Abstract
Seasonal influenza infects approximately 5–20% of the U.S. population every year, resulting in over 200,000 hospitalizations. The ability to more accurately assess infection levels and predict which regions have higher infection risk in future time periods can instruct targeted prevention and treatment efforts, especially during epidemics. Google Flu Trends (GFT) has generated significant hope that “big data” can be an effective tool for estimating disease burden and spread. The estimates generated by GFT come in real-time – two weeks earlier than traditional surveillance data collected by the U.S. Centers for Disease Control and Prevention (CDC). However, GFT had some infamous errors and is significantly less accurate at tracking laboratory-confirmed cases than syndromic influenza-like illness (ILI) cases. We construct an empirical network using CDC data and combine this with GFT to substantially improve its performance. This improved model predicts infections one week into the future as well as GFT predicts the present and does particularly well in regions that are most likely to facilitate influenza spread and during epidemics.
Google Flu Trends (GFT) uses aggregated search query data to estimate influenza activity in the ten U.S. Health and Human Services (HHS) regions and throughout the country. Google's model produces real-time estimates of the percentage of physician visits attributed to influenza-like illness (ILI) using a combination of query terms that best correlated with CDC ILI data from 2003–20081. Initial excitement at the potential of GFT to predict influenza earlier than traditional methods by harnessing “big data” declined when it dramatically erred in February 2013: GFT predicted double the number of doctors' visits from the flu than was later reported by the CDC's traditional sentinel system of hospitals and clinics2,3. An additional limitation is that GFT is significantly less correlated with laboratory-confirmed cases of the flu than with ILI levels1,4. Reducing error in estimates of actual influenza cases as opposed to ILI is critical for prevention and control efforts because ILI captures a multitude of other pathogens and provides a noisy measure of actual flu levels5,6,7.
Following the call by Lazer et al.2, we combine data generated by GFT and the CDC in a model that dynamically recalibrates to produce better estimates of actual cases of the flu using methods borrowed from social network analysis. Influenza spreads from person-to-person via respiratory droplets and requires close physical proximity for infection. As a result, regions with populations that are highly connected to one another (through geographic proximity, air traffic, commuting, etc.) will likely experience highly correlated patterns in influenza levels. This study seeks to improve GFT's accuracy by using historical correlations between influenza outbreaks in different regions to create a network of connected regions that are likely to experience outbreaks at similar times (Figure 1). Other recent work has used empirical models8,9,10 as well as computer simulations11,12 to better understand systematic patterns in the geographic spread of influenza using network analysis. However, to our knowledge, no other studies have used empirical data on connectivity between geographic units in models that assess influenza levels in real time.
Figure 1. Network Map of Influenza Correlations Between HHS Regions.
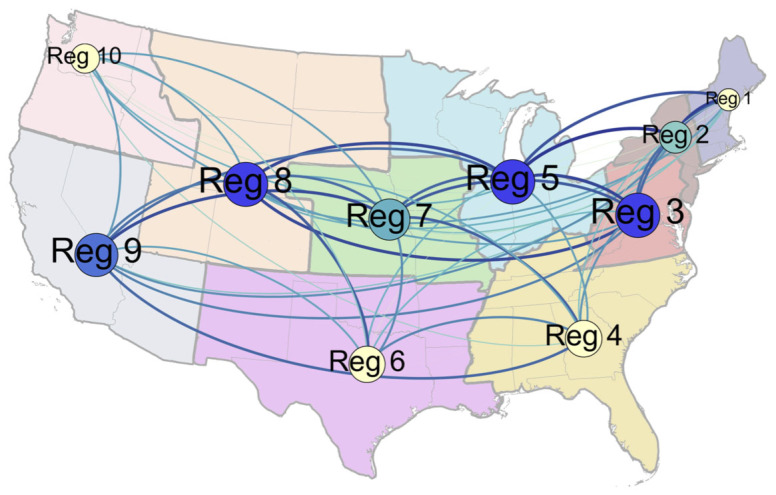
Nodes are sized by weighted degree centrality, which incorporates the number of ties a given region has to other regions (in this case, each region is connected to all other regions) as well as the strength of those ties, which is determined by the strength of the cross-correlation between regions. Nodes are colored by betweenness centrality, which represents the number of shortest paths to other regions that go through a certain node (blue indicates high betweenness and yellow indicates low betweenness). Influenza is likely to pass through regions with high betweenness on its way to other parts of the country. Edges between nodes are colored by the weight of the tie between two regions as measured by the correlation between flu trends in those regions (darker, thicker edges denote stronger ties); only the ties whose weights are in the upper two quartiles are shown. All statistics for this figure are calculated using correlations over the full time period of the data ranging 2003–2012. The nodes and ties were created using GEPHI (version 0.8.2), a social network visualization software, and the background map was added using Adobe Photoshop.
Incorporating information on flu levels of connected regions allows for better assessment of real-time infection levels because knowledge of flu levels in connected regions tempers inflated Google search volumes caused by excess media coverage, especially during epidemics. In addition, incorporating this information on connected regions allows for more accurate predictions of future spread by taking into account how the disease spread in previous years. For example, because flu levels in the mid-Atlantic (Region 3) are historically highly correlated with flu levels in the Midwest (Region 5), observing a flu outbreak in the mid-Atlantic can inform predictions of future flu levels in the Midwest. The model performs particularly well during periods of heightened flu activity, when GFT is most likely to overestimate influenza prevalence. The model also performs best in regions that are most likely to facilitate the spread of influenza (structurally “central” regions). Because these regions are more highly connected to other regions, more information can be gleaned from flu levels in these other regions. Thus, the times and places in which our model provides the biggest improvements on GFT are the most important for prevention and control efforts.
Creating the Network Measure
Incorporating network information into GFT's real-time predictive model followed a two-step process. First, we created a series of weighted ties between regional units defined by the correlation of influenza levels using the CDC data13 on laboratory confirmed cases in every week of the previous year, T-1. In our study, each region had nine weighted ties, one with each of the other regions. A tie between two regions that experience high and low influenza prevalence at the same time was assigned a larger weight on a scale from 0 to 1 than a tie between two regions that experienced peaks and troughs at different times. Cross-correlations between regions are likely a function of factors such as airline traffic, commuting traffic, geographic proximity, vaccination coverage, and climatic patterns, which all impact how influenza spreads between regions.
Step 2 of our method applies the network of weighted ties based on time-series correlations in the previous year T-1 to real-time empirical data in the current year T. Because the factors facilitating the spread of influenza between regions remain relatively constant from year to year, the co-incidence of influenza in the previous year provides a useful framework for predicting the spread of influenza in the current year. At the same time, using only the previous year's correlations (as opposed to correlations over a longer timeframe) allows the model to adapt to smaller changes in the underlying mechanisms facilitating the spread of influenza. In order to incorporate information from the network into the real time model, we multiplied the connectivity factor (the weight representing the strength of the tie between regions i and j) by the estimated levels of influenza produced by GFT in each region j14. Through this method, we produced an influenza network-load measure, ΣWij Pj,t for each region i. In this measure, W is the strength of the tie between regions i and j and P is the current GFT estimate for influenza level in each region j in week t. For example, the influenza-load measure for Region 1 was created by multiplying the weight of the tie between Region 1 and Region 2 by the estimated influenza level in Region 2. This process was repeated for regions 3–10 and the products were summed.
Results
Better Predictions in the Present and Future
Using network data from the previous year to inform our predictions, we hypothesized that incorporating this weighted influenza load measure would allow for (1) better assessment of real-time infection levels and (2) more accurate predictions regarding the future spread of influenza. To test the first prediction, we regress the GFT ILI prediction and our network flu-load measure on laboratory-confirmed cases of influenza in the same week for all weeks 2003–2012 (Table 1). The coefficient on the network measure was positive and highly significant when added to the basic model (column 2). To analyze the substantive significance of this finding, we compared predicted flu levels from the basic GFT model and the network model to actual cases of the flu measured by the CDC. Using this simple network model in real-time reduced error in predicting actual cases by 2.1% relative to the GFT model on its own (Figure 2).
Table 1. Regression on lab-confirmed influenza data in present and future, with and without network statistic. Models one and two regress the actual flu level in time period t on GFT with and without the network statistic, respectively. Models three and four are identical except the dependent variable is the flu level in time period t + 1 (i.e. one week into the future).
| CDC Actual Flu Level (Virologic % Positive) | ||||
|---|---|---|---|---|
| (Present) | (Present) | (Future) | (Future) | |
| Google Flu Trend | 0.839*** | 0.637*** | 0.791*** | 0.607*** |
| (0.022) | (0.028) | (0.022) | (0.029) | |
| Network Statistic | 0.028*** | 0.025*** | ||
| (0.002) | (0.003) | |||
| Constant | −4.053*** | −4.046*** | −3.699*** | −3.690*** |
| (0.159) | (0.156) | (0.161) | (0.159) | |
| Observations | 3,082 | 3,082 | 3,069 | 3,069 |
| R2 | 0.322 | 0.348 | 0.291 | 0.313 |
| Adjusted R2 | 0.322 | 0.348 | 0.291 | 0.312 |
Note: *p < 0.1; **p < 0.05; ***p < 0.01.
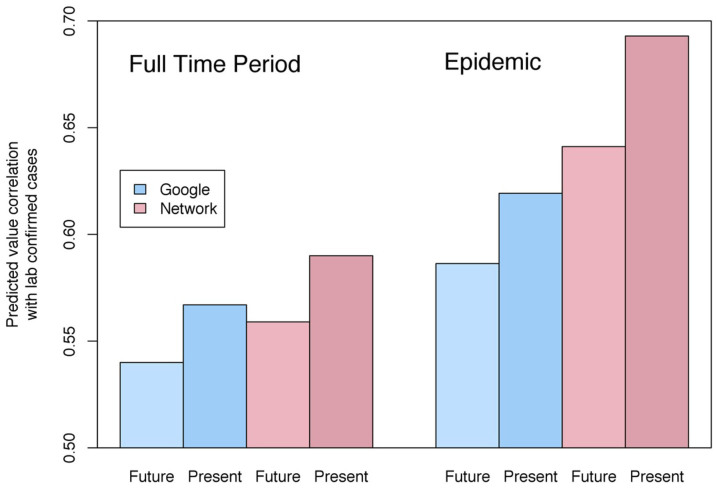
Figure 2. Correlations of Predicted Values with Laboratory-Confirmed Influenza.
Over the full time period of the data, the network model predicted laboratory-confirmed influenza cases significantly better than Google Flu Trends in both the present and one week into the future. Network model predictions for one week into the future were within 1% of GFT predictions for the present. During periods of heightened flu levels above the CDC baseline for each region (seasonal epidemics), estimates influenza levels one week into the future produced by the network model were more than 2% better than GFT predictions for the present.
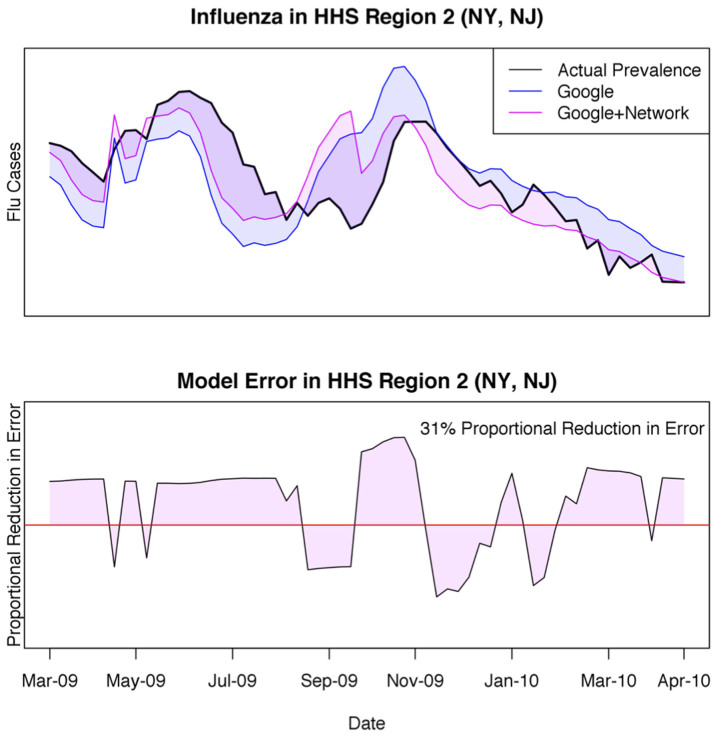
One particularly good example of our model's performance relates to New York and New Jersey (HHS Region 2) during the 2009 H1N1 pandemic, which was first identified in the U.S. and later spread around the globe. Google notoriously underestimated and later overestimated influenza activity during this period, due in part to increased media coverage driving internet search traffic15. Including our network statistic tempered this effect by accounting for estimates of influenza in connected regions and resulted in a 31% proportional reduction in error of estimated influenza levels compared to GFT on its own (Figure 3).
Figure 3. Region 2 Time Trend during the 2009 H1N1 Pandemic.
Google Flu Trends is particularly prone to error during pandemics. The top panel compares the network model (pink) and GFT on its own (blue) to laboratory confirmed influenza levels during the 2009–2010 season. The y-axis on the bottom panel is the residual of the network model subtracted from the residual of Google's model at any given week, with areas above the red line indicating times in which the network model outperformed Google on its own. The proportional reduction in error of the network model compared to GFT during this time span is 31%.
Perhaps more importantly, incorporating the network measure significantly improved predictions of laboratory confirmed influenza cases one week into the future (Hypothesis 2). With the knowledge of where influenza is now, along with an understanding of which regions are likely to experience influenza outbreaks at similar times, we estimated next week's influenza levels in all regions. Columns 3 and 4 of Table 1 show the same regression model but with the dependent variable (actual flu cases) being predicted one week in the future. The proportional reduction in error of the network model compared to the basic GFT model in this case is 1.7%. By including our network variable, we do just as well at predicting virological outcomes one week into the future as GFT on its own does at predicting outcomes in the present (Figure 2).
Epidemics and Centrality
In addition to improving on the general GFT model, the network model performs particularly well in the times and places that are most pivotal for prevention and control efforts. During seasonal epidemics, proportional reduction in error of the network model relative to GFT alone was three times greater than during periods of low or normal flu levels (6.3% compared to 2.1%). Moreover, during these periods, the network model predicted flu levels one week into the future nearly 2% more accurately than GFT predicted influenza levels in the present (Figure 2).
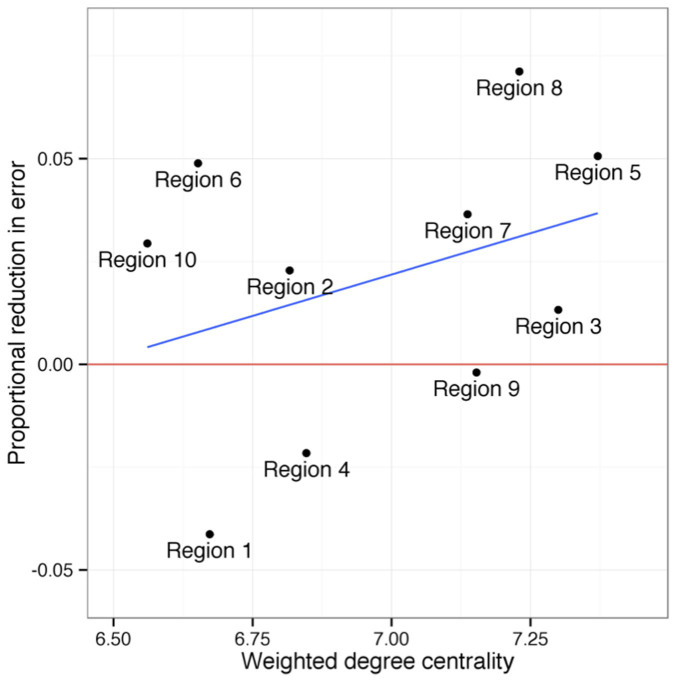
The network model also performed best in regions that were most important for facilitating the spread of influenza. The proportional reduction in error of the network model compared to GFT was greatest in regions that were most highly connected to other regions (Figure 4). We calculated a weighted degree centrality score for each region using correlations between regions over the full time period. In the most central region (Region 5), the network reduced more than twice as much error as the average across all other regions (over 5% compared to an average of just over 2%). Our results show that geography plays an important role in the network: regions with more central geographic locations were more likely to have strong ties to a greater number of regions and consequently have a large influence over influenza spread. However, we also found that regions that are important transportation hubs (e.g. Regions 2 and 9 – New York and California) were more central to the influenza network than geography alone might suggest (Figure 1).
Figure 4. Centrality and Network Model Effectiveness.
The network model does particularly well in more central regions. The y-axis (proportional reduction in error) is a measure comparing the residuals of the network model to the residuals of the Google Flu Trends model, with positive numbers indicating regions in which the network model outperforms Google on its own. All statistics are calculated using the full time period of data ranging from 2003–2012.
Discussion
This study responds to the need to combine novel, modern data sources with time-proven data collection2. By combining sentinel data on laboratory confirmed cases of influenza with GFT, we make strides towards accessing the best of both. There are several reasons why our model improves on either data source alone. Our dynamic network model combines the accuracy of time proven sentinel data collection with the real-time predictions that make GFT valuable. In addition, a network based in empirical trends of connectivity between US regions makes it possible to leverage data on infection levels in adjacent areas when estimating current illness levels for a given area. Information on infection rates in other regions is particularly valuable in predicting future flu incidence because many of the factors that facilitate the spread of disease between areas remain relatively constant from year to year (for example, travel between regions).
The findings of this paper have important implications for prevention and control efforts at the local and national level. Predicting the geographical spread of influenza is critical for informing clinical treatment of disease as well as prioritizing public health interventions such as vaccination. Early and accurate detection of influenza activity can inform efforts to reduce the spread and impact of the disease5,6,16. At the national level, vaccination campaigns can target central regions in the network that are likely to be epicenters for large-scale regional and national outbreaks7. Having more accurate predictions of influenza levels in the most central regions is particularly valuable for prevention and control efforts, as they are likely to facilitate the spread of influenza to other parts of the country. Reducing influenza levels in these regions will have the greatest spillover effects on influenza levels elsewhere.
Our focus on estimating laboratory-confirmed influenza levels (as opposed to ILI) is particularly impactful because targeted prevention efforts will only be successful at culling outbreaks if the proper illness is being tracked. Accurate ILI assessment and prediction can help prepare medical personnel for the influx of patients but is less useful for targeting the future spread of disease. Because ILI is a measure of doctors visits as opposed to actual disease, it is highly sensitive to factors that influence visits and not the disease itself (e.g. media coverage). In addition to the focus on actual cases of the flu as opposed to ILI, this paper takes the important step of assessing real-time models that predict the future spread of the flu. Prevention and control efforts relying on current flu estimates suffer from the tendency to chase the disease rather than anticipate its spread. Models that improve predictions of future disease spread in real-time allow officials to get a leg up on the disease and target efforts in areas that are likely to be affected, thus increasing a potential campaign's effectiveness. Knowing future spread is particularly important during epidemics, time periods in which our model performs particularly well.
More broadly, this paper highlights the advantages of incorporating network measures into real-time models of the spread of disease and of integrating, rather than replacing, traditional data collection with “big data”. Building on these methods may have implications for a wide range of epidemiological models. Given the increasing focus on the structural spread of disease through individual17 and geographic networks8, incorporating these aspects into real-time predictive models is a natural next step.
Methods
Data from GFT and the CDC were available for every week from October 2004 through September 2011 and were joined by week and HHS Region. The CDC reports data on the number of doctor visits attributed to influenza-like-illness (ILI) as well as the percentage of respiratory samples tested for influenza that come back positive13. Virological data comes from state public health laboratories and certain smaller level public health laboratories and participating medical centers. Weeks in which a region experienced heightened flu activity (an epidemic) were determined using the CDC's threshold for epidemics, defined as an increase of 1.645 standard deviations above the seasonal baseline of deaths attributed to influenza and pneumonia.
To construct the weighted network measure, we first calculate a connectivity factor, W, for every pair of regions i and j, where i denotes the region for which the measure is being calculated. The connectivity factor is the cross-correlation in laboratory confirmed-influenza cases for each pair of regions ij in the previous year, T-1 (correlation taken across all weeks). We then multiply the current GFT value in region j by the connectivity factor, Wij. Lastly, we sum the product of these process for region i across all other regions, j.
We report models predicting lab confirmed cases in table 1 and calculate a proportional reduction in error (PRE) for the models with and without the network measure. PRE is calculated by subtracting the sum of prediction errors for the model with the network measures from the sum of errors of the base model and dividing this difference by the summed errors of the base model.
We fit the following linear regression models (OLS):
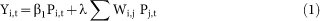 |
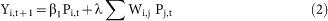 |
OLS regression is used in keeping with Ginsberg et al (2009)1 and other papers assessing the effectiveness of GFT. In each model, the first right-hand-side term is the GFT estimate for each region i in the current week t and the second is our weighted influenza load measure. Model 1 allows us to assess the impact of including our network term in real-time assessments for the current week (t) while Model 2 allows us to assess its usefulness in making predictions one week in the future (t + 1). Following previous studies that evaluate GFT's estimates, we compared these data to CDC data on the percentage of cases that exhibit ILI as well as lab-confirmed cases of influenza (Y)1,3.
To verify the resilience of our main findings, we performed out-of-sample testing through K-fold cross-validation. This method involved splitting the sample into equal sized subsamples, or folds. Over k rounds, the model was recursively fit on a training set, consisting of (k-1)/k folds, and then the dependent variable was predicted for observations in the validation set (1/k). This method included every observation in the testing set only once, helping avoid any testing error that might result from single out of sample predictions on a single fold18. The results confirm the reported reductions in error.
Statistical analyses were done using R (version 3.0).
Acknowledgments
We thank James H. Fowler for critical discussions and reading of the manuscript. We also thank members of the Human Nature Group workshop at the University of California, San Diego for providing comments and guidance. M.D. and D.A.H. thank the Robert Wood Johnson Foundation (RWJF) and the James S. McDonnell Foundation (JSMF).
Footnotes
The authors declare no competing financial interests.
Author Contributions M.D., D.A.H. and J.M.R. designed the study, collected data, analyzed data, and wrote the paper. All authors discussed the results and commented on the manuscript. M.D., D.A.H. and J.M.R. contributed equally to the study.
References
- Ginsberg J. et al. Detecting influenza epidemics using search engine query data. Nature 457, 1012–5 (2009). [DOI] [PubMed] [Google Scholar]
- Lazer D., Kennedy R., King G. & Vespignani A. The parable of Google Flu: traps in big data analysis. Science 343, 1203–5 (2014). [DOI] [PubMed] [Google Scholar]
- Butler D. When Google got flu wrong. Nature 494, 155–6 (2013). [DOI] [PubMed] [Google Scholar]
- Ortiz J. R. et al. Monitoring influenza activity in the United States: a comparison of traditional surveillance systems with Google Flu Trends. PLoS ONE 6, e18687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M. M. et al. Strategies for mitigating an influenza pandemic. Nature 442, 448–52 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N. M. et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 437, 209–214 (2005). [DOI] [PubMed] [Google Scholar]
- Longini I. M. Jr et al. Containing pandemic influenza at the source. Science 309, 1083–7 (2005). [DOI] [PubMed] [Google Scholar]
- Chan J., Holmes A. & Rabadan R. Network Analysis of global influenza spread. PLoS Compu Bio 6, e1001005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget J., Marquet R., Meijer A. & van der Velden K. Influenza activity in Europe during eight seasons (1999–2007): an evaluation of the indicators used to measure activity and an assessment of the timing, length and course of peak activity (spread) across Europe. BMC Infect Dis 7, 1–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C., Nelson M. I., Tan Y. & Holmes E. C. Contrasting the epidemiological and evolutionary dynamics of influenza spatial transmission. Philos Trans R Soc Lond B Biol Sci 368, 20120199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenah E., Chao D. L., Matrajt L., Halloran M. E. & Longini I. M. Jr The global transmission and control of influenza. PLoS One 6, e19515 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simini F., González M. C., Maritan A. & Barabási A. L. A universal model for mobility and migration patterns. Nature 484, 96–100 (2012). [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. FluView. Available at: http://www.cdc.gov/flu/weekly/. (Date of access: 12/05/2013).
- Google Inc. Google Flu Trends. http://www.google.org/flutrends/us/data.txt (Date of access: 12/09/2013).
- Cook S., Conrad C., Fowlkes A. L. & Mohebbi M. H. Assessing Google Flu Trends performance in the United States during the 2009 influenza virus A (H1N1) pandemic. PloS One 6, e23610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Communicable Disease Surveillance and Response. WHO consultation on priority public health interventions before and during an influenza pandemic. World Health Organization. (2004). http://www.afro.who.int/fr/downloads/doc_download/5116-who-consultation-on-priority-public-health-interventions-before-and-during-an-influenza-pandemic.html (Date of access: 01/11/13).
- Christakis N. A. & Fowler J. H. Social Network Sensors for Early Detection of Contagious Outbreaks. PloS One 5, e12948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteller F. A k-sample slippage test for an extreme population. Ann Mat Statist 19, 58–65 (1948). [Google Scholar]