AD/PDR5− appeared to be saturated after 30 min, and 10 μM BM2 stimulated the uptake by two- to threefold throughout the time course of the assay (Fig. 5A). This suggests that 10 μM BM2 modified the cell permeability and/or diminished the membrane potential of the plasma membrane.
FIG. 5.
Effects of BM2 on Rh6G uptake and efflux. (A) Rh6G uptake by S. cerevisiae AD/PDR5+ and AD/PDR5− cells was measured in the presence or the absence of 10 μM BM2, as described in Materials and Methods. (B) Rh6G efflux from S. cerevisiae AD/PDR5+ cells was measured in the presence or the absence of 0.2% glucose, as described in Materials and Methods. Glucose-dependent efflux is defined as the Rh6G efflux detected in the presence of glucose minus the efflux detected in the absence of glucose.
Glucose-dependent Rh6G efflux in strain AD/PDR5+ cells provided a measure of the activity of the Pdr5p ABC transporter. BM2 inhibited glucose-dependent Rh6G efflux from preloaded AD/PDR5+ cells (Fig. 5B). Low concentrations of BM2 (<2.5 to 5 μM) activated glucose-dependent Rh6G efflux by about 25%. Glucose-dependent Rh6G efflux was inhibited 80% with 20 μM BM2 and was fully inhibited by 40 μM BM2. BM2 concentrations >10 μM caused significant efflux of Rh6G in the absence of glucose, indicating that BM2 permeabilized the yeast plasma membrane. In either the presence or the absence of glucose, BM2 (>10 μM) also caused Rh6G efflux from AD/PDR5− cells that was essentially identical to the glucose-independent efflux seen with AD/PDR5+ cells (data not shown). This accounted for about 30, 40, and 50% of the control efflux with 10, 20, and 40 μM BM2, respectively. The activation of Rh6G efflux by 2.5 to 5 μM BM2 is consistent with a loss of cell membrane potential due to Pma1p inhibition. The dose-dependent inhibition of glucose-dependent Rh6G efflux by BM2 at concentrations ≥5 μM may involve direct or indirect effects on the activity of the Pdr5p multidrug efflux pump, which transports Rh6G across the plasma membrane. BM2 was found to inhibit the ATPase activity of the hyperexpressed Pdr5p multidrug efflux pump, with an IC50 of 4 ± 1 μM (n = 3) at pH 7.0.
Chemosensitization of drug resistance.
The functional hyperexpression of the endogenous S. cerevisiae Pdr5p multidrug efflux pump in an FLC-sensitive background from which six other multidrug efflux pumps were deleted confers high-level FLC resistance (MIC = 600 μg/ml) (31). In agar diffusion assays 4 nmol of peptides BM0, BM20, and BM2 gave larger zones of inhibition when the AD/PDR5+ strain was exposed to FLC at a sub-MIC (120 μg/ml) (Fig. 6). Of the group of peptides, BM2 had the greatest toxicity for the yeast in the absence of FLC and was also the most effective chemosensitizer. Checkerboard chemosensitization assays (31) demonstrated that sub-MICs of BM2 chemosensitized the AD/PDR5+ strain to FLC (Table 7).
FIG. 6.
Chemosensitization of multidrug efflux in fungi by BM2. An agarose diffusion assay was used to test the effects of 4 nmol of selected peptides on strain AD/PDR5+ in the presence or the absence of 120 μg of FLC per ml. Disk 0, DMSO control; disk 1, BM0; disk 2, BM20; disk 3, BM2.
TABLE 7.
BM2 chemosensitizes S. cerevisiae constructs and fungal pathogens to azoles
| Drug tested and species | Strain (hyperexpressed protein) | Concn used for chemosensitization/MIC
|
Fold chemosensitization by BM2 (for FLC or ITC MIC) | |
|---|---|---|---|---|
| BM2 (μM) | FLC or ITC (μg · ml−1) | |||
| FLC | ||||
| S. cerevisiae | T48 | 1.25/2.5 | 5/20 | 4 |
| AD/PDR5− | 1.25/2.5 | 0.5/1 | 2 | |
| AD/PDR5+ (Pdr5p) | 2.5/5 | 80/600 | 7.5 | |
| AD1-8u−a | 0.75/3 | 1/2 | 2 | |
| AD/PDR5 (Pdr5p)a | 1.75/3 | 320/40 | 8 | |
| AD/CaCDR1 (CaCdr1p)a | 1.5/2 | 80/320 | 4 | |
| AD/MDR1 (Mdr1p) | 1.5/2 | 3.75/60 | 16 | |
| AD/ERG11 (Erg11p) | 1.5/2 | 0.5/2 | 4 | |
| AD/CaCDR2 (CaCdr2p) | 2/3 | 20/80 | 4 | |
| AD/CgCDR1-1B (CgCdr1p)a | 1.75/3 | 40/320 | 8 | |
| C. albicans | ATCC 10261 | 10/20 | 0.25/>64 | >256 |
| FR2 | 5/20 | 2/>32 | >16 | |
| KB | 20/40 | 10/>160 | >16 | |
| C. glabrata | CBS138 | 5/10 | 20/>80 | >4 |
| 850821 | 10/40 | 80/160 | 2 | |
| 85090 | 5/40 | 2.5/>40 | >16 | |
| C. tropicalis | IF00618 | 1/3 | 1.25/>160 | 128 |
| C. krusei | B2399 | 10/20 | 20/>80 | >4 |
| IFO 0011 | 10/20 | 20/>80 | >4 | |
| 89.102 | 10/20 | 40/>80 | >2 | |
| C. dubliniensis | CD36 | 5/20 | 0.3/>20 | >60 |
| CD43 | 2.5/20 | 1.25/>20 | >16 | |
| C. parapsilosis | 90.493 | 10/40 | 2/8 | 4 |
| 425 | 40/40 | >32/>32 | Not detected | |
| ITC | ||||
| S. cerevisiae | AD1-8u− | 1.5/3 | 0.2/0.8 | 4 |
| AD/PDR5 | 1.75/3 | 10/>40 | >4 | |
| AD/CDR1 | 1.75/3 | 20/>40 | >2 | |
| AD/CgCDR1-1B | 1.75/4 | 20/>40 | >2 | |
| C. glabrata | CBS138 | 5/10 | 0.2/>0.8 | >4 |
| C. krusei | B2399 | 10/20 | 0.05/>0.8 | >16 |
| IFO 0011 | 10/20 | 0.05/0.8 | 16 | |
| 89.102 | 10/20 | 0.05/>0.8 | >16 | |
Incubated for 72 h instead of 48 h to allow full development of resistance phenotype.
We used an S. cerevisiae strain from which seven ABC pumps were deleted to functionally hyperexpress ABC and MFS multidrug efflux pumps or Erg11p (23, 30, 31). Heterologous hyperexpression of the C. albicans ABC multidrug efflux pumps Cdr1p and Cdr2p gave high-level FLC resistance, hyperexpression of the MFS pump Mdr1p gave intermediate-level FLC resistance, while expression of Erg11p gave low-level resistance. Treatment with BM2 at concentrations within a narrow window of sub-MICs chemosensitized each hyperexpressing strain to FLC (Table 7). BM2 also chemosensitized several, but not all, wild-type and drug-resistant strains of C. albicans, C. glabrata, C. tropicalis C. dubliniensis, and C. parapsilosis to FLC. Because the ITC resistance of the S. cerevisiae strains hyperexpressing the ABC transporter exceeded the concentration at which ITC is soluble (<32 μg/ml), it was more difficult to quantitate the BM2-dependent chemosensitization to ITC compared with that to FLC for these strains. Table 7 shows that Pdr5p, C. albicans Cdr1p, and C. glabrata Cdr1p were significantly chemosensitized to ITC by BM2 when they were hyperexpressed in S. cerevisiae.
Both FLC-resistant strains and the trailing tail of resistance obtained with some strains of so-called sensitive organisms like C. albicans were chemosensitized by treatment with BM2 (Table 7). C. krusei is intrinsically resistant to FLC, and this organism was chemosensitized modestly (approximately four- to eightfold) by BM2. Furthermore, C. krusei infections are preferably treated with ITC. BM2 gave at least a 16-fold chemosensitization of C. krusei to ITC. Similar results were obtained with two further C. krusei clinical isolates, and BM2 chemosensitized C. glabrata strain CBS138 to both FLC and ITC more than fourfold.
DISCUSSION
Antimicrobial peptides related to BM0 and BM2.
A model d-hexapeptide containing a C-terminal amidated triarginine motif was excluded from the interior of fungal cells and concentrated at their surface in a reversible association with cell wall phosphomannan. This membrane-impermeant surface-targeting motif was incorporated into a 1.8-million-member d-octapeptide combinatorial library. Screening of the combinatorial library for inhibitors of S. cerevisiae and C. albicans growth and S. cerevisiae Pma1p ATPase activity identified BM0 (d-NH2-RFWWRRR-CONH2) as a broad-spectrum surface-active antifungal. A recent study (39) of short (six- to seven-residue) arginine- and tryptophan-rich antimicrobial peptides has identified several factors important for antimicrobial activity mediated by general membrane permeabilization. These include the presence of three tryptophan and three arginine residues, a free N-terminal group, and an amidated C terminus. BM0 matches these criteria fairly closely. Optimization of the fungicidal and Pma1p-inhibitory activities of BM0 via an N-terminal extension of two arginine residues led to the more potent d-decapeptide BM2 (d-NH2-RRRFWWFRRR-CONH2). Hong et al. (12) previously found that a greater net charge increased antifungal activity without improving the antibacterial activity of their model antimicrobial peptide, NH2-KKVVFKVKFK-CONH2. Similarly, Kundu et al. (15) obtained a 17-fold increase in activity against C. albicans and C. neoformans by using a combinatorial approach to extend and optimize the lead antifungal hexapeptide NH2-l-R-d-W-d-F-l-I-d-F-l-H-CONH2 to a nonapeptide, NH2-l-R-d-W-d-F-l-I-d-F-l-H-l-K-l-K-l-R-CONH2 or NH2-l-R-d-W-d-F-l-I-d-F-l-H-l-K-l-K-l-I-CONH2. The positively charged and aromatic residues of such peptides are thought to preferentially interact with the interfacial region of negatively charged microbial membranes but not with erythrocyte membranes, which are predominantly zwitterionic. BM2 has some similarity to the tryptophan-rich, cationic antimicrobial peptide indolicidin (l-NH2-ILPWKWPWWPWRR-COOH) (8); antifungal hexapeptides such as l-CH3CO-RRWWRR-CONH2, d-CH3CO-RKKWFW-CONH2, and d-CH3CO-RKKWFW-CONH2 that were obtained from combinatorial libraries (13, 17); and the d-enantiomer of the lactoferricin active core (d-NH2-RRWQWRMKK-CONH2) (42). Purified d-, l-, and d-reverse versions of the lactoferricin active core all inhibit S. cerevisiae Pma1p, but with IC50s that are at least 10-fold higher than those of BM2 (B. C. Monk and K. Niimi, unpublished).
Mode of action.
BM2 and its congeners are a novel class of fungicide. They are among the most potent noncovalent inhibitors of Pma1p known at present. BM2 has an IC50 of 0.5 μM for S. cerevisiae Pma1p and inhibits the Pma1ps from several pathogenic Candida species and C. neoformans, with IC50s in the range of 0.5 to 2.5 μM at pH 7.0. Kinetic studies with purified plasma membrane preparations with comparable Pma1p contents from S. cerevisiae and several fungal pathogens indicate that BM2 has little or no effect on the binding of ATP to the cytoplasmic active site of Pma1p. TRITC-labeled BM2, which retained significant antifungal activity and noncompetitively inhibited Pma1p (IC50 = 1.4 μM), was strongly bound to the surfaces of S. cerevisiae cells. This interaction withstood washing, unlike the model hexapeptides used to demonstrate the membrane impermeability conferred by the triarginine motif and its interaction with the phosphomannan of the fungal cell wall. Similarly, S. cerevisiae cells pretreated with ≤10 μM BM2 at neutral pH gave dose-dependent inhibition of glucose-dependent proton pumping at pH 5.0, without substantial leakage of cellular buffering capacity.
The ∼1,600-Da lead d-decapeptide BM2 has an aromatic core (FWWF) bounded by a pair of triarginine motifs. Preliminary low-temperature aqueous solution nuclear magnetic resonance studies indicate that although the N terminus of BM2 is disordered, there is significant order within the aromatic core and the C-terminal triarginine amidated motif (B. C. Monk and A. R. Hayman, unpublished results), consistent with i, i + 4 tryptophan-arginine pi-cation interactions between the core tryptophans and the most C-terminal arginines (where i is the position of an amino acid in the primary sequence); this order makes a favorable energetic contribution to the helix structure (38). The pH dependence of fungal growth inhibition by BM2 suggests the involvement of negative charges located at or near the cell surface, with a pKa of about 7. A likely candidate in intact cells is wall phosphomannan. In vitro enzyme inhibition experiments with purified plasma membranes in the presence or the absence of purified cell walls show that Pma1p has a higher affinity than cell wall phosphomannan for BM2 (B. C. Monk and S. Tsao, unpublished observations).
The precise nature of the interaction between Pma1p and BM2 has yet to be determined. Although BM2 bound to the cell surface did not permeabilize yeast cells at concentrations that affect cell growth or Pma1p-mediated proton pumping (≤5 μM), a more general interaction with the lipid bilayer at high BM2 concentrations (>10 μM) is likely and an anesthetic-like effect at the Pma1p-lipid interface at lower peptide concentrations has not been excluded. Pma1p in yeasts is located in lipid rafts that are enriched for sphingolipids and ergosterol (2). FLC treatment of AD/PDR5+ cells, designed to slightly lower the plasma membrane ergosterol content, reduced glucose-dependent proton pumping activity by 20% without modifying the dose dependence of BM2 inhibition of proton pumping (data not shown). It therefore seems unlikely that low BM2 concentrations perturb a critical interaction at the Pma1p protein-lipid interface, which would also need to be significantly different between S. cerevisiae and C. albicans Pma1p. Could the binding of BM2 to the Pma1p ectodomain inhibit the enzyme? Cell surface amino acid mutations in transmembrane loops can affect the activity of Pma1p (29). Comparative modeling of S. cerevisiae and C. albicans Pma1ps with the program MODELLER (18) and a model of the homologous Neurospora crassa enzyme as the threading template (14) indicate where BM2 might bind to the Pma1ps. Both of our models have analogous ionic and hydrogen bond networks linking the five ectodomain loops at or near the enzyme surface that could be disrupted by BM2 binding. Drug susceptibility experiments, including the use of S. cerevisiae Pma1p chimeras containing C. albicans transmembrane loops TM1-TM2 and/or TM3-TM4, suggest that the accessible amino acid differences (S. cerevisiae→C. albicans) D714→N and D720→N in TM5-TM6, F778→L in TM7-TM8, and E847→Q in TM9-TM10 rather than those in TM1-TM2 and TM3-TM4 might determine the differential affinities of the two enzymes for BM2. This hypothesis can be tested by cross-linking studies, mutational analysis, and structural resolution of inhibitor-enzyme complexes.
BM2 and azole chemosensitization.
Like the related d-octapeptide KN20 (31), ≥10 μM BM2 caused the bidirectional flux of Rh6G, indicating membrane permeabilization that could also chemosensitize S. cerevisiae to azoles. The mechanisms by which BM2 at sub-MICs (≤2.5 μM) causes chemosensitization to azoles are not known. BM2 may directly affect the Pdr5p drug efflux pump, for which the BM2 IC50 is 4 μM in vitro. The BM2-mediated chemosensitization to FLC of S. cerevisiae cells which functionally hyperexpress the FLC resistance determinant Pdr5p, the ABC transporters or MFS transporters, or the FLC target Erg11p from pathogenic fungi might involve indirect inhibition of these enzyme activities via Pma1p (22). Pma1p generates the membrane electrochemical gradient that powers both Mdr1p and the uptake of nutrients that are metabolized to produce the ATP and the reduced NADPH required by the ABC transporters and Erg11p, respectively. BM2 (5 μM) inhibited glucose-dependent proton pumping by 90% and activated the glucose-dependent efflux of the preloaded Pdr5p substrate Rh6G in S. cerevisiae. The increased rate of Rh6G pumping is consistent with a collapsed membrane potential, which would explain the chemosensitization of Mdr1p-mediated FLC resistance in strain AD/MDR1 (Table 7). Further experiments are needed to determine if BM2 inhibition of Pma1p at sub-MICs affects the plasma membrane electrochemical gradient, which would impose metabolic limitations that affect lanosterol 14α-demethylase activity and azole efflux by ABC transporters.
Chemosensitization of the tail of FLC resistance in some but not all C. glabrata and C. dubliniensis strains may involve inhibition of efflux pumps and/or Erg11p. For example, matrix-assisted laser desorption ionization-time of flight mass spectrometric analysis of tryptic fingerprints demonstrated that exposure to FLC induces the expression of the 170-kDa CgCdr1p multidrug efflux pump and Erg11p in C. glabrata strain CBS138 (32). BM2 chemosensitized this strain to both FLC and ITC. Two mechanisms of drug resistance have been identified in the innately FLC-resistant organism C. krusei. Increased drug efflux has been implicated in ITC resistance (40), while the relative insensitivity of C. krusei to FLC compared with that of C. albicans may be due to structural differences in Erg11p (6). Both of these mechanisms may contribute in the superior BM2-dependent chemosensitization of C. krusei to ITC compared with that to FLC. BM2 chemosensitization of yeast to azoles support, but do not prove, our contention (22) that substantial inhibition of Pma1p can enhance the potencies of azoles that are affected by several common resistance mechanisms. Compounds with BM2-like activities could increase the overall potency and commercial lifetimes of drugs like FLC and ITC and possibly avoid the evolution of drug resistance from innate low-level resistance.
Prospects for application.
BM2 and its congeners provide a proof of principle for several concepts. The interaction of the amidated triarginine motif with the fungal cell wall deposits a reservoir of membrane-impermeant peptide at the fungal cell surface but not at the surface of host cells. This motif-based selectivity could be used to deliver libraries and prodrugs to cell surface enzymes of pathogens and cancer cells. Cationic peptides also bind to a variety of artificial surfaces used by microbiologists and surgeons (7), indicating that derivatives of BM2 could be used to sterilize implants and catheters. The identification of fungicidal Pma1p inhibitors extends therapeutic targeting of P-type ATPases from established targets such as the Na+,K+-ATPase and the gastric H+,K+-ATPase of humans to the P-type ATPases of fungal, bacterial, and parasitic systems (28). Such targets could circumvent known drug resistance mechanisms and, by operating at the cell surface, minimize possibilities for the emergence of drug resistance via detoxification and drug efflux pathways. A facet of the activity of BM2 that requires improvement, however, is the achievement of fungicidal potencies against different pathogenic fungi at concentrations in the submicromolar range. The small-scale production of d-peptide inhibitors like BM2 is technically demanding and expensive, and although the inhibitor differentiates between fungal Pma1ps and a surrogate host enzyme (dog kidney Na+,K+-ATPase), it causes low-level hemolysis and detectable toxicity in cultured HEp-2 cells at the concentrations required to rapidly kill several of the major fungal pathogens. While the observation that oral administration of the lactoferricin active core d-enantiomer did not cause lethal toxicity to mice when it was administered at up to 100 mg/kg of body weight (42) is encouraging, significant challenges will likely be retention of a window of fungal specificity and the recovery of bioavailable peptide-based compounds. We expect that the main uses of peptide-based Pma1p inhibitors like BM2 will be in superficial applications at mucosal surfaces or on implants and catheters, as markers for high-throughput screens of pharmacophore libraries, and in the structural resolution of enzyme-inhibitor complexes that will help define the features of Pma1p required for structure-directed drug discovery.
Acknowledgments
We acknowledge the financial support of the Health Research Council of New Zealand, the Marsden Fund, the New Zealand Lottery Grants Board, and the U.S. National Institutes of Health (grant R21DE15075 to R.D.C.) and The Wellcome Trust for a major equipment grant.
We thank Sarah Tsao for excellent technical assistance, Erwin Lamping for providing yeast strains that functionally hyperexpress drug resistance determinants, Susan Cutfield for creating the Pma1p models, A. Brett Mason for his contribution to some early aspects of the paper, and the University of Otago Electron Microscopy Unit for assistance with confocal microscopy.
REFERENCES
- 1.Albertson, G. D., M. Niimi, R. D. Cannon, and H. F. Jenkinson. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnat, M., S. Keranen, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decottignies, A., B. Rogers, M. Kowlaczkowski, E. Carvajal, E. Balzi, G. Conseil, K. Niimi, A. Di Pietro, B. C. Monk, and A. Goffeau. 2002. The pleiotropic drug ABC transporters from Saccharomyces cerevisiae, p. 157-176. In I. T. Paulsen and K. Lewis (ed.), Microbial multidrug efflux. Horizon Scientific Press, Wymondham, United Kingdom.
- 4.De Lucca, A. J., and T. J. Walsh. 1999. Antifungal peptides: novel therapeutic compounds against emerging pathogens. Antimicrob. Agents Chemother. 43:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eraso, P., A. Cid, and R. Serrano. 1987. Tight control of the amount of yeast plasma membrane ATPase during changes in growth conditions and gene dosage. FEBS Lett. 224:193-197. [DOI] [PubMed] [Google Scholar]
- 6.Fukuoka, T., D. A. Johnston, C. A. Winslow, M. J. de Groot, C. Burt, C. A. Hitchcock, and S. G. Filler. 2003. Genetic basis for differential activities of fluconazole and voriconazole against Candida krusei Antimicrob. Agents Chemother. 47:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giacometti, A., O. Cironi, R. Ghiselli, L. Goffi, F. Moccheggiani, A. Riva, G. Scalise, and V. Saba. 2000. Polycationic peptides as prophylactic agents against methicillin-susceptible or methicillin-resistant Staphylococcus epidermidis vascular graft infection. Antimicrob. Agents Chemother. 44:3306-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock, R. E., and G. Diamond. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8:402-410. [DOI] [PubMed] [Google Scholar]
- 9.Hancock, R. E., and M. G. Scott. 2000. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. USA 97:8856-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock, R. E. W., and R. Lehrer. 1998. Cationic peptides; a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 11.Helmerhorst, E. J., W. Van't Hof, E. C. Veerman, I. Simoons-Smit, and A. V. Nieuw Amerongen. 1997. Synthetic histatin analogues with broad-spectrum antimicrobial activity. Biochem. J. 326:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong, S. Y., T. G. Park, and K. H. Lee. 2001. The effect of charge increase on the specificity and activity of a short antimicrobial peptide. Peptides 22:1669-1674. [DOI] [PubMed] [Google Scholar]
- 13.Houghten, R. A., C. Pinilla, S. E. Blondelle, J. R. Appel, C. T. Dooley, and J. H. Cuevo. 1991. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature 354:84-86. [DOI] [PubMed] [Google Scholar]
- 14.Kuhlbrandt, W., J. Zeelen, and J. Dietrich. 2002. Structure, mechanism, and regulation of the Neurospora plasma membrane H+-ATPase. Science 297:1692-1696. [DOI] [PubMed] [Google Scholar]
- 15.Kundu, B., T. Srinivasan, A. P. Kesarwani, A. Kavishwar, S. K. Raghuwanshi, S. Batra, and P. K. Shukla. 2002. Identification of novel antifungal nonapeptides through the screening of combinatorial peptide libraries based on a hexapeptide motif. Bioorg. Med. Chem. Lett. 12:1473-1476. [DOI] [PubMed] [Google Scholar]
- 16.Li, X. S., M. S. Reddy, D. Baev, and M. Edgerton. 2003. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J. Biol. Chem. 278:28553-28561. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Garcia, B., E. Perez-Paya, and J. F. Marcos. 2002. Identification of novel hexapeptides bioactive against phytopathogenic fungi through screening of a synthetic peptide combinatorial library. Appl. Environ. Microbiol. 68:2453-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marti-Renom, M. A., A. C. Stuart, A. Fiser, R. Sanchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291-325. [DOI] [PubMed] [Google Scholar]
- 19.Mason, A. B., T. B. Kardos, and B. C. Monk. 1998. Regulation and pH-dependent expression of a bilaterally truncated yeast plasma membrane H+-ATPase. Biochim. Biophys. Acta 1372:261-271. [DOI] [PubMed] [Google Scholar]
- 20.Mason, A. B., T. B. Kardos, D. S. Perlin, and B. C. Monk. 1996. Functional complementation between transmembrane loops of Saccharomyces cerevisiae and Candida albicans plasma membrane H+-ATPases. Biochim. Biophys. Acta 1284:181-191. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell, D. J., D. T. Kim, L. Steinman, C. G. Fathman, and J. B. Rothbard. 2000. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 56:318-325. [DOI] [PubMed] [Google Scholar]
- 22.Monk, B., and A. Goffeau. 2000. Platforms for antifungal discovery in the 21st century. Biochemist 20:20-25. [Google Scholar]
- 23.Monk, B. C., R. D. Cannon, K. Nakamura, M. Niimi, K. Niimi, D. R. K. Harding, A. R. Holmes, E. Lamping, A. Goffeau, and A. Decottignies. March 2003. Membrane protein expression system and its application in drug screening. International provisional patent PCT/NZ02/00163.
- 24.Monk, B. C., M. B. Kurtz, J. A. Marrinan, and D. S. Perlin. 1991. The cloning and characterization of the plasma membrane H+-ATPase from Candida albicans. J. Bacteriol. 173:6826-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monk, B. C., A. B. Mason, G. Abramochkin, J. E. Haber, D. Seto-Young, and D. S. Perlin. 1995. The yeast plasma membrane proton pumping ATPase is a viable antifungal target. 1. Effects of the cysteine-modifying reagent omeprazole. Biochim. Biophys. Acta 1239:81-90. [DOI] [PubMed] [Google Scholar]
- 26.Monk, B. C., C. Montesinos, C. Ferguson, K. Leonard, and R. Serrano. 1991. Immunological approaches to the transmembrane topology and conformational changes of the carboxyl terminal regulatory domain of the yeast plasma membrane H+-ATPase. J. Biol. Chem. 266:18097-18103. [PubMed] [Google Scholar]
- 27.Monk, B. C., M. Niimi, and M. G. Shepherd. 1993. The Candida albicans plasma membrane and H+-ATPase during yeast growth and germ tube formation. J. Bacteriol. 175:5566-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monk, B. C., and D. S. Perlin. 1994. Fungal plasma membrane proton pumps as promising new antifungal targets. Crit. Rev. Microbiol. 20:209-233. [DOI] [PubMed] [Google Scholar]
- 29.Na, S., D. S. Perlin, D. Seto-Young, G. Wang, and J. B. Haber. 1993. Characterization of yeast plasma membrane H+-ATPase mutant pma1-A135V and its revertants. J. Biol. Chem. 268:11792-11797. [PubMed] [Google Scholar]
- 30.Nakamura, K., M. Niimi, K. Niimi, A. R. Holmes, J. E. Yates, A. Decottignies, B. C. Monk, A. Goffeau, and R. D. Cannon. 2001. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob. Agents Chemother. 45:3366-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niimi, K., D. R. K. Harding, R. Parshot, A. King, D. J. Lun, A. Decottignies, M. Niimi, S. Lin, R. D. Cannon, A. Goffeau, and B. C. Monk. 2004. Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a d-octapeptide derivative. Antimicrob. Agents Chemother. 48:1256-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niimi, M., Y. Nagai, K. Niimi, S. Wada, R. D. Cannon, Y. Uehara, and B. C. Monk. 2002. Identification of two proteins induced by exposure of the pathogenic fungus Candida glabrata to fluconazole. J. Chromatogr. B 782:245-252. [DOI] [PubMed] [Google Scholar]
- 33.Ostresh, J. M., S. E. Blondelle, B. Dorner, and R. A. Houghten. 1996. Generation and use of nonsupport-bound peptide and peptidomimetic combinatorial libraries. Methods Enzymol. 267:220-234. [DOI] [PubMed] [Google Scholar]
- 34.Pinilla, C., J. R. Appel, P. Blanc, and R. A. Houghten. 1992. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. BioTechniques 13:901-905. [PubMed] [Google Scholar]
- 35.Portillo, F., and R. Serrano. 1989. Growth control strength and active site of yeast plasma membrane ATPase studied by site-directed mutagenesis. Eur. J. Biochem. 186:501-507. [DOI] [PubMed] [Google Scholar]
- 36.Serrano, R., M. C. Kielland-Brandt, and G. R. Fink. 1986. Yeast plasma membrane ATPase is essential for growth and has homology with (Na++K+), K+- and Ca2+-ATPase. Nature 319:689-693. [DOI] [PubMed] [Google Scholar]
- 37.Seto-Young, D., B. Monk, A. B. Mason, and D. S. Perlin. 1997. Exploring an antifungal target region in the plasma membrane H+-ATPase of fungi. Biochim. Biophys. Acta 1326:249-256. [DOI] [PubMed] [Google Scholar]
- 38.Shi, Z., C. A. Olson, and N. R. Kallenbach. 2002. Cation-pi interaction in model alpha-helical peptides. J. Am. Chem. Soc. 124:3284-3291. [DOI] [PubMed] [Google Scholar]
- 39.Strom, M. B., O. Rekdal, and J. S. Svendsen. 2002. Antimicrobial activity of short arginine- and tryptophan-rich peptides. J. Pept. Sci. 8:431-437. [DOI] [PubMed] [Google Scholar]
- 40.Venkateswarlu, K., D. W. Denning, N. J. Manning, and S. L. Kelly. 1996. Reduced accumulation of drug in Candida krusei accounts for itraconazole resistance. Antimicrob. Agents Chemother. 40:2443-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada, S., M. Niimi, K. Niimi, A. R. Holmes, B. C. Monk, R. D. Cannon, and Y. Uehara. 2002. Candida glabrata ABC transporters Cdr1p and Pdh1p expressed in a Saccharomyces cerevisiae strain deficient in membrane transporters show phosphorylation-dependent pumping properties. J. Biol. Chem. 277:46809-46821. [DOI] [PubMed] [Google Scholar]
- 42.Wakabayashi, H., H. Matsumoto, S. Terguchi, M. Takase, and H. Hayasawa. 1999. N-acylated and d-enantiomer derivatives of a nanomer core peptide of lactoferrin B showing improved antimicrobial activity. Antimicrob. Agents Chemother. 43:1267-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White, T., K. Marr, and R. Bowden. 1998. Clinical, cellular and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeaman, M. R., and N. Y. Yount. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27-55. [DOI] [PubMed] [Google Scholar]