Abstract
We have generated a human induced pluripotent stem cell (iPSC) line under feeder-free culture conditions using the urine derived cells (UCs) collected from non-affected control subjects to use as a comparison group for the iPSC lines containing a Plasminogen Activator Inhibitor-1 (PAI-1 null) mutation. The Sendai virus (SeV) vector encoding pluripotent Yamanaka transcription factors was used at a low multiplicity of infection to reprogram the UCs.
Resource Table
| Name of Stem Cell line | iPAI-005-B |
| Institution | Department of Medicine and Cardiovascular Center, Medical College of Wisconsin, 8701 Watertown Plank Road, Milwaukee, WI-53226 |
| Person who created resource | Muhammad Zeeshan Afzal and Melanie Gartz |
| Contact person and email | Jennifer Strande, jstrande@mcw.edu |
| Date archived/stock date | September, 2015 |
| Origin | Human urine-derived cells |
| Type of resource | Induced pluripotent stem cells (iPSC) generated from non-affected control subjects |
| Sub-type | Human induced pluripotent stem cells |
| Key transcription factors | Oct3/4, Sox2, Klf4, and cMyc |
| Authentication | Identity and purity of the cell line confirmed (Fig 2) |
| Link to related literature | |
| Information in public databases | |
| Ethics | Patient informed consent obtained and procedures were approved by the Institutional Review Boards (IRBs) at the Medical College of Wisconsin (Milwaukee, WI) and St. Vincent’s Health (Indianapolis, IN). |
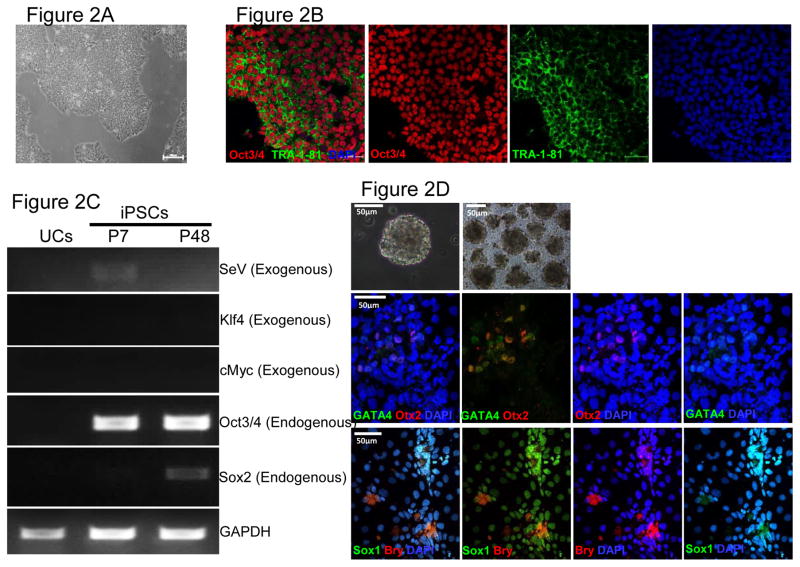
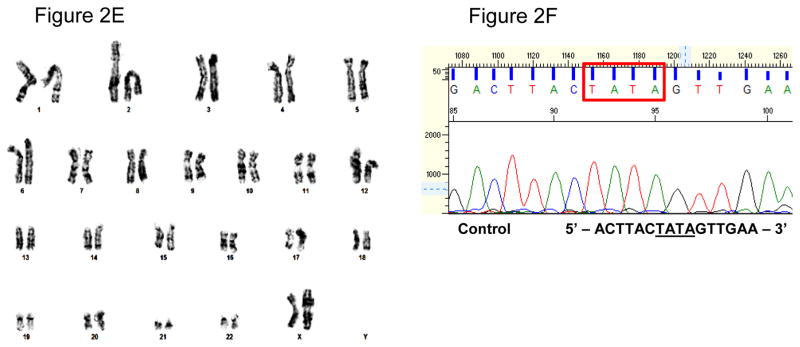
Figure 2. Characterization of iPSCs and genotypic evaluation.
(A) Phase contrast micrograph of the cultured iPSC clone, plated under feeder-free conditions on matrigel. Scale bar, 50μm. (B) Pluripotency of iPSCs confirmed by immunostaining with Oct3/4 (Red) and TRA-1-81 (green) markers and counterstained nuclei with blue DAPI stain. Scale bar, 50μm. (C) RT-PCR gene expression analysis show upregulation of endogenous pluripotent genes (Oct3/4 and Sox2) and elimination of the non-integrating viral genome and exogenous genes (SeV, Klf4 and cMyc). (D) Differentiation into three embyonic germ layers was confirm after processing iPSCs to form embryoid bodies (EBs) and allowing spontaneous in vitro differentiation. Ectoderm, mesoderm and endoderm characterized by immunostaining with Otx 2 (Red)/SOX1 (Green), Brachyury (Red), and GATA4 (Green), respectively. Scale bar, 50μm. (E) Karyogram of iPSCs demonstrate normal ploidy of a female subject. (G) Electropherogram confirms the wild-type gene sequence on the exon 4 of PAI-1 gene in control iPSCs (iPAI-005-B).
Resource Details
We generated human iPSC clones from urine cells of non-affected subjects (iPAI-005-B) to serve as control for iPSCs containing homozygous or heterozygous dinucleotide (TA) insertion mutations within exon 4 of the PAI-1 gene(Fay et al. 1992, Fay et al. 1997) (see companion lab resource articles). iPSC clones were established under feeder-free culture conditions using the Cytotune® iPSReprogramming Kit (Life Technologies, Carlsbad, CA) which employs the non-integrating Sendai virus to deliver reprogramming factors, OCT3/4, SOX2, KLF4 and cMYC (Takahashi et al. 2007). The morphologic iPAI-005-B iPSC clones displayed typical pluripotent cell morphology (Fig 2A). The iPSC clones stained positive for the pluripotency markers Oct3/4 and TRA-1-81 (Fig 2B). The presence of endogenous pluripotency markers OCT3/4 and SOX2 and the absence of exogenous reprogramming transgenes KLF4, cMYC and the SeV was confirmed by RT-PCR (Fig 2C). Demonstration of three germ layer differentiation capacity was confirmed by immunofluorescence analysis of SOX1, OTX2, BRY and GATA4 to identify ectoderm, mesoderm and endoderm in spontaneously differentiated embryoid bodies (Fig 2D). The iPSC lines showed normal karyotype (46, XX) (Fig 2E) and the exon 4 of the PAI-1 gene was confirmed to be the wild-type sequence (Fig 2F).
Materials and Methods
Urine sample collection and urine cell isolation
All procedures described were approved by the Institutional Review Boards (IRBs) at the Medical College of Wisconsin (Milwaukee, WI) and St. Vincent’s Health (Indianapolis, IN). Subject enrollment and sample collection occurred at a distant site. After informed consent was obtained, subjects were instructed on to collect a sterile mid-stream catch urine sample in a sterile urine cup available commercially. The samples were aseptically processed under a portable laminar flow hood and were transferred to sterile 50 mL centrifuge tubes and spun at 400× g for 10 minutes. The urine supernatant was discarded after aspiration and the remaining pellet with particulate portion was resuspended with 2 mL of collection/storage media (1 volume Keratinocyte Serum Free (KSF) Medium, 1 volume Progenitor Cell Medium + 1 volume FBS). Samples were stored on ice for 24–48 hrs until transported back to the laboratory where the urine cell isolation and culturing continued as described by Afzal and Strande (Afzal and Strande 2015). The cells were expanded for a maximum of 5 passages when they were either directly reprogrammed or banked in liquid nitrogen for future experiments.
Non-integrative reprogramming of PAI-1 mutant UCs into iPSCs
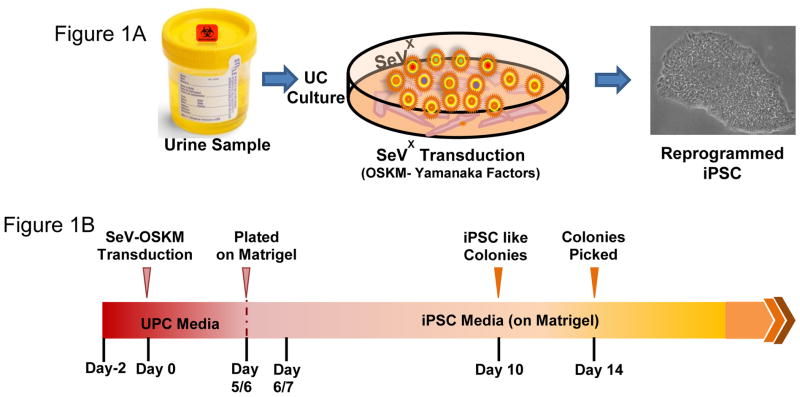
Urine cells grown to 80–90 percent confluency, were exposed to the Sendai virus (SeV) vectors included in the CytoTune® iPS Reprogramming Kit (Life Technologies, Carlsbad, CA) at a multiplicity of infection of 1.5. After 24 hours, the media was replaced with fresh UPC medium daily. Once iPSC-like clones with cobblestone morphology appear on day 5 or 6 post SeV transduction, the reprogrammed UCs were dissociated with trypsin-EDTA, resuspended in UPC medium (supplemented with 5 μM Y27632, RhO Kinase inhibitor) and seeded onto fresh matrigel (10 μg/cm2) coated 12 well plates in the ratio ranging from 1:5 – 1:10. The next day, culture medium was replaced with TeSR-E8 supplement (Stem Cell Technologies, Vancouver, BC, Canada). The media was replaced daily until individual iPSC-like clones were large enough to be selected for picking and expansion, usually by day 14. (Figure 1, A–B)
Figure 1. Schematic representation of the protocol.
(A) Urine derived cells are cultured and reprogrammed using Sendai Virus (SeV) transfection with OSKM-Yamanaka Factors. (B) Timeline for reprogramming UCs.
Embryoid body formation and differentiation
Dissociated iPSC clones were suspended in embryoid body induction (EB) Medium (KO-DMEM, 20% KSR, penicillin/streptomycin, Glutamax, β-mercaptoethanol and non-essential amino acids) and transferred to 6 well suspension culture plates (CELLSTAR, Greiner Bio-One, Monroe, NC) under hypoxic conditions. After 4–5 days, EBs were plated onto gelatin coated chamber slides and cultured under normoxic conditions in differentiation medium (DMEM/F12 with 20% FBS, penicillin/streptomycin, Glutamax, β-mercaptoethanol and non-essential amino acids) for 6–7 days until confluent.
Immunofluorescence staining
Cells grown on matrigel-coated chamber slides were fixed with 4% paraformaldehyde, permeabilized with PBS containing 0.1% Triton X-100, and incubated with CAS-Block™ (Thermo Fischer Scientific Inc., Waltham, MA) for 30 minutes. Unconjugated primary antibodies to Oct3/4 (Santa Cruz Biotechnology, Dallas, Texas) and TRA-1-81 (Thermo Fischer Scientific Inc.), or fluorochrome conjugated antibodies to SOX1, Otx-2, Brachyury, and GATA4 (Human Three Germ Layer 3-Color Immunocytochemistry Kit, R&D Systems Inc, Minneapolis, MN) were incubated overnight at 4°C. When necessary, cells were incubated with fluorochrome conjugated secondary antibodies (Thermo Fischer Scientific Inc.) for 4 hrs at room temperature under humid conditions. Coverslips were mounted with fluorshield mounting medium containing nuclear counterstain DAPI (Sigma Aldrich, St. Louis, MO). Cells were visualized with confocal imaging (Nikon A1-R confocal microscope; Nikon Instruments Inc., Melville, NY).
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA) and complementary DNA (cDNA) was synthesized from 1μg of total mRNA using iScript cDNA synthesis kit (BioRad Laboratories, Hercules, CA). RT-PCR was performed with the primers listed in Table 1. Amplification followed an initial denaturation at 95°C for 5 minutes, and 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, and final extension at 72°C for 7 minutes.
Table 1.
List of primer sequences used for RT-PCR or genotyping analysis.
| SeV (Exogenous) | Forward | GGATCACTAGGTGATATCGAGC |
| Reverse | ACCAGACAAGAGTTTAAGAGATATGTATC | |
| Klf4 (Exogenous) | Forward | TTCCTGCATGCCAGAGGAGCCC |
| Reverse | AATGTATCGAAGGTGCTCAA | |
| cMyc (Exogenous) | Forward | TAACTGACTAGCAGGCTTGTCG |
| Reverse | TCCACATACAGTCCTGGATGAT | |
| Oct3/4 (Endogenous) | Forward | CAGTGCCCGAAACCCACAC |
| Reverse | GGAGACCCAGCAGCCTCAAA | |
| Sox2 (Endogenous) | Forward | CAAGATGCACAACTCGGAGA |
| Reverse | GTTCATGTGCGCGTAACTGT | |
| GAPDH | Forward | GTGGACCTGACCTGCCGTCT |
| Reverse | GGAGGAGTGGGTGTCGCTGT | |
| PAI-1 (Exon-4) | Forward | CCTGACTGCAGCCCTTTGACATACA |
| Reverse | ACATCTAGAGCATTCCCTGTGGTCTTCCTC |
Karyotype Analysis
Chromosomes of at least 20 proliferating cells per line were counted and fully analyzed using G-banding by Wisconsin Diagnostic Services (Milwaukee, WI).
Assessment of PAI-1 exon 4 sequence
Genomic DNA from iPSCs was extracted using QIAamp DNA Mini Kit (Qiagen) and a genomic 753 bp fragment flanking the dinucleotide (TA) insertion site within exon 4 of PAI-1 gene was PCR amplified. The PAI-1 primer set used for the amplification is listed in Table 1. PCR products were purified using QIAquick PCR purification kit (Qiagen) and sent to Retrogen, Inc (San Diego, CA) for DNA sequencing.
Acknowledgments
This work was supported by the National Institutes of Health K08 Grant Number HL111148 (J.L.S.) and R01HL051387 (D.E.V.). We are especially indebted to patients and their families for participating in this study and donating samples.
Footnotes
Author disclosure statement:
There are no competing financial interests in this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afzal Muhammad Z, Strande Jennifer L. Generation of Induced Pluripotent Stem Cells from Muscular Dystrophy Patients: Efficient Integration-free Reprogramming of Urine Derived Cells. Journal of Visualized Experiments. 2015;(95):e52032. doi: 10.3791/52032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay William P, Parker Andrew C, Condrey Lorraine R, Shapiro Amy D. Human Plasminogen Activator Inhibitor-1 (PAI-1) Deficiency: Characterization of a Large Kindred With a Null Mutation in the PAI-1 Gene. Blood. 1997;90(1):204–208. [PubMed] [Google Scholar]
- Fay William P, Shapiro Amy D, Shih Judy L, Schleef Raymond R, Ginsburg David. Complete Deficiency of Plasminogen-Activator Inhibitor Type 1 Due to a Frame-Shift Mutation. New England Journal of Medicine. 1992;327(24):1729–1733. doi: 10.1056/NEJM199212103272406. [DOI] [PubMed] [Google Scholar]
- Takahashi Kazutoshi, Tanabe Koji, Ohnuki Mari, Narita Megumi, Ichisaka Tomoko, Tomoda Kiichiro, Yamanaka Shinya. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. http://dx.doi.org/10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]