Significance
Deleterious mutations in different copies of a duplicated gene pair have the potential to cause hybrid incompatibility between diverging subpopulations, contributing to reproductive isolation and speciation. This study demonstrates a case of epigenetic gene silencing rather than pseudogene creation by mutation, contributing to a lethal gene combination on hybridization of two ecotypes of Arabidopsis thaliana. Our findings provide direct evidence that naturally occurring epigenetic variation can contribute to incompatible hybrid genotypes, reducing gene flow between subtypes of the same species.
Keywords: gene silencing, epigenetic inheritance, DNA methylation, histone deacetylation, hybrid incompatibility
Abstract
Hybrid incompatibility resulting from deleterious gene combinations is thought to be an important step toward reproductive isolation and speciation. Here, we demonstrate involvement of a silent epiallele in hybrid incompatibility. In Arabidopsis thaliana accession Cvi-0, one of the two copies of a duplicated histidine biosynthesis gene, HISN6A, is mutated, making HISN6B essential. In contrast, in accession Col-0, HISN6A is essential because HISN6B is not expressed. Owing to these differences, Cvi-0 × Col-0 hybrid progeny that are homozygous for both Cvi-0 HISN6A and Col-0 HISN6B do not survive. We show that HISN6B of Col-0 is not a defective pseudogene, but a stably silenced epiallele. Mutating HISTONE DEACETYLASE 6 (HDA6), or the cytosine methyltransferase genes MET1 or CMT3, erases HISN6B's silent locus identity, reanimating the gene to circumvent hisn6a lethality and hybrid incompatibility. These results show that HISN6-dependent hybrid lethality is a revertible epigenetic phenomenon and provide additional evidence that epigenetic variation has the potential to limit gene flow between diverging populations of a species.
Mutations that accumulate in separate subpopulations of a species can facilitate reproductive isolation by engendering hybrid incompatibility, a reduction in fitness observed among hybrid progeny at the F1 or F2 generation (1–3). Bateson (4), Dobzhansky (5), and Muller (6) independently proposed that gene mutations made benign by compensatory mutations in interacting genes within isolated subpopulations prove deleterious when subpopulations or ecotypes interbreed, thereby contributing to speciation by reducing gene flow. Examples of this include the lethal consequences of a Saccharomyces cerevisiae protein directing improper splicing of essential Saccharomyces bayanus mRNAs in S. cerevisiae × S. bayanus hybrids (3) or the hybrid necrosis that results from expression of incompatible innate immune receptors in Arabidopsis thaliana (1).
Lynch and Force (7) envisioned an alternative scenario by which hybrid incompatibilities might arise, as a result of gene duplication. Gene duplication initially results in gene redundancy, thereby relaxing constraints on sequence and functional divergence, but mutations most often transform one paralog into a nonfunctional pseudogene (8–10). Lynch and Force recognized that asymmetric resolution of gene duplicates in this manner could result in either paralog becoming the sole operational copy in different subpopulations of a species, such that hybrid progeny inheriting nonfunctional alleles of both subpopulations would suffer reduced fitness (7).
Nonmutational (epigenetic) gene silencing could potentially contribute to hybrid incompatibility via the scenario envisioned by Lynch and Force. In plants, silent epialleles segregating in Mendelian fashion can be stably inherited over many generations, and are known to affect a number of well-studied traits, including flower morphology (11, 12), fruit ripening (13), and sex determination (14). Silent epialleles are inherited via the perpetuation of repressive chromatin states, and various plant chromatin-modifying enzymes are known to participate in epigenetic inheritance, including the Rpd3-like histone deacetylase HDA6, the ATP-dependent chromatin remodeler DDM1, and the DNA methyltransferases MET1 and CMT3 (15-25). These enzymes are important for maintaining patterns of cytosine methylation following each round of DNA replication, such that newly replicated daughter strands of DNA inherit the methylation status of mother strands.
Multicopy transgenes frequently become methylated and silenced, particularly when inserted into the genome as inverted repeats that can give rise to double-stranded RNAs (26). Such double-stranded RNAs can be diced into small interfering RNAs (siRNAs) that guide the cytosine methylation of homologous DNA sequences, a process known as RNA-directed DNA methylation (RdDM) (27, 28). Known cases of silencing that involve duplicated endogenous genes, such as the phosphoribosylanthranilate isomerase (PAI) or folate transporter (AtFOLT) genes of A. thaliana, resemble transgene silencing in that complex sequence rearrangements coincide with duplication of a gene to create a novel (nonancestral) locus engendering siRNA production and homology-dependent DNA methylation (29, 30). Silencing of the ancestral AtFOLT1 gene, directed by siRNAs derived from the novel AtFOLT2 locus present in some ecotypes, causes reduced fertility in ecotype-specific hybrid combinations (29). This interesting case study has shown that naturally occurring RdDM, involving a new paralog that inactivates the ancestral paralog in trans, can be a cause of hybrid incompatibility.
Here, we demonstrate an epigenetic basis for a previously identified case of hybrid incompatibility (31) involving a gene pair, HISN6A and HISN6B. The underlying mechanism of HISN6-based hybrid incompatibility was previously unknown. We show that the HISN6B gene of ecotype Col-0, and hundreds of other A. thaliana natural accessions, is hypermethylated in its promoter region and is epigenetically silent, making its paralog, HISN6A, essential. Inheritance of the silent HISN6B epiallele requires HDA6-, MET1-, and CMT3-dependent cytosine methylation, but is unaffected by mutations disrupting RdDM. Although methylated HISN6B epialleles can be stably inherited for at least 30 generations, they can revert to an active state if the epigenetically silent state is erased by passage through an hda6 mutant background. This allows HISN6B to now rescue hisn6a null mutations in ecotype Col-0 and to restore compatibility with ecotype Cvi-0, in which HISN6A is defective owing to an internal deletion. Collectively, our results demonstrate that HISN6-dependent hybrid lethality is a previously unrecognized epigenetic phenomenon.
Results
HISN6A and HISN6B are duplicated, paralogous genes (Fig. 1A and Fig. S1) encoding histidinol-phosphate aminotransferase, which converts imidazole-acetol phosphate to histidinol-phosphate in the histidine biosynthesis pathway (32, 33) (Fig. 1B). The gene duplication occurred after A. thaliana diverged from a common ancestor with Arabidopsis lyrata (34, 35), resulting in duplication of a segment of chromosome 1 containing HISN6B (At1g71920) on chromosome 5, yielding the HISN6A locus, At5g10330 (33, 36). Although HISN6A and HISN6B coding regions are 100% identical at the amino acid level, homozygous hisn6a null mutations are embryo-lethal in ecotype Col-0, because HISN6B is not expressed (Fig. 1 B and C) (31, 32). This differential expression of HISN6A and HISN6B can be observed by RT-PCR amplification followed by digestion with RsaI, which cuts only HISN6A cDNA (Fig. 1C). In wild-type (WT) Col-0, only HISN6A is expressed (Fig. 1D); however, in hda6-6 or hda6-7 mutants (37, 38), HISN6B is expressed as well (Fig. 1D), which is also true in mutants for MET1 or CMT3, which encode enzymes responsible for cytosine maintenance methylation in the CG and CHG sequence contexts (39), respectively (Fig. 1D and Fig. S2A). In contrast, HISN6B is not derepressed in mutants deficient for RNA-directed de novo cytosine methylation, such as nrpd1-3 (Pol IV), nrpe1-11 (Pol V), and drm2 (Fig. 1D and Fig. S2A).
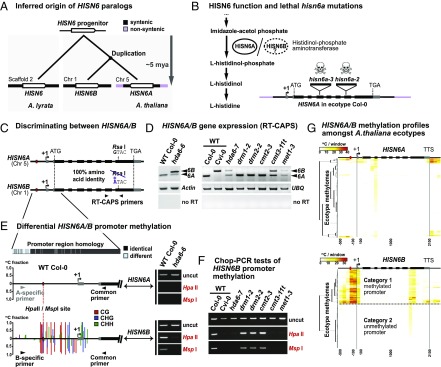
Fig. 1.
HDA6, CMT3, and MET1 silence HISN6B via promoter region DNA methylation. (A) Inferred phylogenetic origin of HISN6 paralogs based on synteny between HISN6B flanking regions on A. thaliana chromosome 1 and sequences including the single copy HISN6 gene of A. lyrata scaffold 2 (Fig. S1 A and B). (B, Left) HISN6A/B protein function in histidine biosynthesis. Steps upstream of Imidazole-acetol phosphate are omitted. (B, Right) Gene structure of HISN6A in ecotype Col-0: UTRs, exons, and T-DNA insertion positions in mutant alleles are indicated by gray boxes, black boxes, and inverted triangles, respectively. (C) RT-CAPS assay for discrimination of HISN6A and HISN6B mRNAs using primers flanking a polymorphic RsaI site present only in HISN6A. (D) HISN6A/B expression analysis via RT-CAPS in hda6-6, hda6-7, drm1-2, drm2-2, cmt2-3, cmt3-11t, and met1-3 mutants compared with WT Col-0 or Cvi-0. Actin and ubiquitin (UBQ) reactions served as loading controls. Reactions omitting reverse transcriptase (no RT) are controls for genomic DNA contamination. (E) Analysis of DNA methylation in HISN6A and HISN6B promoter regions. Bar plots show WT Col-0 methylation profiles, color-coded by sequence context (CG, CHG, and CHH), tabulated as fractional cytosine methylation (y-axis), based on methylome data of Stroud et al. (40). No methylation was detected in the HISN6A promoter. Gel images show Chop-PCR assays of cytosine methylation status at a HISN6A/B promoter HpaII/MspI site (red dotted line) in WT Col-0 or the hda6-6 mutant. Reactions omitting restriction enzymes (uncut) demonstrate equivalent DNA input. HISN6A/B primer specificity was verified by sequencing PCR products. (F) Analysis of HISN6B promoter methylation in the mutant series of D, using Chop-PCR. (G) Hierarchical clustering of cytosine methylation profiles for HISN6A (Top) or HISN6B (Bottom) genes in 892 ecotype methylomes of A. thaliana. Methylated cytosines, based on data of Kawakatsu et al. (41), were tallied within 100-bp nonoverlapping windows starting 500 bp upstream of the transcription start site (+1) and stopping 300 bp downstream of the TTS.
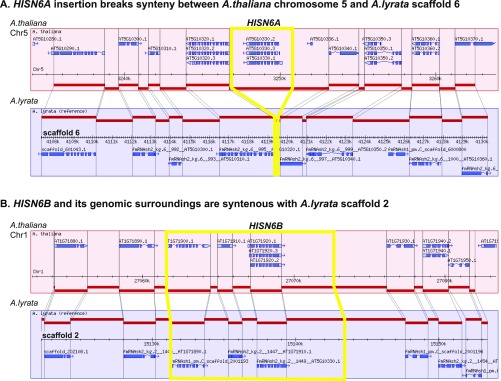
Fig. S1.
Chromosomal contexts of HISN6A and HISN6B. (A) The HISN6A locus of A. thaliana interrupts a region displaying extensive synteny between A. thaliana chromosome 5 and A. lyrata scaffold 6, suggesting insertion at this site after the divergence of the two species from a common progenitor (33, 36). (B) HISN6B is present within a region displaying extensive synteny with A. lyrata scaffold 2, the location of the sole HISN6 gene of A. lyrata. Diagrams generated using the Synteny Viewer tool (https://www.arabidopsis.org/servlets/tools/synteny).
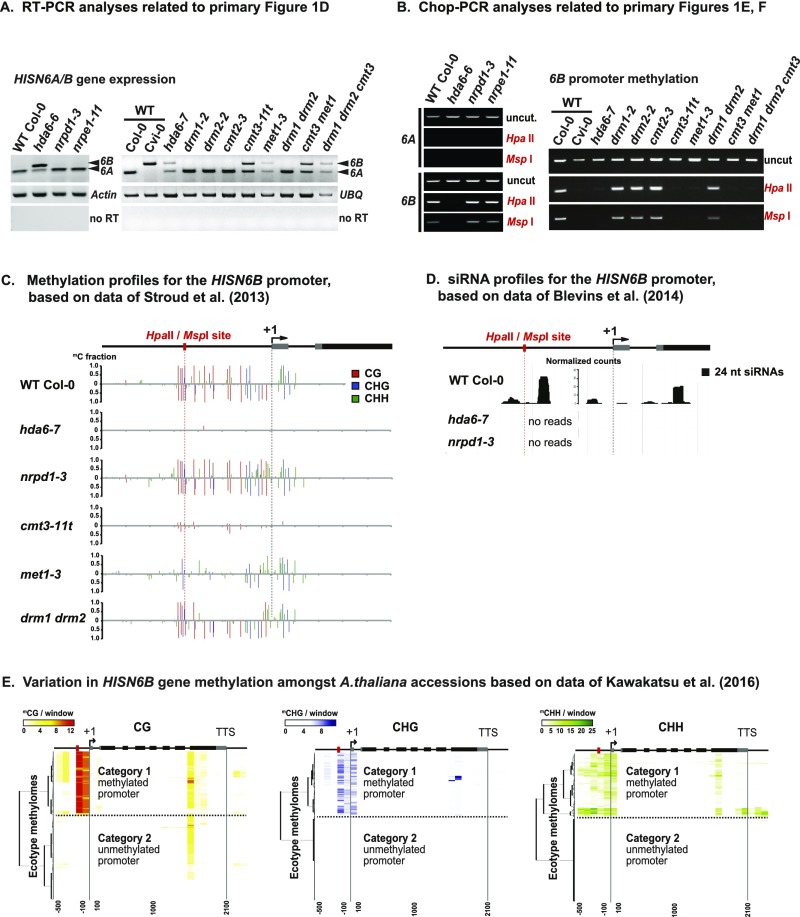
Fig. S2.
HISN6A and HISN6B expression and methylation in WT Col-0 and in mutants affecting chromatin modification and gene silencing. (A) RT-PCR products were digested with RsaI to discriminate HISN6A/B expression in WT Col-0 or Cvi-0 and the indicated Col-0 mutants (complete panel corresponding to Fig. 1D). (B) HISN6A and HISN6B promoter methylation in WT and various Col-0 mutants was analyzed by Chop-PCR at the promoter region HpaII/MspI restriction site (complete panel corresponding to Fig. 1 E and F). Reactions omitting restriction enzymes (uncut) control for equivalent DNA input in all reactions. (C) Cytosine methylation profiles are shown for a 600-bp region encompassing the HISN6B promoter and transcription start site (+1). Methylomes of 3-wk-old leaves isolated from WT and mutant lines of the Col-0 ecotype were analyzed [data from Stroud et al. (40)]. Bars represent fractional methylation per cytosine position, color-coded by sequence context (CG, CHG, and CHH). The HpaII/MspI restriction enzyme site used for Chop PCR assays is highlighted by a red dotted line. (D) A genome browser shot showing small RNA profiles of 2-wk-old leaves of WT, hda6-7, or nrpd1-3 mutants of the Col-0 ecotype. Read counts corresponding to 24-nt siRNAs are plotted in black on the y-axis, normalized by total mapped reads [data from Blevins et al. (15)]. (E) Hierarchical clustering analysis of HISN6A/B methylation, as in Fig. 1G, but divided by CG, CHG, or CHH sequence context [data from Kawakatsu et al. (41)].
HISN6 promoter CG and CHG methylation is easily assayed using Chop-PCR, a test in which genomic DNA is first digested (chopped) with methylation-sensitive restriction endonuclease HpaII or MspI before PCR amplification of a region that includes a HpaII/MspI recognition site, CCGG. In our tests, we assayed a HpaII/MspI site in the promoter region (Fig. 1E, red dotted line), where Col-0 methylome data (40) showed that dense CG and CHG methylation and scattered CHH methylation occur in HISN6B, but not in HISN6A (Fig. 1E gene diagrams). Using the Chop-PCR assay, PCR products were detected for HISN6B (Fig. 1E, Bottom Right), but not for HISN6A (Fig. 1E, Top Right), indicating CG and CHG methylation of the HISN6B promoter CCGG site, but not of the corresponding HISN6A site. HISN6B promoter CG and CHG methylation was lost in hda6-6, met1-3, and cmt3-11t mutants, but not in Pol IV (nrpd1-3) and Pol V (nrpe1-11) mutants or mutants defective for the DNA methyltransferases DRM1, DRM2, or CMT2 (Fig. 1 E and F and Fig. S2 B and C). Although 24-nt siRNAs matching the HISN6B promoter were detected by RNA-seq (Fig. S2D) (15), their absence in the nrpd1-3 mutant was not correlated with HISN6B reactivation. Collectively, the data of Fig. 1 D-F show that HISN6B silencing in Col-0 correlates with HDA6-, MET1-, and CMT3-dependent CG and CHG methylation. In ecotype Cvi-0, in which HISN6A has suffered a deletion mutation, HISN6B lacks promoter methylation (Fig. 1F) and is expressed (Fig. 1D). Analysis of publicly available methylome data (41) reveals HISN6B promoter hypermethylation in 43% (387 of 892 datasets) of the A. thaliana ecotype methylomes analyzed (Fig. 1G and Fig. S2E), suggesting that differential methylation, and likely silencing, of HISN6B is not unique to Col-0. The basis for the differential methylation of HISN6B among these accessions is unclear, however.
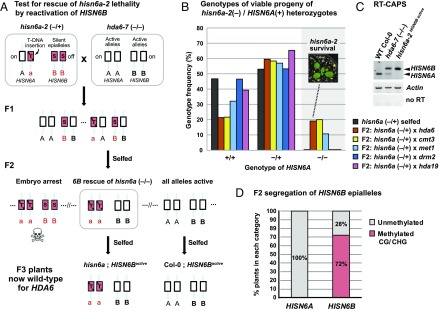
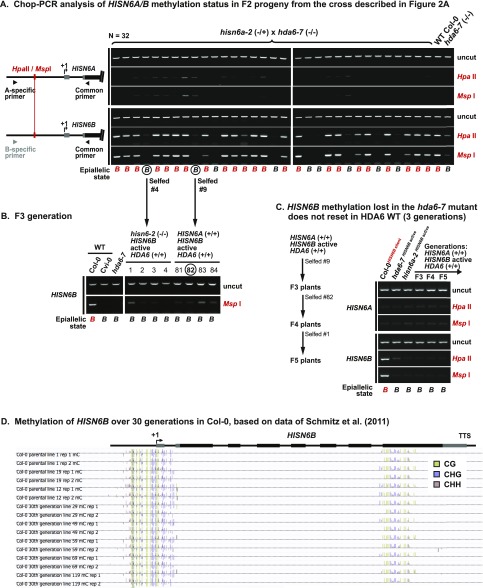
In Col-0, homozygous hisn6a-2 progeny of hisn6a-2/HISN6A heterozygotes arrest as preglobular embryos (32). To test whether HISN6B derepression in the hda6-7 mutant background rescues hisn6a-2 lethality, we crossed a hisn6a-2 heterozygote (−/+) with homozygous hda6-7 (Fig. 2A). One-half of the resulting F1 plants harbored one hisn6a-2 mutant allele (red “a”), one functional HISN6A allele (black “A”), one silent HISN6B allele from the hisn6a-2 parent (red “B”), and one active, derepressed HISN6B allele (black “B”) from the hda6-7 parent (Fig. 2A). Among 89 of their F2 progeny, 17 (19%) hisn6a-2 homozygous mutant (−/−) plants were recovered (Fig. 2B, red bars). In contrast, zero hisn6a-2 (−/−) plants were among the 64 progeny of self-fertilized hisn6a-2 (−/+) plants WT for HDA6 (Fig. 2B, black bars). Whereas WT Col-0 plants expressed only HISN6A, and hda6-7 mutants expressed both HISN6A and HISN6B, the viable hisn6a-2 homozygotes expressed only HISN6B (Fig. 2C; hisn6a-2;HISN6B active).
Fig. 2.
Reactivating HISN6B via elimination of symmetric DNA methylation rescues hisn6a-2 lethality. (A) Strategy for rescue of hisn6a-2 lethality. The red “a” indicates the hisn6a-2 null mutant allele, the red “B” indicates transcriptionally silent HISN6B, and the black “A” and “B” indicate transcriptionally active HISN6A and HISN6B, respectively. HDA6 genotypes are omitted for simplicity. (B) Tests of hisn6a-2 rescue by passage of HISN6B alleles through null mutants affecting gene silencing. Heterozygous hisn6a-2 (−/+) was selfed (black) or crossed to hda6-7 (orange), cmt3-11t (yellow), met1-7 (light blue), drm2-2 (blue), or hda19-1t (purple). Selfed hisn6a-2 (−/+) progeny (n = 64 plants) or F2 progeny resulting from the crosses to other mutants (n = 89, 60, 28, 60, and 30 plants, respectively) were genotyped for hisn6a-2 and HISN6A alleles, the frequencies of which were plotted. (C) RT-CAPS analysis of HISN6A/B gene expression in the hda6-7 mutant and in a rescued hisn6a-2 mutant line now harboring active HISN6B alleles. Actin reactions served as loading controls. Reactions omitting reverse transcriptase (no RT) served as controls for genomic DNA contamination. (D) Mendelian segregation of hypomethylated HISN6B epialleles. F2 progeny of hisn6a-2 × hda6-7 were assayed for the presence (red) or absence (gray) of cytosine methylation at the HpaII/MspI restriction site of HISN6A/B promoters. The percentage of plants in each category is plotted (n = 32). Unlike HISN6B, HISN6A did not show significant levels of promoter methylation (Fig. S3 A and C).
We also crossed hisn6a-2 (−/+) to the cytosine methyltransferase mutants cmt3-11t or met1-7. Viable hisn6a-2 homozygotes represented 20% of the F2 plants resulting from the cmt3-11t cross and 11% of the F2 plants resulting from the met1-7 cross (Fig. 2B). In contrast, crosses to drm2-2 or hda19-1t, a histone deacetylase that is functionally distinct from HDA6 (42, 43), yielded no viable hisn6a-2 homozygous F2 progeny (Fig. 2B). We conclude that HISN6B can be converted from a silent epiallele to an active allele in hda6, cmt3, or met1 mutants, allowing the reanimated gene to rescue plants lacking the normally essential HISN6A gene.
Analysis of F2 individuals in the hisn6a-2 (−/+) × hda6-7 segregating population revealed HISN6B promoter methylation in 72% of the plants, consistent with a 3:1 ratio owing to Mendelian inheritance of methylated or unmethylated alleles (Fig. 2D and Fig. S3A). Because the unmethylated HISN6B allele segregated independently of the hda6-7 mutation, we were able to identify an F3 line (Col-0HISN6B active) that is homozygous for active HISN6B epialleles in an otherwise genetically WT background (Fig. 2A; F3 individual 82 in Fig. S3B). After self-fertilization of this line for three generations, no resetting to the silent, methylated state was observed (Fig. S3C); thus, this line, designated Col-0HISN6B active, was used for further genetic comparisons to WT plants, designated Col-0HISN6B silent, homozygous for silent, methylated HISN6B epialleles. Analysis of methylome data obtained by Schmitz et al. (44) showed that methylated HISN6B epialleles were stably inherited over a span of 30 generations (Fig. S3D).
Fig. S3.
Stable HISN6B methylation is lost in hda6 mutants and not regained upon restoration of WT HDA6 genes. (A) Chop-PCR DNA methylation analysis of F2 progeny from the cross in Fig. 2A testing HISN6A/B methylation at the promoter HpaII/MspI site. Reactions omitting restriction enzymes serve to control for equivalent DNA input to all reactions (uncut). PCR specificity was verified by sequencing uncut PCR products. (B) Chop-PCR analysis of HISN6B promoter methylation in F3 progeny from the cross in Fig. 2A. (C) Chop-PCR analysis of HISN6A/B promoter methylation in a HISN6B active lineage derived from an hda6 mutant background, in three self-fertilized generations after restoration of HDA6 function (F3, F4, and F5 generation from the cross in Fig. 2A). WT Col-0 (HISN6B silent) and hda6-7 and hisn6a-2 mutants (both HISN6B active) served as controls. (D) Browser shot of HISN6B methylation profiles in WT Col-0 lines propagated by single-seed descent for 30 generations. Profiles of independent parental lines and 30th generation lines are shown, with the methylcytosine sequence context (CG, CHG, or CHH) indicated using different colors. Data are from Schmitz et al. (44), displayed using the genome browser tool described in that paper (neomorph.salk.edu/30_generations/browser.html).
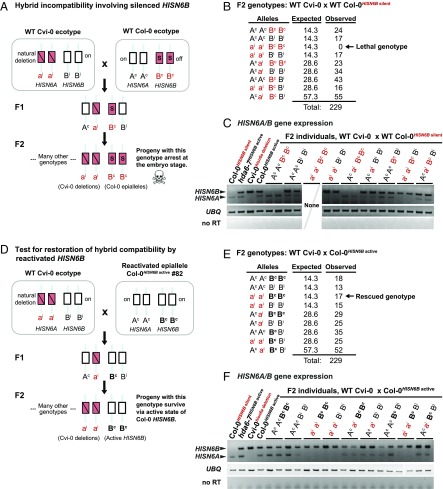
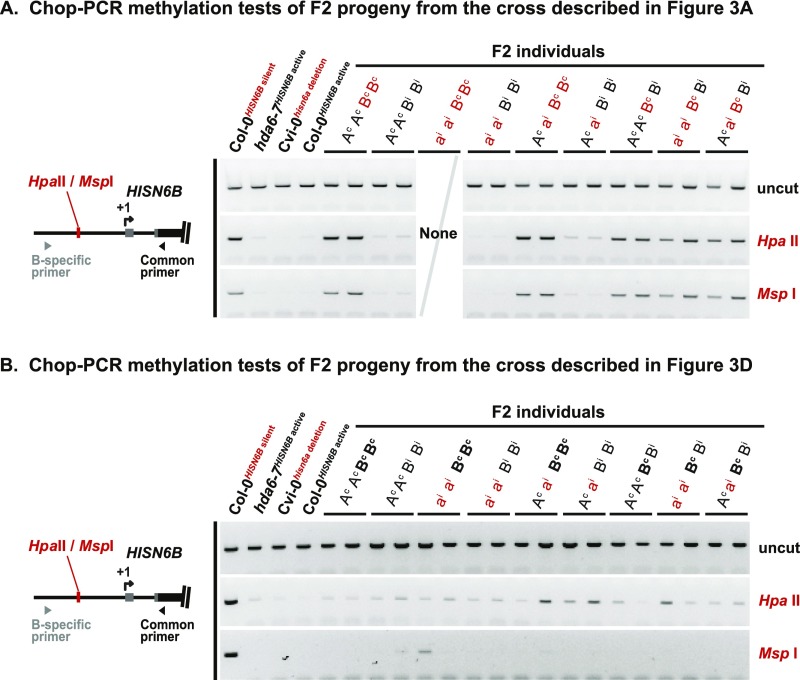
In Cvi-0 × Col-0 F2 hybrids, homozygosity for both Col-0 HISN6B and Cvi-0 hisn6a is lethal (Fig. 3A) (31). We confirmed this among 229 Cvi-0hisn6a deletion × Col-0HISN6B silent F2 progeny, using PCR to detect Col-0 HISN6A (abbreviated Ac), Col-0 HISN6B (Bc), Cvi-0 hisn6a (ai), and Cvi-0 HISN6B (Bi) alleles. Although ∼14 individuals (1/16) could be expected to have the genotype aiai BcBc, none were observed (Fig. 3B, arrow), a significant deviation (P = 4.8 × 10−8, Pearson’s χ2 test) indicating hybrid incompatibility. Reverse-transcription–cleaved amplified polymorphic sequence (RT-CAPS) analyses revealed that hybrid individuals that are homozygous for WT Col-0 (BcBc) HISN6B alleles failed to express HISN6B, whereas individuals that inherited at least one HISN6B allele from Cvi-0 (Bi or BiBi) showed HISN6B expression (Fig. 3C). This indicates that silent or active HISN6B alleles of the parental ecotypes are faithfully transmitted. The promoter methylation marks of Col-0 HISN6B (Bc) alleles were also faithfully inherited among F2 hybrid individuals (Fig. S4A).
Fig. 3.
Active HISN6B epialleles circumvent lethality in A.thaliana Cvi-0 x Col-0 hybrids. (A) Schematic diagram of hybrid incompatibility between A. thaliana ecotypes Col-0 and Cvi-0. The red “ai” indicates Cvi-0 hisn6a deletion alleles, the red “Bc” indicates silent Col-0 HISN6B, and the black “Ac” and “Bi” indicate active Col-0 HISN6A and Cvi-0 HISN6B, respectively. Note that alleles inherited from Col-0 carry the superscript “c”, and alleles from Cvi-0 carry the superscript “i”. All combinations of HISN6A and HISN6B alleles result in viable plants, except the double-homozygous combination of Cvi-0 hisn6a alleles (aiai) and Col-0 HISN6B alleles (BcBc). (B) F2 genotypes resulting from the cross: WT Cvi-0 × WT Col-0 (with silent HISN6B). A total of 229 individuals were genotyped. Expected allele frequencies were calculated assuming Mendelian segregation. Comparison of observed to expected frequencies using Pearson’s χ2 test resulted in a P value of 4.8 × 10−8, indicating hybrid incompatibility (arrow). (C) RT-PCR analysis of HISN6A/B gene expression in F2 progeny of the WT Cvi-0hisn6a deletion × WT Col-0HISN6B silent cross. Two individuals for each of the eight observed genotypes were assayed. Ubiquitin (UBQ) amplification products served as loading controls. Reactions without reverse transcriptase (no RT) served as controls for DNA contamination. Col-0HISN6B silent, Cvi-0hisn6a deletion, hda6-7, and Col-0 carrying reactivated HISN6B alleles served as HISN6 expression controls. (D) The schematic depicts the genetic mechanism underlying rescue of hybrid compatibility following reanimation of Col-0 HISN6B alleles. The red “ai” indicates Cvi-0 hisn6a deletion alleles, the black “Bc” indicates active Col-0 HISN6B, and the black “Ac” and “Bi” indicate active Col-0 HISN6A and Cvi-0 HISN6B, respectively. Note that alleles inherited from Col-0 carry the superscript “c”, and alleles from Cvi-0 carry the superscript “i”. All combinations of HISN6A and HISN6B alleles result in viable plants, including the previously lethal combination of two Cvi-0 hisn6a alleles (aiai) and two Col-0 HISN6B alleles (BcBc). (E) F2 genotypes of progeny resulting from the cross of WT Cvi-0 × Col-0 (active HISN6B). Allele frequencies among 229 F2 individuals are shown. Expected allele frequencies were calculated assuming Mendelian segregation. Comparison of observed and expected statistics using Pearson’s χ2 test resulted in a P value of 0.78, indicating restored hybrid compatibility (arrow). (F) HISN6A/B expression in the F2 progeny of the Cvi-0hisn6a deletion × Col-0HISN6B active cross. Two individuals from each of the nine observed genotypes were assayed. UBQ reactions served as loading controls. The interruption in this panel corresponds to the end of one agarose gel row and the beginning of the next row. Reactions without reverse transcriptase (no RT) served as controls for DNA contamination. Col-0HISN6B silent, Cvi-0hisn6a deletion, hda6-7, and Col-0 carrying reactivated HISN6B served as controls.
Fig. S4.
Methylation status of HISN6A/B alleles inherited from WT Col-0 or Cvi-0, or a Col-0 hda6-7 mutant. (A) Chop-PCR DNA methylation analysis of F2 progeny from the cross in Fig. 3A, assessing methylation at the promoter region HpaII/MspI site. The double-homozygous combination Cvi-0 hisn6a (aiai) and Col-0 HISN6B (BcBc) was not recovered in 229 plants and thus was not available for analysis. Reactions omitting restriction enzymes (uncut) reveal equivalent DNA input in all reactions. Primer specificity was verified by sequencing uncut PCR products. (B) Chop-PCR analysis of F2 progeny from the cross in Fig. 3D.
We next tested whether derepression of Col-0 HISN6B alleles would now allow the survival of F2 hybrids homozygous for both Col-0 HISN6B and Cvi-0 hisn6a (Fig. 3D). Indeed, the Cvi-0hisn6a deletion × Col-0HISN6B active cross yielded 17 healthy aiai BcBc individuals in an F2 population of 229 plants (Fig. 3E), which is statistically indistinguishable from the expected ∼14 plants (P = 0.78, Pearson’s χ2 test). Col-0 HISN6B allele expression in the F2s (Fig. 3F) correlates with the near absence of HISN6B methylation (Fig. S4B). Collectively, these genetic tests show that reversion of HISN6B epialleles from a methylated, silent state to a hypomethylated, active state eliminates HISN6-based hybrid incompatibility between the Col-0 and Cvi-0 ecotypes of A. thaliana.
Discussion
Our study shows that ecotype-specific silencing of duplicated HISN6 genes occurs in A. thaliana and is maintained by symmetric CG and CHG methylation involving HDA6, MET1, and CMT3. MET1- and CMT3-dependent DNA methylation can maintain silent epialleles over numerous meiotic generations, independently of initial silencing signals (16, 17, 22–24, 45). Mutation of MET1, CMT3, or HDA6 converts silent HISN6B epialleles into active, unmethylated alleles that are stably transmitted according to Mendelian rules of segregation. Moreover, reactivation of HISN6B circumvents the normal lethality of hisn6a mutations in Col-0 and prevents the occurrence of HISN6 allele-dependent hybrid lethality among Cvi-0 × Col-0 hybrid progeny.
Hybrid incompatibility involving HISN6A and HISN6B alleles fits the model of Lynch and Force in that alternative HISN6A vs. HISN6B expression states lead to deleterious gene combinations in Cvi-0 × Col-0 F2 progeny (7, 31). Unlike the model of Bateson, Dobzhansky, and Muller, the Lynch and Force model for hybrid incompatibility does not require gene neofunctionalization; instead, differential loss of function of one member of a duplicated gene pair in different subpopulations or ecotypes spawns incompatibilities if the ecotypes hybridize. Examples in plants include the DPL1 and DPL2 genes of Oryza sativa subspecies indica and japonica (46), and the AtFOLT1 and AtFOLT2 genes of A. thaliana (29). The latter report showed that Col-0 × C24 and Col-0 × Sha incompatibilities correlate with the duplication, and additional complex rearrangement, of AtFOLT1 in the C24 and Sha ecotypes. These mutations trigger methylation of AtFOLT1, leaving AtFOLT2 as the sole active copy in C24 or Sha and causing hybrid incompatibility with Col-0, which lacks AtFOLT2 altogether. Our study demonstrates that transgenerationally heritable (but fully revertible) epialleles that have not undergone inverted duplication, rearrangement, or mutation also contribute to hybrid incompatibility, providing additional evidence that epigenetic variation can foster reproductive isolation (29, 47, 48) in a manner consistent with the hypothesis of Lynch and Force.
Materials and Methods
Plant Materials.
The A. thaliana ecotype Col-0 used in this study was a laboratory stock of the C.S.P. laboratory. Ecotype Cvi-0 (CS22614) was obtained from the Arabidopsis Biological Resource Center at Ohio State University. The hda6-6 (axe1-5) and hda6-7 (rts1-1) mutants were described by Murfett et al. (38) and Aufsatz et al. (37). hda19-1t is the athd1-t1 (Ws) mutant allele of Tian et al. (42) introgressed into Col-0 by three backcrosses. The RNA polymerase mutants pol IV (nrpd1-3) and pol V (nrpe1-11) were described by Onodera et al. (49) and Pontes et al. (2006) (50). The mutants drm2-2, cmt2-3 (SALK_012874), cmt3-11t (SALK_148381), hisn6a-2 (SAIL_750), and hisn6a-3 (SALK_089516), and the triple mutant drm1-2 drm2-2 cmt3-11t, were obtained from the Arabidopsis Biological Resource Center. met1-3 was described by Saze et al. (16), and met1-7 (SALK_076522) was obtained from the Nottingham Arabidopsis Stock Center.
RNA Analyses.
Total RNA was extracted from 2-wk-old rosettes or from inflorescences using TRI Reagent (Molecular Research Center). For semiquantitative RT-PCR, 1.5 µg of DNase I-treated total RNA was used for random-primed cDNA synthesis by SuperScript III reverse transcriptase (Invitrogen). Standard PCR was performed on cDNA aliquots (∼100 ng of RNA input) using GoTaq Green (Promega) and the primers listed in Table S1. PCR products were analyzed either directly by agarose gel electrophoresis (Actin and UBQ controls) or following restriction enzyme digest (RsaI) and then agarose gel electrophoresis (RT-CAPS for HISN6A/B).
Table S1.
Oligonucleotides used for genotyping, Chop-PCR, and RT-PCR
| Primer use | Primer ID | Primer sequence |
| Genotyping | ||
| hda6-7 genotyping | hda6-7_geno_F | GATTCTGAGTGAGAGACGGAG |
| hda6-7_geno_R | AGCCATACGGATCCGGTGAGG | |
| hisn6a-2 (SAIL_750) genotyping | HISN6A_Chop_F | GTTCCTTTTAGATCGCCGGGAAATCGATC |
| SAIL_750_F01-RP | AAGGCGTCTCTCACATCTTCC | |
| hisn6aCvi-0 (deletion) genotyping | SEQ503246F3 | GGTTTGCACAGACAACTATCATATTGC |
| SEQ503254R4 | CATTCGATCTGAATTTGTTCCGATCTAATTC | |
| HISN6ACol-0 (intact) genotyping | HISN6_RsaI-CAPS_F1 | TATCATCAGCGAGGACGATCTGTTGAAG |
| HISN6_RsaI-CAPS_R1 | ACCAAGGCGTCTCTCACATCTTC | |
| dCAPS assay for HISN6BCol-0 vs. HISN6BCvi-0 | HINS6B_dCAPS_BspHI_F | ATCGAAGATTCTAAACTTTAGTCTATAATCCCTCG |
| HINS6B_dCAPS_BspHI_R | CGATAATTGGACCAGACTTTTGACATTGAAATGTTTCTCATTTCACATGTTACTATAGAATCATG | |
| Chop-PCR | ||
| HISN6A promoter | HISN6A_Chop_F | GTTCCTTTTAGATCGCCGGGAAATCGATC |
| HISN6_Chop_comR | GAGCCCTGAACATTGATCACACCC | |
| HISN6B promoter | HISN6B_Chop_F | GGAATCATTGGGAGAAAAATTAGGTG |
| HISN6_Chop_comR | GAGCCCTGAACATTGATCACACCC | |
| RT-PCR | ||
| HISN6A/B (RT-CAPS) | HISN6_RsaI-CAPS_F1 | TATCATCAGCGAGGACGATCTGTTGAAG |
| HISN6_RsaI-CAPS_R1 | ACCAAGGCGTCTCTCACATCTTC | |
| Actin | Actin_RT_F | ACCAGATAAGACAAGACACAC |
| Actin_RT_R | AAGTCATAACCATCGGAGCTG | |
| UBQ | UBQ_RT_F | GATCTTTGCCGGAAAACAATTGGAGGATGGT |
| UBQ_RT_R | CGACTTGTCATTAGAAAGAAAGAGATAACAGG | |
DNA Analyses.
For genotyping, genomic DNA was purified from 2-wk-old seedlings using a CTAB extraction protocol. GoTaq Green Master Mix (2×; Promega) was mixed with ∼100 ng of genomic DNA and particular genotyping primer pairs. PCR products were scored either directly by agarose gel electrophoresis or following restriction enzyme digest (BspHI) and then agarose gel electrophoresis (CAPS). For DNA methylation analyses, genomic DNA was isolated from inflorescence tissue using the Nucleon PhytoPure DNA extraction kit (Amersham). Chop PCR assays were performed using 100 ng of restriction enzyme-digested (“chopped”) genomic DNA as in Earley et al. (51). Primers used for genotyping and Chop PCR are listed in Table S1.
Bioinformatic Analyses.
Analysis of WT and mutant methylomes of ecotype Col-0 (Fig. 1E and Fig. S2C) were performed on data from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) accession no. GSE39901 (six datasets, wiggle format) (40). Methylation profiles were based on HISN6A gene model AT5G10330.8 and HISN6B gene model AT1G71920.2 (Araport11), whose similar intron/exon structures facilitated comparison of the two genes. Fractional cytosine methylation in CG, CHG, and CHH sequence contexts were converted from wiggle to bigWig format, extracted using the bwtool software, and then plotted in Microsoft Excel (52). For the comparison of A. thaliana ecotype methylomes (Fig. 1G and Fig. S2E), datasets were obtained from NCBI GEO accession no. GSE43857 (927 ecotype datasets, tabular format) (41). After removing redundant or unidentified ecotypes, 892 methylomes remained for analysis. For each methylome, CG, CHG, and CHH methylation were tallied separately over 100-bp nonoverlapping windows in HISN6A or HISN6B, respectively, starting 500 bp upstream of the transcription start site (+1) and stopping 300 bp downstream of the transcription termination site (TTS). Hierarchical clustering was then performed using Euclidean distance and Ward's method to regroup ecotypes with similar patterns of cytosine methylation along HISN6A or HISN6B. Heatmaps were drawn using the “heatmap.2” function of R. For HISN6B, ecotypes were assigned to two visually evident categories: methylated promoter versus unmethylated promoter.
Statistical Tests.
Chi-square goodness-of-fit tests were performed on genotype data shown in Fig. 3 B and E to test the hypothesis of HISN6B methylation-dependent hybrid incompatibility and generate P values in each case. The GPower software (53) estimated that sample sizes of at least 158 F2 plants would be needed to determine whether the observed allele frequencies deviate from Mendelian expectations, a number far exceeded by the 229 F2 plants genotyped in each experiment.
Acknowledgments
C.S.P. is an Investigator at the Howard Hughes Medical Institute (HHMI) and the Gordon and Betty Moore Foundation (GBMF). This work was supported by National Institutes of Health (NIH) Grant GM077590 and GBMF Grant GBMF3036 (to C.S.P.). T.B. was supported by an NIH Ruth L. Kirschstein National Research Service Award, funding from the HHMI, as well as LabEx consortium ANR-10-LABX-0036_NETRNA, with resources managed by the French Agence Nationale de la Recherche as part of the “Investissements d'Avenir” program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 3558.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700368114/-/DCSupplemental.
References
- 1.Chae E, et al. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell. 2014;159(6):1341–1351. doi: 10.1016/j.cell.2014.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5(9):e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Annu Rev Genet. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- 4.Bateson W. Heredity and variation in modern lights. In: Seward AC, editor. Darwin and Modern Science. Cambridge Univ Press; Cambridge, UK: 1909. pp. 85–101. [Google Scholar]
- 5.Dobzhansky T. Studies on hybrid sterility, II: Localization of sterility factors in Drosophila pseudo-obscura hybrids. Genetics. 1936;21(2):113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- 7.Lynch M, Force AG. The origin of interspecific genomic incompatibiity via gene duplication. Am Nat. 2000;156:590–605. doi: 10.1086/316992. [DOI] [PubMed] [Google Scholar]
- 8.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 9.Gossmann TI, Schmid KJ. Selection-driven divergence after gene duplication in Arabidopsis thaliana. J Mol Evol. 2011;73(3-4):153–165. doi: 10.1007/s00239-011-9463-2. [DOI] [PubMed] [Google Scholar]
- 10.Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19(5):859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401(6749):157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen SE, Meyerowitz EM. Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science. 1997;277(5329):1100–1103. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- 13.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38(8):948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 14.Martin A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461(7267):1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 15.Blevins T, et al. A two-step process for epigenetic inheritance in Arabidopsis. Mol Cell. 2014;54(1):30–42. doi: 10.1016/j.molcel.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34(1):65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, et al. Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci Rep. 2015;5:9192. doi: 10.1038/srep09192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1(3):E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22(1):94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 20.Kankel MW, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163(3):1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittelsten Scheid O, Afsar K, Paszkowski J. Release of epigenetic gene silencing by trans-acting mutations in Arabidopsis. Proc Natl Acad Sci USA. 1998;95(2):632–637. doi: 10.1073/pnas.95.2.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindroth AM, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292(5524):2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 23.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15(14):1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quadrana L, Colot V. Plant transgenerational epigenetics. Annu Rev Genet. 2016;50:467–491. doi: 10.1146/annurev-genet-120215-035254. [DOI] [PubMed] [Google Scholar]
- 25.Teixeira FK, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323(5921):1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 26.Matzke MA, Aufsatz W, Kanno T, Mette MF, Matzke AJ. Homology-dependent gene silencing and host defense in plants. Adv Genet. 2002;46:235–275. doi: 10.1016/s0065-2660(02)46009-9. [DOI] [PubMed] [Google Scholar]
- 27.Wendte JM, Pikaard CS. The RNAs of RNA-directed DNA methylation. Biochim Biophys Acta. 2017;1860(1):140–148. doi: 10.1016/j.bbagrm.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15(6):394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 29.Durand S, Bouché N, Perez Strand E, Loudet O, Camilleri C. Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr Biol. 2012;22(4):326–331. doi: 10.1016/j.cub.2011.12.054. [DOI] [PubMed] [Google Scholar]
- 30.Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83(5):725–734. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 31.Bikard D, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323(5914):623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- 32.Muralla R, Sweeney C, Stepansky A, Leustek T, Meinke D. Genetic dissection of histidine biosynthesis in Arabidopsis. Plant Physiol. 2007;144(2):890–903. doi: 10.1104/pp.107.096511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingle RA. Histidine biosynthesis. Arabidopsis Book. 2011;9:e0141. doi: 10.1199/tab.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berr A, et al. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J. 2006;48(5):771–783. doi: 10.1111/j.1365-313X.2006.02912.x. [DOI] [PubMed] [Google Scholar]
- 35.Schmickl R, Jørgensen MH, Brysting AK, Koch MA. The evolutionary history of the Arabidopsis lyrata complex: A hybrid in the amphi-Beringian area closes a large distribution gap and builds up a genetic barrier. BMC Evol Biol. 2010;10:98. doi: 10.1186/1471-2148-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore RC, Purugganan MD. The early stages of duplicate gene evolution. Proc Natl Acad Sci USA. 2003;100(26):15682–15687. doi: 10.1073/pnas.2535513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21(24):6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001;13(5):1047–1061. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1-2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawakatsu T, et al. 1001 Genomes Consortium Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166(2):492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian L, et al. Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics. 2003;165(1):399–409. doi: 10.1093/genetics/165.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao MJ, et al. SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nat Commun. 2015;6:7243. doi: 10.1038/ncomms8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334(6054):369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130(5):851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Mizuta Y, Harushima Y, Kurata N. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc Natl Acad Sci USA. 2010;107(47):20417–20422. doi: 10.1073/pnas.1003124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lafon-Placette C, Köhler C. Epigenetic mechanisms of postzygotic reproductive isolation in plants. Curr Opin Plant Biol. 2015;23:39–44. doi: 10.1016/j.pbi.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Richards EJ. Inherited epigenetic variation: Revisiting soft inheritance. Nat Rev Genet. 2006;7(5):395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 49.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120(5):613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 50.Pontes O, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126(1):79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 51.Earley KW, et al. Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 2010;24(11):1119–1132. doi: 10.1101/gad.1914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pohl A, Beato M. bwtool: A tool for bigWig files. Bioinformatics. 2014;30(11):1618–1619. doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]