SUMMARY
The decision of stem cells to proliferate and differentiate is finely controlled. The Caenorhabditis elegans germ line provides a tractable system to study the mechanisms that control stem cell proliferation and homeostasis [1–4]. Autophagy is a conserved cellular recycling process crucial for cellular homeostasis in many different contexts [5], but its function in germline stem cell proliferation remains poorly understood. Here, we describe a function for autophagy in germline stem cell proliferation. We found that autophagy genes, such as bec-1/Beclin1, atg-16.2/ATG16L, atg-18/WIPI1/2, and atg-7/ATG7 are required for the late larval expansion of germline stem cell progenitors in the C. elegans gonad. We further show that BEC-1/Beclin1 acts independently of the GLP-1/Notch or DAF-7/TGFβ pathways, but together with the DAF-2/insulin IGF-1 receptor (IIR) signaling pathway to promote germline stem cell proliferation. Similar to DAF-2/IIR, BEC-1/Beclin1, ATG-18/WIPI1/2 and ATG-16.2/ATG16L all promote cell cycle progression, and are negatively regulated by the phosphatase and tensin DAF-18/PTEN. However, whereas BEC-1/Beclin1 acts through the transcriptional regulator SKN-1/Nrf1, ATG-18/WIPI1/2 and ATG-16.2/ATG16L exert their function through the DAF-16/FOXO transcription factor. In contrast, ATG-7 functions in concert with the DAF-7/ TGFβ pathway to promote germline proliferation, and is not required for cell cycle progression. Finally, we report that BEC-1/Beclin1 functions cell non-autonomously to facilitate cell cycle progression and stem cell proliferation. Our findings demonstrate a novel non-autonomous role for BEC-1/Beclin1 in the control stem cell proliferation, and cell cycle progression, which may have implications for the understanding, and development, of therapies against malignant cell growth in the future.
Keywords: C. elegans, germ line, autophagy, stem cell
RESULTS AND DISCUSSION
BEC-1 is required for the normal accumulation of germline progenitor cells during larval development
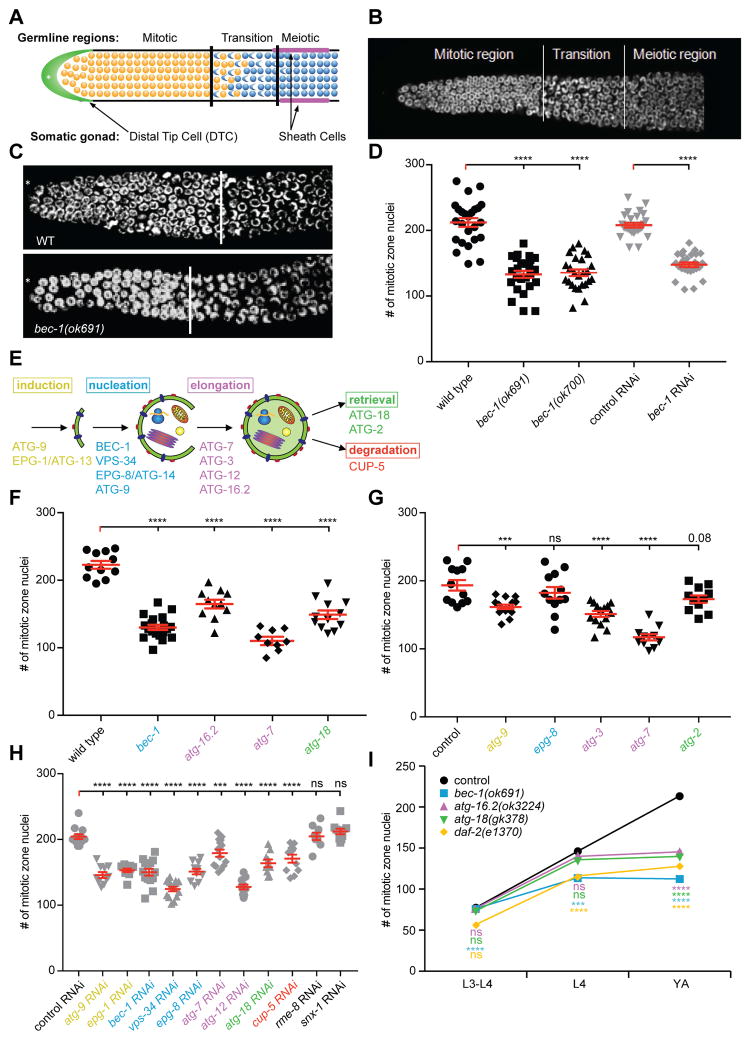
In C. elegans, loss-of-function mutations in the autophagy gene bec-1/beclin1 result in a sterile phenotype [6], suggesting a role for BEC-1/Beclin1 in gonadogenesis or in germline development. We found that compromising BEC-1/Beclin1 function resulted in a significant reduction (up to 50%) in the number of stem cells, in the distal proliferative mitotic zone, when compared to wild-type animals of the same stage (Figure 1A–D). Moreover, the proliferating mitotic zone was shortened, from an average length of 20 cell diameters in wild-type animals, to an average of 15 cell diameters in bec-1 mutants (Figure 1C). bec-1 homozygous mutants that segregated from a heterozygous parent, are maternally rescued from the lethal phenotype of the bec-1 complete loss of function [6]. The germline phenotypes of homozygous bec-1 mutant and bec-1 RNAi depleted animals are indistinguishable (Figure 1D). Since RNAi targets both the maternal and zygotic mRNA, and the phenotype of bec-1 mutants subjected to RNAi against bec-1 was not enhanced (data not shown), these results suggest that there is no significant maternal rescue of the bec-1 mutant germline phenotype.
Figure 1. BEC-1-mediated autophagy controls germ cell population in the distal gonad.
(A) Schematic, and representative DAPI stained image (B) of the distal part of the gonad from a wild-type animal. (C) Representative images of wild-type and bec-1(ok691) null mutant animals. (D) Quantification of nuclei in the mitotic proliferative region of animals with the indicated genotypes (black symbols) or RNAi-treated (gray symbols). (E) Schematic of the step-wise autophagy pathway with relevant genes indicated. (F, G, H) Quantification of mitotic nuclei in the proliferative zone upon loss of autophagy genes (by genomic mutation [black] or RNAi [grey]). (I) Quantification of mitotic nuclei at the indicated developmental stages. In F-I, genes are color coded according to (E), retromer genes are in black. In G, all animals carry him-5(e1490) in the background. Animals were grown at 15°C, and for (C) , (D), (E), and (H), shifted to 20°C as L3 larvae, and analyzed as young adults. For panel (G), animals were shifted to 20ºC as L1 larvae. Results reflect the average of at least three biological replicates shown as the mean ± SEM (error bars). Statistical significance compared to control was determined by one-way ANOVA with Dunnett’s correction in all panels, and indicated as *** P≤0.001, **** P≤0.0001; ns - not significant. Number of analyzed gonads N≥20 for all experiments, except for (H), where N≥15. See also Figure S1 and S4E.
BEC-1-mediated autophagy, not retromer function, controls germline proliferation
In addition to its role in the nucleation of autophagosomes [7], BEC-1/Beclin1 has been shown to function in a complex with VPS-34/PI3K in endocytosis, and as part of the retromer, in the transport from endosomes to the Golgi network [6]. We first inhibited genes required at different steps of autophagy, a stepwise process mediated by different protein complexes for all of which orthologs have been identified in C. elegans (Figure 1E) [8–10]. The steps include: induction (e.g. ATG-9, EPG-1/ATG-13), nucleation of the pre-autophagosomal structure (e.g. BEC-1/Beclin1/Atg6/Vps30, VPS-34/PI3K, EPG-8/ATG14, and VPS-15), elongation of the isolation membrane (e.g., ATG-7, ATG-3, LGG-3/Atg12, ATG-16.2/ATG16L), docking and fusion with the lysosome (eg. SNAP29), and retrieval of membrane or membrane proteins (e.g. ATG-18/WIPI1/2, ATG-2)(Figure 1E). Loss of function of autophagy genes at different steps resulted in a reduced number of germ cell progenitors in both hermaphrodites (Figure 1F-G) and males (data not shown). Importantly, we found that loss of CUP-5/ MCOLN1, the C. elegans ortholog of human mucolipin 1, a protein important for normal lysosomal degradation [11, 12], also resulted in a reduced number of germ cell progenitors in the mitotic zone (Figure 1G). In contrast, RNAi against genes that encode core retromer proteins, including the sorting nexin factor snx-1/SNX1 or the receptor mediated endocytosis protein rme-8/RME8, resulted in no change of germline stem cell number (Figure 1G). We conclude that autophagy genes functioning at different steps of the autophagic process, including lysosomal degradation, are required for the proliferation of stem cells during development, whereas retromer transport may not serve a major function in this process.
Under normal growth conditions, wild-type germline stem cell progenitors expand during the third and fourth larval stages to establish an adult pool of germ cells [13]. We determined that autophagy genes are not required for early proliferation, but are essential for the late larval accumulation of germline stem cell progenitors (from this point on we refer to this phenotype as a germline proliferation defect) (Figure 1H). This germline phenotype of autophagy mutants is similar to that of mutants in the sole insulin-like growth factor receptor daf-2/IIR (Figure 1H) [14], suggesting that autophagy proteins, such as BEC-1/Beclin1, ATG-16.2/ATG16L and ATG-18/WIPI1/2, may function in a similar manner as the DAF-2/IIR receptor.
To obtain insight into the germline defects in bec-1 loss of function animals, we examined the plasma membrane of the germ line by using a GFP::PLC1 (phospholipase C delta) reporter (Figure S1). PLC1 (derived from rat PLC1) comprises a PH domain that specifically binds to plasma membrane associated phosphatidylinositol 4,5-biphosphate [15]. In wild-type hermaphrodites, the membranes of the germline syncytium show uniformly arranged hexagons, and line up at the periphery surrounding the shared cytoplasm (rachis), in the shape of a T-letter (Figure S1 and S4E). In contrast, bec-1 loss of function animals contained germ cells with discontinuous, randomly shaped membranes, and appeared disorganized, with T-shapes no longer visible (Figure S1 and S4E). Surprisingly, atg-7 depleted animals did not display the same level of membrane disorganization as bec-1 mutants (Figure S1), despite the fact that atg-7 RNAi animals have a decrease in stem cell progenitors (Figure 1E and G). However, we failed to observe the characteristic division patterns of wild-type germ cells in atg-7 mutants (Figure S1). Together, our results suggest that BEC-1 is necessary for membrane formation, recruitment and/or stability, to ensure proper division patterns during germ line stem cell proliferation. A function for autophagy in membrane trafficking, or nonconventional secretion, could underlie the observed phenotypes (reviewed in [16]). Alternatively, but not mutually exclusive, autophagy could provide metabolites, such as nucleosides, fatty acids, sugars, or amino acids, and may have a role in the polarity or orientation of cell division [17].
BEC-1 acts independently of GLP-1/Notch signaling to control germ cell proliferation
GLP-1/Notch, DAF-7/TGFβ and DAF-2/insulin-like IGF-1 receptor (IIR) signaling have been shown to promote germ cell proliferation [14, 18–20]. To determine in which pathways autophagy genes act, we tested GLP-1/Notch signaling, which acts from the distal tip cell (DTC) to create a stem cell niche, and maintains the proliferative fate of the germ line [19]. We found that loss of bec-1 gene function, either by RNAi or by genomic mutation significantly decreased the number of stem/progenitor cells in the weak gain of function allele glp-1(ar202gf) (Figure S2A, S2B), an allele that is characterized by lack of germ cells differentiation, and overproliferation [21]. Similarly, genetic removal of atg-16.2(ok3224) or atg-18(gk378) significantly suppressed the glp-1(ar202gf) overproliferation phenotype (Figure S2B). Thus, autophagy gene function is required for the excessive proliferation of progenitor/stem cells in glp-1(ar202gf) mutants. Interestingly, the glp-1(ar202gf) allele suppressed the mitotic germline proliferation defect of mutations in bec-1/Beclin1, atg-16.2/ATG16L, or atg-18/WIPI1/2 to wild-type levels, but not to the level of glp-1(ar202gf) alone (Figure S2B), suggesting that autophagy has a GLP-1 independent activity in promoting germline stem cell proliferation. In contrast, loss of BEC-1 function further decreased germ cell proliferation in the glp-1(e2141lf) loss of function allele (Figure S2C), which is characterized by a reduction in germline progenitors, and a severe Glp (Germline abnormal proliferation) phenotype, where germ cells do not proliferate and differentiate prematurely [19]. Therefore, reducing bec-1 activity enhanced the phenotype associated with a reduction in glp-1 function, and suppressed the phenotype associated with elevated glp-1 activity. One possibility was that autophagy proteins, such as BEC-1/Beclin1, act to modulate the production or localization of a DTC-expressed Notch ligand or ligands. To examine this possibility, we depleted bec-1 in a gld-2(q497) gld-1(q1485); glp-1(q175) triple mutant [1], where proliferative germ cells are observed, despite the complete absence of GLP-1/Notch activity. However, we found that bec-1/beclin1 RNAi still reduced the number of mitotic germ cells in the gld-2(q497) gld-1(q1485); glp-1(q175) triple mutants to a similar extent as in wild-type animals (Figure S2D). Taken together, our glp-1/Notch gain and loss of function experiments suggest that BEC-1/Beclin1 functions, at least in part, independently of GLP-1/Notch activity to promote germ cell proliferation.
BEC-1 functions in a DAF-18/PTEN and SKN-1/Nrf dependent but DAF-16/FOXO independent manner to promote germline proliferation
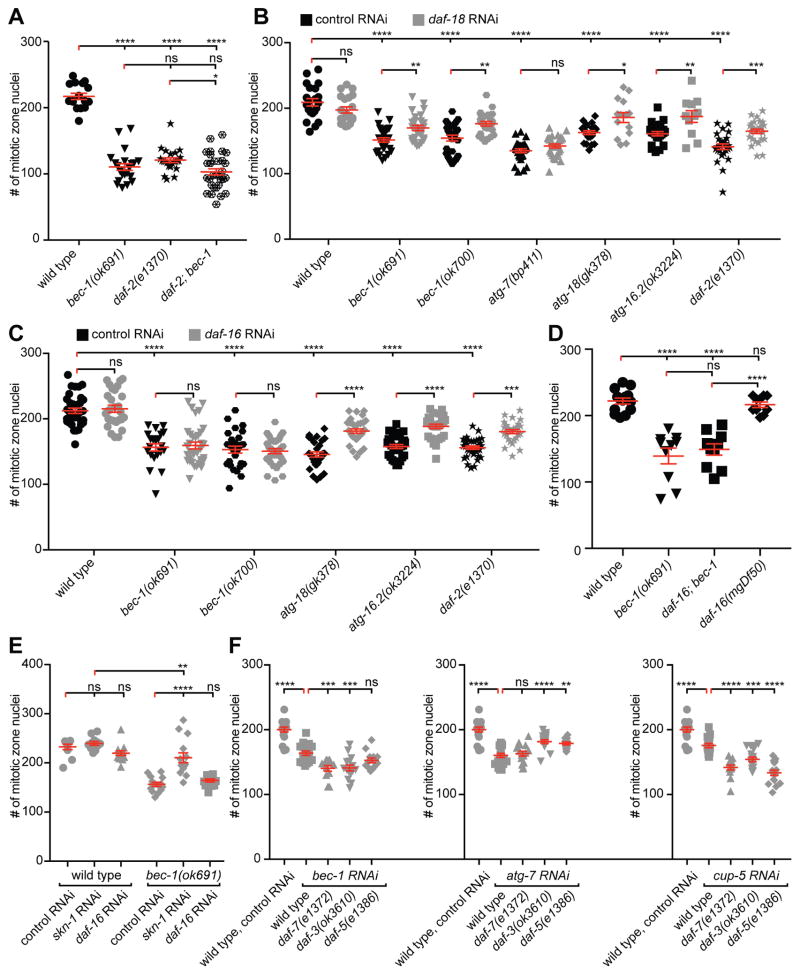
The reduction of germline proliferation in autophagy mutants is similar to that of loss of function mutants in daf-2/IIR signaling (Figure 1H) [14]. Thus, we investigated possible interactions between the DAF-2/IIR pathway and autophagy genes. The reduction of germline progenitor cells in bec-1(ok691); daf-2(e1370) double mutants was indistinguishable from that of bec-1 single mutants, however slightly enhanced when compared to daf-2/IIR single mutants (P≤0.05) (Figure 2A). We conclude that bec-1 acts in a pathway with daf-2/IIR signaling to promote germline proliferation.
Figure 2. BEC-1 functions in a DAF-18/PTEN and SKN-1/Nrf dependent but DAF-16/FOXO independent manner to promote germline proliferation.
(A) Quantification of mitotic nuclei in the proliferative zone of wild type, bec-1(ok691), daf-2(e1370) single, and daf-2(e1370); bec-1(ok691) double mutants. (B, C) Quantification of mitotic nuclei in wild-type, bec-1(ok691), bec-1(ok700), atg-7(bp411), atg-18(gk378), atg-16.2(ok3224) or daf-2(e1370) single mutants treated with RNAi against daf-18/PTEN (B), or daf-16/FOXO (C); plasmid (L4440) served as a negative control in all RNAi experiments. (D) Quantification of mitotic nuclei in animals of the indicated genotype. (E) Quantification of mitotic nuclei in wild type or bec-1(ok691) animals, treated with RNAi against skn-1/Nrf or daf-16/FOXO. (F) Quantification of mitotic nuclei in wild-type, daf-7(e1372), daf-3(ok3610), daf-5(e1386) single mutants, RNAi depleted against control, bec-1, atg-7 or cup-5. For (A)-(E), animals were raised at 15ºC, shifted to 20ºC as L3 larvae, and analyzed as young adults. Results reflect the average of at least 2 trials and shown as mean ± SEM (shown as error bars). The number of analyzed gonads was N≥21 for all experiments. Statistical significance was determined by one-way ANOVA with corrections for multiple comparisons using the method of Tukey (A,E) or Šídák (B,C,D,F), and indicated as * P ≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.0001, ns - not significant. See also Figure S2.
DAF-2/IIR signaling promotes germline stem cell proliferation through the canonical class I PI3K pathway, which inactivates the transcription factor DAF-16/FOXO, and is negatively regulated by DAF-18/PTEN [22, 23]. Interestingly, we found that daf-18/PTEN RNAi depletion suppressed the decrease in the number of mitotic nuclei of bec-1 (ok691 and ok700)(P≤0.01), atg-18(gk378) (P≤0.05), or atg-16.2(ok3224) (P≤0.01), mutant animals (Figure 2B). In contrast, daf-16/FOXO RNAi depletion had no effect on bec-1 (ok691 or ok700) mutants, but suppressed the decrease in the number of mitotic nuclei in atg-16.2(ok3224) or atg-18(gk378) mutant animals (Figure 2C). To further corroborate the notion that bec-1/beclin1 acts independently of daf-16/FOXO, we analyzed a daf-16(mgDf47); bec-1(ok691) double mutant, and found that complete loss of daf-16/FOXO also failed to suppress the bec-1/beclin1 mutant phenotype (Figure 2D). However, RNAi depletion of skn-1, a gene that encodes a bZip transcription factor orthologous to mammalian Nuclear factor-erythroid-related factor (Nrf), an alternative transcription factor to DAF-16/FOXO [24], suppressed the reduction of mitotic nuclei phenotype of bec-1 mutants (Figure 2E). Thus, autophagy genes display complex interactions with the DAF-2/IIR signaling pathway to control stem cell proliferation. Whereas bec-1 function is dependent on DAF-18/PTEN and SKN-1/Nrf, but independent of DAF-16/FOXO, atg-16.2/ATG16L and atg-18/WIPI1/2 function is dependent on DAF-18/PTEN and DAF-16/FOXO. The mechanism for germ cell proliferation for BEC-1 we suggest here is reminiscent of the mechanisms that control entry into the L1 diapause arrest in response to starvation. This process is also regulated by DAF-2/IIR in a DAF-18/PTEN-dependent, but DAF-16/FOXO independent manner [25]. In contract, the loss of function phenotype of atg-7 was not suppressed by RNAi-mediated knock down of daf-18/PTEN under the same conditions. This suggests that ATG-7 may not be modulated by DAF-18/PTEN to control germline stem cell proliferation (Figure 2B), a finding consistent with the observation that ATG-7 acts in a pathway with TGFβ signaling (see below). Alternatively, but not mutually exclusive, different autophagy genes may display different sensitivity to reducing the function of the DAF-18/PTEN negative regulator.
BEC-1 acts independently of DAF-7/TGFβ signaling, whereas ATG-7 promotes germline proliferation together with DAF-7/TGFβ signaling
A third pathway shown to control germline proliferation is the DAF-7/TGFβ signaling pathway, which negatively regulates the daf-3/CoSMAD and daf-5 genes to maintain the balance between proliferation and differentiation in response to sensory cues that report on food availability and population density [18]. When we tested the impact of autophagy genes on TGFβ signaling, we found neither enhancement nor suppression of the germline proliferation defect after depletion of bec-1 or cup-5 by RNAi in daf-3/CoSMAD or daf-5 mutants (Figure 2F). This observation contrasts with the phenotype of daf-7/TGFβ, which is readily suppressible by genetic removal of the negative regulators of TGFβ-signaling, daf-3/CoSMAD or daf-5 [18]. Interestingly, we found that the atg-7 RNAi loss of function phenotype was suppressed by loss of daf-3/CoSMAD or daf-5. Since the daf-7/TGFβ loss of function phenotype was not enhanced by atg-7 RNAi depletion (Figure 2F), our results suggest that atg-7 acts in a pathway with daf-7/TGFβ to promote proliferation of the germ cell population, and provide another example of a genetic distinction between the autophagy genes bec-1/Beclin1 and atg-7/ATG7 (see above). In contrast, the daf-7/TGFβ loss of function phenotype was enhanced upon bec-1 or cup-5/MCOLN1 RNAi depletion (Figure 2F), suggesting that BEC-1/Beclin1 and CUP-5/MCOLN1 act in a parallel pathway with DAF-7/TGFβ. Lastly, the reduction in adult germline stem cell proliferation observed as a consequence of dietary restriction, in eat-2 mutants, was exacerbated by bec-1 RNAi (Figure S2E). We conclude that BEC-1/Beclin1 acts in an independent pathway from DAF-7/TGFβ signaling, whereas ATG-7 may act together with DAF-7/TGFβ to promote the expansion of germline stem cell progenitors.
BEC-1/Beclin1 regulates cell cycle progression
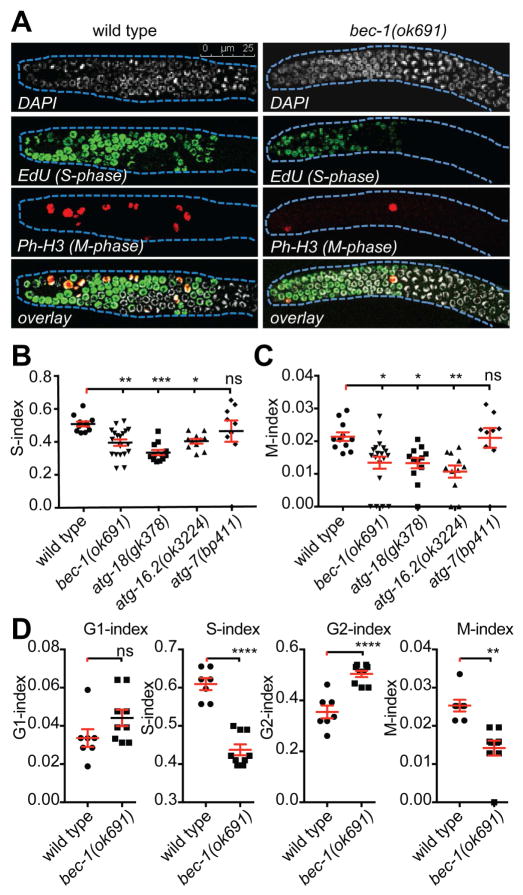
Our results showed that autophagy genes are necessary for the stem cell proliferation in the developing germ line that occurs during the L4 larval stage and thereafter (Figure 1H). We considered three possible mechanisms for how BEC-1/Beclin1 mediated autophagy may be required to maintain a mitotic population of stem cells: (1) by ensuring cell survival, (2) by promoting the transition from proliferation to differentiation, and/or (3) by controlling cell cycle progression. We found no increase in the number of apoptotic corpses (or cellular debris) in the gonad at the L4 larval stage (Figure S3A), even when the decrease in stem cell proliferation due to autophagy gene knock down (bec-1, vps-34, or atg-18) was already visible. Thus, we conclude that the reduction of progenitor/stem cell number in young autophagy gene mutants is not due to a decrease in cell survival.
Precocious transition into meiosis can decrease the stem/progenitor cell pool. The GLD-1 and GDL-3 RNA binding proteins are important for the transition of germ cells from proliferation to differentiation [1, 26, 27]. Lack of GLD-1 in gld-1(q485) mutants decreases the length of the mitotic zone due to precocious differentiation, whereas lack of GLD-3 in gld-3(q741) mutants extends the mitotic zone, due to a delay in the decision to differentiate [1, 27]. We found that RNAi against bec-1, vps-34, atg-18 or atg-7 in gld-1(q485) or gld-3(q741) single mutants resulted in no change in the length of the proliferative zone (Figure S3B). These results suggest that autophagy genes are not involved in the decision to transition from mitosis to meiosis (i.e. proliferation vs differentiation).
Finally to investigate the role of autophagy genes in cell cycle progression, we first evaluated the fraction of germ cells in the actively dividing Mitotic-phase (M-index), and in the DNA-Synthesis phase (S-index), in wild type animals, bec-1, atg-16.2, atg-18 or atg-7 mutants (Figure 3A–C). We found a significant reduction in S-index (Figure 3B), as well as a reduced M-phase index (Figure 3C) in bec-1, atg-18 and atg-16.2 mutants compared to wild type animals. In contrast, the loss of atg-7 activity appeared to have no effect on either S-phase or M-phase (Figure 3B–C), consistent with our genetic analysis, demonstrating that atg-7 acts in a pathway with daf-7/TGFβ, which is not required for cell cycle progression [18]. In addition to the reduced M- and S-indices, bec-1 mutants displayed an increase in the number of cells in G2 and G1, although G1 failed to reach statistical significance (Figure 3D), indicating that the cell cycle is slowed by an extension of the G2 phase. Furthermore, autophagy loss of function mutants exhibited no visible cell cycle arrest (data not shown), providing additional evidence for the conclusion that autophagy functions to promote cell cycle progression.
Figure 3. BEC-1 is important for the cell cycle progression.
(A) Confocal images of gonads stained with DAPI, and EdU for S- phase, and with anti-Ph-H3 for M-phase. (B) Quantification of S-phase index and (C) M-phase index in wild type, bec-1(ok691), atg-18(gk378), atg-16.2(ok3224), and atg-7(bp411) mutants. (D) Quantification of M-phase, S-phase, G1 and G2 indices in wild type and bec-1(ok691). For (A)–(D), animals were grown at 15°C, shifted to 20°C as L3 larvae, and st ained for analysis as young adults. Results reflect the average of at least three biological replicates shown as the mean ± SEM (shown as error bars). Significance was determined by one-way ANOVA with corrections for multiple comparisons using the method Dunnett’s: * P ≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.0001, ns - not significant; number of analyzed gonads N≥20. See also Figure S3.
BEC-1/Beclin1 promotes germline stem cell proliferation cell non-autonomously from somatic tissues
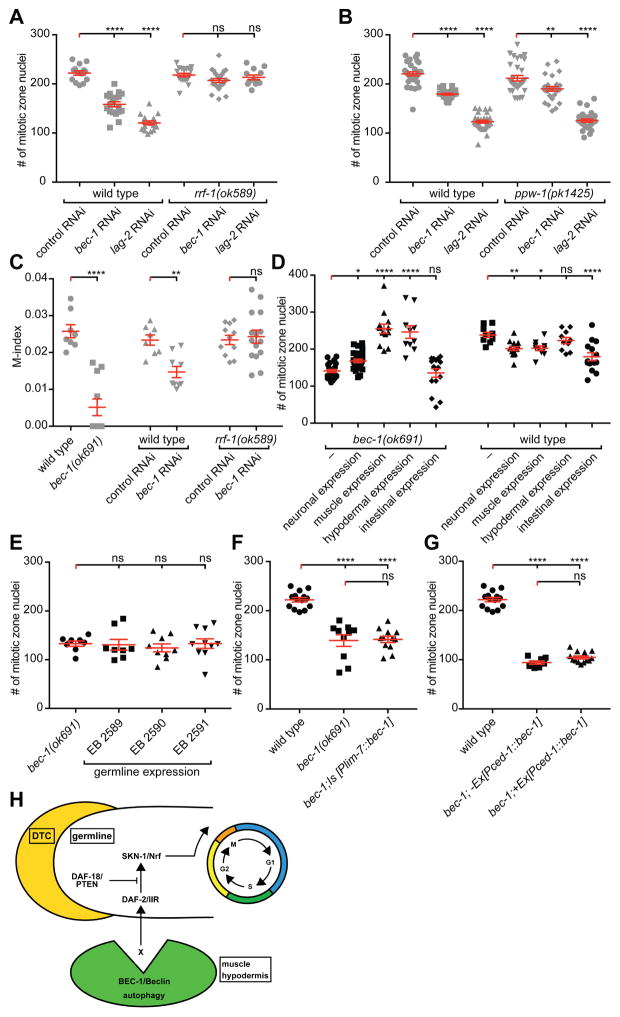
Germline stem cell proliferation could be regulated cell non-autonomously from the surrounding somatic tissue, or cell autonomously within the germ line. Several lines of evidence indicate that BEC-1/Beclin1 can act non-autonomously to control germline stem cell proliferation. First, RNAi depletion of bec-1 in ppw-1 but not in rrf-1 mutants resulted in a reduced germ cell progenitor pool (Figure 4A–B). Since RNAi in rrf-1 and ppw-1 mutants compromise RNAi effectiveness in somatic tissues [28], or in the germ line [29], respectively, these results suggest that BEC-1/Beclin1 functions cell non-autonomously to control germ cell proliferation. Moreover, the significant reduction in M-phase observed upon bec-1 depletion by RNAi was not observed in rrf-1 mutants (Figure 4C). Similarly, the decrease in germ cell proliferation upon RNAi depletion of vps-34 or atg-18 was not observed in rrf-1 germline sensitive mutants (Figure S4C), suggesting that VPS-34/Vps34 and ATG-18/WIPI1/2 are also required cell non-autonomously to control germline proliferation.
Figure 4. BEC-1 acts cell non-autonomously to control proliferation (A, B).
Quantification of the mitotic nuclei number in the proliferative zone of wild-type or rrf-1(ok589) (A), or ppw-1(pk1425) (B) mutants, treated with RNAi against bec-1 or lag-2 (GLP-1/Notch ligand as a positive control); plasmid (L4440) served as a negative control in all RNAi experiments. (C) Quantification of the mitotic index in wild-type, and bec-1(ok691) mutants, compared to wild-type or rrf-1(ok589) animals treated with RNAi against bec-1. (D) Quantification of mitotic nuclei in bec-1 mutants that express the BEC-1 cDNA from a pan-neuronal (Prgef-1), muscle (Pmyo-3), hypodermal (Pdpy-7), intestinal (Pglo-1) promoter, or with no transgene (–). (E) Quantification of the number of mitotic nuclei in animals expressing the BEC-1 cDNA in the germ line as single copy insertions (miniMos). Three independent miniMos lines were assayed. (F) Quantification of mitotic nuclei in bec-1 mutants expressing BEC-1 in sheath cells (Plim-7), or (G) with a ced-1 promoter (Pced-1). For (A)–(C), animals were grown at 20°C and analyzed as young adults. For (D)–(G), animals were raised at 15ºC, shifted to 20ºC as L3 larvae, and analyzed as young adults. Results reflect the average of at least three biological replicates shown as the mean ± SEM (shown as error bars). Statistical significance was determined by one-way ANOVA with corrections for multiple comparisons using the method of Dunnett (A, B, D, E) or Šídák (F, G). Statistical significance of individual comparisons in (C) was determined with the Mann-Whitney test. * P ≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.0001, ns - not significant; number of analyzed animals N≥20, except for (G), where the N≥10. (H) Current model of the cell non-autonomous function of BEC-1 and autophagy during germline proliferation. BEC-1-mediated autophagy interacts with components of the DAF-2/IIR signaling pathway, to directly or indirectly facilitate cell cycle progression of mitotic cell cycle in germline stem cells. See Figure S4.
A second line of evidence for the cell non-autonomous function of BEC-1 is provided by cell specific rescue experiments. We found that BEC-1 expression from a hypodermal [Pdpy-7::BEC-1], a muscle-specific [Pmyo-3::BEC-1], its own promoter [Pbec-1::BEC-1], and less so from a pan-neuronal promoter [Prgef-1::BEC-1], rescued the germline proliferation defects in bec-1 mutant animals (Figure 4D, S4A–B). In addition, all cell cycle defects (the decrease in S- and M- index, as well as the increase in G2) in bec-1 mutant animals were rescued upon hypodermal expression of bec-1 (Figure S4D). In contrast, expression of bec-1 from an intestinal promoter [Pglo-1::BEC-1] did not rescue the proliferation defect of bec-1 mutants (Figure 4D). Similarly, expression of bec-1 in the germ line, through miniMos-mediated single copy transgenes [30], failed to rescue the bec-1 mutant phenotype (Figure 4E), as did bec-1 expression in the somatic gonad [Pced-1, or Plim-7] (Figure 4F–G). Since expression of BEC-1 under the ced-1 promoter partially rescued defects in cell corpse clearance of bec-1/beclin1 mutants [31], our results demonstrate that BEC-1/Beclin1 functions in different tissues to control germline proliferation and cell corpse clearance. Interestingly, tissue specific over-expression of bec-1 in neurons, muscles and intestine in wild-type animals also decreased proliferation (Figure 4D). Thus, too much or too little bec-1/Beclin activity is detrimental, suggesting that bec-1 activity, and by inference, autophagy has to be finely controlled in surrounding tissues to ensure germline stem cell homeostasis. Our observations corroborate a rheostat model to control stem cell progenitor proliferation, which has been previously proposed for Beclin1 activity in mammals, where too little or too much autophagy may have negative consequences [32, 33].
In contrast to previous studies that established Beclin1 or autophagy genes as cell autonomous regulators of Notch signaling [34–36], we find BEC-1 to act non-autonomously, and in parallel to both GLP-1/Notch signaling and TGFβ to promote germline stem cell proliferation. Instead, BEC-1/Beclin acts in a DAF-2/IIR-dependent non-canonical pathway through the transcriptional regulator SKN-1/Nrf (rather than DAF-16/FOXO), and is negatively regulated by DAF-18/PTEN (Figure 4H). In contrast, ATG-16.2/ATG16L and ATG-18/WIPI1/2 function through DAF-16/FOXO to promote germline stem cell proliferation, while also being negatively regulated by DAF-18/PTEN. Intriguingly, ATG-7 functions in a pathway with DAF-7/TGFβ to promote stem cell proliferation, suggesting that ATG-7 may function independently of autophagy in C. elegans germ line proliferation, possibly in analogy to its function in cell cycle arrest of starved mouse embryonic fibroblasts [37]. Thus, although atg-16.2/ATG16L, atg-18/WIPI1/2, atg-7 and bec-1/Beclin1 mutants display a similar decrease in proliferation of the stem cell population, they appear to show different genetic interactions, revealing a previously underappreciated complexity of function.
BEC-1/Beclin1 can function non-autonomously from some but not all tissues to promote the expansion of stem cells in the late larval and adult stages. Similarly, VPS-34/Vps34 and atg-18/WIPI1/2 may also act cell non-autonomously. A possible explanation could be that BEC-1/Beclin1-mediated autophagy functions as part of a secretory pathway that provides signals that control germ line proliferation. Alternatively, autophagy may function to supply metabolites such as amino acids, nucleosides, lipids, or membrane to the developing germ line. Since autophagy genes are conserved from C. elegans to humans, understanding the molecular mechanisms by which autophagy modulates the proliferation and/or maintenance of the stem progenitor cell population in vivo may advance our understanding of the etiology of cancers. Lastly, we cannot exclude that BEC-1 serves additional functions in the germline. For example, we found that the expression of the BEC-1 cDNA in the germline from the pie-1 promoter appeared to at least partially rescue the defects in germline membrane integrity of bec-1 mutants (Figure S4E). Future experiments will distinguish between these possibilities.
EXPERIMENTAL PROCEDURES
Strains, plasmids and imaging
Strains were maintained using standard methods [38]. For a complete strain, plasmid and transgene list, see Supplemental Experimental Procedures.
Counting mitotic nuclei and measurement of proliferative zone length
The number of mitotic nuclei was counted in the proliferative mitotic zone, the region between the distal tip and the edge of the transition zone, in DAPI stained gonads. The edge of the transition zone was determined by the presence of two or more nuclei that had entered meiosis, and adopted the characteristic crescent shape [39]. The distance from the distal tip to the transition zone was measured as the number of cell diameters from the distal tip to the transition zone.
RNA interference
HT115 dsRNA expressing bacteria were grown overnight and seeded on carbenicillin plates. The IPTG/carbenicillin mix was added to induce production of dsRNA, 12 hours prior to placing the animals on the plate. L4 animals were fed the dsRNA-expressing bacteria, and their progeny were synchronized and analyzed. HT115 bacteria with the control empty vector (L4440), or bacteria expressing dsRNA against gfp (gfp RNAi), were used as negative controls. All RNAi experiments were repeated and results were pooled.
Time Course experiment
Individual mid/late L4 stage animals were placed onto plates at 15°C and allowed to self-fertilize. For the analysis of the bec-1(ok691) homozygous animals, F1 progeny from bec-1(ok691)/nT1 heterozygous parents were selected as L3-stage animals, and shifted to 20°C. The germline cell proliferation phenotype was analyzed at larval stages L3, L3/L4, L4, and young adult, using DAPI staining on whole animals.
Cell death
The functional CED-1::GFP fusion protein expression in somatic sheath cells, was used to identify engulfed apoptotic cell corpses [40].
Statistics
Statistical comparisons were performed using the GraphPad Prism 7. For multiple comparisons, we used one-way ANOVA with appropriate corrections for multiple comparisons. In cases that required pairwise comparisons of all means, we used Tukey’s test, whereas we used Dunnett’s correction when all means were compared to a control mean. For cases in which we compared only select pairs of means, the corrections for multiple comparisons were conducted using the method of Šídák. Comparisons of two individual means were performed using the Mann-Whitney test.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (R15 GM102846 to A.M., and R01 GM01313 to H.E.B.), and the National Science Foundation (NSF 0818802 to A.M.). D.S.D.C. is the recipient of a Ph.D. exchange fellowship from CAPES, Brazil. H.E.B. is an Irma T. Hirschl/Monique Weill-Caulier Research Fellow and A.M. is an Ellison Medical Foundation New Scholar in Aging. We thank the Caenorhabditis Genetics Center (GCG, funded by NIH, P40 OD010440), Drs. Kimble, Zetka, Kipreos, Hubbard, Hobert, Greenwald, and Schedl, for strains and reagents. We thank Alex Ruck, Ameer Mourad, Lizbeth Nuñez, David Hoffman, and John Attonito for initial work on tissue specific rescue. We also thank the Queens College Core Facility, Dr. Holtzmann, as well as Shawn Jordan and Dr. Canman, for assistance with imaging, and the Savage-Dunn, Bülow and Meléndez labs for advice and discussions.
Footnotes
AUTHOR CONTRIBUTIONS
A.M. and K.A. conceived and K.A., D.S., D.C., B.G., M.K., F.L., G.S., L.S., and S.W. performed experiments. K.A., H.E.B. and A.M. analyzed the data and prepared the manuscript with editorial input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hansen D, Hubbard EJ, Schedl T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol. 2004;268:342–357. doi: 10.1016/j.ydbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 2.Korta DZ, Hubbard EJ. Soma-germline interactions that influence germline proliferation in Caenorhabditis elegans. Dev Dyn. 2010;239:1449–1459. doi: 10.1002/dvdy.22268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard EJ. Caenorhabditis elegans germ line: a model for stem cell biology. Dev Dyn. 2007;236:3343–3357. doi: 10.1002/dvdy.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubbard EJ, Korta DZ, Dalfo D. Physiological control of germline development. Adv Exp Med Biol. 2013;757:101–131. doi: 10.1007/978-1-4614-4015-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 6.Ruck A, Attonito J, Garces KT, Nunez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7:386–400. doi: 10.4161/auto.7.4.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 8.Melendez A, Levine B. Autophagy in C. elegans. WormBook. 2009:1–26. doi: 10.1895/wormbook.1.147.1. [DOI] [PubMed] [Google Scholar]
- 9.Melendez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Chang JT, Guo B, Hansen M, Jia K, Kovacs AL, Kumsta C, Lapierre LR, Legouis R, Lin L, et al. Guidelines for monitoring autophagy in Caenorhabditis elegans. Autophagy. 2015;11:9–27. doi: 10.1080/15548627.2014.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Natl Acad Sci U S A. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hersh BM, Hartwieg E, Horvitz HR. The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc Natl Acad Sci U S A. 2002;99:4355–4360. doi: 10.1073/pnas.062065399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Killian DJ, Hubbard EJ. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev Biol. 2005;279:322–335. doi: 10.1016/j.ydbio.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Michaelson D, Korta DZ, Capua Y, Hubbard EJ. Insulin signaling promotes germline proliferation in C. elegans. Development. 2010;137:671–680. doi: 10.1242/dev.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, 3rd, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 17.Lobert VH, Stenmark H. Cell polarity and migration: emerging role for the endosomal sorting machinery. Physiology (Bethesda) 2011;26:171–180. doi: 10.1152/physiol.00054.2010. [DOI] [PubMed] [Google Scholar]
- 18.Dalfo D, Michaelson D, Hubbard EJ. Sensory regulation of the C. elegans germline through TGF-beta-dependent signaling in the niche. Curr Biol. 2012;22:712–719. doi: 10.1016/j.cub.2012.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 20.Crittenden SL, Eckmann CR, Wang L, Bernstein DS, Wickens M, Kimble J. Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos Trans R Soc Lond B Biol Sci. 2003;358:1359–1362. doi: 10.1098/rstb.2003.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepper AS, Killian DJ, Hubbard EJ. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics. 2003;163:115–132. doi: 10.1093/genetics/163.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 23.Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 24.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuyama M, Rougvie AE, Rothman JH. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol. 2006;16:773–779. doi: 10.1016/j.cub.2006.02.073. [DOI] [PubMed] [Google Scholar]
- 26.Francis R, Maine E, Schedl T. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics. 1995;139:607–630. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckmann CR, Crittenden SL, Suh N, Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 29.Tijsterman M, Okihara KL, Thijssen K, Plasterk RH. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr Biol. 2002;12:1535–1540. doi: 10.1016/s0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- 30.Frokjaer-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, Pozniakovsky A, Moerman DG, Jorgensen EM. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods. 2014;11:529–534. doi: 10.1038/nmeth.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang S, Jia K, Wang Y, Zhou Z, Levine B. Autophagy genes function in apoptotic cell corpse clearance during C. elegans embryonic development. Autophagy. 2013;9:138–149. doi: 10.4161/auto.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 33.Thorburn A. Autophagy and its effects: making sense of double-edged swords. PLoS Biol. 2014;12:e1001967. doi: 10.1371/journal.pbio.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee G, Liang C, Park G, Jang C, Jung JU, Chung J. UVRAG is required for organ rotation by regulating Notch endocytosis in Drosophila. Dev Biol. 2011;356:588–597. doi: 10.1016/j.ydbio.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barth JM, Hafen E, Kohler K. The lack of autophagy triggers precocious activation of Notch signaling during Drosophila oogenesis. BMC Dev Biol. 2012;12:35. doi: 10.1186/1471-213X-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu X, Fleming A, Ricketts T, Pavel M, Virgin H, Menzies FM, Rubinsztein DC. Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis. Nat Commun. 2016;7:10533. doi: 10.1038/ncomms10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu N, Yu X, He X, Zhou Z. Detecting apoptotic cells and monitoring their clearance in the nematode Caenorhabditis elegans. Methods Mol Biol. 2009;559:357–370. doi: 10.1007/978-1-60327-017-5_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.