Summary
The importance of transgenerationally inherited epigenetic states to organismal fitness remains unknown as well-documented examples are often not amenable to mechanistic analysis or rely on artificial reporter loci. Here we describe an induced silenced state at an endogenous locus that persists, at 100% transmission without selection, for up to 13 generations. This unusually persistent silencing enables a detailed molecular genetic analysis of an inherited epigenetic state. We find that silencing is dependent on germline nuclear RNAi factors and post-transcriptional mechanisms. Consistent with these later observations, inheritance does not require the silenced locus, and we provide genetic evidence that small RNAs embody the inherited silencing signal. Notably, heritable germline silencing directs somatic epigenetic silencing. Somatic silencing does not require somatic nuclear RNAi but instead requires both maternal germline nuclear RNAi and chromatin modifying activity. Coupling inherited germline silencing to somatic silencing may enable selection for physiologically important traits.
Keywords: Transgenerational inheritance, epigenetics, piRNAs, nuclear RNAi, C. elegans
Graphical Abstract
Introduction
Most inherited traits arise from pre-existing DNA sequence variation which, for recessive traits, must become sufficiently frequent in the population to produce homozygotes. In contrast, dominantly transmitted epigenetic traits can appear within a single generation and can spread rapidly in the population in response to environmental conditions. Mitotically propagated stable epigenetic traits have been described in microbes, plants and animals (Jablonka and Raz, 2009). However, few transgenerational epigenetic phenomena have been mechanistically characterized and the precise nature of the trait-specific information that is transmitted between generations remains elusive.
Several well characterized examples of inherited epigenetic silencing in C. elegans involve RNA interference (RNAi) (Fire et al., 1998). Silencing of the endogenous germline expressed gene oma-1 by exogenously introduced double stranded RNA (dsRNA) persists for up to four generations with selection (Alcazar et al., 2008), while dsRNA-initiated silencing of a germline expressed gfp transgene can persist, with selection, for 80 or more generations (Ashe et al., 2012; Vastenhouw et al., 2006). Furthermore, endogenous piRNAs can initiate silencing of a single copy germline gfp transgene in RNAe (Shirayama et al., 2012). Once initiated, transgenerational transgene silencing requires the germline specific nuclear RNAi pathway as well as several putative histone modifying enzymes (Ashe et al., 2012; Buckley et al., 2012; Shirayama et al., 2012). In germline nuclear RNAi, PRG-1-stabilized piRNAs or dsRNA-derived short interfering RNAs (siRNAs) direct the production of 22G siRNAs that are stabilized by the nuclear localized Argonaute HRDE-1 (Bagijn et al., 2012; Batista et al., 2008; Buckley et al., 2012; Lee et al., 2012; Sapetschnig et al., 2015). RNAi pathways can inhibit gene expression transcriptionally (Buckley et al., 2012; Burkhart et al., 2011; Burton et al., 2011; Guang et al., 2008; 2010) and post-transcriptionally (Montgomery et al., 1998; Sapetschnig et al., 2015; Tsai et al., 2015).
piRNA pathway-silenced loci are often paramutagenic (Ashe et al., 2012; de Vanssay et al., 2012; Hermant et al., 2015; Sapetschnig et al., 2015; Shirayama et al., 2012). In paramutation, an allele’s expression state can stably alter the expression of a homologous locus in trans (Chandler, 2010). Silencing associated with a transgenic piRNA sensor can be transmitted to naïve alleles and single copy transgenes silenced in the germline produce a paramutagenic silencing signal in RNAe (Sapetschnig et al., 2015; Shirayama et al., 2012).
The import of RNAi-triggering dsRNA into cells and between generations (“systemic RNAi”) requires the transmembrane protein SID-1 (Winston et al., 2002). Here we show that multiple copies of the sid-1 upstream region initiate stable transgenerational silencing of the endogenous sid-1 locus, resulting in animals that are resistant to RNAi in germline and somatic cells. The ability of a promoter and 5′ UTR transgene to silence a coding region has not been reported previously. In co-suppression, multi-copy transgenes must include the coding sequence to silence the endogenous gene (Dernburg et al., 2000; Ketting and Plasterk, 2000). Furthermore, epigenetic silencing at this endogenous locus is remarkably stable after loss of the initiating transgene, as silencing persists in the absence of selection and at near 100% penetrance for up to 13 generations in the germline and for at least four generations in the soma. Our study shows that the silenced state is only transmitted maternally, and provides evidence that small RNAs embody the inherited silencing signal. In addition, and in contrast to previous reports, germline silencing does not require the chromatin modifying enzymes implicated in the piRNA pathway. Instead, these two histone modifying methyltransferase homologs are required to silence sid-1 specifically in the soma, suggesting that the mechanisms for transgenerational silencing in the germline and soma are distinct.
Results
sid-1 promoter arrays silence the sid-1 locus
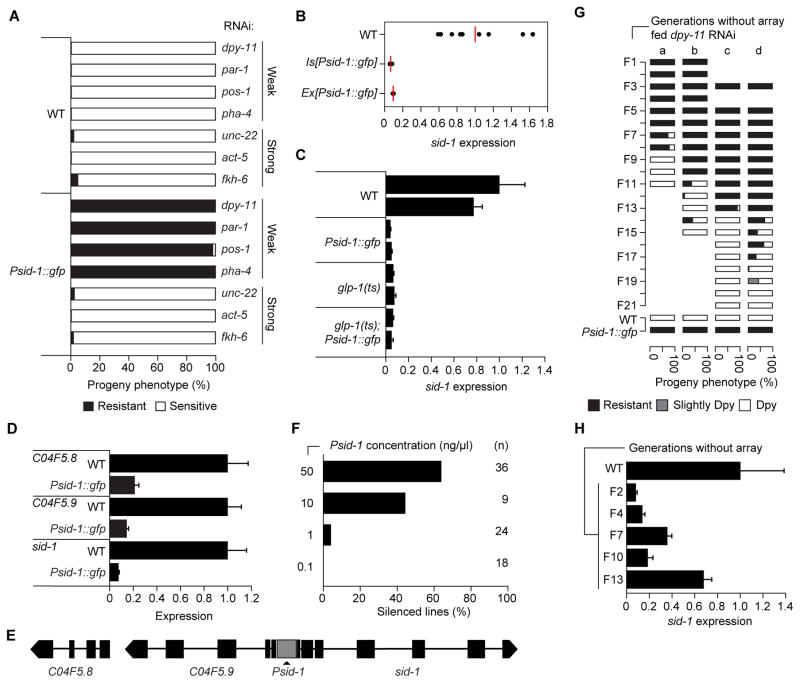
We previously constructed sid-1 transcriptional reporter worms (Winston et al., 2002). Surprisingly, we found that animals harboring the multi-copy Psid-1::gfp array were defective for RNAi induced by ingested dsRNA (feeding RNAi) (Figure 1A). Quantitative reverse transcription (qRT)-PCR showed a ten-fold reduction in sid-1 mRNA levels in both extrachromosomal (Ex) and chromosomally integrated (Is) Psid-1::gfp array lines (Figure 1B). The majority of sid-1 mRNA expression is confined to the germline in wild-type C. elegans. Consistent with gene expression analyses of isolated gonads and pre-transcriptional embryos, which indicate abundant germline-derived sid-1 transcripts (Arnold et al., 2014; Baugh et al., 2003; Levin et al., 2012), sid-1 mRNA levels are reduced ten-fold in germline-depleted (glp-1) young adults with or without the Psid-1::gfp array (Figure 1C). Thus, in adults, sid-1 is primarily expressed in the germline and the Psid-1::gfp array strongly reduces this expression.
Figure 1. Transgenerational epigenetic sid-1 silencing.
(A) RNAi sensitivity of >100 progeny of L4 worms. mRNA expression in (B) mixed stage worms (biological replicate average in red), (C) single young adults (25°C, each bar represents a single worm and two technical replicates), and (D) synchronized young adults. (E) Silenced region. (F) Fraction of dpy-11 RNAi resistant F2 Psid-1 array lines (n). (G) Inherited RNAi resistance of average 103 (a, b) or 870 (c, d) progeny from 3 (a, b) or 20 (c, d) L4 larvae fed dpy-11 RNAi. (H) sid-1 expression in mixed stage line b. Expression is relative to gpd-2/3, wild-type is set to 1.0. Average ± SD of two technical replicates, unless otherwise noted. See also Figure S1.
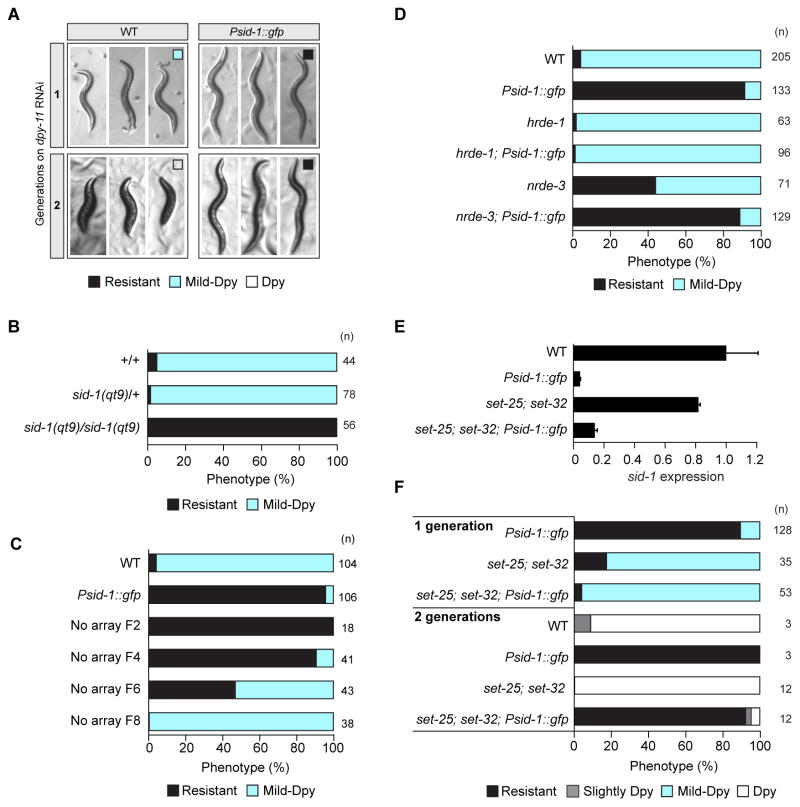
Psid-1::gfp-induced sid-1 silencing results in feeding RNAi defects for some RNAi foods (Figure 1A). While Psid-1::gfp worms remain sensitive to potent RNAi foods that can silence the targeted gene in directly exposed animals, they are resistant to weak RNAi triggers that require two continuous generations of exposure for wild-type worms to show the expected phenotype (Figure S1A). The specific resistance of Psid-1::gfp worms to weak RNAi foods that require maternal transmission of dsRNA is consistent with reduced sid-1 germline expression.
To characterize the specificity of this unusual silencing, we sequenced all mRNA from synchronized wild-type and Psid-1::gfp young adults (Figure S1B). Only three genes showed significant silencing: sid-1 and its two immediate upstream (5′) neighbors (Figure 1D, 1E). Thus, the Psid-1::gfp array specifically silences multiple genes in the vicinity of the endogenous sid-1 locus.
The Psid-1::gfp transcriptional fusion contains the entire 716bp intergenic region starting 10 base pairs upstream of the sid-1 start codon. To determine whether the sid-1 promoter is sufficient to silence sid-1, we injected into wild-type worms this 716bp region along with a co-injection marker and DNA ladder to make complex arrays. 23/36 Psid-1 lines placed on dpy-11 RNAi food produced only resistant progeny, indicating that the sid-1 promoter array is sufficient to silence sid-1 (Figure 1F, S1C). In a separate experiment, a similar proportion of lines were resistant after two generations, but six generations after injection nearly all lines produced only resistant progeny (Figure S1D). Additionally, diluting the sid-1 promoter decreased the proportion of silenced lines (Figure 1F, S1C). These results confirm that sid-1 promoter arrays induce sid-1 silencing.
In feeding RNAi experiments, even the progeny that did not inherit the array from Ex[Psid-1::gfp] worms were resistant to dpy-11 RNAi. Non-array worms remained resistant to dpy-11 RNAi food for 8–13 generations after loss of the Psid-1::gfp array (Figure 1G). qRT-PCR analysis confirmed that sid-1 expression correlated with resistance and sensitivity to dpy-11 RNAi (Figure 1H). The upstream genes can also remain silenced over multiple generations after Psid-1::gfp array loss (Figure S1E). Therefore, exposure to the Psid-1::gfp array initiates a robust and stable epigenetic silenced state at the sid-1 locus.
Transmission of the silenced sid-1 state
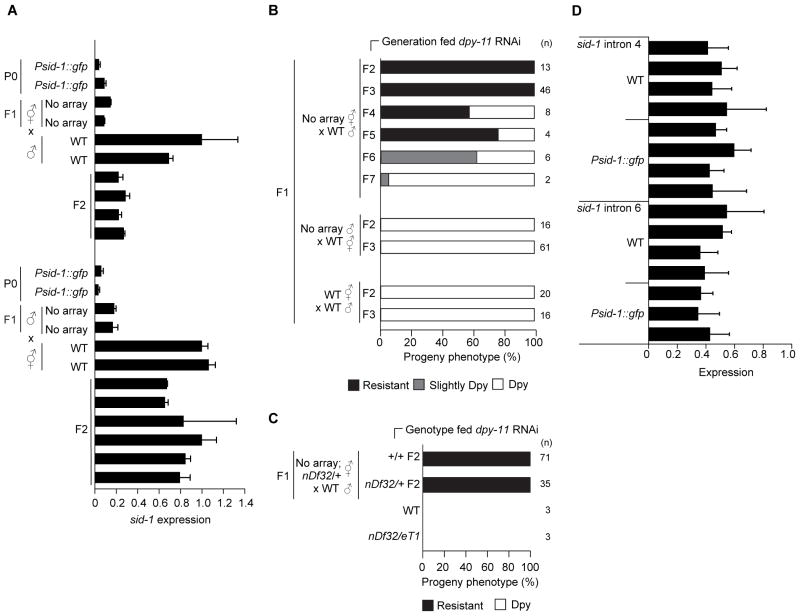
To characterize the inheritance of the silenced sid-1 state, we crossed sid-1 silenced animals that no longer carry the Psid-1::gfp array to wild-type animals. The cross progeny of wild-type males and silenced hermaphrodites all silenced sid-1, and all F2 and F3 worms produced only resistant progeny on dpy-11 RNAi food (Figure 2A, 2B, S2A). Thus, when an expressed locus from a male is crossed to a silenced hermaphrodite, the naïve locus from the male becomes stably silenced, indicating that the silenced state is dominant. Paramutation-like silencing can be transmitted via sperm and oocytes in C. elegans (Alcazar et al., 2008; Shirayama et al., 2012). However, we found that the cross progeny of wild-type hermaphrodites and silenced males all expressed sid-1 and all F3 and F4 progeny were sensitive to dpy-11 RNAi (Figure 2A, 2B). Thus, the silenced state at the sid-1 locus is paramutagenic but transmitted only maternally (Figure S2C).
Figure 2. Maternally transmitted epigenetic silencing.
(A) Normalized sid-1 mRNA levels (relative to cpf-1) in single silenced and non-silenced parents and progeny (see Figure S2B for non-normalized data). Average ± SD of at least two technical replicates. (B) RNAi sensitivity of progeny (average of 62 per worm) from (n) F2 cross progeny described in (A) and their subsequent self-progeny fed dpy-11 RNAi. (C) RNAi sensitivity of progeny (average 68 per worm) from (n) F2 sid-1+/nDf32 hemizygous or sid-1+/sid-1+ cross progeny fed dpy-11 RNAi. (D) qRT-PCR measurement of sid-1 intron 4 and intron 6 expression in single worm adults relative to cpf-1 intron 5 expression. cDNA used for qRT-PCR was generated using a gene specific primer amplifying sid-1 and cpf-1. Average ± SD of at least two technical replicates is shown. See also Figure S2.
Epigenetic silencing in C. elegans is often associated with changes to chromatin structure (Ashe et al., 2012; Buckley et al., 2012; Burton et al., 2011; Guang et al., 2010; Vastenhouw et al., 2006). However, a heritable transgene-silencing signal can be transmitted in the absence of the silenced transgene (Sapetschnig et al., 2015). To determine whether the sid-1 locus is required for transmission of the sid-1 silenced state, we generated Psid-1::gfp/+;sid-1+/nDf32 worms that contain a large deficiency (nDf32) that deletes over three megabases (>700 genes) including the sid-1 locus (Winston et al., 2002). We then crossed wild-type males to silenced sid-1+/nDf32 progeny (Figure S2D). All resulting cross progeny, whether they inherited the intact sid-1 locus (sid-1+/sid-1+) or the deleted sid-1 locus (sid-1+/nDf32), produced resistant progeny when placed on dpy-11 RNAi food (Figure 2C, S2C). Thus, the sid-1 locus and any associated chromatin marks are not required for transmission of silencing signals.
Epigenetic silencing in C. elegans is often associated with reduced transcription (Buckley et al., 2012; Burton et al., 2011; Guang et al., 2010; Shirayama et al., 2012). For example, comparison of pre-mRNA and mRNA levels indicates that both transcriptional and post-transcriptional mechanisms may contribute to dsRNA-mediated heritable silencing of a gfp transgene (Ashe et al., 2012), while silencing of gfp in RNAe is primarily transcriptional (Shirayama et al., 2012). If the 10-fold decrease in sid-1 mRNA in Psid-1::gfp worms is due to transcriptional silencing, we also expect a 10-fold decrease in sid-1 pre-mRNA in Psid-1::gfp worms. However, sid-1 pre-mRNA levels, inferred by qRT-PCR of two sid-1 introns, were indistinguishable in wild-type and Psid-1::gfp worms (Figure 2D). Although pre-mRNA levels are often difficult to measure, we found that our single-worm qRT-PCR protocol (see STAR Methods) results in reproducibly detectable levels of sid-1 pre-mRNA. Thus, although we cannot rule out a minimal contribution of transcriptional silencing that may be undetectable by qRT-PCR or more complex mechanisms of regulation, this result suggests that silencing does not affect sid-1 transcription or RNA splicing rates and is likely post-transcriptional.
22G siRNAs target the silenced sid-1 locus
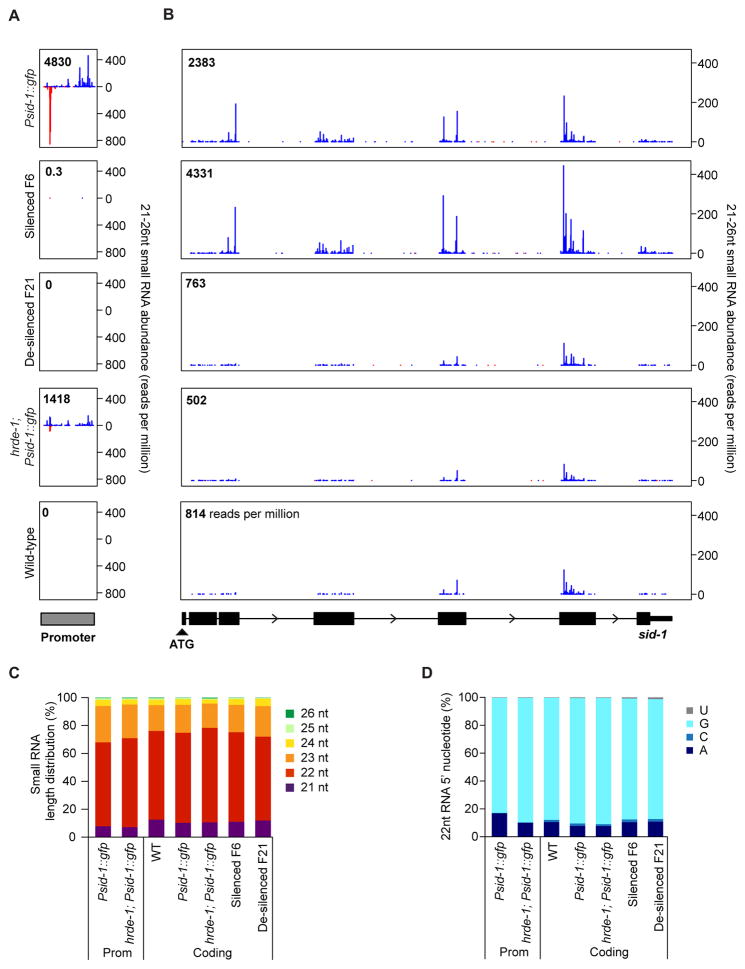
Small RNA pathways can regulate gene expression. Therefore, we sequenced two independent wild-type and four independent Psid-1::gfp small RNA libraries prepared from synchronized young adults. Normalized to library size, small RNAs antisense to the sid-1 coding sequence in Psid-1::gfp worms increased three-fold compared to wild type (Figure 3B). In all libraries, the small RNAs targeting sid-1 were highly enriched for 22 nucleotide RNAs with a 5′ guanine, indicating that they are likely 22G secondary siRNAs (Figure 3C, 3D). The number of sid-1-aligned 22G siRNAs was dramatically reduced in Psid-1::gfp small RNA sequencing libraries prepared without phosphatase treatment, confirming that these small RNAs have a 5′ triphosphate, as would be expected of 22G siRNAs (Pak and Fire, 2007) (Figure S3A). Thus, Psid-1::gfp-dependent silencing is associated with an increase in secondary 22G siRNAs antisense to the sid-1 mature transcript. Importantly, small RNA profiles are not globally altered in sid-1 silenced worms, indicating that the increased small RNAs at sid-1 are not simply due to the perturbation of sid-1 (Figure S3B).
Figure 3. Transgenerational sid-1 silencing is associated with an increase in HRDE-1-dependent small RNAs.
(A–B). Frequency and distribution of sense (red) and antisense (blue) 21–26 nucleotide small RNAs over the (A) sid-1 promoter and (B) the sid-1 gene (start codon to 3′ UTR). Total read counts (upper corner) are normalized to all 21–26nt reads that map to genes. (C–D) Reads (both strands) are highly enriched for (C) 22 nucleotide RNAs with (D) a 5′ guanine in all libraries. Four Psid-1::gfp libraries and two libraries for all other genotypes were combined as replicates. See also Figure S3.
In addition, in Psid-1::gfp libraries, small RNAs enriched for 22G RNAs also aligned to the sid-1 promoter region (Figure 3A, 3C, 3D, S3A). Promoter-aligned small RNAs were never detected in wild-type libraries. Our RNA-seq data indicates that Psid-1::gfp worms contain abundant transcripts that map to the sid-1 promoter while no similar transcripts were detected in wild-type worms (Figure S3C), suggesting that the small RNAs that align to the sid-1 promoter are templated by Psid-1::gfp array-associated transcripts.
To determine whether these sid-1 associated 22G RNAs persist in the absence of the Psid-1::gfp array, we sequenced small RNAs from lines c and d (Figure 1G) at six (silenced) and 21 (de-silenced) generations removed from a Psid-1::gfp array ancestor. No small RNAs aligned to the sid-1 promoter in either line at either generation (Figure 3A). In contrast, in silenced worms (F6) we detected an increase in sid-1 antisense small RNAs (Figure 3B), while in de-silenced (F21) worms, small RNA levels were at wild type levels (Figure 3B). These results extend the correlation between the coding region 22G RNAs and persistent sid-1 silencing.
The PRG-1/HRDE-1 pathway silences sid-1
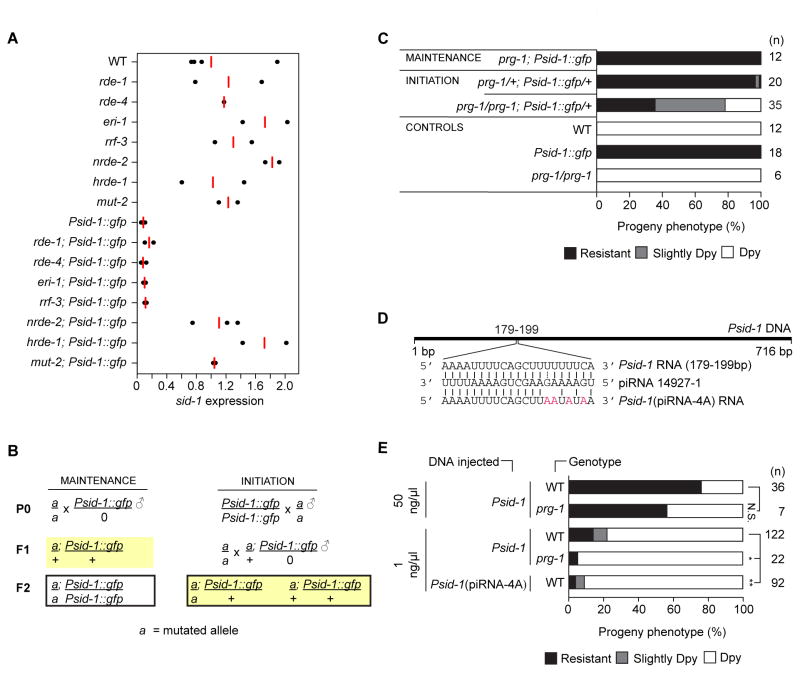
We hypothesized that the small RNAs targeting the sid-1 locus are generated and stabilized by one of the known C. elegans small RNA pathways. Previous cases of heritable transgene silencing in C. elegans require the nuclear RNAi pathway (Ashe et al., 2012; Buckley et al., 2012; Sapetschnig et al., 2015; Shirayama et al., 2012). We found that mutations in the nuclear RNAi factors hrde-1 and nrde-2, and a mutation in mut-2, a putative nucleotidyltransferase required for siRNA accumulation (Zhang et al., 2011), also prevent promoter-mediated sid-1 silencing (Figure 4A).
Figure 4. Genetic requirements for sid-1 silencing.
(A) sid-1 mRNA expression in mixed stage worms. Average (red bar) of biological replicates relative to gpd-2/3. (B) Maintenance and initiation crosses (silencing competent hermaphrodite germline is highlighted). (C) RNAi sensitivity of progeny of (n) F2 L4 larvae produced by crosses in (B). (D) Putative piRNA 14927-1 binding site and mutant Psid-1(piRNA-4A) sequence. (E) RNAi sensitivity of progeny of (n) F2 lines produced by injected wild-type or piRNA-4A Psid-1 DNA. N.S. = Not significant, * = p < 0.05, ** = p < 0.002 (Mann-Whitney test). Resistant and slightly Dpy values were combined for statistics. In A–C, the Psid-1::gfp array is integrated on the X chromosome. See also Figure S4.
In addition to nuclear RNAi, the PIWI Argonaute PRG-1 has been implicated in heritable silencing of transgenes. While RNAe requires PRG-1 only to initiate silencing (Shirayama et al., 2012), another case of heritable transgene silencing also requires PRG-1 to maintain silencing (Bagijn et al., 2012). We found that PRG-1 is only partially required to initiate sid-1 silencing. Specifically, prg-1(n4357); Psid-1::gfp animals that segregate from silenced prg-1/+ heterozygotes (“Maintenance cross”) remained fully silenced, showing that prg-1 is not required to maintain established silencing (Figure 4B, 4C). In contrast, crossing the array from a prg-1(n4357)/+; Psid-1::gfp/0 male into prg-1(n4357) homozygotes (“Initiation cross”), produced prg-1(n4357); Psid-1::gfp animals that incompletely silence sid-1 (Figure 4B, 4C). The dsRNA-dependent exogenous (rde-1, rde-4) and 26G small RNA-dependent endogenous (rde-4, eri-1, rrf-3) RNAi pathways were not required to initiate (Figure S4B–D) or maintain (Figure 4A) sid-1 silencing. Thus, prg-1 uniquely participates in the initiation of promoter-mediated sid-1 silencing, while hrde-1 maintains silencing.
To characterize the prg-1-dependent initiation of sid-1 silencing, we injected the Psid-1 DNA fragment into wild-type and prg-1(n4357) mutant worms. High concentrations of the Psid-1 DNA fragment caused prg-1-independent sid-1 silencing, but at lower concentrations, the Psid-1 DNA fragment was significantly better at silencing sid-1 in wild-type worms than in prg-1 mutants (Figure 4E, S4E). We then identified a 21 nucleotide sequence in the sid-1 promoter region that could be targeted by the endogenous piRNA 14927-1 (Bagijn et al., 2012; Batista et al., 2008) and disrupted the putative piRNA binding site by four nucleotide changes (“Psid-1(piRNA-4A)”, Figure 4D). The efficiency of silencing induced by Psid-1(piRNA-4A) DNA fragment injection in wild-type worms was similar to that observed by injecting the wild-type fragment into prg-1 mutant worms (Figure 4E, S4E). Thus, the requirement for prg-1 to initiate efficient sid-1 silencing likely reflects a requirement for piRNA 14927-1.
siRNAs embody the inherited silencing signal
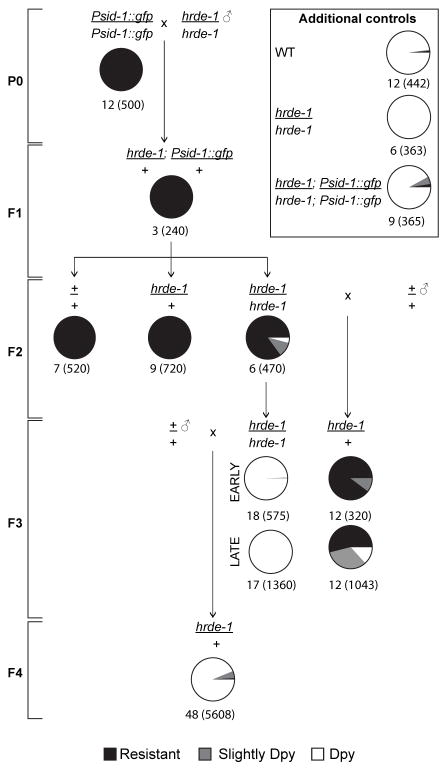
HRDE-1 is a nuclear Argonaute required to silence sid-1 in Psid-1::gfp worms (Figure 4A). To further characterize the contribution of HRDE-1 to transgenerational epigenetic inheritance of sid-1 silencing, we generated hrde-1(tm1200)/+; Psid-1::gfp/+ F1s and placed their F2 non-array progeny on dpy-11 RNAi (Figure 5, S5A). All hrde-1(tm1200)/hrde-1(tm1200) F2s produced non-Dpy F3 progeny, demonstrating that sid-1 remained silenced. This likely represents maternal F1-produced, HRDE-1-deposited small RNAs in F2 embryos. In contrast, F3 worms fed dpy-11 RNAi produced only Dpy F4 progeny (Figure 5, S5A), indicating that HRDE-1 and/or silencing signal depletion results in the re-expression of sid-1 in the F3 generation. If 22G RNAs targeting sid-1 persist in the F3 but cannot execute silencing due to the depletion of HRDE-1, then re-introduction of HRDE-1 into the F3 progeny should restore sid-1 silencing. Indeed, all tested F3 hrde-1/+ worms placed on dpy-11 RNAi food produced non-Dpy F4 progeny (Figure 5, S5A). Thus, sequence-specific silencing information was transmitted from the heterozygous F1 grandparent, through the homozygous F2 mother, and into the heterozygous F3 grand-progeny, where the re-introduced wild-type HRDE-1 was sufficient to execute silencing. Importantly, introduction of a wild-type HRDE-1 a generation later cannot restore sid-1 silencing, indicating that the silencing signal is impermanent and requires HRDE-1 for its production and/or maintenance. The restoration of silencing by wild-type HRDE-1 in the heterozygous F3 generation is strong evidence that small RNAs embody the inherited epigenetic information. Temporal analysis of silencing strength in F3 worms provides further evidence for this model. Cross progeny F3 worms laid earlier more efficiently silenced sid-1 than cross progeny laid later (Figure 5, S5A). This likely reflects dilution of silencing signals in the F2 germline such that early oocytes have more anti-sid-1 siRNAs than do the late oocytes.
Figure 5. HRDE-1 is required to execute transgenerational silencing.
Crosses to test hrde-1 necessity and sufficiency for transgenerational silencing. Pie charts show RNAi sensitivity of progeny of singled L4 larvae on dpy-11 RNAi food. Below each pie chart is the number of L4 parents and the number of progeny scored (in parentheses). Early progeny are the first progeny laid, late progeny are progeny laid subsequently by F3 parent. See also Figure S5.
HRDE-1 is required for the transmission of the silencing signal across generations (Figure 5) and sid-1 promoter and exon associated small RNAs decrease significantly in hrde-1(tm1200); Psid-1::gfp worms compared to Psid-1::gfp worms (Figure 3A, 3B). Thus, in the presence of the Psid-1::gfp array, HRDE-1 is required for the accumulation and spread of 22G RNAs from the sid-1 promoter region to the coding region.
How could small RNAs targeting array-produced promoter mRNA sequences direct HRDE-1-dependent accumulation of small RNAs to the sid-1 coding region? Psid-1::gfp animals contain many small RNAs complementary to the abundant array-transcribed RNA corresponding to the sid-1 upstream region (Figure 3A, S3C). Like many C. elegans transcripts, sid-1 is trans-spliced, meaning that a 5′ UTR present in pre-mRNA is replaced by a splice leader (SL1) sequence in the mature transcript (Saito et al., 2013). While RNA-seq analysis detects only the abundant SL1 trans-spliced mature sid-1 transcript, experiments designed to detect transcription start sites identify sites within the promoter region of sid-1 (Chen et al., 2013; Saito et al., 2013) (Figure S5B). Thus the abundant array-dependent small RNAs could target the sid-1 primary transcript enabling HRDE-1-dependent spread and accumulation of small RNAs targeting the sid-1 exons.
Transgenerational somatic sid-1 silencing
Wild-type animals exposed to dpy-11 RNAi food for two consecutive generations are strongly Dpy. This strongly Dpy phenotype requires both maternal sid-1 expression in the germline and somatic sid-1 expression. However, wild-type animals exposed to dpy-11 RNAi food for a single generation have a readily detectable and highly penetrant mild-Dpy phenotype (Figure 6A). To determine whether maternally inherited sid-1 activity contributes to the single-generation mild-Dpy phenotype, we tested the progeny of sid-1(qt9)/+ parents in the single generation dpy-11 RNAi assay. While sid-1(qt9)/+ progeny developed the mild-Dpy phenotype as adults, sid-1(qt9)/sid-1(qt9) progeny were completely resistant adults (Figure 6B). Thus, maternal sid-1 expression is not sufficient to produce the mild-Dpy phenotype. Further, the cross progeny of a non-array silenced worm and a wild-type male, which should express the non-silenced paternal sid-1 allele, are mildly Dpy (Figure S6A). These results indicate that somatic sid-1 expression is necessary and sufficient for the mild-Dpy phenotype. Therefore, the absence of this phenotype is a reliable indirect measure for somatic sid-1 silencing. Because Psid-1::gfp worms are completely resistant in the single generation dpy-11 RNAi assay (Figure 6A), sid-1 must be silenced in the soma in Psid-1::gfp worms.
Figure 6. Transgenerational somatic silencing.
(A) Progeny of wild-type or Psid-1::gfp larvae (two generations) or embryos (one generation) placed on dpy-11 RNAi food. (B–D) RNAi sensitivity of (n) worms hatched on dpy-11 RNAi food, scored as adults. To determine sid-1 genotype in (B), adults were fed act-5 RNAi (L1 arrest). (E) qRT-PCR analysis of sid-1 mRNA levels (normalized, relative to gpd-2/3) in young adults. Average ± SD of at least two technical replicates. (F) RNAi sensitivity of methyltransferase mutants after dpy-11 dsRNA exposure for one (n worms scored) or two generations (average of 116 progeny from (n) L4 larvae scored). See also Figure S6.
The single generation dpy-11 RNAi assay accurately, sensitively, and specifically detects silencing of somatic sid-1 expression. RT-PCR experiments comparing wild-type and Psid-1::gfp L1 larvae (~550 somatic cells and two germline precursors) also detect reduced sid-1 transcripts (data not shown). However, unlike the single generation dpy-11 RNAi assay, these experiments cannot distinguish between maternal and zygotic sid-1 transcripts. That is, if maternal sid-1 transcripts persist in larval stages, then this measured decrease could reflect reduced maternal contribution rather than reduced zygotic expression. Thus, we used the single generation dpy-11 RNAi assay to specifically measure somatic sid-1 expression.
Heritable silencing of a somatically expressed gene in C. elegans in response to exogenous RNAi has been reported. However, the silencing is transmitted at reduced penetrance (30%) and maternal (germline) contributions to the silencing were not investigated (Vastenhouw et al., 2006). To determine whether somatic sid-1 silencing can occur and be inherited in the absence of the array, we tested the progeny of non-array silenced animals in the single-generation dpy-11 RNAi assay. Not only did we find that sid-1 somatic silencing can occur in the absence of the array, but the silencing is remarkably stable; somatic sid-1 silencing is inherited, at nearly 100% penetrance, for four generations (Figure 6C, S6B).
The stability of somatic sid-1 silencing allowed us to determine the genetic basis for heritable silencing in the soma. Unexpectedly, sid-1 silencing in the soma did not require the somatically expressed HRDE-1 paralog NRDE-3 (Guang et al., 2008). nrde-3(gg66); Psid-1::gfp and Psid-1::gfp worms are equally and completely resistant to dpy-11 RNAi (Figure 6D). In contrast, hrde-1(tm1200); Psid-1::gfp worms fail to silence sid-1 in the soma (Figure 6D). We conclude that sid-1 silencing in the soma is dependent on previous silencing in the maternal germline.
In contrast to other examples of heritable silencing (Ashe et al., 2012), promoter-mediated sid-1 silencing in the germline does not require the putative histone H3 lysine 9 (H3K9) methyltransferases set-25 and set-32 (hereafter referred to as methyltransferases): set-25; set-32; Psid-1::gfp worms have reduced sid-1 expression and when placed on dpy-11 RNAi produce resistant progeny (Figure 6E, 6F). However, in single generation dpy-11 RNAi, set-25; set-32; Psid-1::gfp worms are mildly Dpy, indicating that both methyltransferases contribute to somatic sid-1 silencing (Figure 6F). Thus, the requirement for HRDE-1 in both germline and somatic silencing, together with the requirement for SET methyltransferases exclusively in somatic silencing suggests that small RNA-dependent silencing in the germline directs chromatin-dependent silencing in the soma.
Discussion
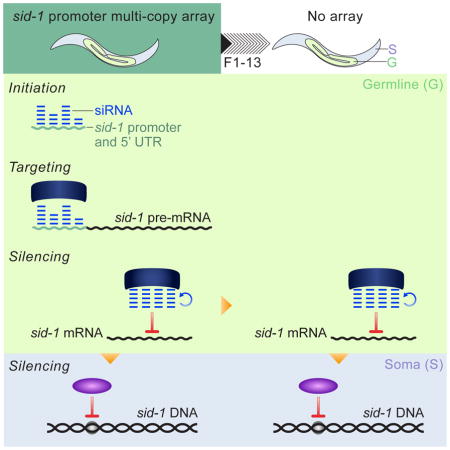
Here we provide the first molecular and genetic analysis of heritable RNAi silencing at an endogenous locus in C. elegans. Silencing at the sid-1 locus is initiated by multiple copies of the promoter and 5′ UTR in a dose-dependent process that is partially dependent on the piRNA-stabilizing protein PRG-1. Once initiated, the silencing is remarkably stable in the absence of the promoter array, persisting at 100% transmission for up to 13 generations without selection. The silencing is dependent on components of the germline heritable RNAi pathway and HRDE-1-dependent small RNAs antisense to sid-1 exons are associated with the silenced state. Our genetic analysis reveals that the silenced locus is not required for inheritance and that HRDE-1-responsive silencing information persists in the absence of HRDE-1 function over two generations.
Silencing in the context of an endogenous locus is more likely to reflect characteristics of evolutionarily relevant silencing than silencing of custom designed transgenes. In fact, comparison of sid-1 silencing to previously described multigenerational transgene silencing identifies numerous differences (Table S1). First, previously described cases of heritably silenced loci are initiated by either dsRNA or by endogenous piRNAs (Alcazar et al., 2008; Ashe et al., 2012; Buckley et al., 2012; Sapetschnig et al., 2015; Shirayama et al., 2012; Vastenhouw et al., 2006). In contrast, sid-1 silencing is initiated by a multi-copy array containing only its promoter and 5′ UTR, which has not been reported for any other gene. An array-derived dsRNA intermediate is unlikely to be required for sid-1 silencing because RDE-1 and RDE-4 are not required for silencing. Furthermore, although an endogenous piRNA can contribute to sid-1 silencing, piRNAs are not required to initiate sid-1 silencing. Second, sid-1 is the first stably silenced endogenous locus, both in the germline and in the soma. In all other examples of endogenous silenced loci, silencing is passed on to only a fraction of progeny even one generation after removal of the initiating trigger (Alcazar et al., 2008; Vastenhouw et al., 2006). In contrast, after removal of the Psid-1 array, sid-1 silencing continues at 100% transmission, for up to 13 generations in the germline and for four generations in the soma. Third, while transgene silencing and silencing of the endogenous oma-1 gene can be inherited through both the male and female germlines (Alcazar et al., 2008; Shirayama et al., 2012), silencing of sid-1 is inherited exclusively through the maternal germline. As in other cases of heritable silencing (Ashe et al., 2012; Buckley et al., 2012; Sapetschnig et al., 2015; Shirayama et al., 2012), the nuclear Argonaute HRDE-1 is required for sid-1 silencing in the germline. More surprisingly, two putative histone H3K9 methyltransferases that are required to silence transgenes in the germline (Ashe et al., 2012) are not required to silence sid-1 in the germline. Instead, these methyltransferases are required to silence sid-1 in the soma, suggesting a transition from post transcriptional gene silencing (PTGS) in the germline to transcriptional gene silencing (TGS) in the soma.
The initiation of sid-1 silencing likely reflects a two-step process: first, the accumulation of a persistent population of small RNAs that target the sid-1 promoter region on the multi-copy array, followed by spreading of small RNAs from the endogenous promoter to the coding exons. There is precedent for spreading of siRNAs 3′ of the original trigger (Pak and Fire, 2007; Sapetschnig et al., 2015) and the efficient spreading of siRNAs along a spliced transcript is dependent on nuclear silencing (Zhuang et al., 2013; Sapetschnig et al., 2015). We also note that the presence of siRNAs at the expressed sid-1 locus may facilitate the spreading of promoter siRNAs to the gene body. The delay in initial silencing likely reflects a below threshold level of array promoter small RNAs. Consistent with this, injecting the promoter fragment showed dose-dependent efficiency of silencing (Figure 1F). At the lowest dose, the establishment of silencing is partially dependent on prg-1 and the presence of a binding site for a specific piRNA (Figure 4E). Analysis of double mutants would be required to determine if any of the other known small RNA pathways are sufficient to initiate silencing in the absence of the piRNA pathway. Additionally, factors required for maintenance of silencing likely participate in the initiation of silencing, but this is not possible to test, as we cannot genetically isolate an initiation function for such factors.
Where tested, the Argonaute HRDE-1 is required for all RNAi-dependent transgenerational silencing described in C. elegans. Experiments comparing wild-type to hrde-1 or nrde-3 mutants undergoing RNAi support a role for TGS in nuclear RNAi (Buckley et al., 2012; Burkhart et al., 2011; Guang et al., 2008; 2010). However, we did not detect the expected TGS associated decrease in sid-1 pre-mRNA levels in worms that silence sid-1 (Figure 2D) nor did we detect a requirement for the putative histone methyltransferases set-25 and set-32 in sid-1 silencing in the germline (Figure 6E, 6F), supporting a PTGS mechanism for transgenerational sid-1 silencing in the germline. Thus, hrde-1-dependent silencing can act through TGS and PTGS mechanisms in different contexts, as has been previously proposed due to the role of nuclear RNAi Argonautes in transitive RNAi (Sapetschnig et al., 2015; Zhuang et al., 2013).
Several groups have shown correlation between the presence or absence of siRNAs and silencing and non-silencing. These siRNAs could be a byproduct of an RNAi-independent silencing mechanism. For example, marked chromatin can trigger siRNA production in yeast (Bühler et al., 2006). Alternatively, these siRNAs could be the parentally-provided sequence-specific cause of silencing in the progeny. In mammals, diet and stress influences the abundance of sperm associated small RNAs that can alter gene expression and behavior in progeny (Chen et al., 2016; Gapp et al., 2014; Sharma et al., 2016). Our small RNA sequencing and multi-generational genetic analysis of hrde-1 showed that hrde-1 is required to stabilize siRNAs and execute sid-1 silencing. hrde-1 dependent silencing signals can persist in hrde-1 homozygotes for two generations, but cannot execute silencing unless wild-type hrde-1 is re-introduced. Further, we showed that the sid-1 physical locus is not required for transmission of silencing. Together, these results strongly support the supposition that siRNAs physically embody the transgenerationally transmitted silencing information. Although our genetic analysis cannot discriminate between direct transmission of grandparental siRNAs acting in the germline of the grandprogeny versus a re-synthesis of siRNAs after the reintroduction of wild-type hrde-1, our results provide direct evidence for sustained RNA-directed transgenerational inheritance.
This study also provides the first example of stable, highly penetrant, transgenerational silencing in somatic cells. Multiple features of this somatic silencing were unexpected. First, not only was nrde-3, the somatically expressed homolog of hrde-1, not required for somatic sid-1 silencing, but maternal hrde-1 was required. Second, the putative methyltransferases set-25 and set-32 are specifically required to silence sid-1 in the soma (Figure 6E, 6F). Low somatic sid-1 expression levels relative to the germline expression levels likely masked any measurable effect on pre-mRNA in the soma (Figure 1C). Since both hrde-1 and set-25/set-32 are required for somatic silencing, they do not act redundantly and likely act in series. Thus, siRNA mediated hrde-1-dependent trans-acting germline silencing likely establishes chromatin modifying machinery-dependent cis-acting somatic silencing in the progeny.
Many multi-copy reporter construct arrays containing promoters have been made and analyzed and none are reported to epigenetically silence the endogenous locus. sid-1 silencing is distinct from co-suppression, in which multi-copy arrays that include coding sequences can silence endogenous loci in the C. elegans germline (Dernburg et al., 2000; Ketting and Plasterk, 2000). First, co-suppression requires that the coding sequence be present in the multi-copy array; for example, a multicopy array of the fem-1 gene that includes the promoter, 5′UTR and coding sequence silences germline fem-1 expression. However, a multicopy array that contains the promoter and 5′UTR only does not silence fem-1 (Dernburg et al., 2000; Saito et al., 2013). The Psid-1 fragment does not contain the sid-1 coding sequence. The inclusion of a trans-spliced 5′ UTR in our Psid-1::gfp reporter construct does not sufficiently differentiate this array from other reporter constructs. 70% of C. elegans transcripts are trans-spliced (Zorio et al., 1994) and transcriptional fusions often contain 5′ UTRs. However, they do not silence. Second, co-suppression-induced silencing is transmitted in the absence of the array weakly or not at all (Dernburg et al., 2000), whereas sid-1 silencing can be transmitted for more than a dozen generations at 100% penetrance. Third, only germline genes can be silenced by co-suppression, whereas sid-1 is also heritably silenced in the soma.
It is unlikely that sid-1 is the only gene that can exhibit stable promoter-mediated epigenetic silencing. Indeed, the two germline-enriched genes upstream of sid-1 can also be epigenetically silenced (Figure 1D, S1E) by a HRDE-1 dependent process, although we note that statistically significant changes in small RNAs targeting these genes are not consistently detected in silenced worms, perhaps because these small RNAs are present at very low levels (data not shown). Instead, sid-1 may be special in our ability to readily detect epigenetic silencing. Unlike many germline-expressed genes, sid-1 is not required for viability or fertility. If essential genes are silenced by a promoter multi-copy array, any silenced segregants would die out and only non-silenced progeny would persist. Additionally, like the two genes upstream of sid-1, loss of other epigenetically silenced germline genes may be undetectable unless measured intentionally. Loss of systemic RNAi is not only easily scored, but the strength, sensitivity, and target tissue(s) can be readily modified by the choice of RNAi food. This is particularly important as the level of sid-1 silencing by the multi-copy promoter array is incomplete (90%). Had we only tested potent RNAi foods, we never would have detected the silencing. Additionally, the ability to infer small changes in sid-1 expression levels from RNAi food phenotypes was crucial to recognize that sid-1 was transgenerationally silenced in the soma.
Shared sequence-specific silencing between the germline and the soma potentially enables selection for advantageous phenotypes beyond fecundity. Several recent studies in C. elegans implicate hrde-1-dependent processes in physiologically-induced transgenerational changes in gene expression (Rechavi et al., 2014; Schott et al., 2014). It remains unknown whether or how changes in expression of these genes contribute to a stress response. Because hrde-1 is involved, there is presumably selection for genes like sid-1 that are expressed both in the germline, to initiate and transmit heritable signals, and in somatic cells to effect physiological phenotypes.
It remains unknown how silencing of endogenous genes is triggered in response to environmental stress. The initiation of sid-1 silencing that we describe likely involves accumulation of the Psid-1::gfp array transcribed sid-1 promoter and 5′ UTR RNA. We hypothesize that accumulation and export of this piRNA-targeted 5′ UTR exon to the cytoplasm initiates production of antisense 22G RNAs targeting the 5′ UTR, which can then be transported to the nucleus to initiate nuclear RNAi dependent silencing. The endogenous sid-1 5′ UTR is efficiently trans-spliced prior to mRNA export to the cytoplasm, thus the sid-1 locus is not readily silenced in wild-type animals. However, it is plausible that sid-1 or other loci could be similarly targeted for silencing if alternative splicing produces a transcript with an exon that contains a piRNA binding site. Because splicing is regulated by environmental conditions (Biamonti and Caceres, 2009), stress responsive alternative splicing could trigger gene-selective epigenetic silencing in response to specific environmental conditions.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Craig P. Hunter, Harvard University, cphunter@g.harvard.edu.
The Lead Contact holds responsibility for responding to requests and providing reagents and information.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were maintained as previously described (Brenner, 1974). All experiments were performed at 20°C unless otherwise indicated. See Table S2 for strains and alleles.
METHOD DETAILS
Worm synchronization
To obtain synchronized young adult worms, young adults laid embryos for 3 hours. Adults were subsequently removed and staged embryos developed into synchronized young adult worms that were collected 64.5–65.5 hours after embryos were laid. For strains with more varied growth than N2, improperly staged worms were manually removed prior to collection.
RNAi assays
E. coli carrying IPTG-inducible vectors expressing dpy-11, par-1, pos-1, pha-4, unc-22, act-5 and fkh-6 dsRNA from the Ahringer RNAi library (Kamath and Ahringer, 2003) were grown for 15–17 hours in LB media with 100μg/mL carbenicillin. Cultures were seeded onto NGM plates containing IPTG and carbenicillin, and left at room temperature for 24 hours before use. Worms placed on RNAi food as embryos were scored as adults. The progeny of L4 larvae placed on RNAi food were continuously on RNAi and scored as adults. To avoid scoring the first progeny that may have received a lesser dose of dsRNA, L4 larvae placed on RNAi food were moved as adults the next day to a new RNAi plate. Only progeny laid for a day on this plate were scored unless otherwise noted. If all worms scored on RNAi were the same phenotype, the number of worms was often estimated to be greater than n worms, and n was used to calculate average number of worms scored. In the two generation assay on dpy-11 RNAi, mild-Dpy worms were scored as resistant, since the two generation assay is used as an indirect measure for sid-1 expression in the germline rather than the soma.
Transgenesis
Worms carrying the extrachromosomal and integrated Psid-1::gfp arrays were generated previously (Winston et al., 2002). To generate Psid-1 worms, the sid-1 promoter was amplified from N2 genomic DNA (primers: 5′-GGTCATGAGAGGGTCGAGAG-3′, 5′-GGAAAAATGAGGAGTTTTAATTTC-3′) and gel purified (QIAquick Gel Extraction Kit, Qiagen, 28704). To make complex extrachromosomal array lines, the germline of N2 (wild-type) worms was injected with 0.1–75ng/μl Psid-1, 15ng/μl pHC183 (myo-3::dsRed2) (Winston et al., 2002) and 25ng/μl DNA ladder (New England Biolabs, N3232S).
The Psid-1(piRNA-4A) fragment was amplified from genomic DNA in two pieces using a site directed mutagenesis strategy to introduce the appropriate mutations (See Table S3 for primers) and cloned (pHC516). Psid-1(piRNA-4A) fragments were amplified, gel purified and injected at a concentration of 50ng/μl or 1ng/μl into N2 or prg-1(n4357) worms with 15ng/μl pHC183 (myo-3::dsRed2) (Winston et al., 2002) and 25ng/μl DNA ladder (New England Biolabs, N3232S).
Genetics: maintenance and initiation crosses
To test for a requirement in the maintenance of silencing, Psid-1::gfp males were crossed to mutant hermaphrodites, resulting in mutant/mutant; Psid-1::gfp/Psid-1::gfp F2 or F3 worms. Mixed stage worms were collected for expression measurements, unless otherwise stated. To test for a requirement for prg-1 in the initiation of silencing, Psid-1::gfp hermaphrodites were crossed to prg-1;prg-1 males. Resulting prg-1/+; Psid-1::gfp/0 males were crossed to prg-1/prg-1 hermaphrodites, and F2 cross progeny carrying the Psid-1::gfp array were placed on dpy-11 RNAi. prg-1/prg-1 worms were back-crossed prior to starting the experiment due to their mortal germline, and maintained at 15°C. The experiment was performed at 20°C. To test for a requirement for rrf-3 and rde-1 in the initiation of silencing, Psid-1::gfp/0 males were crossed to mutant/mutant; Psid-1::gpf/Psid-1::gfp hermaphrodites. F1 mutant/+; Psid-1::gfp/0 males were crossed to mutant/mutant hermaphrodites. For rrf-3, resulting F2s were placed on dpy-11 RNAi as L4 larvae and their progeny were scored. F2 rde-1/rde-1; Psid-1::gfp/+ cross progeny self-fertilized and resulting rde-1/rde-1; Psid-1::gfp/Psid-1::gfp mixed staged worms were collected for RNA extraction and sid-1 mRNA expression measurements because rde-1 mutant animals are RNAi deficient. See Table S2 for strains and alleles.
Genetics: paramutation experiments
To test for maternal transmission, non-array hermaphrodites from Ex[Psid-1::gfp] parents were crossed to Is[Podr-1::rfp] males. To test for paternal transmission, non-array males from Ex[Psid-1::gfp] parents were crossed to Is[Podr-1::rfp] hermaphrodites. In both crosses, Pod-1::rfp was used as a marker to identify cross progeny. sid-1 expression was measured in resulting single worm cross progeny directly as described below or by singling cross progeny onto dpy-11 RNAi as described above.
To determine whether modified chromatin at the sid-1 locus is required for transmission, dpy-11(e224) nDf32 V/eT1 (III;V) worms were crossed to N2 males. Resulting males were crossed to Is[Psid-1::gfp] worms. Progeny were singled (called “P0” for consistency with Figure 2) and nDf32/+; Psid-1::gfp/+ worms were identified by F1 phenotype (25% dead progeny, no Unc worms). Singled non-array F1 worms from nDf32/+; Psid-1::gfp/+ P0 parents were crossed to Is[Podr-1::rfp] males. F2 L4 cross progeny were placed on dpy-11 RNAi and progeny were scored for the Dpy phenotype. Embryonic lethality in F3 was used to determine if F2 worms carried nDf32 deficiency. See Figure S2D for further details.
Genetics: Non-array silenced and de-silenced worms
To directly compare non-array worms that silence sid-1 to non-array de-silenced sid-1 worms, two independent lines were established and maintained three generations after loss of the Ex[Psid-1::gfp] array. Synchronized sid-1 silenced worms were collected three generations later, at the 6th generation, and synchronized de-silenced sid-1 worms were collected 18 generations later, at the 21st generation. To determine whether sid-1 remained effectively silenced at each intervening generation, 20 L4 worms were placed on dpy-11 RNAi food and their progeny were scored. Resistant worms indicate that sid-1 is silenced, while Dpy worms indicate that sid-1 is expressed.
Confirmation that non-array worms do not contain the Psid-1::gfp array
Four observations indicate that non-array worms do not surreptitiously carry a silenced array that maintains silencing. First, since the extrachromosomal array is normally maintained in only a fraction of progeny, to persist in 100% of the progeny for 8–13 generations it would need to be integrated in the genome and then when sid-1 expression is restored coordinately lost in all or nearly all progeny over the course of 1–3 generations (Figure 1G). Second, while we can detect the array by PCR amplification in Ex[Psid-1::gpf] worms, we never detect this array by PCR from wild-type worms or from non-array F1 worms (picked as “non-array” from Ex[Psid-1::gfp] parent based on lack of gfp and rol-6D co-marker). Third, if the silencing was due to an undetected array, then we would expect “silenced non-array” males crossed to wild-type hermaphrodites to result in progeny that silence sid-1. However, silenced males cannot transmit the silencing signal (Figure 2A, B). Fourth, we detect very abundant promoter associated siRNAs in Psid-1::gfp array worms, but do not detect these siRNAs in silenced non-array animals (Figure 3A).
RNA extraction and first strand cDNA synthesis
To purify RNA from mixed stage or synchronized young adult worms, frozen worm pellets were extracted in Trizol/chloroform. The aqueous fraction was precipitated, resuspended in water, DNaseI treated for 1 hour (Roche, 04716728001) and purified (RNeasy Mini Kit, Qiagen, 74106 or RNA Clean & Concentrator™-5, Zymo Research, R1015 if small RNAs were required in downstream applications). To prepare RNA for 5′-independent small RNA sequencing, 5μg of RNA was treated with 5′ polyphosphatase (Epicentre, RP8092H) and re-purified (RNA Clean & Concentrator™-5, Zymo Research, R1015).
First strand cDNA was synthesized using ThermoScript™ RT-PCR System (Invitrogen, 11146-024) with an OligodT primer and 125ng-1μg total RNA. Control cDNA synthesis reactions without the reverse transcriptase enzyme were included for each sample.
cDNA synthesis from single worms
A single adult worm was frozen in 5μl 10 mM Tris-Cl containing 90μg/mL proteinase K at −80°C for at least 10 minutes. Worms were lysed at 65°C for 10 minutes, followed by 95°C for 1 minute to inactivate proteinase K. 2μl of lysis was used directly in a 20μl OligodT-primed cDNA synthesis reaction with and without reverse transcriptase (control) for measuring mRNA expression (Invitrogen, 11146-024). If measuring pre-mRNA levels, the lysis was treated with DNase I (Roche, 04716728001) for 10 minutes at 37°C and DNase I was inactivated for 10 minute at 75°C prior to cDNA synthesis using a gene specific primer. See Table S3 for primers.
qRT-PCR
Quantitative RT-PCR analysis (QuantiTect SYBR Green PCR Kit, Qiagen, 204145) was performed on an Eppendorf Mastercycler ep realplex4. For expression measurements in a population of worms, 1/20th of the cDNA reaction was used in each qRT-PCR. qRT-PCRs were incubated at 95°C for 15 min followed by 35–40 cycles of 94°C 15 sec, 50°C 30 sec, 72°C 30 sec. For single worm expression, 1/10th of the cDNA was used in each qRT-PCR and the extension time (72°C) was decreased to 15 seconds. qRT-PCR primers were designed and verified to amplify only cDNA and not genomic DNA, which was especially important for single worm expression in which the genomic DNA was not degraded prior to cDNA synthesis. Ct values were determined using noiseband quantification. Error bars represent standard deviation for at least two technical replicates unless otherwise stated. See Table S3 for primers used in qRT-PCR.
mRNA and small RNA library preparation
mRNA sequencing libraries were made from 1μg total RNA from synchronized young adult worms. RNA was PolyA purified using the Apollo 324™ NGS Library Prep System with the PrepX™ PolyA 8 Protocol (Beta v1, Wafergen) and stranded mRNA sequencing libraries were made using the PrepX™ mRNA 8 Protocol (Beta v1, Wafergen). Resulting libraries were PCR amplified for 15 cycles and purified using the PCR Cleanup 8 Protocol (Apollo 324™ NGS Library Prep System). Non-stranded mRNA sequencing libraries were prepared using the TruSeq RNA Library Preparation Kit v2 (Illumina, RS-122-2001). Libraries were fragmented after dsDNA synthesis on the Covaris instrument as described in Alternate Fragmentation Protocol (Illumina, RS-122-2001). Two biological replicates for each genotype were pooled and single end (stranded) or paired end (non-stranded) libraries were sequenced on the Illumina NextSeq 500 Mid output flow cell for 150 cycles.
Small RNA libraries were prepared from 1μg total RNA from synchronized young adult worms (5′ polyphosphatase treated for 5′-independent libraries as described above) using the PrepX™ Small RNA 8 Protocol on the Apollo 324™ NGS Library Prep System. Libraries were amplified for 12 cycles and purified (QIAquick PCR Purification Kit, Qiagen, 28106). Small RNAs were size selected on the Pippin Prep 3% dye free cassette (Sage Science, CDP3010) by collecting 126–160bp fragments. Size selected RNA libraries were purified and concentrated to 10μl (DNA Clean & Concentrator™-5, Zymo Research, D4013). Libraries were pooled and sequenced on the Illumina NextSeq 500 High output flow cell for 75 cycles.
Library data processing
mRNA libraries were aligned to C. elegans genome WS235 using Tophat (v2.0.1) (Trapnell et al., 2012) with all parameters set to default except for minimum and maximum intron length (-i 20, -I 10000). Differential expression analysis was performed using Cufflinks (v2.2.1) (Trapnell et al., 2012) with default parameters, allowing a false discovery rate (FDR) of 0.05.
21–26 nucleotide reads were filtered from small RNA libraries and aligned to C. elegans genome WS235 using Bowtie2 (Langmead et al., 2009), allowing 0 mismatches (--score-min L,0,0) and up to four alignments per read (-k 4). Small RNA counts per gene were generated using HTseq (Anders et al., 2015) and normalized to total number of 21–26 nucleotide reads mapped to genes. Genes differentially targeted by small RNAs were identified with edgeR (Robinson et al., 2009) using the exact test and allowing an FDR of 0.05.
QUANTIFICATION AND STATISTICAL ANALYSIS
The Mann-Whitney test was used in Figure 4E because the data are independent of each other and the test does not assume a normal distribution. Values and n from Figure 4E, S4E are used in the statistical test, with Slightly Dpy and Dpy values combined. P<0.05 was defined as significant.
Differential expression analysis for mRNA-seq data was performed using Cufflinks (v2.2.1) (Trapnell et al., 2012) with default parameters, allowing a false discovery rate (FDR) of 0.05. Genes differentially targeted by small RNAs were identified with edgeR (Robinson et al., 2009) using the exact test and allowing an FDR of 0.05.
All error bars represent Standard Deviation, as stated in Figure legends.
Supplementary Material
Acknowledgments
We thank members of the Hunter lab for helpful discussion throughout the project, Phil Shiu and John Calarco for comments on this manuscript, Renate Hellmiss for help with graphics and Kareem Carr for bioinformatics consultation. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by NIH grant GM089795.
Footnotes
DATA AND SOFTWARE AVAILABILITY
mRNA and small RNA sequence data have been deposited in the Gene Expression Omnibus under GEO Accession: GSE81708.
Author contributions
O.M. performed all experiments and analyzed the data. O.M. and C.P.H. designed experiments and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcazar RM, Lin R, Fire AZ. Transmission Dynamics of Heritable Silencing Induced by Double-Stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A, Rahman MM, Lee MC, Muehlhaeusser S, Katic I, Gaidatzis D, Hess D, Scheckel C, Wright JE, Stetak A, et al. Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes. Nucleic Acids Res. 2014;42:13353–13369. doi: 10.1093/nar/gku1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Molecular Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem Sci. 2009;34:146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S. A Pre-mRNA–Associating Factor Links Endogenous siRNAs to Chromatin Regulation. PLoS Genet. 2011;7:e1002249. doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Verdel A, Moazed D. Tethering RITS to a Nascent Transcript Initiates RNAi- and Heterochromatin-Dependent Gene Silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Chandler VL. Paramutation’s Properties and Puzzles. Science. 2010;330:628–629. doi: 10.1126/science.1191044. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan W, Duan E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nature Reviews Genetics. 2016;17:733–743. doi: 10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RAJ, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Research. 2013;23:1339–1347. doi: 10.1101/gr.153668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bougé AL, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Zalevsky J, Colaiacovo MP, Villeneuve AM. Transgene-mediated cosuppression in the C. elegans germ line. Genes & Development. 2000;14:1578–1583. [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nature Publishing Group. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute Transports siRNAs from the Cytoplasm to the Nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermant C, Boivin A, Teysset L, Delmarre V, Asif-Laidin A, van den Beek M, Antoniewski C, Ronsseray S. Paramutation in Drosophila Requires Both Nuclear and Cytoplasmic Actors of the piRNA Pathway and Induces Cis-spreading of piRNA Production. Genetics. 2015;201:1381–1396. doi: 10.1534/genetics.115.180307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol. 2009;84:131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Developmental Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Hashimshony T, Wagner F, Yanai I. Developmental Milestones Punctuate Gene Expression in the Caenorhabditis Embryo. Developmental Cell. 2012;22:1101–1108. doi: 10.1016/j.devcel.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct Populations of Primary and Secondary Effectors During RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Park EC, Horvitz HR. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics. 1986;113:821–852. doi: 10.1093/genetics/113.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Rechavi O, Houri-Ze’evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, Hobert O. Starvation-Induced Transgenerational Inheritance of Small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TL, Hashimoto SI, Gu SG, Morton JJ, Stadler M, Blumenthal T, Fire A, Morishita S. The transcription start site landscape of C. elegans. Genome Research. 2013;23:1348–1361. doi: 10.1101/gr.151571.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapetschnig A, Sarkies P, Lehrbach NJ, Miska EA. Tertiary siRNAs Mediate Paramutation in C. elegans. PLoS Genet. 2015;11:e1005078. doi: 10.1371/journal.pgen.1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D, Yanai I, Hunter CP. Natural RNA interference directs a heritable response to the environment. Sci Rep. 2014;4:7387. doi: 10.1038/srep07387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Chen CCG, Conte D, Jr, Moresco JJ, Chaves DA, Mitani S, Yates JR, III, Tsai M-D, Mello CC. A Ribonuclease Coordinates siRNA Amplification and mRNA Cleavage during RNAi. Cell. 2015;160:407–419. doi: 10.1016/j.cell.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RHA. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SEJ, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JJ, Banse SA, Hunter CP. The nuclear argonaute NRDE-3 contributes to transitive RNAi in Caenorhabditis elegans. Genetics. 2013;194:117–131. doi: 10.1534/genetics.113.149765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Cheng NN, Blumenthal T, Spieth J. Operons as a common form of chromosomal organization in C. elegans. Nature. 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.