Abstract
In 2002, the first two clinical isolates of vancomycin-resistant Staphylococcus aureus (VRSA) containing vanA were recovered in Michigan and Pennsylvania. Tn1546, a mobile genetic element that encodes high-level vancomycin resistance in enterococci, was present in both isolates. With PCR and DNA sequence analysis, we compared the Tn1546 elements from each isolate to the prototype Tn1546 element. The Michigan VRSA element was identical to the prototype Tn1546 element. The Pennsylvania VRSA element showed three distinct modifications: a deletion of nucleotides 1 to 3098 at the 5′ end, which eliminated the orf1 region; an 809-bp IS1216V-like element inserted before nucleotide 3099 of Tn1546; and an inverted 1,499-bp IS1251-like element inserted into the vanSH intergenic region. These differences in the Tn1546-like elements indicate that the first two VRSA isolates were the result of independent genetic events.
The first two clinical isolates of vancomycin-resistant Staphylococcus aureus (VRSA) were recovered from patients in Michigan and Pennsylvania in June and September of 2002, respectively (2, 3). The MIC of vancomycin was 1,028 μg/ml for the Michigan VRSA (MI-VRSA) isolate and 32 μg/ml for the Pennsylvania VRSA (PA-VRSA) isolate by broth microdilution (7). The vanA gene was detected in both isolates by PCR amplification and localized to a plasmid of either 58 (Michigan) or 127 (Pennsylvania) kb by Southern hybridization (2, 3, 11, 12). The MI-VRSA vanA plasmid was transferable to S. aureus COL by filter mating (12), but conjugal transfer of the PA-VRSA plasmid to other staphylococcal or enterococcal recipients has been unsuccessful. The SmaI pulsed-field gel electrophoresis patterns of both isolates fall within the USA100 lineage (New York/Japan clone), which is the most common staphylococcal pulsed-field type found in U.S. hospitals. However, the patterns are clearly distinguishable, indicating that the MI-VRSA and PA-VRSA isolates are not epidemiologically linked (11, 12).
The prototype vanA gene is present on transposon Tn1546, a mobile genetic element containing genes responsible for high-level glycopeptide resistance among enterococci and several other gram-positive organisms (1). Acquisition of vancomycin resistance usually involves horizontal transfer of a Tn1546-containing plasmid. Tn1546 is composed of nine genes: vanR, vanS, vanH, vanA, and vanX are required for the expression of resistance; orf1 and orf2 encode transposase and resolvase enzymes, respectively; vanY encodes a carboxypeptidase; and vanZ has an unknown function, although expression of this gene is associated with teicoplanin resistance (1, 5). In 1992, Noble et al. reported in vitro transfer of glycopeptide resistance from Enterococcus faecalis to S. aureus (8), but natural transfer was not documented until 2002. This report describes the similarities and differences of the genetic elements encoding vancomycin resistance in the MI-VRSA and PA-VRSA isolates and compares them with the prototype Tn1546 transposon from enterococci.
(These data were presented in part at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., September 2003 [abstract C1-1303].)
PCR amplification of the individual genes comprising Tn1546 was performed with an Applied Biosystems 9600 thermocycler (Applied Biosystems, Foster City, Calif.) in accordance with previously described protocols (4). PCR primers included P1 to P19, described by Arthur et al. (1), and additional primers selected from the sequence of Tn1546 with Oligo 6 software (Molecular Biology Insights, Inc., Cascade, Colo.) (Table 1). The DNA sequences were determined by dRhodamine Dye Terminator Cycle Sequencing and an ABI 377 automated DNA sequencer (Applied Biosystems). DNA sequences were aligned and compared with DNAsis for Windows (version 2.5; MiraiBio, Inc., San Francisco, Calif.).
TABLE 1.
Additional primers used for PCR and sequencing
| Primere | Sequence (5′→3′) | Reference |
|---|---|---|
| Tn1546R | GGAAAATGCGGATTTACAACGCTAAG | 14 |
| VanR4 (+) | (3907)a ATGCTTATAAATTCGGCCCTA | This study |
| VanS3 (+) | (4771)a CCGAGGGAAACTTGGGGATTG | This study |
| VanA3 (−) | (7002)a TATTGCAACTTTTATTCTATTCATG | This study |
| VanA4 (+) | (7984)a TTGATCGTATTAGCGTTAAAGGGG | This study |
| IS1251-1 (+) | (106)b AACCCAAAAGGAGGAATCAAG | This study |
| IS1251-2 (−) | (1057)b CTTTGAAGCCAGGTCGC | This study |
| IS1251-4 (+) | (1041)b GCGACCTGGCTTCAAAG | This study |
| 6113R (−) | (6113)a TATCGTTGCCATAACGC | 13 |
| 7875F (+) | (7875)a CCGCATTGTACTGAACG | 13 |
| 8544F (+) | (8544)a GCATATAGCCTCGAATGG | 13 |
| 9519F (+) | (9519)a ACCAGCAGGTTATAGTGAGC | 13 |
| 10687F (+) | (10687)a CTCGCCCGTAGGTGTGAAGTG | This study |
| 10778F (+) | (10778)a TTTAGTGCTGAGGAATTGG | 13 |
| IS1216V.A (+) | (90)c GGAAAGCAATTTCAGCAG | 13 |
| IS1216V.C (−) | (495)c CACTTGTAATAGAGGGGGC | 13 |
| IS1216V.E (+) | (749)c AGCTTAAATCATAGATACCGTAAGG | 13 |
| UP211 (+) | (211)d CCATCGATTATGAGGCTAGACA | This study |
| DN645 (−) | (663)d ATTCTTGCACCCAACGATA | This study |
The primer positions correspond to the sequence of Tn1546 (GenBank accession no. M97297).
The primer positions correspond to the sequence of IS1251 (GenBank accession no. AF148130).
The primer positions correspond to the sequence of IS1216V (GenBank accession no. AF093508).
The primer positions correspond to the sequence of the Tn1546-like element reported in this study.
+, forward primer; −, reverse primer (relative to the respective sequence).
DNA sequence analysis revealed that the vanA sequences from the two strains were identical to the vanA sequence of Tn1546 (GenBank accession no. M97297). However, PCR amplification of additional transposon elements from the MI-VRSA and PA-VRSA isolates suggested significant differences. By Long PCR (14), the entire 10.8-kb transposon was amplified from the MI-VRSA isolate and Tn1546-containing E. faecalis A256 (4, 9). The restriction patterns of the MI-VRSA and A256 Long PCR products obtained with EcoRI, EcoRV, HindIII, and XbaI were identical (data not shown) (14). With the same oligonucleotide primers, the complete transposon could not be amplified from the PA-VRSA isolate.
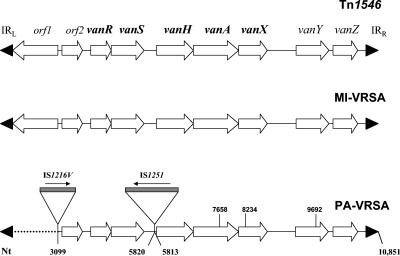
The PCR products obtained from the MI-VRSA isolate and E. faecalis A256 with all of the primer pairs reported by Arthur et al. from orf1 through vanZ were of the expected sizes. The DNA sequences of the MI-VRSA products were identical to those of the prototype Tn1546 element. Although amplification products from the vanR, vanS, vanH, vanA, vanX, vanY, and vanZ regions of the PA-VRSA isolate were of the predicted sizes, no products were obtained for the orf1 gene. However, PCR products that were larger than expected were generated from the orf1-orf2 and vanS-vanH intergenic regions. These data suggested that insertion sequences were present. This was confirmed by DNA sequence analysis, which revealed truncation of the 5′ region of the Tn1546-like element resulting in the loss of nucleotides 1 to 3098, which eliminated the orf1 region. Upstream from the truncated Tn1546 element was a 419-bp sequence with homology to bases 58293 to 58711 of E. faecalis V583 pTEF1 (GenBank accession no. AE016833). This sequence, represented by the dotted line in Fig. 1, was followed by an 809-bp sequence designated an “IS1216V-like” element (GenBank accession no. AF093508) that is in the same 5′→3′ orientation as the transposon (6). For confirmation of this result, we generated a 1,417-bp product with a primer (UP211) within the pTEF1 sequence that preceded the IS1216V-like element and the primer P6 (1), located in the orf2 region.
FIG. 1.
Comparison of the Tn1546-like elements in the MI-VRSA and PA-VRSA isolates to the prototype Tn1546 element. Numbers correspond to the nucleotide (Nt) positions in the prototype Tn1546 element. Deletions are indicated by dotted lines. IR, inverted repeat.
The Tn1546-like element sequence from the PA-VRSA isolate began at nucleotide 3099 of the prototype Tn1546 sequence (located between orf1 and orf2) and continued through nucleotide 5820 (in the vanS-vanH intergenic region). The elimination of the orf1 (transposase) region of the element may affect the mobility of the truncated transposon (13). The expression of vancomycin resistance may also be affected by the presence of the transposon on such a large plasmid in S. aureus (11). DNA sequence analysis also revealed a 1,499-bp sequence designated “IS1251-like” (GenBank accession no. AF148130) (10). This element was inserted downstream from position 5820 in the opposite orientation relative to the transposon but was in the same position and orientation as an insertion described by Handwerger et al. (5) in a Tn1546-like element from an E. faecium isolate. IS1251 and IS1251-like elements have been reported almost exclusively in U.S. isolates. The exceptions are two isolates from Ireland and one from Norway (5, 10). The IS1251-like element in the PA-VRSA isolate was also flanked by an 8-bp duplication of the target sequence (ATAATTTT) corresponding to bases 5813 to 5820 of Tn1546 (5, 13). The remainder of the Tn1546-like element (5,049 bp) had 99% nucleotide homology with the reported prototype sequence. Nucleotide substitutions identified at positions 7658 (T→C), 8234 (G →T), and 9692 (C →T) (Fig. 1) were the same as those described by Willems et al. (13) for Tn1546 type F2. The PA-VRSA isolate differs in that the IS1216V-like element is not in the reverse orientation and there is a larger deletion at the 5′ end of the PA-VRSA transposon (3,098 versus 889 bp). Tn1546 type F2 isolates have only been found in hospitalized patients in the United States (13).
Ligation of PA-VRSA EcoRI plasmid fragments into plasmid vector pUC19 enabled us to determine the sequence of the 3′ inverted repeat. The ligation mixture was used as the template for PCR amplification with the M13 reverse sequencing primer (GGAAACAGCTATGACCATG) [Strategies 6(1):15, 1993; Stratagene, La Jolla, Calif.] and a Tn1546-specific primer (10687F or 10778F). The PA-VRSA inverted repeat was identical to that reported in Tn1546.
In summary, vancomycin resistance in the MI-VRSA and PA-VRSA isolates is due to acquisition of vanA on plasmid-borne Tn1546-like elements. The MI-VRSA isolate acquired a Tn1546 element on 58-kb plasmid pLW1043 (12). The Tn1546-like element on the 127-kb plasmid (11) in the PA-VRSA isolate is quite different, lacking orf1 because of a truncation at the 5′ end and containing two insertion sequences; an IS1216V-like insertion sequence preceded the truncated Tn1546 sequence, and an IS1251-like insertion sequence was located in the vanS-vanH intergenic region. These differences in the Tn1546-like elements indicate that the first two VRSA isolates arose from independent genetic events.
REFERENCES
- 1.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 3.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, S. K. Fridkin, and the Vancomycin-Resistant Staphylococcus aureus Investigative Team. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 4.Clark, N. C., R. C. Cooksey, B. C. Hill, J. M. Swenson, and F. C. Tenover. 1993. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob. Agents Chemother. 37:2311-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Handwerger, S., J. Skoble, L. F. Discotto, and M. J. Pucci. 1995. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob. Agents Chemother. 39:362-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen, L. B. 1998. Internal size variations in Tn1546-like elements due to the presence of IS1216V. FEMS Microbiol. Lett. 169:349-354. [DOI] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 5th edition. NCCLS document M7-A5.National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 8.Noble, W. C., Z. Virani, and R. G. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195-198. [DOI] [PubMed] [Google Scholar]
- 9.Shlaes, D., A. Bouvet, C. Devine, J. H. Shlaes, S. Al-Obeid, and R. Williamson. 1989. Inducible, transferable resistance to vancomycin in Enterococcus faecalis A256. Antimicrob. Agents Chemother. 33:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonsen, G. S., M. R. Myhre, K. H. Dahl, O. Olsvik, and A. Sundsfjord. 2000. Typeability of Tn1546-like elements in vancomycin-resistant enterococci using long-range PCRs and specific analysis of polymorphic regions. Microb. Drug Resist. 6:49-57. [DOI] [PubMed] [Google Scholar]
- 11.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Characterization of a vancomycin-resistant clinical isolate of Staphylococcus aureus from Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 13.Willems, R. J., J. Top, N. van den Braak, A. van Belkum, D. J. Mevius, G. Hendriks, M. van Santen-Verheuvel, and J. D. van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodford, N., and J. M. Stigter. 1998. Molecular investigation of glycopeptide resistance in gram-positive bacteria, p. 579-615. In N. Woodford and A. P. Johnson (ed.), Methods in molecular medicine, vol. 15. Molecular bacteriology: protocols and clinical applications. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]