Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) isolates have previously been classified into major epidemic clonal types by pulsed-field gel electrophoresis in combination with multilocus sequence typing (MLST) and staphylococcal cassette chromosome mec typing. We aimed to investigate whether genetic variability in potentially polymorphic domains of virulence-related factors could provide another level of differentiation in a diverse collection of epidemic MRSA clones. The target regions of strains representative of epidemic clones and genetically related methicillin-susceptible S. aureus isolates from the 1960s that were sequenced included the R domains of clfA and clfB; the D, W, and M regions of fnbA and fnbB; and three regions in the agr operon. Sequence variation ranged from very conserved regions, such as those for RNAIII and the agr interpromoter region, to the highly polymorphic R regions of the clf genes. The sequences of the clf R domains could be grouped into six major sequence types on the basis of the sequences in their 3′ regions. Six sequence types were also observed for the fnb sequences at the amino acid level. From an evolutionary point of view, it was interesting that a small DNA stretch at the 3′ clf R-domain sequence and the fnb sequences agreed with the results of MLST for this set of strains. In particular, clfB R-domain sequences, which had a high discriminatory capacity and with which the types distinguished were congruent with those obtained by other molecular typing methods, have potential for use for the typing of S. aureus. Clone- and strain-specific sequence motifs in the clf and fnb genes may represent useful additions to a typing methodology with a DNA array.
Methicillin-resistant Staphylococcus aureus (MRSA) isolates have been classified into major clonal types by pulsed-field gel electrophoresis (PFGE) in combination with multilocus sequence typing (MLST) and staphylococcal cassette chromosome mec (SCCmec) typing (18, 23). Certain MRSA clones have been shown to become rapidly dominant in an outbreak situation in several hospitals or display the capacity for long-distance geographic spread over the years and have been called epidemic. In the context of this work, a clone has been considered epidemic when it was shown to comprise isolates which have been recovered repeatedly in geographically distant hospitals over a period of several years. Examples of epidemic clonal types are Iberian (21) (ST247-I), Brazilian (74) (ST239-III), NY/Japan (62) (ST5-II), Pediatric (65) (ST5-IV), Berlin (79) (ST45-IV), EMRSA-15 (60) (ST22-IV), and EMRSA-16 (14) (ST36-II). (The nomenclature in parentheses refers to the clone's genotype, as defined by MLST sequence type [ST], followed by the roman number associated with its SCCmec type [23].) MLST is based on the sequences of fragments of seven housekeeping genes which accumulate genetic variation relatively slowly and is therefore useful for global epidemiological and evolutionary studies. In contrast to MLST, spa typing, which was also used in this study, takes advantage of the changes in the numbers and sequences of repeat units occurring in the gene coding for protein A and has been shown to be discriminatory enough for outbreak investigations but has also been shown to be efficient for global epidemiological studies (37, 67). The primary purpose of this study was to investigate whether genetic variability in the potentially polymorphic repeat domains of the clumping factor (clf) and fibronectin-binding protein (fnb) genes could provide another level of differentiation to a collection of S. aureus isolates, which included representatives of epidemic MRSA clones with diverse isolation dates and geographic origins. The genes included in our study are associated with staphylococcal virulence, and new information on patterns of sequence variation may also provide useful clues for future studies on the virulence of epidemic clones if variability in the repeat domains suggests that these genes may have sequence polymorphisms that translate into functionally relevant modifications at the protein level. Previous studies have linked the genetic background of S. aureus isolates to the sequence types of a global regulator of virulence gene expression, the accessory gene regulator operon agr (32, 33, 68). As a further contribution to the genotypic characterization of major MRSA epidemic clones, we also sought to determine the agr sequence types of the strains in this study.
Several microbial surface proteins (adhesins) mediate the adherence of S. aureus to host proteins, such as fibrinogen and fibronectin. These plasma proteins coat indwelling medical devices, and the ability of the bacteria to adhere to the deposited proteins is believed to be an important factor in the pathogenesis of wound and foreign body infections (25). In S. aureus, the adhesin genes which have been characterized in detail include clf (45, 48) and fnb (36, 69), which encode the fibrinogen- and the fibronectin-binding proteins, respectively. Although the clumping factor genes, clfA and clfB, encode proteins whose structural organizations resemble those of other cell wall-anchored surface proteins, one distinctive feature is the presence of a repeat region, the R domain, that connects the cell wall-spanning domain to the A domain, which contains the ligand-binding site. It has been proposed that the R domain may function as a “stalk” that allows the exposure of the A domain at the bacterial surface for ligand interaction or that acts in the attachment of the protein to the cell wall (44). The R domain, both in ClfA and in ClfB, is mainly composed of repeats of the dipeptide serine-aspartate encoded by an 18-bp variable repeat, GAYTCNGAYTCNGAYAGY (45, 48). Owing to its potential for genetic variability, the R domain of the clumping factor genes was one of the target regions for sequencing in this study.
The fibronectin-binding protein genes, fnbA and fnbB, encode proteins which, in addition to fibronectin, also bind to fibrinogen (78). Besides their role in the pathogenesis of medical device-related infections, the fibronectin-binding proteins of S. aureus have been shown to be sufficient for the invasion of human cell lines (43, 71) and factors that contribute to the colonization of the mammary gland in a mouse model of mastitis (10). The fnb genes are in tandem in the S. aureus chromosome and share sequence homology, in particular at the 3′ end: in the region containing the D repeats, in the wall-spanning WR repeats, and in hydrophobic membrane-spanning domain M (36). Originally, the fibronectin-binding activity was placed in the D region of FnbA (69), but recently it has been shown that this protein has multiple fibronectin-binding regions which include the B and C domains (35, 43). In FnbB, an additional fibronectin-binding region has been observed upstream to the D domain (36). In this study the DNA region containing the D, W, and M domains in both fnbA and fnbB has been sequenced.
Expression of these and other virulence factors is, at least in part, differentially regulated by the agr operon (58) and several other global regulator loci, which is likely to be important for the adaptation and survival of the microorganism in the host. In our study, in addition to regions of the clf and fnb genes, the following regions were also sequenced in the agr operon: (i) the 5′ region of agrC; (ii) the DNA region between the P2 and P3 promoters containing the binding site for SarA (11), another transcription regulator related to virulence factor expression; and (iii) the complete DNA sequence coding for the effector molecule, RNAIII.
MATERIALS AND METHODS
Bacterial strains.
The collection of strains analyzed included a total of 33 S. aureus isolates representing previously characterized epidemic clones, as well as methicillin-susceptible S. aureus (MSSA) isolates with the same genetic backgrounds (defined by PFGE clonal type and MLST type) as some of the MRSA clones (Table 1). All strains originated from infection sites. Most genetic backgrounds were represented by at least three strains, preferably with diverse isolation dates and geographic origins. The MSSA strains recovered from bloodstream infections in the 1960s in Denmark during the period when the first MRSA strains were identified (15) have been included in the study in order to test if genetic polymorphisms were consistent within a genetic background over time. Sequence data for strain MW2 were downloaded from the genome sequence available in the GenBank database (genome accession no. NC_003923).
TABLE 1.
Molecular data for the representative collection of MRSA and MSSA strains used in this study
| Strain | Yr of isolation | City, state, or country | Referencea | Clonal typeb | spa typec | MLST profilec | ST | SCCmec typed | Investigator source |
|---|---|---|---|---|---|---|---|---|---|
| E2125 | 1964 | Denmark | 17 | Archaic | YHFGFMBQBLO | 3-3-1-12-4-4-16 | 247 | I | H. Westh |
| E2453 | 1965 | Varde, Denmark | 17 | Archaic | YHFGFMBQBLO | 3-3-1-12-4-4-16 | 247 | MSSA | H. Westh |
| 10395 | 1961 | Colindale, United Kingdom | 15 | Archaic | YHGFMBQBLO | 3-3-1-1-4-4-16 | 250 | I | J. Hamilton-Miller |
| E213 | 1957 | Denmark | 15 | Archaic | YHGFMBQBLO | 3-3-1-1-4-4-16 | 250 | MSSA | H. Westh |
| HPV107 | 1992 | Portugal | 66 | Iberian | YHFGFMBQBLO | 3-3-1-12-4-4-16 | 247 | IA | ITQB/RUe |
| PER34 | 1989 | Barcelona, Spain | 21 | Iberian | YHFGFMBQBLO | 3-3-1-1-4-4-16 | 250 | IA | ITQB/RU |
| BK1953 | 1995 | New York, N.Y. | 62 | Iberian | YHFGFMBQBLO | 3-3-1-12-4-4-16 | 247 | IA | ITQB/RU |
| HSJ216 | 1997 | Portugal | 4 | Brazilian | WGKAOMQ | 2-3-1-1-4-4-3 | 239 | IIIA | ITQB/RU |
| HU25 | 1993 | Rio de Janeiro, Brazil | 74 | Brazilian | XKAOMQ | 2-3-1-1-4-4-3 | 239 | IIIA | ITQB/RU |
| PLN104 | 1997 | Poland | 40 | Brazilian | WGKAOMQ | 2-3-1-1-4-4-3 | 239 | IIIA | ITQB/RU |
| CPS22 | 1985 | Portugal | 18 | Portuguese | WGKAOM | 2-3-1-1-4-4-3 | 239 | III variant | ITQB/RU |
| CPS68 | 1985 | Portugal | 18 | Portuguese | WGKAOM | 2-3-1-1-4-4-3 | 239 | III variant | ITQB/RU |
| ICP5011 | 1993 | Portugal | 13 | Portuguese | WGKAOM | 2-3-1-1-4-4-3 | 239 | III variant | ITQB/RU |
| HU101 | 1996 | Kaposvar, Hungary | 53 | Hungarian | XKAOKAOKAOMQ | 2-3-1-1-4-4-3 | 239 | III | ITQB/RU |
| HUSA304 | 1993 | Dunaujvaros, Hungary | 20 | Hungarian | WGKAKAOKAOM | 2-3-1-1-4-4-3 | 239 | III | ITQB/RU |
| TAW9 | 1998 | Taipei, Taiwan | 1 | Hungarian | WGKAOMK | 2-3-1-1-4-4-30 | 241 | III | ITQB/RU |
| HDE288 | 1996 | Lisbon, Portugal | 65 | Pediatric | TJMBDMGMK | 1-4-1-4-12-1-10 | 5 | IV-like | ITQB/RU |
| HDE1 | 1992 | Lisbon, Portugal | 65 | Pediatric | TJMBDMGMK | 1-4-1-4-12-1-10 | 5 | IV-like | ITQB/RU |
| BM18 | 1989 | New York, N.Y. | 19 | Pediatric | TJMBMDMGMK | 1-4-1-4-12-1-10 | 5 | IV | ITQB/RU |
| COB3 | 1996 | Bogota, Colombia | 28 | Pediatric | TMDMGK | 1-4-1-4-12-1-10 | 5 | IV | ITQB/RU |
| BK2464 | 1990 | New York, N.Y. | 62 | NY/Japan | TJMBMDMGMK | 1-4-1-4-12-1-10 | 5 | II | ITQB/RU |
| JP1 | 1997 | Tokyo, Japan | 3 | NY/Japan | TJMBMDMGMK | 1-4-1-4-12-1-10 | 5 | II | ITQB/RU |
| CN1 | 1996 | Connecticut | 61 | NY/Japan | TMDMGMK | 1-4-1-4-12-1-10 | 5 | II | ITQB/RU |
| E2104 | 1964 | Denmark | 15 | NY/Japan | TJMBMDMGMK | 1-4-1-4-12-1-10 | 5 | MSSA | H. Westh |
| E3001 | 1964 | Denmark | 15 | NY/Japan | TJMBMDMMK | 1-4-1-4-12-1-10 | 5 | MSSA | H. Westh |
| N315 | 1982 | Japan | 39 | TJMBMDMGMK | 1-4-1-4-12-1-10 | 5 | II | K. Hiramatsu | |
| HAR24 | 1993 | United Kingdom | 14 | EMRSA-16 | WGKAKAOMQQQ | 2-2-2-2-3-3-2 | 36 | II | Harmony collection |
| E1410 | 1962 | Denmark | 15 | EMRSA-16 | WGKAKAOMQ | 2-2-2-2-6-3-2 | 30 | MSSA | H. Westh |
| HAR22 | 1991 | United Kingdom | 60 | EMRSA-15 | TJEJNF2MNF2MOMOKR | 7-6-1-5-5-8-6 | 22 | IV | Harmony collection |
| PLN49 | 1997 | Poland | 40 | Berlin | XKAKBEMBKB | 10-14-8-6-10-3-2 | 45 | IV | ITQB/RU |
| CA04f | 1998 | United States | 16 | Berlin | NDh | 10-14-8-6-10-3-2 | 45 | IV | R. Daum |
| E3812 | 1969 | Denmark | A2AKBEMBKMBKB | 10-14-8-6-10-3-2 | 45 | MSSA | H. Westh | ||
| MW2f | 1998 | North Dakota | 6 | UJJFKBPE | 1-1-1-1-1-1-1 | 1 | IVa | NARSA collectiong |
Reference for the first molecular characterization of the strain.
Clonal types were defined by ClaI polymorphisms of mecA and Tn554 and by PFGE patterns.
spa types and MLST profiles for strains E2125, E2453, 10395, E213, HPV107, PER34, HDE288, BM18, COB3, BK2464, E2104, E3001, and E1410 were described previously (15), as were the spa types and MLST profiles for strains BK1953, HSJ216, HU25, JP1, and N315 (55); TAW9 (1); PLN49 (2); CA04 and E3812 (M. Chung et al., unpublished results); CPS22, CPS68, ICP5011, HU101, and HDE1 (M. I. Crisóstomo et al., unpublished results); the spa types for strains HUSA304 (53) and CN1 (Crisóstomo et al., unpublished results); and the MLST profiles for strains HUSA304 (Crisóstomo et al., unpublished results) and HAR22 and HAR24 (22).
SCCmec types for strains E2125, HPV107, PER34, BK1953, HSJ216, HU25, HUSA304, HDE288, BM18, COB3, BK2464, JP1, and N315 were described previously (55), as were the SCCmec types for strains PLN49 (2), CA04 (16), and MW2 (6). Most strains of type IV determined by multiplex PCR (HDE288, HDE1, BM18, COB3, HAR22, and PLN49) had ccrAB allele 2; strains HDE288 and HDE1 had ccrAB allele 4. Strains HDE288 and HDE1 were originally assigned SCCmec type IV (55), but since both strains had ccrAB allele 4, their SCCmec types are now designated IV-like. Type III variant strains (CPS22 and ICP5011) had ccr4B allele 3; strain CPS68 was nontypeable for the ccrAB allele.
ITQB/RU, Instituto de Tecnologia Química e Biológica/Rockefeller University strain collection.
Community-acquired MRSA strains.
NARSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
ND, not determined.
PCR amplification.
Chromosomal DNA for PCR was prepared as described previously (5). Primers for amplification and sequencing were designed on the basis of available published sequences and were purchased from MWG Biotech (Ebersberg, Germany) and Invitrogen Life Technologies (Barcelona, Spain). The primer sequences and their target sites are listed in Table 2. Target regions were amplified from approximately 10 ng of template DNA in a 100-μl reaction mixture containing 2.5 U of AmpliTaq DNA polymerase (Applied Biosystems, Foster City, Calif.), 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin), each deoxynucleoside triphosphate at a concentration of 200 μM, and each primer (forward and reverse primers) at a concentration of 400 nM. Amplifications were carried out in a Perkin-Elmer thermocycler (GeneAmp PCR System 9600) with the following parameters: predenaturation for 4 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C (50°C for agrC), and 60 s at 72°C; and postextension for 10 min at 72°C.
TABLE 2.
Oligonucleotide primers used for amplification and sequencing of virulence-related loci in this study
| Product | Primer | Sequence (5′-3′) | GenBank accession no. | Position |
|---|---|---|---|---|
| clfA R domain | clfAF1 | ATG GGA CAA CGA AGT AGC A | Z18852 | 1867-1885 |
| clfAR1 | GCT TCA TCT TCA GAA CCT G | 2997-3015 | ||
| clfB R domain | clfBF2 | GTT ATG GTG GTG GAA GTG CTG | AJ224764 | 1613-1633 |
| clfBR1 | CGC TCT TAT CTC CTG TTT CTG G | 2650-2671 | ||
| fnbA D, W, and M regions | fnbF1 | TAG GAA CTG AAA ATG GTC AC | J04151 | 2255-2274 |
| fnbAR1 | GAA GCA ATC AGA AAA CAC TC | 3262-3281 | ||
| fnbB D, W, and M regions | fnbF1 | TAG GAA CTG AAA ATG GTC AC | X62992 | 2465-2484 |
| fnbBR1 | GAG TAT GTA ATT ATT TCT TGG | 3417-3437 | ||
| agrC 5′ region | agrCF1 | GAA TTA ACD CAA TTA CAC GA | AF001783 | 685-704 |
| agrCF1 | CAA TTT CTT CTT GAT TAC G | AF001783 | 1366-1384 | |
| agrCF2 | CCA TTG AAA TCA CTC CTT CC | X52543 | 1483-1502 | |
| agrCR2 | GAT AGA CCT AAA CCA CGA CC | AF001783 | 1906-1925 | |
| RNAIII and P2-P3 interpromoter | RNAIIIF1 | GAC CTT TTC CAA CAT TAG AC | X52543 | 972-991 |
| RNAIIIR1 | ACA CCA CTC TCC TCA CTG TC | 1752-1771 | ||
| RNAIIIR2 | AGA TAC GTG GCA AAC TGG TC | 1800-1819 |
(i) clf genes.
The R domains of clfA and clfB were amplified with primers designed to be specific for the flanking nonrepeat sequence of each gene. The clfA R domains of strains HAR24 and E1410 could not be amplified with this set of primers for clfA. The R-domain sequence representative of clone EMRSA-16 was obtained from the genome sequence, available on the Internet (www.sanger.ac.uk/Projects/S_aureus/).
(ii) fnb genes.
A 3′ fragment of the fnb genes that included the repeated D region, the wall-spanning W region, membrane-spanning domain M, and the translation stop codon was amplified with one forward primer common to both fnbA and fnbB (primer fnbF1) and one reverse primer specific for each gene located in the intergenic region (primer fnbAR1) or downstream of fnbB (primer fnbBR1).
(iii) agr operon.
The 5′ variable region of the agrC gene was amplified with primers agrCF1 and agrCR1. For strains HDE288, BK2464, BM18, COB3, JP1, CN1, N315, E2104, and E3001, however, for which no amplifications were obtained, primers agrCF2 and agrCR2 were used instead, with an extension time of 3′ at 72°C and sequencing with primer agrCR2 only. (After sequencing, these strains were all shown to belong to agr group II.) The DNA fragment containing the complete RNAIII sequence and the region between the P2 and P3 promoters was amplified with primers RNAIIIF1 and RNAIIIR1; however, for the strains mentioned above, as well as strains HAR24 and E1410, the same forward primer and primer RNAIIIR2 were used under the same amplification conditions.
DNA sequencing.
PCR products were purified with a Wizard PCR Preps purification system (Promega) and used as templates for automated sequencing with a BigDye Terminator cycle sequencing kit (Applied Biosystems) run on an ABI Prism 3700 DNA analyzer at the DNA Sequencing Resource Center, The Rockefeller University. Sequence data analysis was performed with the DNAStar package (Lasergene).
Representation of clf sequences.
The following encoding was devised in order to represent the unexpectedly high degree of variation in the units composing the R domains of the clf sequences. We represented each of the 4 nucleotides by a three-dimensional floating-point vector in a way that all pairwise distances between these points were equal; we thus obtained for the 18-nucleotide repeat units of the form GAYTCNGAYTCNGAYAGY a set of 54-dimensional floating-point vectors, with 1 vector corresponding to one occurrence of the pattern in a clf sequence. We included in our analysis all occurrences of a repeat which differed by, at the most, four positions from the consensus sequence presented above. While this encoding can be used to specify any sequence of nucleotides precisely, it is impossible to visualize. In order to try and achieve a graphic representation of the R-domain sequences, we used the method of singular value decomposition to represent the points in a space with fewer dimensions and color coded the coordinates of the lower-dimensional space. We projected the 54-dimensional space onto a 3-dimensional space and used these three coordinates to color code each occurrence of the repeat in terms of hue, brightness, and saturation. Since hue is the most striking feature of color, it was used for the first coordinate, which reflects the highest variance of the scatter of points. We mapped the second coordinate to brightness and the third coordinate to saturation. As a result of this representation, repeats which were close to each other in the 54-dimensional space remained close to each other in the 3-dimensional space and displayed similar shades of color, and, thus, differences in color reflect differences in the nucleotide sequence of the repeat units. The sequences of the clf genes were displayed as strings of colored segments representing the repeat units of the R domain and black fragments representing sequences which did not follow the repeat unit consensus pattern defined above.
Construction of dendrograms from sequence data.
Dendrograms were generated from the amino acid sequence data for the fnb genes on the basis of the percent identity of optimal pairwise alignments obtained with the program lalign (29) and with a BLOSUM-50 score matrix and default gap opening and extension penalties of −14 and −4. Sequences were clustered by the unweighted pair group method with arithmetic means (UPGMA). The dendrogram for the DNA sequences of agrC was generated from a distance matrix obtained from a multiple alignment by use of the CLUSTALX program (75), and sequences were also clustered by UPGMA.
PFGE.
PFGE of SmaI digests of chromosomal DNA from the strains shown in Table 1 was performed as described previously (12). Relatedness among the PFGE profiles was evaluated with Bionumerics software (version 3.0; Applied Maths, Ghent, Belgium). The dendrogram was generated from a similarity matrix calculated with the Jaccard coefficient, and patterns were clustered by UPGMA.
MLST and spa typing.
MLST was performed as described previously (22), with the exception that primer arcCF2 (5′-CCT TTA TTT GAT TCA CCA GCG-3′) (15) was used. MLST alleles and STs were identified by using the MLST database, available at http://www.mlst.net. Molecular typing based on the sequence of the polymorphic region of protein A (spa typing) was performed as described previously (67).
SCCmec typing.
SCCmec types were determined by a multiplex PCR strategy which establishes a specific amplification pattern for each structural type (54). The exceptions were strain CA04, whose SCCmec type was previously assigned by PCR analysis of the ccrAB genes and the mec complex (16, 31, 42), and strain MW2, whose SCCmec element has been fully sequenced (6). In strains of SCCmec type IV, as determined by the multiplex strategy, the ccrAB allele type was also determined by PCR with primer sets specific for alleles 1, 2, and 3 and a control set of primers specific for the ccrAB locus, as described elsewhere (31, 52). According to the criteria defined by these previous investigators, SCCmec types are defined by the combination of the ccrAB allele with the genetic organization of the mecA regulon: the presence (class A mec, mecI-mecR1-mecA) or the absence (class B mec, IS1272-ΔmecR1-mecA) of the mecA transcription repressor (the mecI gene), information also provided by the multiplex strategy. SCCmec types I and IV are negative for mecI and have ccrAB alleles 1 and 2, respectively; and SCCmec types II and III are mecI positive and have ccrAB alleles 2 and 3, respectively. Since SCCmec type IV is defined by the multiplex strategy by the presence of only two bands (one corresponding to the dcs region and the other corresponding to the mecA gene), all SCCmec type IV strains were further characterized by ccrAB allele determination for confirmation purposes. Most SCCmec type IV strains in this collection characterized by the multiplex strategy had ccrAB allele 2. The exceptions were strains HDE288 and HDE1, which had ccrAB allele 4, as described previously (55); and although they were originally assigned to SCCmec type IV, they are now designated IV-like. In strains CPS22, CPS68, and ICP5011, which are variants of SCCmec type III (mecI positive), ccrAB alleles were also characterized: strains CPS22 and ICP5011 have ccrAB allele 3, while strain CPS68 was nontypeable for the ccrAB allele.
Comparison of virulence loci sequencing with typing methods.
Discriminatory power was measured with Simpson's index of diversity, which calculates the probability that two unrelated strains sampled from the test population will be placed into different typing groups (30). Cross-classification concordance levels between clf sequence-based clustering and the results of known typing methods for this collection of isolates were determined by comparing pairwise matches and calculating the percentage of classification agreement (37).
RESULTS
clf genes.
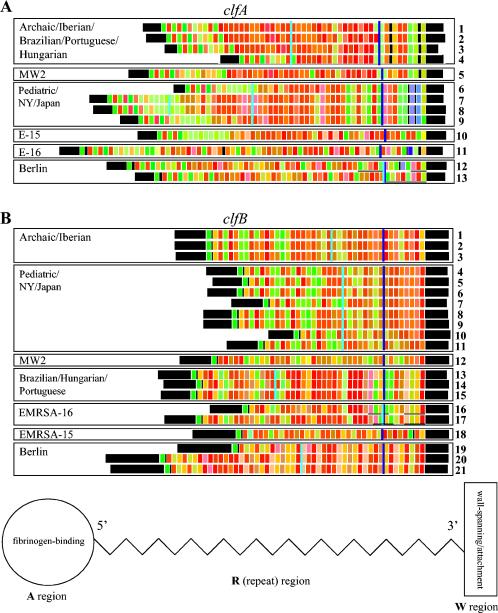
The R domains of clfA and clfB of a collection of strains representing epidemic clones and other MRSA and MSSA isolates were sequenced. The R domain is a potentially polymorphic DNA sequence composed of repeat units of the form GAYTCNGAYTCNGAYAGY. In order to try and understand the structural organization of the R domain, we color coded each repeat unit in a way that similar repeat sequences had similar shades of color. The results, depicted in Fig. 1, showed that the 13 unique clfA sequences were more divergent in the extremities of the R-domain sequence. In particular, the last eight repeat units at the 3′ end displayed the greatest sequence dissimilarity and defined six classes of sequences with the same (or very similar) set of 3′ repeat sequences (Fig. 1A). The Archaic-Iberian class comprised sequences of the Archaic, Brazilian, Hungarian, Portuguese, and Iberian clones. The sequences of strain MW2 and representative strains of the EMRSA-15, EMRSA-16, and Berlin clones defined four individual classes. A sixth group included all sequences which belonged to the Pediatric and the NY/Japan clones. Sequences of MSSA strains of the Archaic, NY/Japan, and Berlin clonal types also fit within the respective clfA R-domain 3′ classes. While the last eight repeats at the 3′ end of the R domain defined six classes of clfA sequences (navy blue line in Fig. 1A), if the sequences were read farther upstream (up to the perpendicular turquoise line dissecting the sequences in Fig. 1), one was able to distinguish some individual clonal types within each class, such as the Iberian clone (sequence 4) from all other clones in the Archaic-Iberian group and the American isolates of the NY/Japan clone (sequence 6) from the strains isolated in Japan (sequence 8).
FIG. 1.
Clumping factor sequences represented by color-coded repeats of the R domain. (A) clfA R-domain sequences 1 to 13 are unique sequences representative of the following strains: sequence 1, strains E2125, E2453, 10395, and E213 (Archaic clone); strains HSJ216, HU25, and PLN104 (Brazilian clone); and strains CPS22, CPS68, and ICP5011 (Portuguese clone); sequence 2, strain TAW9 (Hungarian clone); sequence 3, strains HU101 and HUSA304 (Hungarian clone); sequence 4, strains HPV107, PER34, and BK1953 (Iberian clone); sequence 5, strain MW2; sequence 6, strains BK2464 and CN1 (NY/Japan clone); sequence 7, strains HDE1 and HDE288 (Pediatric clone); sequence 8, strain N315; strains BM18 and COB3 (Pediatric clone); and strains JP1 and E3001 (NY/Japan clone); sequence 9, strain E2104 (NY/Japan clone); sequence 10, strain HAR22 (EMRSA-15 clone); sequence 11, clone EMRSA-16 (www.sanger.ac.uk/Projects/S_aureus/); sequence 12, strains PLN49 and E3812 (Berlin clone); sequence 13, strain CA04 (Berlin clone). (B) clfB R-domain sequences 1 to 21 are unique sequences representative of the following strains: sequence 1, strains E2125 and E2453 (Archaic clone); sequence 2, strains 10395 and E213 (Archaic clone); sequence 3, strains HPV107, PER34, and BK1953 (Iberian clone); sequence 4, strain N315 and strain HDE288 (Pediatric clone); sequence 5, COB3 (Pediatric clone); sequence 6, strain CN1 (NY/Japan clone); sequence 7, strain BK2464 (NY/Japan clone); sequence 8, strain JP1 (NY/Japan clone); sequence 9, strain E3001 (NY/Japan clone); sequence 10, strain BM18 (Pediatric clone); sequence 11, strain E2104 (NY/Japan clone); sequence 12, strain MW2; sequence 13, strains HSJ216 and PLN104 (Brazilian clone); sequence 14, strains HU101 and HUSA304 (Hungarian clone); sequence 15, strain HU25 (Brazilian clone); strain TAW9 (Hungarian clone); and strains CPS22, CPS68, and ICP5011 (Portuguese clone); sequence 16, strain HAR24 (EMRSA-16 clone); sequence 17, strain E1410 (EMRSA-16 clone); sequence 18, strain HAR22 (EMRSA-15 clone); sequence 19, strain PLN49 (Berlin clone); sequence 20, strain CA04 (Berlin clone); sequence 21, strain E3812 (Berlin clone). Lines perpendicular to the sequences delimit the 3′-end eight-repeat region (navy blue lines), which groups the sequences into classes circumscribed by rectangles and the minimum amount of additional sequence which is necessary to differentiate sequences within each class (turquoise lines). Regions of homology are underlined in instances in which the 3′-end eight-repeat region is interrupted.
As was observed for clfA, the sequences of the 3′ end of the R domain of clfB also allowed the grouping of clones into classes, and the number of unique clfB sequences was even larger (n = 21) than the 13 sequence types identified in clfA. The 21 unique sequences were grouped into seven classes on the basis of the 3′ R-domain repeats (Fig. 1B). The discriminatory power of the clfB sequences was greater than those of the clfA and spa sequences (Table 3): it could resolve the Brazilian, Hungarian, and Portuguese clones as a group separate from the Archaic and Iberian clones. The clfB polymorphisms allowed a wider level of discrimination among clones and between strains of the same clone on the basis of the sequence upstream to the last eight repeats (up to the turquoise line in Fig. 1A). Examples were the identification not only of the Brazilian clone but also of strains of this clone with different geographic origins (sequence 13 from Europe and sequence 15 from Brazil) and the specific identification of nearly all MSSA strains isolated in the 1960s (sequences 9, 11, 17, and 21). However, the clfB 3′ region was not as divergent from the rest of the sequence for each strain, nor was it as divergent between the 3′ sequences of different classes when compared with the clfA 3′ region. For instance, only one among the eight 3′ clfB repeats in the EMRSA-16 class was different from the 3′ repeats of the strains in the Brazilian, Hungarian, and Portuguese class. Moreover, the similarity of the 3′ clfB sequences within clusters was not as consistent as that of the 3′ clfA sequences, as shown by the 3′ sequence variation on a pattern displayed by the Pediatric and NY/Japan classes.
TABLE 3.
Discriminatory abilities of typing methods and sequencing results for virulence-related loci
| Method | No. of classes | Index of diversity (%) |
|---|---|---|
| PFGE | 27 | 98.7 |
| clfB sequencing | 21 | 96.4 |
| spa typing | 20 | 95.1 |
| clfA sequencing | 13 | 87.5 |
| MLST | 10 | 83.9 |
| agr sequencing | 10 | 75.4 |
| fnbB sequencing | 7 | 67.9 |
| fnbA sequencing | 5 | 62.8 |
fnb genes.
The 3′ regions of the fnb genes, which encode the D, W, and M domains of the fibronectin-binding proteins, of representative strains of all clones, with the exception of the fnbA genes of HAR22 (EMRSA-15) and HAR24 and E1410 (both of which have the EMRSA-16 genetic background), were amplified and sequenced. Homology searches did not result in any similar sequence in tandem with fnbB for strain MRSA 252 (EMRSA-16 clone) or elsewhere in the available sequence of this strain's genome. In a study of variance in the fnb locus, Rice et al. (59) reported that the epidemic CMRSA-4 strain, which was later shown to be indistinguishable from EMRSA-16 by PFGE (70), contains only one fnb gene. We now show that fnbB is the gene present in clone EMRSA-16 (both in a contemporary MRSA strain and in an MSSA strain from 1962) and also in clone EMRSA-15, which likely possesses a single fnb gene as well.
Pairwise comparisons of translated fnb sequences resulted in identities between 85.2 and 96% for fnbA and 86.1 and 94.6% for fnbB when MRSA strains with distinct genetic backgrounds were analyzed. For both genes the most similar sequences were those of strain MW2 and those of the Archaic, Iberian, Brazilian, Hungarian, and Portuguese clones; and the most divergent ones were those of the Berlin clone and the NY/Japan clone (in fnbA) or strain MW2 (fnbB) (Fig. 2A and 3A). As expected, the fnb sequences of MSSA strains with genetic backgrounds similar to those of MRSA strains were nearly identical to (identity, 98.7 to 99%) or the same as the corresponding sequences of MRSA strains. Similar to what was observed for the clf genes, seven unique fnbB sequences defined six classes of clonal types: Archaic-Iberian, Pediatric-NY/Japan, EMRSA-15, EMRSA-16, MW2, and Berlin (Fig. 3A). These sequence types have an equivalent unique sequence in fnbA; EMRSA-15 and EMRSA-16 clones, however, do not appear to have this gene (Fig. 2A).
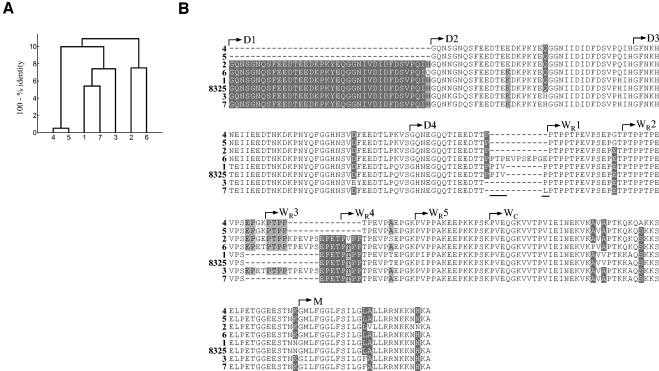
FIG. 2.
fnbA amino acid sequences derived from nucleotide sequences encoding the D, W, and M domains. Unique sequences 1 to 5 represent the following strains: sequence 1, strains E2125, E2453, 10395, and E213 (Archaic clone); strains HPV107, PER34, and BK1953 (Iberian clone); strains HSJ216, HU25, and PLN104 (Brazilian clone); strains HU101, HUSA304, and TAW9 (Hungarian clone); and strains CPS22, CPS68, and ICP5011 (Portuguese clone); sequence 2, strain N315; strains HDE288, BM18, and COB3 (Pediatric clone); and strains BK2464, JP1, CN1, and E3001 (NY/Japan clone); sequence 3, strain E2104 (NY/Japan clone); sequence 4, strains PLN49, CA04, and E3812 (Berlin clone); sequence 5, strain MW2. (A) Dendrogram based on percent identities of fnbA amino acid sequences; (B) fnbA amino acid sequences aligned with the corresponding published sequence for strain 8325-4 (GenBank accession no. J04151) with the CLUSTALX program (75); D1 to D5, repeats of the D domain; WR1 to WR7, repeats of the repetitive region of the cell wall-spanning (W) domain; WC, nonrepetitive region of the W domain; M, membrane-spanning domain (69). The GIDFVED and PIVP motifs are underlined.
FIG. 3.
fnbB amino acid sequences derived from nucleotide sequences encoding the D, W, and M domains. Unique sequences 1 to 7 represent the following strains: sequence 1, strains E2125, E2453, 10395, and E213 (Archaic clone); strains HPV107, PER34, and BK1953 (Iberian clone); strains HSJ216, HU25, and PLN104 (Brazilian clone); strains HU101, HUSA304, and TAW9 (Hungarian clone); and strains CPS22, CPS68, and ICP5011 (Portuguese clone); sequence 2, strains N315; strains HDE288, BM18, and COB3 (Pediatric clone); and strains BK2464, JP1, CN1, E2104, and E3001 (NY/Japan clone); sequence 3, strain HAR22 (EMRSA-15 clone); sequence 4, strain HAR24 (EMRSA-16 clone); sequence 5, strain E1410 (EMRSA-16 clone); sequence 6, strains PLN49, CA04, and E3812 (Berlin clone); sequence 7, strain MW2. (A) Dendrogram based on percent identities of fnbB amino acid sequences; (B) fnbB amino acid sequences aligned with the corresponding published sequence of strain 8325-4 (GenBank accession no. X62992) with the CLUSTALX program (75); D1 to D4, repeats of the D domain; WR1 to WR5, repeats of the repetitive region of the cell wall-spanning (W) domain; WC, nonrepetitive region of the W domain; M, membrane-spanning domain (36). The PIVP motif is underlined.
The differences in this set of fnb sequences among clonal types were mostly due to different structural organizations of the D and W repeats. The Berlin clone had some of the longest D and WR fnb regions: five D repeats and five W repeats in fnbA (Fig. 2B, sequence 4) and a sixth incomplete repeat in the beginning of the WR region in fnbB (Fig. 3B, sequence 6). The GIDFVED motif described for Canadian clone CMRSA-1 by Rice et al. (59) instead of SVDFEED was observed at the end of the fourth D repeat in the fnbA sequence of the Berlin clone (Fig. 2B). On the other hand, EMRSA-16 has the shortest fnb 3′ sequence, with only three D repeats and four W repeats (Fig. 3B, sequence 4). Both the fnbA and the fnbB sequences of the Archaic, Iberian, Brazilian, Hungarian, and Portuguese clones (Fig. 2B and 3B, sequences 1) were exactly identical to the sequences of strain 8325-4; and all strains had a PIVP motif not present in other sequences at the end of the last D repeat.
agr.
Three DNA regions in the agr operon were sequenced: the 5′ region of agrC, the region between the P2 and P3 promoters, and the region coding for RNAIII. On the basis of the agrC 5′ sequence, all strains in this study could be grouped into previously identified agr groups. Most clones belonged to agr group I: the Archaic, Iberian, Brazilian, Portuguese, Hungarian, EMRSA-15, and Berlin clones had agrC sequences with 98.3 to 99.8% identities to the prototype sequence for agr group I. Interestingly, in strain HUSA304 (Hungarian clone) the tnp gene from IS256 was inserted at nucleotide 472 from the agrC translation start site, which raises issues about the functionality of the agr receptor protein in this strain. The remaining clones belonged to groups II (NY/Japan and Pediatric clones and strain N315) and III (EMRSA-16 clone and strain MW2) (Fig. 4).
FIG. 4.
Dendrogram based on a multiple alignment of agrC 5′ DNA sequences performed with the CLUSTALX program (75). Unique sequences 1 to 13 represent the following strains: sequence 1, strain HUSA304 (Hungarian clone); sequence 2, prototype agrC 5′ variable sequence for agr group I (50); sequence 3, strains E2125, E2453, 10395, and E213 (Archaic clone), strains HPV107, PER34, and BK1953 (Iberian clone); strains HSJ216, HU25, and PLN104 (Brazilian clone): strains CPS22, CPS68, and ICP5011 (Portuguese clone); and strain HU101 (Hungarian clone); sequence 4, strain TAW9 (Hungarian clone); sequence 5, strain HAR22 (EMRSA-15 clone); sequence 6, strains PLN49, CA04, and E3812 (Berlin clone); sequence 7, prototype agrC 5′ variable sequence for agr group IV (GenBank accession no. AF288215); sequence 8, strain HAR24 (EMRSA-16 clone), prototype agrC 5′ variable sequence for agr group III (GenBank accession no. AF001783); sequence 9, strain E1410 (EMRSA-16 clone); sequence 10, strain MW2; sequence 11, strains BK2464, JP1, CN1, E2104, and E3001 (NY/Japan clone) and strains HDE288, BM18, and COB3 (Pediatric clone); sequence 12, prototype agrC 5′ variable sequence for agr group II (GenBank accession no. AF001782); sequence 13, strain N315.
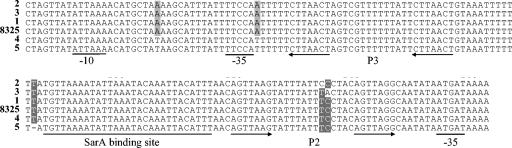
The sequences of RNAIII and the agr interpromoter region were conserved. Only three, nearly identical, sequence types were identified for the sequence coding for RNAIII: there was a consensus sequence for all clones except for an additional A nucleotide at position 406 (according to the RNAIII start site described by Kornblum et al. [38]) for strains with the EMRSA-16 background (strains HAR24 and E1410) and a T-to-C substitution at nucleotide 62 for strains of the Iberian clone (strains HPV107, PER34, and BK1953). The consensus RNAIII sequence was identical to the published sequence of a derivative of strain 8325 (GenBank accession no. X52543). Five nearly identical sequence types were also identified for the agr interpromoter region (Fig. 5). The SarA-binding sites of all sequences had the same composition. The unique sequence types for the agr interpromoter region corresponded to clones EMRSA-15, EMRSA-16, Berlin, and Portuguese and a fifth sequence type common to all other strains.
FIG. 5.
Nucleotide sequences of the agr interpromoter region were aligned with the published sequence for a derivative of strain 8325 (GenBank accession no. X52543). Unique sequences 1 to 5 represent the following strains: sequence 1, all strains of the Archaic, Iberian, Brazilian, Hungarian, NY/Japan, and Pediatric clonal types (Table 1) and strains N315 and MW2; sequence 2, strains CPS22, CPS68, and ICP5011 (Portuguese clone); sequence 3, strain HAR22 (EMRSA-15 clone); sequence 4, strains HAR24 and E1410 (EMRSA-16 clone); sequence 5, strains PLN49, CA04, and E3812 (Berlin clone). The annotation is according to Novick et al. (50), Morfeldt et al. (47), and Chien and Cheung (11). Arrows underline direct repeats of the P2 and P3 promoters. The putative −35 and −10 boxes and the SarA-binding site are also underlined.
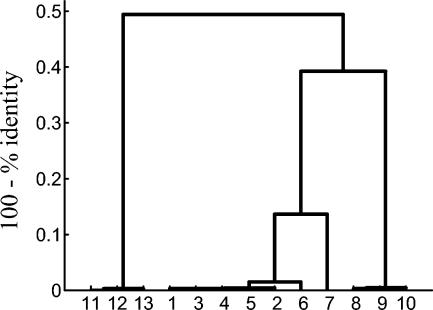
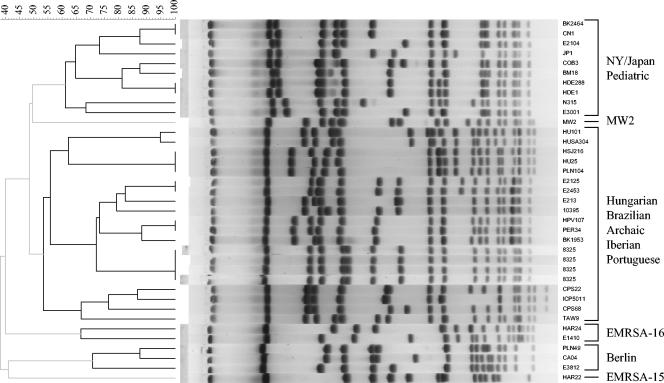
PFGE.
When the PFGE patterns of the epidemic MRSA strains and the other MRSA and MSSA strains in this study were analyzed, the clustering of strains into six classes was also observed (Fig. 6). One large cluster comprised the Archaic, Iberian, Brazilian, Hungarian, and Portuguese clones; a second cluster included strains of the Pediatric and NY/Japan clones and strain N315; and the four remaining clusters corresponded to the EMRSA-16, Berlin, and EMRSA-15 clones and strain MW2.
FIG. 6.
Comparison of PFGE patterns of the representative epidemic MRSA strains and the other MRSA and MSSA strains used in this study (Table 1). The dendrogram was generated from a similarity matrix calculated with the Jaccard coefficient, and patterns were clustered by UPGMA. Band comparisons were performed with a tolerance of 1.06. The scale on the top of the dendrogram represents similarity. Shaded branches are below the cluster cutoff value calculated with Bionumerics software.
DISCUSSION
Sequence analysis of the clf, fnb, and agr loci provided new perspectives on the variability of the S. aureus genome: it has allowed a detailed analysis of the units composing the highly polymorphic R domains of the clf genes, identification of motifs specific to certain clonal types provided by the fnb genes, and a broad genotypic classification of the MRSA clones included in this study on the basis of the sequences of three regions of the agr operon. Such diverse perspectives stem from the different evolutionary trajectories of different regions of the genome and may contribute to a better understanding of the genetic relationships among these clones.
Our studies documenting the clone- and strain-specific sequence variations in S. aureus genes, which are important for interactions with the human host, may also provide useful information for future studies on the mechanisms responsible for the superior epidemicity and geographic dominance of globally spread MRSA clones.
Sequence variations in clf genes.
The genetic relatedness of the MRSA clones evaluated in this study was classified from the sequences for the R domains of the clumping factor genes. The R-domain sequences themselves provided two levels of analysis: clustering into six classes of clones on the basis of the last eight 3′ repeats of clfA (Fig. 1) and identification of individual clones within each cluster or even specific strains of each clone when the sequence was read farther upstream (i.e., to the left from the navy blue perpendicular line markers dissecting the sequences in Fig. 1). The results of the same analysis performed with clfB were concordant with the data for clfA albeit more discriminating; a seventh class was identified, and a larger number of specific sequence types was observed for individual strains from each clone. Interestingly, the six classes of clones which were identified on the basis of the 3′ clf sequences, the Archaic-Iberian, MW2, Pediatric-NY/Japan, EMRSA-15, EMRSA-16, and Berlin clones, are in agreement with the clonal complex classifications of the same strains defined by MLST: CC8, CC1, CC5, CC22, CC30, and CC45, respectively (23, 24). They also coincide with the sequence data for the fnb genes. Therefore, by sequencing of a single locus it was possible to identify not only the clonal complex corresponding to, for instance, the Pediatric and NY/Japan clones but also to distinguish between the NY/Japan strains isolated in the United States from one isolated in Japan. Owing to this dual capability, clf sequencing may have potential as a complementary typing method. Sequencing of the R domains of the clf genes needs to be performed with a larger number of isolates, and the in vitro and in vivo stabilities of these DNA regions need to be assayed in order to establish meaningful comparisons with other sequence typing data.
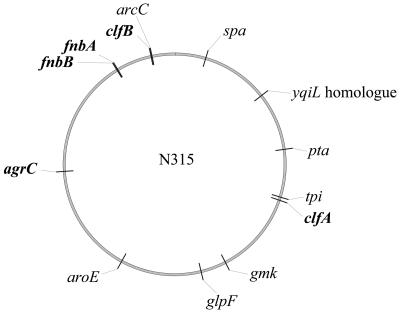
Some clfB sequences seem to register a genetic event already documented by spa typing and other sequencing data: the recombination of a large fragment of 557 kb from ST30 (EMRSA-16 background) into the branch of ST8 (Archaic-Iberian background) which gave rise to ST239 (Brazilian, Portuguese, and Hungarian lineage) (63). According to our data, the recombined fragment common to the EMRSA-16 clone and the Brazilian, Portuguese, and Hungarian cluster includes clfB, in addition to the spa and arcC genes, but did not include the fnb genes, which classified these clones into separate sequence types (Fig. 7). Further evidence for the relatedness between EMRSA-16 and the Brazilian, Portuguese, and Hungarian cluster in this stretch of the genome is the striking similarity between the clfB sequence in strain E1410, the MSSA strain from the 1960s which has the same MLST background as EMRSA-16, and the clfB sequences in strains belonging to the Brazilian, Portuguese, and Hungarian cluster. Yet, nucleotide mutations within the repeats allowed differentiation of strains with the EMRSA-16 clonal background from those belonging to the Brazilian, Portuguese, and Hungarian cluster. The importance of the background rate of nucleotide mutation within repeats, along with the extent of repeat number variation, has been documented for spa typing, which was recently been proposed as a means to address both long- and short-term epidemiological issues (37).
FIG. 7.
Map locations of virulence-related loci sequenced in this study (in boldface), as well as those of the spa and MLST genes. Sequence data for S. aureus strain N315 were downloaded from www.ncbi.nlm.nhi.gov/genomes.
The occurrence of recombination also cannot be ruled out for the region of the genome which contains clfA. However, for the set of strains examined here, the 3′ clfA stretch of eight repeat units is rather specific for previously defined clonal complexes. One exception seems to be strain MW2, whose clfA R-domain sequence is quite similar to those of the Archaic- Iberian sequence types. Nevertheless, from these data, together with the sequence data for fnbA and fnbB, which are located at a distant region of the chromosome and yet displayed similar sequence types for the two lineages, one is inclined to think rather about some genetic relatedness between MW2 and the Archaic and Iberian group of clones which have distinct MLST profiles and which have been thought of as unrelated clonal complexes. The usefulness of various typing methods resides, therefore, in the complementarity of these methods which is brought to the characterization of S. aureus strains. Recently, the clumping factor genes have been included with other loci with variable numbers of tandem repeats in a typing methodology based on PCR analysis of repeat polymorphisms which had a discriminatory power comparable to that of PFGE (64). The clumping factor B gene, which may be a major determinant in S. aureus nasal colonization due to its role in binding to host keratin (51), has also been recently used for microepidemiological typing based on sequence variations in the repeat region (L. Koreen, S. Ramaswamy, S. Naidich, E. A. Graviss, and B. N. Kreiswirth, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. C-429, 2003).
Sequence variations in fnb genes.
Sequence data for the fnb genes provided another source of information concerning the relationships between the S. aureus strains evaluated in this study. One fnbA sequence and one fnbB sequence corresponded to each class of clones defined by clfA sequencing, and these were the same for all clones within the class, thus confirming the classification obtained by clf sequencing and MLST. Most clones possessed two fnb genes, the only exceptions being clones EMRSA-16 and EMRSA-15, which had fnbB only. This is in agreement with previous studies in which not all but only between 77 and 91% of S. aureus clinical isolates possessed both fnbA and fnbB (57, 59). It was reported that isolates which differed in the number of fnb genes did not exhibit significant differences in fibronectin binding. On the other hand, fibronectin binding was negatively correlated with protease activity, which, in turn, is regulated by agr expression (59). This was documented in some detail for Canadian strain CMRSA-4, which had only one fnb gene (fnbB), was indistinguishable from EMRSA-16 by PFGE (70), and exhibited a high level of protease activity and a low-level capacity to bind to fibronectin.
Analysis of the fnb sequences in the strains that we tested resulted in the observation of a closer sequence similarity between the sequences of strain MW2 and those of the group comprising the Archaic, Iberian, Brazilian, Hungarian, and Portuguese clones (the Archaic-Iberian group) and complete identity between the sequences of this group and those of strain 8325-4. Previously reported motifs, such as the PIVP motif at the end of the fourth D repeat described for strain 8325-4 (36, 69), which is unique to the fnb sequences of this strain and the Archaic-Iberian group, or the GIDFVED motif described for Canadian epidemic CMRSA-1 (59), were also observed. In CMRSA-1 the GIDFVED motif has been reported to replace the SVDFEED epitope, which is essential for fibronectin binding, in one of the D repeats of FnbA as a possible strategy of immune evasion, but with a cost in terms of fibronectin binding. In our study, the GIDFVED motif replaced the SVDFEED motif in the fourth D repeat of the fnbA sequence for the Berlin clone, which, like CMRSA-1, possesses an additional D repeat in this gene. Are CMRSA-1 and the Berlin clone from the same lineage? Indeed, it appears to be so, although the Berlin clone was not included in the survey which compared Canadian MRSA isolates with international epidemic clones (70). However, the investigators referred to a previous report (7) in which CMRSA-1, under the designation of OE-MRSA, clustered with epidemic strains from Belgium, Switzerland, and Germany after randomly amplified polymorphic DNA analysis. In yet two other studies, the Canadian OE clone (8), also referred to as the Ontario epidemic clone (9), was shown by PFGE to belong to the same clonal type as the Berlin clone. The PIVP motif of fnb and the GIDFVED motif of fnbA may be useful as specific markers for clonal classification.
Sequence variations in some regions of agr.
All strains in this study could be assigned to one of three major agr specificity groups. Most clones belonged to agr group I; but this reflected the fact that the Archaic, Iberian, Brazilian, Hungarian, and Portuguese clones (the Archaic-Iberian cluster of clones) have similar genetic backgrounds. If agr grouping is looked at as a genomic classification, the clones in this study were distributed more evenly: the Archaic-Iberian cluster, EMRSA-15, and Berlin clones in group I; the Pediatric and NY/Japan clones and strain N315 in group II; and the EMRSA-16 clone and strain MW2 in group III. Novick (49) has proposed that agr groups may represent ancient evolutionary divisions in terms of the organism's fundamental biology, and subsequent studies have linked the agr type to the genetic background of S. aureus from both disease (32, 33) and colonization (68) isolates. As observed previously (26), agr group I was the interference group which showed more genetic variation at the agr locus and which displayed the largest number of agrC sequence variants (which may again reflect the fact that more strains belonging to group I were analyzed). In particular, in one strain of the Hungarian clone, the tnp gene from IS256 was inserted into the 5′ variable region of agrC. Instability in the agrC-coding region that leads to a truncated or a mutated protein has been documented (46, 72, 76). The mutations which occurred in AgrC during in vitro serial passage of S. aureus decreased the levels of production of secreted virulence factors and increased the growth yields of the bacteria, suggesting that the fitness of agr variants may be increased in certain ecological niches (72). On the other hand, the sequences of RNAIII and the region between the P2 and the P3 promoters were highly conserved in the set of clones analyzed in the present study. Yet, it would be interesting to clarify whether the point mutations in the P3 promoter in the Berlin and EMRSA-16 clones and in the P2 promoter in the Portuguese and EMRSA-15 clones have an effect on the overall regulation of agr in these clones.
Geographic dominance of clonal types and agr type.
The form of bacterial interference mediated by inhibition of the synthesis of virulence factors and other extracellular proteins in S. aureus strains of different agr groups has been well documented in vitro (32, 34). Although different agr groups have been observed for several years in strains isolated from S. aureus carriers (76), only one agr type was detected at a single time in each healthy individual's nasal flora (27, 41). The observation that the clones with different agr types in our study correspond to distinct geographic areas in which these clones are dominant (i.e., are most frequently recovered) again raises the question of whether some type of agr-associated interference exists in vivo: strains of agr group I, represented by the Iberian, Brazilian, Portuguese, Hungarian, Berlin, and EMRSA-15 clones, are predominant in Europe and some South American countries; strains of group II, represented by the Pediatric and NY/Japan clones, have mainly been isolated in Japan and North America (but also in some European countries); and strains of group III, which were represented only by the EMRSA-16 clone, are also mainly isolated in Europe (see references 23 and 56 and the references therein). Although the agr types showed geographic overlap, some paradigmatic instances are noteworthy, such as the one in which two different agr types have coexisted for years in the same hospital but in different wards and in which one type does not overtake the other, as described for the Iberian (group I) and Pediatric (group II) clones in a Portuguese hospital (65). A similar case was recorded in Colombia, in which the Brazilian clone (group I) widely disseminated in other Southern American countries was totally absent and instead the dominant clone was the Pediatric clone of MRSA (group II) (28). On the other hand, the displacement of one major local clone with another was also observed in Portuguese hospitals: the Portuguese clone which was most frequently recovered during the 1985 surveillance study (18) was replaced by the Iberian clone (66) in 1992-1993, followed most recently by further replacement of the Iberian clone by the Brazilian clone (4). Each of these three clones belongs to the same agr type (group I). Similarly, clones belonging to agr group I were dominant in German hospitals in the 1990s (the Northern German [ST247] and Hannover [ST254] clones), and agr group I was still predominant in 2002, even though it was represented by different clones (the Berlin [ST45] and Barnim [ST22] clones). The rise of the Southern German (ST228) clone from agr group II observed in 2000 was not sustained in the following years, and it would be interesting to monitor the evolution of the recently emerged Rhine-Hesse (ST5) clone, also from group II (73). We hypothesize that, due to differences in genomic characteristics associated with a given agr type, MRSA epidemic clones belonging to three agr types may be competing for dominance in the hospital setting throughout the world.
In conclusion, the sequence polymorphisms observed in virulence-related loci may be associated with differential regulation by a global regulator of virulence genes (agr interpromoter region) or mechanisms that interfere with epidemiological dynamics (agr receptor variable region) in a collection of isolates representative of MRSA epidemic clones. Further experiments are warranted in order to investigate these issues. The sequences of the fnb genes also presented polymorphisms at the amino acid level in a region which is important for fibronectin binding. However, recent findings that the fibronectin-binding proteins have multiple, substituting fibronectin-binding regions (35, 43) suggest that the polymorphisms in the D region alone may not reflect a functional difference in fibronectin-binding capability and, consequently, may not have an effect on the capacity to initiate infection. Nevertheless, the fnb sequences together with the clf sequences have provided useful tools for genotypic characterization of MRSA isolates at a resolution higher than that provided by MLST. A specific motif in fnbA (GIDFVED) allowed the identification of a clonal type (Berlin) previously reported under several unrelated designations. In broad evolutionary terms, the types detected by fnb and clf sequencing were in agreement with those obtained by MLST and allowed the recognition of six lineages among the collection of MRSA isolates evaluated. In particular, the results of clfB sequencing, which had a discriminatory capacity greater than that of spa typing (Table 3), were also highly congruent with those of PFGE and spa typing, and if the last eight repeats (144 bp) of the R domain of this single locus are considered, the results of clfB sequencing are also congruent with those of MLST (Table 4). Thus, sequences from the R domain of clfB have strong potential for use in the typing of S. aureus strains.
TABLE 4.
Cross-classification concordance levels for the collection of S. aureus isolates
| Sequence | % Concordance with:
|
||
|---|---|---|---|
| MLST | spa | PFGE | |
| clfBa | 85.9 | 95.0c | 96.4 |
| clfAa | 83.3 | 87.5 | 88.9 |
| clfB 3′b | 95.8 | 85.5 | 81.7 |
| clfA 3′b | 83.3 | 71.4 | 67.5 |
| spa | 87.3 | 95.3 | |
Sequence of the R domain of the clf genes.
Sequence of the last eight repeats (144 bp) at the 3′ end of the R-domain of the clf genes.
Underscores indicate the highest concordance values.
Sequence information, applied as MLST or spa types, has been useful for both evolutionary studies and global epidemiological analyses, as well as, to a lesser extent, for short-term or local epidemiological analyses. Recently, an oligonucleotide array suited for use with the loci detected by MLST has been developed (77). The results of clf and fnb sequencing described in this study, which have identified clonal types that agree with those detected by MLST and spa typing, as well as clone- and strain-specific sequence motifs, may represent useful additions to a DNA array sequence typing methodology.
Acknowledgments
We thank the investigators who kindly provided some of the bacterial strains used in this study: Henrik Westh for the eight MRSA and MSSA strains isolated in the 1960s, Keiichi Hiramatsu for strain N315, Robert Daum for strain CA04, J. Hamilton-Miller for strain 10395, Barry Cookson for strains 90/10685 (strain HAR22) and 96/32010 (strain HAR24) from the Harmony collection, and the Network on Antimicrobial Resistance in Staphylococcus aureus for strain MW2. We also thank Erik van Nimwegen for collaboration with the color representation of clf sequences and João Carriço for preparing the dendrogram from the PFGE data. We thank Alexander Tomasz for helpful discussions as well as partial support to this work during A. R. Gomes' stay at the Laboratory of Microbiology, The Rockefeller University.
This work was partially supported by Project POCTI/1999/ESP/34872, Fundação para a Ciência e a Tecnologia, Lisbon, Portugal. A. R. Gomes was supported by grants PRAXIS XXI/BPD/16375/98 and SFRH/BPD/9373/2002; and S. Vinga was supported by grant SFRH/BD/3134/2000, Fundação para a Ciência e a Tecnologia, Lisbon, Portugal.
REFERENCES
- 1.Aires de Sousa, M., M. I. Crisóstomo, I. S. Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., and H. de Lencastre. 2003. Evolution of sporadic isolates of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals and their similarities to isolates of community-acquired MRSA. J. Clin. Microbiol. 41:3806-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires de Sousa, M., H. de Lencastre, I. S. Sanches, K. Kikuchi, K. Totsuka, and A. Tomasz. 2001. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6:253-258. [DOI] [PubMed] [Google Scholar]
- 4.Aires de Sousa, M., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aires de Sousa, M., I. S. Sanches, A. van Belkum, W. van Leeuwen, H. Verbrugh, and H. de Lencastre. 1996. Characterization of methicillin-resistant Staphylococcus aureus isolates from Portuguese hospitals by multiple genotyping methods. Microb. Drug Resist. 2:331-341. [DOI] [PubMed] [Google Scholar]
- 6.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 7.Blanc, D. S., A. L. Banuls, P. M. Hauser, P. Moreillon, P. Francioli, and M. Tibayrenc. 2000. Methicillin-resistant Staphylococcus aureus: phylogenetic relatedness between European epidemic clones and Swiss sporadic strains. Microb. Drug Resist. 6:231-238. [DOI] [PubMed] [Google Scholar]
- 8.Blanc, D. S., C. Petignat, P. Moreillon, J. M. Entenza, M. Eisenring, H. Kleiber, A. Wenger, N. Troillet, C. Blanc, and P. Francioli. 1999. Unusual spread of a penicillin-susceptible methicillin-resistant Staphylococcus aureus clone in a geographic area of low incidence. Clin. Infect. Dis. 29:1512-1518. [DOI] [PubMed] [Google Scholar]
- 9.Blanc, D. S., D. Pittet, C. Ruef, A. F. Widmer, K. Muhlemann, C. Petignat, S. Harbarth, R. Auckenthaler, J. Bille, R. Frei, R. Zbinden, P. Moreillon, P. Sudre, and P. Francioli. 2002. Molecular epidemiology of predominant clones and sporadic strains of methicillin resistant Staphylococcus aureus in Switzerland and comparison with European epidemic clones. Clin. Microbiol. Infect. 8:419-426. [DOI] [PubMed] [Google Scholar]
- 10.Brouillette, E., B. G. Talbot, and F. Malouin. 2003. The fibronectin-binding proteins of Staphylococcus aureus may promote mammary gland colonization in a lactating mouse model of mastitis. Infect. Immun. 71:2292-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien, Y., and A. L. Cheung. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273:2645-2652. [DOI] [PubMed] [Google Scholar]
- 12.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sá-Leão, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 13.Couto, I., J. Melo-Cristino, M. L. Fernandes, T. Garcia, N. Serrano, M. J. Salgado, A. Torres-Pereira, I. S. Sanches, and H. de Lencastre. 1995. Unusually large number of methicillin-resistant Staphylococcus aureus clones in a Portuguese hospital. J. Clin. Microbiol. 33:2032-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, R. A., C. Conquest, C. Mallaghan, and R. R. Marples. 1995. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 29:87-106. [DOI] [PubMed] [Google Scholar]
- 15.Crisóstomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 17.de Lencastre, H., M. Chung, and H. Westh. 2000. Archaic strains of methicillin-resistant Staphylococcus aureus: molecular and microbiological properties of isolates from the 1960s in Denmark. Microb. Drug Resist. 6:1-10. [DOI] [PubMed] [Google Scholar]
- 18.de Lencastre, H., I. Couto, I. Santos, J. Melo-Cristino, A. Torres-Pereira, and A. Tomasz. 1994. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur. J. Clin. Microbiol. Infect. Dis. 13:64-73. [DOI] [PubMed] [Google Scholar]
- 19.de Lencastre, H., A. de Lencastre, and A. Tomasz. 1996. Methicillin-resistant Staphylococcus aureus isolates recovered from a New York City hospital: analysis by molecular fingerprinting techniques. J. Clin. Microbiol. 34:2121-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lencastre, H., E. P. Severina, H. Milch, M. K. Thege, and A. Tomasz. 1997. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin. Microbiol. Infect. 3:289-296. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez, M. A., H. de Lencastre, J. Liñares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin- resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 26.Gilot, P., G. Lina, T. Cochard, and B. Poutrel. 2002. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goerke, C., M. Kummel, K. Dietz, and C. Wolz. 2003. Evaluation of intraspecies interference due to agr polymorphism in Staphylococcus aureus during infection and colonization. J. Infect. Dis. 188:250-256. [DOI] [PubMed] [Google Scholar]
- 28.Gomes, A. R., I. S. Sanches, M. Aires de Sousa, E. Castañeda, and H. de Lencastre. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Colombian hospitals: dominance of a single unique multidrug-resistant clone. Microb. Drug Resist. 7:23-32. [DOI] [PubMed] [Google Scholar]
- 29.Huang, X., and W. Miller. 1991. A time efficient, linear space local similarity algorithm. Adv. Appl. Math. 12:337-357. [Google Scholar]
- 30.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarraud, S., G. J. Lyon, A. M. Figueiredo, L. Gerard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 35.Joh, D., P. Speziale, S. Gurusiddappa, J. Manor, and M. Hook. 1998. Multiple specificities of the staphylococcal and streptococcal fibronectin-binding microbial surface components recognizing adhesive matrix molecules. Eur. J. Biochem. 258:897-905. [DOI] [PubMed] [Google Scholar]
- 36.Jonsson, K., C. Signas, H. P. Muller, and M. Lindberg. 1991. Two different genes encode fibronectin-binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202:1041-1048. [DOI] [PubMed] [Google Scholar]
- 37.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornblum, J., B. N. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373-402. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, Inc., New York, N.Y.
- 39.Kuwahara-Arai, K., N. Kondo, S. Hori, E. Tateda-Suzuki, and K. Hiramatsu. 1996. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2′ production. Antimicrob. Agents Chemother. 40:2680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leski, T., D. Oliveira, K. Trzcinski, I. S. Sanches, M. Aires de Sousa, W. Hryniewicz, and H. de Lencastre. 1998. Clonal distribution of methicillin-resistant Staphylococcus aureus in Poland. J. Clin. Microbiol. 36:3532-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lina, G., F. Boutite, A. Tristan, M. Bes, J. Etienne, and F. Vandenesch. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massey, R. C., M. N. Kantzanou, T. Fowler, N. P. Day, K. Schofield, E. R. Wann, A. R. Berendt, M. Hook, and S. J. Peacock. 2001. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell Microbiol. 3:839-851. [DOI] [PubMed] [Google Scholar]
- 44.McDevitt, D., and T. J. Foster. 1995. Variation in the size of the repeat region of the fibrinogen receptor (clumping factor) of Staphylococcus aureus strains. Microbiology 141:937-943. [DOI] [PubMed] [Google Scholar]
- 45.McDevitt, D., P. François, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 46.McNamara, P. J., and J. J. Iandolo. 1998. Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KSI9051. J. Bacteriol. 180:2609-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 48.Ni Eidhin, D., S. Perkins, P. François, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 49.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 50.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 51.O'Brien, L. M., E. J. Walsh, R. C. Massey, S. J. Peacock, and T. J. Foster. 2002. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol. 4:759-770. [DOI] [PubMed] [Google Scholar]
- 52.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliveira, D. C., I. Crisóstomo, I. Santos-Sanches, P. Major, C. R. Alves, M. Aires-de-Sousa, M. K. Thege, and H. de Lencastre. 2001. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 56.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 57.Peacock, S. J., N. P. Day, M. G. Thomas, A. R. Berendt, and T. J. Foster. 2000. Clinical isolates of Staphylococcus aureus exhibit diversity in fnb genes and adhesion to human fibronectin. J. Infect. 41:23-31. [DOI] [PubMed] [Google Scholar]
- 58.Projan, S., and R. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In G. Archer and K. Crossley (ed.), Staphylococci in human diseases. Churchill Livingstone, New York, N.Y.
- 59.Rice, K., M. Huesca, D. Vaz, and M. J. McGavin. 2001. Variance in fibronectin binding and fnb locus polymorphisms in Staphylococcus aureus: identification of antigenic variation in a fibronectin binding protein adhesin of the epidemic CMRSA-1 strain of methicillin-resistant S. aureus. Infect. Immun. 69:3791-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richardson, J. F., and S. Reith. 1993. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hosp. Infect. 25:45-52. [DOI] [PubMed] [Google Scholar]
- 61.Roberts, R. B., M. Chung, H. de Lencastre, J. Hargrave, A. Tomasz, D. P. Nicolau, J. F. John, Jr., and O. Korzeniowski. 2000. Distribution of methicillin-resistant Staphylococcus aureus clones among health care facilities in Connecticut, New Jersey, and Pennsylvania. Microb. Drug Resist. 6:245-251. [DOI] [PubMed] [Google Scholar]
- 62.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 63.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sá-Leão, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanches, I. S., M. Ramirez, H. Troni, M. Abecassis, M. Padua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shopsin, B., B. Mathema, P. Alcabes, B. Said-Salim, G. Lina, A. Matsuka, J. Martinez, and B. N. Kreiswirth. 2003. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J. Clin. Microbiol. 41:456-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Signas, C., G. Raucci, K. Jonsson, P. E. Lindgren, G. M. Anantharamaiah, M. Hook, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simor, A. E., M. Ofner-Agostini, E. Bryce, A. McGeer, S. Paton, and M. R. Mulvey. 2002. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: results of 5 years of National Surveillance, 1995-1999. J. Infect. Dis. 186:652-660. [DOI] [PubMed] [Google Scholar]
- 71.Sinha, B., P. François, Y. A. Que, M. Hussain, C. Heilmann, P. Moreillon, D. Lew, K. H. Krause, G. Peters, and M. Herrmann. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Somerville, G. A., S. B. Beres, J. R. Fitzgerald, F. R. DeLeo, R. L. Cole, J. S. Hoff, and J. M. Musser. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 184:1430-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strommenger, B., C. Cuny, G. Werner, and W. Witte. 2004. Obvious lack of association between dynamics of epidemic methicillin-resistant Staphylococcus aureus in central Europe and agr specificity groups. Eur. J. Clin. Microbiol. Infect. Dis. 23:15-19. [DOI] [PubMed] [Google Scholar]
- 74.Teixeira, L. A., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Leeuwen, W., W. van Nieuwenhuizen, C. Gijzen, H. Verbrugh, and A. van Belkum. 2000. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J. Bacteriol. 182:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Leeuwen, W. B., C. Jay, S. Snijders, N. Durin, B. Lacroix, H. A. Verbrugh, M. C. Enright, A. Troesch, and A. van Belkum. 2003. Multilocus sequence typing of Staphylococcus aureus with DNA array technology. J. Clin. Microbiol. 41:3323-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wann, E. R., S. Gurusiddappa, and M. Hook. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 79.Witte, W., C. Cuny, E. Halle, and J. Wagner. 1994. Methicillin resistance in an epidemic Staphylococcus aureus strain with genomic fingerprints corresponding to those of sensitive strains in the community. Med. Microbiol. Lett. 3:388-395. [Google Scholar]