Significance
Oncogenic KRAS underlies 30–90% of lung, colon, and pancreatic cancers, but despite more than 30 y of research, clinical inhibitors of KRAS—and potential resistance mechanisms—remain elusive. Using CRISPR-mediated genome editing of oncogenic Kras, we show that some lung cancer cells can survive Kras knockout, indicating the existence of mechanisms that allow tumors to escape Kras oncogene addiction. We identify genes highly expressed in Kras knockout cells, including the Fas receptor gene. Antibodies that activate Fas receptor selectively induced apoptosis in Kras-independent lung cancer cells, suggesting a potential strategy for combinatorial therapies against Kras-driven tumors. These findings have direct translational implications for the treatment of lung cancer and other KRAS mutant cancer types.
Keywords: Kras, lung cancer, Fas, apoptosis
Abstract
Genetic lesions that activate KRAS account for ∼30% of the 1.6 million annual cases of lung cancer. Despite clinical need, KRAS is still undruggable using traditional small-molecule drugs/inhibitors. When oncogenic Kras is suppressed by RNA interference, tumors initially regress but eventually recur and proliferate despite suppression of Kras. Here, we show that tumor cells can survive knockout of oncogenic Kras, indicating the existence of Kras-independent survival pathways. Thus, even if clinical KRAS inhibitors were available, resistance would remain an obstacle to treatment. Kras-independent cancer cells exhibit decreased colony formation in vitro but retain the ability to form tumors in mice. Comparing the transcriptomes of oncogenic Kras cells and Kras knockout cells, we identified 603 genes that were specifically up-regulated in Kras knockout cells, including the Fas gene, which encodes a cell surface death receptor involved in physiological regulation of apoptosis. Antibodies recognizing Fas receptor efficiently induced apoptosis of Kras knockout cells but not oncogenic Kras-expressing cells. Increased Fas expression in Kras knockout cells was attributed to decreased association of repressive epigenetic marks at the Fas promoter. Concordant with this observation, treating oncogenic Kras cells with histone deacetylase inhibitor and Fas-activating antibody efficiently induced apoptosis, thus bypassing the need to inhibit Kras. Our results suggest that activation of Fas could be exploited as an Achilles’ heel in tumors initiated by oncogenic Kras.
Lung cancer is a leading cause of cancer death, accounting for ∼1.3 million deaths worldwide each year (1). Non–small-cell lung cancer (NSCLC), the most common subtype, is associated with frequent mutations in KRAS (∼30%). KRAS is also frequently mutated in other tumor types, including pancreatic (>90%) and colon (∼30%) cancer (2). Although various pharmacological inhibitors are being developed for RAS, especially for the mutant KRASG12C (3–5), these small molecules have not been tested in the clinic (6, 7). As a result, advanced oncogenic KRAS lung cancers are usually treated with conventional therapy such as radiation and chemotherapy, often with limited success (1, 8).
Controlled expression of oncogenic RAS cDNA in mouse models of melanoma, lung, breast, and pancreatic cancer has shown that the withdrawal of oncogenic RAS results in complete tumor regression (9–11). This phenomenon, known as “oncogene addiction,” suggests that oncogenic KRAS alleles (e.g., KRASG12D) not only initiate tumorigenesis but also play a crucial role in tumor maintenance. To recapitulate KRAS oncogene addiction in a mouse model of lung cancer, we developed a conditional Kras shRNA system (shKras) to knock down Kras in KrasG12D/+;p53−/− (KP) cell lines derived from a mouse lung tumor (12–14). When we orthotopically transplanted shKras KP cells into immunocompromised mice, we found that Kras-driven lung tumors can escape oncogene addiction and become independent of Kras signaling (termed Kras independence) (15, 16).
Because shRNAs targeting Kras do not completely eliminate Kras expression, residual Kras in cells could contribute to Kras-independent tumor growth. The best way to rule out this possibility is to genetically delete Kras altogether. Unfortunately, the Kras knockout mouse is embryonically lethal (17), and genetic disruption of KRAS—or other oncogenes—in human cells remains a challenge due to the low efficiency of homologous recombination using traditional gene-targeting technology. We recently showed that CRISPR (18) can be used to efficiently and specifically edit cancer genes in adult mice in a fraction of the time and cost of traditional mouse models (19–21). CRISPR therefore provides a flexible genetic system to manipulate the function of cancer genes (22, 23).
Previous work has shown that oncogenic KRAS epigenetically silences Fas expression (24, 25). In addition, RAS directs epigenetic silencing of Fas through a highly ordered pathway that culminates in methylation of the Fas promoter (26, 27). It remains unexplored whether Fas can be restored by genetic inactivation of oncogenic RAS.
Here, we use CRISPR to establish viable Kras knockout (Kras−/−) lung cancer cell lines from parental oncogenic Kras (KrasG12D/+) cells. While investigating the mechanism of Kras-independent tumorigenesis in this model, we identified Fas among the genes most highly regulated by Kras. Fas (also known as CD95, APO-1, and TNFRSF6) encodes a cell surface death receptor that triggers apoptosis upon binding by its cognate ligand, Fas ligand (FasL) (or CD95L), and plays critical roles in the immune elimination of cancer cells (28, 29). In both mouse and human lung cancer cells, genetic disruption of Kras elevated Fas expression on the cell surface and increased sensitivity to Fas-mediated apoptosis, thereby demonstrating a selective vulnerability of Kras-independent cells. Consistent with previous work showing that oncogenic KRAS epigenetically silences Fas expression (24, 25), we show that Fas is activated in Kras−/− cells by loss of both Dnmt1 and Ezh2 recruitment and repressive epigenetic marks of the Fas promoter. Remarkably, treatment of parental KrasG12D/+ cells with pharmacological histone deacetylase (HDAC) inhibitors not only increased Fas levels but also sensitized cells to Fas-mediated apoptosis. These results suggest a combinatorial strategies for targeted elimination of Kras-independent and oncogenic Kras lung cancer cells.
Results
Kras Knockout Murine Lung Cancer Cells Are Viable and Can Form Tumors in Mice.
Our previous study showed that shRNAs targeting Kras do not completely eliminate Kras in cells (16), hence the residual Kras might contribute to Kras independence. We therefore used CRISPR-based method to genetically disrupt oncogenic Kras in two independent mouse KrasG12D/+;p53−/− lung cancer cell lines (termed KP1 and KP2) (30–32). Lentiviral vector (lentiCRISPR) with puromycin selection marker, as described previously (33), was used to deliver Cas9 and a sgRNA targeting KrasG12D (sgKras) into the target cells (Fig. 1A). The puromycin-resistant single-cell clones expressing sgKras were screened for Kras elimination and specific indel mutations at target loci. Immunoblot analysis of single clones expressing sgKras, using anti-Kras antibody showed the total absence of endogenous Kras protein (Fig. 1B). Furthermore, deep-sequencing analysis revealed 2-nt deletions at the G12D allele (Fig. 1C). Although we designed sgKras to target the G12D sequence (Fig. 1A), a 1-nt deletion was detected at the wild-type allele (Fig. 1C). This confirmed that CRISPR can tolerate mismatch between the sgRNA and target site, consistent with known CRISPR off-target effects (34). These biallelic deletions shift the reading frame of Kras mRNA, likely resulting in premature termination of translation and nonsense-mediated decay of the Kras mRNA, as shown in immunoblots of Fig. 1B. We therefore defined these clones as Kras−/− (Kras knockout) cells.
Fig. 1.
CRISPR-mediated Kras knockout in Kras-driven mouse lung adenocarcinoma cells. (A) Schematic diagram of CRISPR sgRNA design targeting exon 2 of the mutant Kras allele (KrasG12D/+). Codon 12 is underlined. “GAT” encodes G12D mutation. Red arrowhead indicates the Cas9 cutting site. (B) Immunoblot demonstrating total Kras protein levels in KP1 and KP2 clone pairs. Each pair included two clones: one parental (KP1/KP2) and one Kras knockout (KP1 clone/KP2 clone). Hsp90 was used as a loading control. (C) Deep-sequencing analysis showing small deletions induced by CRISPR in a representative clone. (Upper) Representative IGV plots. Black bars denote deletions (purple circle). The red “T” is the G12D mutation. The HindIII site (“C” SNP) was engineered in the original KrasG12D/+ mouse model. (Lower) CRISPR induced a 2-nt deletion in the KrasG12D allele and a 1-nt deletion in the wild-type Kras allele. (D) Colony formation assay to examine the ability of KrasG12D/+ and Kras−/− cells to form colonies. Cells were seeded in six-well plates at 1,000 cells per well, cultured for 5 d and stained with crystal violet. Inset shows small Kras−/− colonies.
The Kras−/− clones show markedly reduced proliferation and colony-forming ability compared with KrasG12D/+ (Fig. 1D), further validating the role of Kras in cell proliferation in lung cancer. Nevertheless, Kras−/− cells are viable and form small colonies in an in vitro colony formation assay (Fig. 1D).
To test whether Kras−/− cells can form tumors in vivo, we transplanted KrasG12D/+ and Kras−/− cells s.c. in immunosuppressed nude mice. Kras−/− cells formed tumors (n = 3 mice), but detection of tumor was much slower (60 d) compared with KrasG12D/+ cells (20 d) (Fig. S1A). In addition, we observed gland-like structure and more cytoplasm of Kras−/− tumor by H&E staining (Fig. S1B), providing availability to further investigate Kras−/− tumor pathology. Taken together, these data show that cancer cells derived from oncogenic Kras tumors can indeed escape complete genetic disruption of Kras, and the resulting Kras-independent cancer cells can still form tumors in nude mice.
Fig. S1.
Tumor xenograft in nude mice. (A) KrasG12D/+ and Kras−/− cells were transplanted s.c. in immunocompromised nude mice (n = 3) and monitored tumor growth over time, as indicated. (B) Tumors were excised and H&E stained. Arrows denote gland-like structure (blue) and increased cytoplasm (red).
Transcriptome Analysis of Kras Knockout Cells Revealed Distinct Gene Signatures.
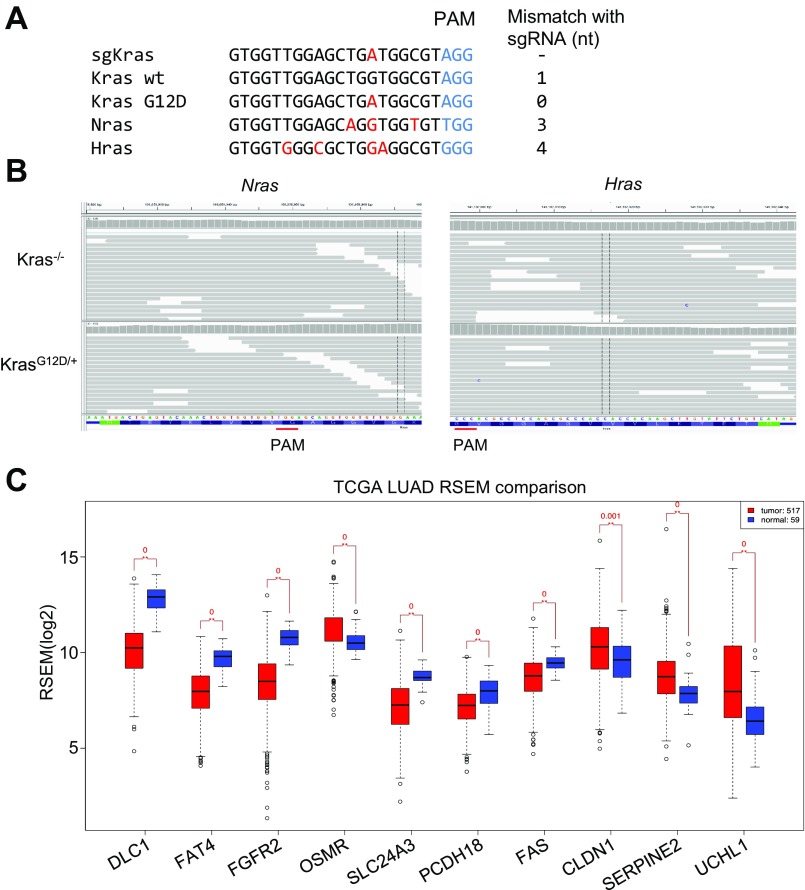
To explore the mechanisms of Kras independence, RNA sequencing (RNA-seq) was performed in KrasG12D/+ and Kras−/− cells. Bioinformatic analysis of RNA-seq data identified 603 up-regulated genes and 216 down-regulated genes in Kras−/− compared with KrasG12D/+ cells (absolute fold change, >2; value of P < 0.05; Fig. 2 A and B), which were consistently up-regulated (down-regulated) in both pairs of KrasG12D/+ and Kras−/− cells (KP1 and KP2, Fig. 2 A and B). These genes are listed in Dataset S1 (“DEG differentially expressed genes”). We then analyzed whether these differentially expressed genes were enriched in any gene ontology or pathways, using the Gene Set Enrichment Analysis (GSEA) and the Molecular Signature Database (MSigDB) (35). The 603 up-regulated genes were enriched in many cancer-related pathways (Fig. 2C), most notably epithelial to mesenchymal transition (q value = 7.25E-53), YAP (q value = 8.30E-18) as well as sets of genes that are down-regulated upon activation of an oncogenic form of KRAS (q value = 1.08E-29). Conversely, the 216 down-regulated genes are enriched in the sets of genes up-regulated by KRAS activation (q value = 3.27E-9). The entire list of enriched gene sets and pathways are provided in Dataset S1 (“Panther or GSEA”). We further confirmed that mRNA levels of RAS ortholog genes (Hras and Nras) did not change in Kras−/− cells (Fig. 2D). In addition, the sgKras did not induce indel mutations at Hras and Nras genomic loci (Fig. S2 A and B). These data suggest that Hras and Nras do not compensate for the inactivation of oncogenic Kras in this model.
Fig. 2.
Identification of KRAS independence genes by RNA-seq. (A) RNA-seq in KrasG12D/+ and Kras−/− cell pairs to identify Kras-independent genes. Heat map shows the clustering of differentially expressed genes. n = 2 for KP2 Kras−/−, n = 3 for other groups. (B) Volcano plot of differentially expressed genes in Kras−/− vs. KrasG12D cells. A total of 603 genes showed increased expression in both KP1 and KP2 pairs; 216 genes showed decreased expression. (C) Selected GSEA dataset. GSEA analysis identified gene sets enriched in up-regulated genes (up in Kras−/− cells) or in down-regulated genes (down in Kras−/− cells). (D) RNA-seq reads showing the relative expression of Ras family genes Hras and Nras in Kras−/− cells. (E) Top 10 candidate genes in the “membrane protein” category.
Fig. S2.
(A and B) Kras sgRNA does not induce indels at Nras or Hras genomic loci. (A) Sequence of sgKras designed to match the G12D sequence. It had 3- and 4-nt mismatches to Nras or Hras, respectively. (B) Deep-sequencing analysis showing that no indels were detected in Nras and Hras genomic loci in Kras−/− cells. (C) Expression analysis of top 10 candidate gene in TCGA datasets, human lung adenocarcinoma (red) vs. normal (blue).
To narrow down the list of differentially expressed genes and identify a possible secondary target in Kras−/− cells, we made the following assumptions: (i) the gene should be up-regulated upon Kras knockout, (ii) the gene should encode a plasma membrane protein, allowing it to be more easily targeted. Out of the 603 up-regulated genes, 189 were annotated as plasma membrane proteins (Dataset S1). To identify up-regulated genes with the most significant P values and largest fold changes, we ranked each of the 189 genes by its average rank of P value. Fold change of the top 10 genes in Kras−/− clones is shown in Fig. 2E. We further analyzed the expression profile of these top 10 candidate genes using The Cancer Genome Atlas (TCGA) datasets (lung adenocarcinoma vs. normal), and found that 7 candidate genes were significantly down-regulated in tumor samples (Fig. S2C). This analysis indicated that these candidates may have a putative role in development or progression of human lung cancer. Moreover, some of these of the candidates, such as DLC1 (36), FAT4 (37), and FAS (28), are well-known tumor suppressors.
Fas Is Up-Regulated in Kras-Independent Cancer Cells.
Among the top 10 genes highly expressed in Kras−/− cells from the RNA-seq, we focused to investigate Fas (which encodes the Fas receptor) for two reasons. First, the Fas receptor triggers apoptosis through cell-intrinsic pathway (38, 39), and it has been implicated in various malignancies (28), hence an attractive therapeutic target. Second, based on previous work (26, 27), Fas expression has been shown to be regulated by Ras-dependent pathways. Following validation of RNA-seq data, the quantitative RT-PCR (qRT-PCR) and immunoblot analysis in parental KrasG12D/+ and Kras−/− cells revealed that KrasG12D/+ cells expressed very low levels of Fas, whereas Kras−/− cells showed a marked increase in Fas mRNA and protein levels (Fig. 3 A and B). Oncogenic RAS activates several downstream signaling pathways, including the MAPK and phosphoinositide 3-kinase (PI3K)/AKT pathways (2). Consistent with previous reports, that Kras-dependent silencing of Fas is partly mediated by MAPK pathway (26), we also observed an increased level of Fas mRNA in KrasG12D/+ cells treated with MEK inhibitor U0126 (Fig. 3C).
Fig. 3.
The Fas receptor is up-regulated in Kras−/− cells. (A) qPCR showing up-regulation of Fas mRNA in Kras−/− cells. (B) Immunoblot demonstrating Fas protein levels in KrasG12D/+and Kras−/− cells. Mouse 3T3 fibroblast cells were used as a positive control for Fas expression. (C) qPCR showing an increase in Fas mRNA upon treatment of MEK inhibitor U0126 in KrasG12D/+ cells at 48 h. (D–F) ChIP assays demonstrating the relative binding of EZH2, DNMT1, and enrichment of H3K27Me3 on Fas promoter in KrasG12D/+ and Kras−/− cells. (G) Methylated DNA immunoprecipitation (MeDIP) assays demonstrating the relative enrichment of 5-methyl cytosine on Fas promoter in KrasG12D/+ and Kras−/− cells. Error bars represent SD (n = 3). **P < 0.01.
Previous studies have shown that expression of oncogenic RAS in mouse NIH 3T3 cells transcriptionally silenced Fas, thereby preventing Fas ligand-induced apoptosis (25). Subsequent studies using genome-wide RNAi screens identified cofactors required for RAS-mediated epigenetic silencing of Fas (27). Kras directs epigenetic silencing of Fas through an ordered pathway that culminates in methylation of the Fas promoter and recruitment of corepressor complex (26, 27). Because KrasG12D/+ cells expressed relatively lower levels of Fas mRNA, we hypothesized that Fas could be transcriptionally silenced in these cells. The Kras-mediated epigenetic silencing of Fas requires two major events, first is the methylation of promoter by DNA methyltransferase, DNMT1, and then trimethylation of histone H3 at lysine 27 (H3K27me3) catalyzed by the histone methyltransferase Ezh2, a component of polycomb repressive complex 2 (26, 27). shRNA-mediated knockdown of either Ezh2 or Dnmt1 significantly increased Fas expression in KrasG12D/+ cells, suggesting that both factors mediate Kras-dependent silencing of Fas (Fig. S3A).
Fig. S3.
(A) qPCR showing Fas expression upon Dnmt1 or Ezh2 knockdown using two independent shRNAs in KrasG12D/+ cells. (B) ChIP assay showing enrichment of H3K4Ac marks on Fas promoter in KrasG12D/+ and Kras−/− cells. Error bars represent SD (n = 3).
We then measured the levels of Dnmt1 and EZH2 and their corresponding epigenetic marks at the Fas promoter in KrasG12D/+ and Kras−/− cells. Chromatin immunoprecipitation (ChIP) revealed a significant enrichment of EZH2 and H3K27me3 at the Fas promoter in KrasG12D/+ cells compared with Kras−/− cells (Fig. 3 D and E). Consistent with previous studies (27), Dnmt1 and DNA methylation were also significantly enriched at the Fas promoter in KrasG12D/+ cells compared with Kras−/− cells (Fig. 3 F and G). Thus, the high levels of Fas mRNA in Kras−/− cells is consistent with low levels of Dnmt1 and EZH2 and their corresponding epigenetic marks at the Fas promoter. Moreover, we found that H3K4 acetylation was enriched at the Fas promoter in Kras−/− cells (Fig. S3B), consistent with its transcriptionally active state. These data indicate that genetic inactivation of oncogenic Kras leads to transcriptional activation and restoration of Fas expression.
Fas-Mediated Apoptosis Can Eliminate Kras-Independent Cells.
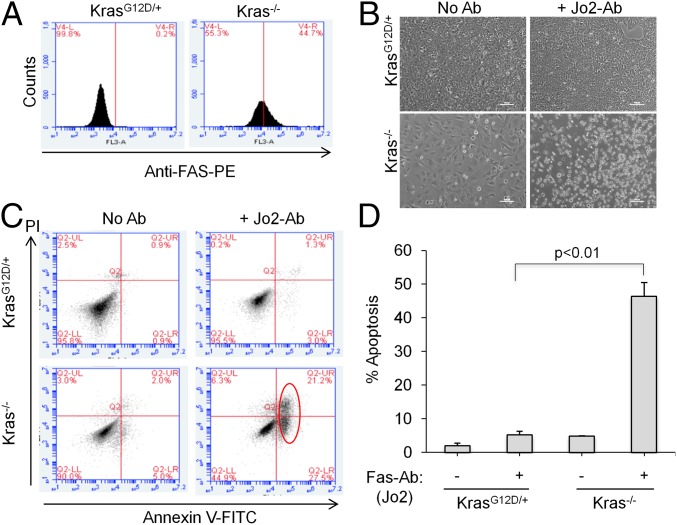
Because Fas is a major mediator of apoptosis on the cell surface, we hypothesized that activation of Fas might selectively trigger apoptosis of Kras−/− cells. Using fluorescence-activated cell sorting (FACS), we confirmed that the Fas receptor is highly expressed on the surface of Kras−/− cells compared with KrasG12D/+ cells (Fig. 4A). As reported before, binding of Fas ligand or Fas-activating antibodies to Fas receptor triggers apoptosis through cell-intrinsic pathway (28). We tested whether a Fas-activating antibody can induce Fas-mediated cell death in Kras−/− cells. When incubating cells with an antibody (Jo2), which binds and activates mouse Fas receptor (28), we observed that many Kras−/− cells became round in shape and detached from the plate, compared with few dead KrasG12D/+ cells (Fig. 4B). Furthermore, we measured the apoptotic cells by Annexin V staining using FACS and observed that ∼50% of Jo2-treated Kras−/− cells were Annexin V positive compared with ∼5% of Jo2-treated KrasG12D/+ cells (Fig. 4 C and D), demonstrating that Fas activation can selectively induce apoptosis in Kras−/− cells over the oncogenic KrasG12D/+ cells. These results demonstrated that, in the absence of Kras, Fas expression is significantly restored, conferring sensitivity to Fas antibody-induced apoptosis.
Fig. 4.
Kras knockout mouse NSCLC cells are sensitive to Fas-mediated apoptosis. (A) FACS histogram showing the Fas receptor expression in Kras−/− and KrasG12D/+ cells. (B) Fas-activating antibodies (Jo2) induces apoptosis in Kras−/− but not KrasG12D/+ cells. Cells were incubated with Jo2 for 24 h. Bright-field images show floating apoptotic Kras−/− cells (Lower Right). (C) Dot plot showing the percentage of Annexin V- or propidium iodide (PI)-positive Kras−/− and KrasG12D/+ cells. Red circle denotes Annexin V-positive cells. x and y axes denote Annexin V and PI signals. (D) Quantitation of Annexin V-positive cells represented as percentage of apoptotic cells. Error bars are SD (n = 3).
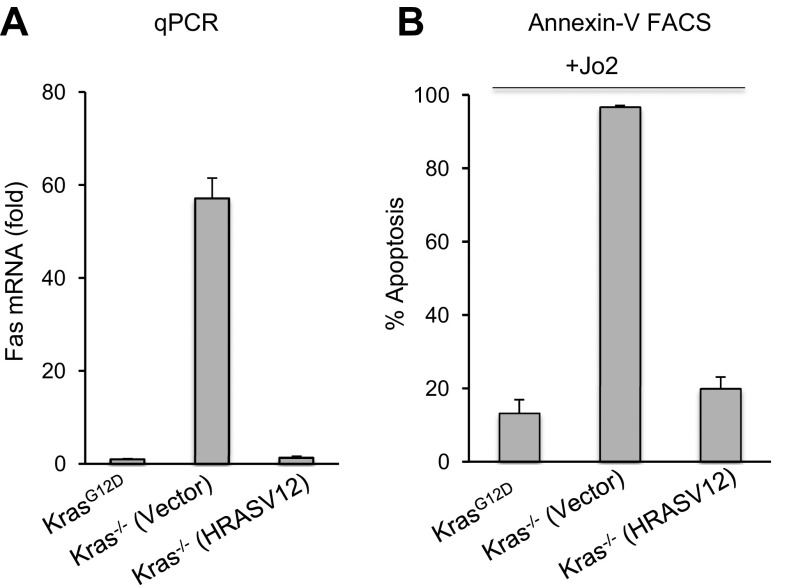
To investigate whether the Fas restoration can be rescued by oncogenic RAS, we reintroduced an oncogenic RAS cDNA in Kras−/− cells. Because Kras−/− cells constitutively expressed Cas9 and Kras sgRNA, these cells are refractory to a KrasG12D cDNA. However, our deep-sequencing data showed that the Hras allele is not affected by the Kras sgRNA. We introduced oncogenic HRAS in Kras−/− cells through retrovirus expressing a HRAS-V12 cDNA (40). Consistent with our previous results of Fas restoration upon loss of Kras (Fig. 3), overexpression of HRAS-V12 in Kras−/− cells led to dramatic decrease in Fas mRNAs (Fig. S4A). Moreover, Fas receptor-mediated apoptosis triggered by the Jo2 antibody was fully rescued in Kras−/− cells expressing HRAS-V12 (Fig. S4B). Thus, reintroduction of oncogenic RAS rapidly suppresses Fas expression in nearly all Kras−/− cells. Together, our data suggest Fas activating antibody as a potential therapeutic strategy to kill Kras-independent cancer cells following Kras silencing.
Fig. S4.
Oncogenic Ras suppresses Fas expression in Kras−/− cells. (A) Fas mRNA levels in Kras−/− cells transduced with retrovirus expressing HRASV12 or empty vector. (B) Kras−/− cells expressing vector or HRASV12 were treated with Jo2 for 24 h, and analyzed for apoptosis using Annexin V staining. The Annexin V-positive cells were quantitated and represented as percentage apoptosis. Error bars are SD (n = 3).
KRAS Knockout Human Cancer Cells Are Sensitive to FAS-Mediated Apoptosis.
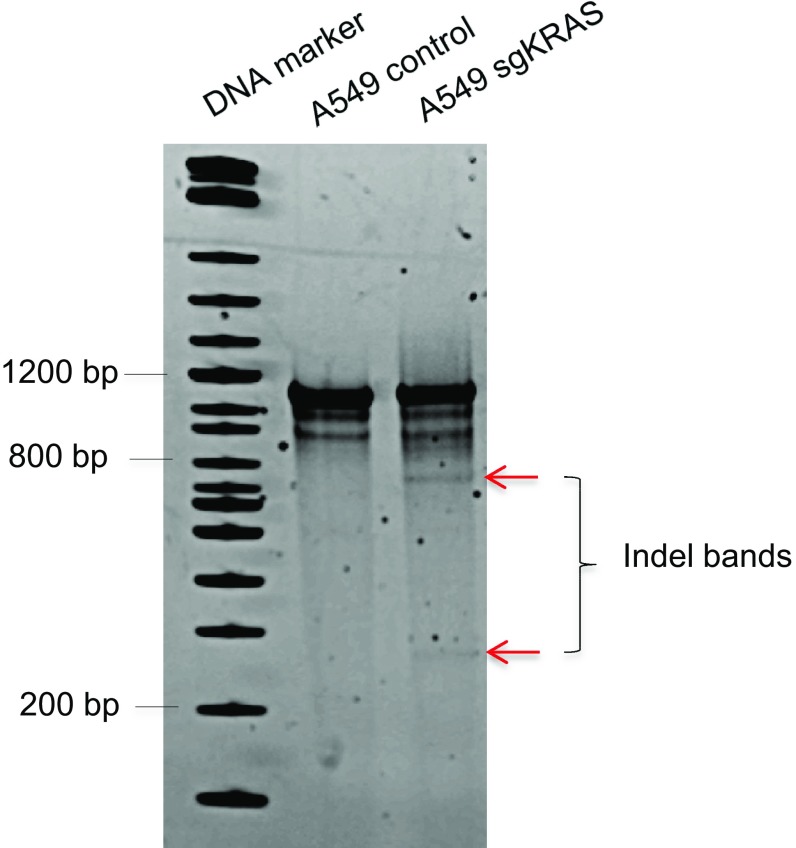
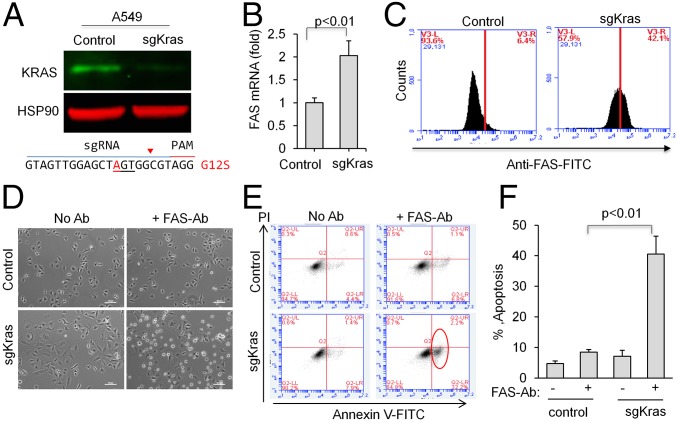
To assess whether CRISPR can edit KRAS in human NSCLC cells carrying KRAS mutations, we chose A549 human lung adenocarcinoma cells, which harbor a homozygous KRASG12S allele (41). Using similar approach as described above, A549 cells were infected with lentivirus expressing Cas9 and a guide RNA that targets human KRAS (sgKRAS). sgKRAS induced genome editing (Fig. S5) and significantly reduced total KRAS protein level in a cell population (Fig. 5A). Concordant with our results in mouse lung cancer cells, we found that some A549 cells survive KRAS knockout. Likewise, we also observed an increase in FAS mRNA (Fig. 5B) and receptor level (Fig. 5C) in sgKRAS-expressing A549 cells.
Fig. S5.
Surveyor nuclease assay demonstrated CRISPR-mediated KRAS genome editing in A549 cells. Arrows denote the indel bands.
Fig. 5.
KRAS knockout human NSCLC cells are sensitive to FAS-mediated apoptosis. (A) CRISPR-mediated KRAS editing in human NSCLC cell line A549 with a homozygous G12S mutation. Pooled cells expressing sgKRAS were analyzed by immunoblot to detect total KRAS protein level. HSP90 was used as a loading control. (B) qPCR measurement of FAS mRNA in control and sgRNA-expressing cells. (C) FACS histogram showing the levels of human FAS receptor in A549 control or sgKRAS cells. (D) Representative bright-field images showing floating apoptotic cells, in the presence of an activating antibody recognizing human FAS receptor. (E) Dot plot showing percentage of Annexin V- or PI-positive cells. Red circle denotes Annexin V-positive cells. (F) Quantification of E. Error bars represent SD (n = 3).
To explore methods to overcome resistance to KRAS knockout in human cells, we hypothesized that activation of FAS could also selectively induce apoptosis in KRAS knockout A549 cells. By incubating cells with an activating antibody for human FAS (clone EOS9.1), we observed that KRAS knockout cells are sensitive to FAS-mediated apoptosis (Fig. 5 D–F). These results indicate a potential strategy to overcome resistance for KRAS inhibition in human NSCLC by activating FAS.
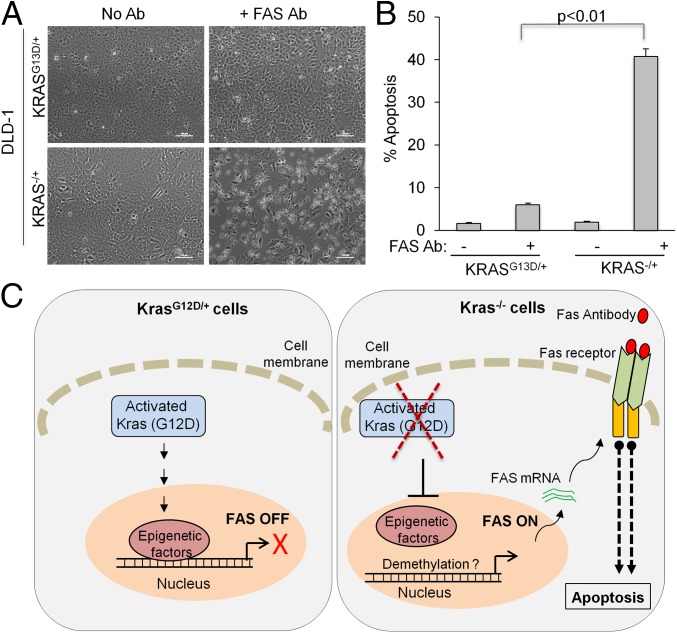
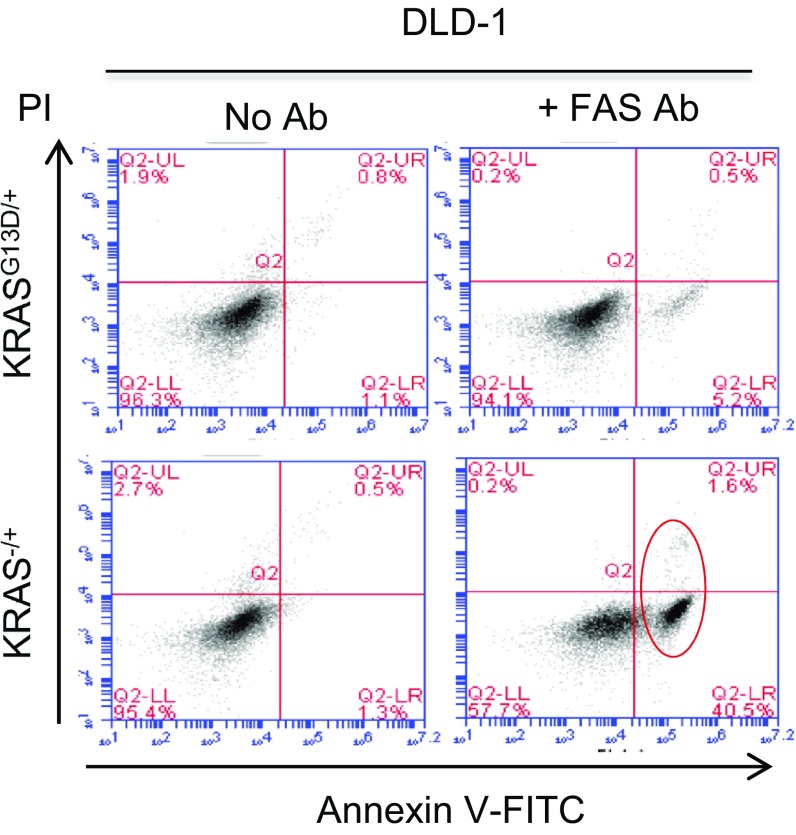
To ensure that the increased sensitivity to FAS activation is indeed a phenotype of CRISPR-mediated KRAS knockout, we used previously reported DLD1 colon cancer cells (KRASG13D/+ vs. KRAS−/+) generated by targeting G13D allele by traditional homologous recombination (42). As expected, KRAS−/+ DLD1 cells also exhibited increased sensitivity to FAS antibody-induced apoptosis. As shown in Fig. 6 A and B and Fig. S6, incubation of FAS-activating antibody with KRAS knockout (KRAS−/+) DLD1 cells, markedly increased Annexin V-positive apoptotic cells. This suggests that KRAS knockout leading to restored FAS expression is a general phenomenon, and that KRAS silencing confers sensitivity to FAS-induced apoptosis both in lung cancer cells edited by CRISPR and in colon cancer cells generated by traditional homologous recombination. Therefore, this conserved mechanism may provide a therapeutic strategy to suppress tumor relapse alongside future generation of KRAS inhibitors (Fig. 6C).
Fig. 6.
KRAS knockout DLD1 cells are sensitive to FAS-mediated apoptosis. (A) KRAS knockout (KRAS−/+) and parent KRASG13D/+ DLD1 cells were incubated with an activating antibody against human FAS. Representative bright-field images show floating/apoptotic KRAS−/+ cells. (B) Quantitation of apoptosis measured by Annexin V/PI staining and FACS analysis. Error bars are SD (n = 3). (C) A simplified model of how KRAS silencing may increase FAS expression and sensitize cells to FAS receptor-mediated apoptosis.
Fig. S6.
FACS analysis demonstrating Annexin V-positive apoptotic cells in FAS antibody-treated KRAS knockout (KRAS−/+) DLD1 compared with parental (KRASG13D/+) cells.
A Combination of HDAC Inhibitor and Fas Antibody Induces Apoptosis in Oncogenic Kras Cells.
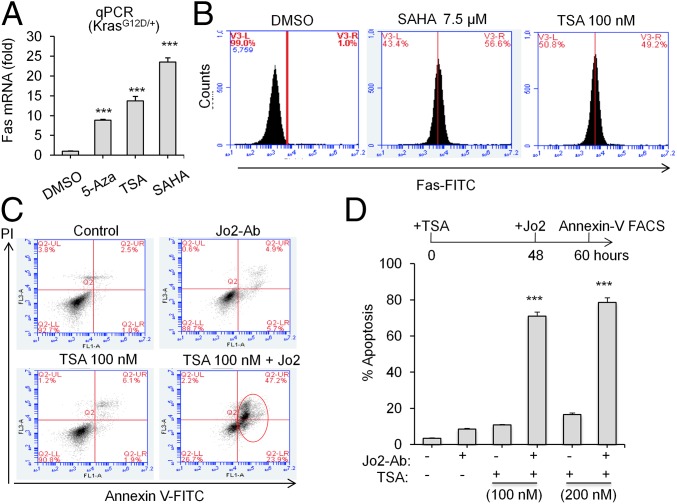
DNA methylation on CpG motifs in gene promoters is often accompanied by the recruitment of HDACs to the transcriptional regulatory site, thereby altering local chromatin structure and inhibiting transcription. General inhibitors of class I and II HDACs, alone or in combination with the methylation inhibitor 5-azacitydine, or its congener 5-aza-deoxycytidine (5Aza) are being studied in clinical trials for treatment of diverse types of tumors (43). Furthermore, HDAC inhibitors alone can induce promoter de-methylation and de-repression (44). Because Kras-directed epigenetic silencing of Fas also requires HDACs (45), we asked whether DNA methylation inhibitors 5Aza or HDAC inhibitors such as Trichostatin A (TSA) or suberoylanilide hydroxamic acid (SAHA) can restore Fas expression in KrasG12D/+ cells. All of these inhibitors significantly increased Fas mRNA expression in KrasG12D/+ mouse lung cancer cells (Fig. 7A). Furthermore, two pan-HDAC inhibitors TSA and SAHA both significantly up-regulated Fas receptor level on the cell surface (Fig. 7B). Finally, we observed that TSA or SAHA pretreatment sensitized KrasG12D/+ cells to Fas-mediated apoptosis in combination with Jo2 antibody (Fig. 7 C and D, and Fig. S7). These results suggest that exploiting the Fas activation as an Achilles’ heel in oncogenic Kras cells using HDAC inhibitors can potentially provide a complementary approach to Kras inhibitors.
Fig. 7.
A combination of HDAC inhibitor and Fas antibody induces apoptosis in oncogenic Kras cells. (A) qPCR showing reactivation of Fas in KrasG12D/+ mouse KP lung cancer cells treated with 5-Aza (5 μM), TSA (10 nM), and SAHA (10 μM) for 48 h. DMSO control is set to 1. (B) Representative FACS histogram showing Fas expression on cell surface in the presence of TSA or SAHA. (C) FACS dot plot showing induction of apoptosis by Jo2 antibody in KrasG12D/+ cells pretreated with TSA. (D) Quantitation of apoptosis measured by Annexin V/PI staining and FACS analysis. Error bars are SD (n = 3). ***P < 0.001.
Fig. S7.
The HDAC inhibitor SAHA sensitizes KrasG12D/+ cells to Fas-mediated apoptosis. (A) FACS analysis demonstrating Annexin V-positive apoptotic cells in presence of SAHA and Jo2. (B) Statistics of percentage apoptosis. Error bars are SD (n = 3). ***P < 0.001.
Discussion
Herein, we report CRISPR/Cas9-mediated genome editing of Kras in mouse and human lung cancer cells.
CRISPR Can Effectively Model Genetic Deletion of Oncogenes.
Traditional technologies have limited ability to model complete silencing of oncogenes because of the low efficiency of homologous recombination without DNA breaks (18, 22). The CRISPR system is flexible and provides a rapid and facile genetic system to functionally investigate mechanisms of both KRAS and other “undruggable” oncogenes in lung cancer pathology. Using CRISPR to model genetic disruption of oncogenes will pave the road for identifying therapeutic targets (22, 23).
Cancer Cells Can Escape KRAS Oncogene Addiction and Survive Independent of KRAS.
Cancer therapy can be improved by specifically modulating genes in resistance pathways. However, resistance to KRAS-targeted therapy is unknown due to the lack of effective small-molecule inhibitors for KRAS (16). Our data uncover KRAS independence upon complete KRAS knockout, which is consistent with our previous report that Kras mutant lung tumor can escape from RNAi-mediated Kras knockdown (16). Although KRAS knockout cells exhibited decreased colony formation ability, these cells are viable, implying mechanisms of resistance to KRAS inhibition or depletion. The recovery of tumors in the absence of Kras activity indicates potential resistance mechanisms to Ras inhibitors. Inhibiting KRAS alone, therefore, might be insufficient for treating KRAS-driven cancer in humans.
RNA-seq revealed a number of differentially expressed genes between oncogenic Kras and Kras knockout cells (Fig. 2). Understanding additional mechanisms of KRAS independence will be critical for tailoring treatment decisions in future generations of KRAS inhibitors being developed by the RAS initiative at the National Cancer Institute (3–5). Moreover, because we used KrasG12D/+;p53−/− mouse lung cancer cells, it remains unclear how additional genetic lesions (e.g., p53−/−) contribute to Kras independence. Future work is needed to investigate how Kras independence is affected by various genetic contexts.
Fas as an Achilles’ Heel in Kras-Independent Cells.
Because KRAS-independent cells are selectively sensitive to FAS-mediated apoptosis, understanding how KRAS regulates FAS might unveil treatment targets. Potential therapeutic options (such as siRNA) might exploit these targets to improve the efficacy of KRAS inhibitors. Moreover, a better understanding of the epigenetic regulation of FAS might allow us to turn FAS on and exploit it in other cancer types. Additional studies are also needed to determine whether Fas activation could mediate rejection of Ras-deficient tumor cells during early stages of lung tumor or organ development.
Tumor-Targeted Fas Activation Is Required in Vivo.
Our results suggest that a Fas-activating antibody can selectively induce apoptosis in Kras knockout cells. However, normal somatic cells also express Fas receptor. For example, hepatocytes are highly sensitive to FAS-mediated apoptosis; the systemic injection of high-dose Jo2 antibody induces apoptosis in the liver (28). Thus, FAS-activating antibody needs to be specifically delivered to tumor cells to avoid liver damage. Future studies will explore ways to tackle this challenge together with KRAS silencing. It may also be that endogenous FAS ligand excreted from tumor microenvironment can modulate apoptosis upon KRAS silencing in vivo.
In conclusion, our study has pinpointed FAS activation as a potential strategy to improve the efficacy of future KRAS inhibitors. Because KRAS mutations are prevalent in lung, colon, and pancreatic cancer and are associated with poor patient outcomes, these findings will be critical for developing effective ways to inhibit KRAS and prevent tumor relapse.
Materials and Methods
Vectors and Cloning.
sgRNAs targeting mouse KrasG12D or human KRAS were designed using Broad Institute online tool (https://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design). The following sgRNA sequences were used: mouse KrasG12D, 5′-GTGGTTGGAGCTGATGGCGT-3′, and human KRAS, 5′-GTAGTTGGAGCTGATGGCGT-3′. Oligos were annealed and cloned into the lenti.U6sgRNA.Cas9-2A-Puro vector using a standard BsmBI protocol. WZL-Hygro (Addgene; 18750) or WZL-HRAS-V12 (40) were gifts from Scott Lowe, Memorial Sloan Kettering Cancer Center, New York.
ChIP Assays.
ChIP assays were performed as previously described (26) using DNMT1 (Abcam), EZH2 (CST) and H3K27me3 (Millipore), and H3K4Ac (Abcam) antibodies. ChIP products were analyzed by qRT-PCR using Fas promoter primer sets (Tables S1 and S2) corresponding to transcription start site (TSS) for EZH2, H3K27me3, and ∼1 kb upstream of TSS for DNMT1. Samples were normalized to input DNA, results were analyzed using the ΔΔCt method, and fold enrichment at target site was calculated with respect to IgG control.
Table S1.
Primer sequences used in this study
| Assay | Gene | Primer sequence, 5′–3′ |
| Quantitative RT-PCR | M-Fas | Forward: GATGCACACTCTGCGATGAAG |
| Reverse: CAGTGTTCACAGCCAGGAGAAT | ||
| Hu-Fas | TaqMan probe: Hs00163653_m1 | |
| Surveyor assay | Hu-KRAS | Forward: GAAGTACAGTTCATTACGATACACGTCTGC |
| Reverse: TGTTGAGAAGAAGATAGGAAAATACTGCTG | ||
| ChIP assay | Fas-TSS (−234 to +20 bp) | Forward: GCCGCCTGTGCAGTGGTGA |
| Reverse: CTGTGTGTGGGCAGCCTGCGGC | ||
| Fas (∼1 kb) (−967 to −722) | Forward: GGCTATAGATCACCTTCATGTA | |
| Reverse: GCAGTTAACTCAGGGACCAAG | ||
| MeDIP assay | Fas-TSS | Forward: CAGCCCAGAGTAACTCACTTC |
| Reverse: CATACCCACAGGCAGTCTAGA | ||
| Fas (2.3–2.6 kb) | Forward: GAAGTAGAAACAGAAGCTGAG | |
| Reverse: TTGCTACATCCCAACTGTAAC |
Table S2.
Oligo ID numbers for shRNAs (Open Biosystem/Thermo Scientific)
| Gene | Oligo ID |
| Dnmt1 | V2MM_46797 |
| V2LMM_43170 | |
| Ezh2 | V2MM_35988 |
| V2MM_30422 |
Quantitative PCR.
Total RNA was purified using an RNeasy Mini Kit (74104; Qiagen). Reverse transcription was performed and diluted cDNA was used as template for real-time PCR. TaqMan probes or SYBG primers were used to measure expression of mouse or human Fas (Tables S1 and S2).
Immunoblot Analysis.
Cells were harvested and lysed in RIPA buffer with proteinase and phosphatase inhibitors. Cell lysate was quantified and equal amounts of protein were loaded into a 4–12% NuPage Bis-Tris gel (Life Technologies). The proteins isolated by the gel was then transferred to nitrocellulose membrane, blocked with blocking buffer (Odyssey), and then incubated with antibody against KRAS (sc-30; Santa Cruz) and Fas (Upstate; 05-351) overnight at 4 °C. An immunofluorescent secondary antibody (LICOR) was used for the Odyssey Imaging machine.
Statistical Analysis.
All quantitation data were collected from at least three independent experiments, and the difference between groups were determined using two-tailed Student’s t test, with P < 0.05 considered to be significant. All animal study protocols were approved by the University of Massachusetts Institutional Animal Care and Use Committee.
SI Materials and Methods
Deep Sequencing, RNA-Seq, and Bioinformatics Analysis.
Genomic regions flanking the Kras gene-editing site were PCR amplified using Herculase II high-fidelity polymerase and PCR purified using a QIAquick Gel Extraction Kit (46). RNA-seq was performed as described recently (16). Data were processed according to standard Illumina sequencing analysis procedures. For bioinformatics analysis, reads were trimmed, and PCR primers were removed using Trimmomatic (version 0.30). We then aligned reads to the mm10 genome using STAR (version 2.3.0e) with the default parameters and selected only for uniquely mapping reads. The redundant read pairs were removed using Samtools (version 0.0.19). Using Gencode M7 annotations, the number of reads per gene was calculated using HTSeq. The differential expression was determined using DESeq and accounted for possible batch effects using a generalized linear model. To be considered differentially expressed, the required gene's expression was considered to be greater than twofold and the adjusted P < 0.05. For additional analysis, we used all genes that were differentially expressed in the same direction between the KP1 and KP2 datasets.
Gene ontology analysis was performed using Panther database using a background of all mouse genes. We also converted our list of genes to their human homologs using The Jackson Laboratory's homolog master list (www.informatics.jax.org/downloads/reports/HOM_MouseHumanSequence.rpt). Human gene lists were analyzed using Gene Set Enrichment Analysis (GSEA). We investigated the following gene sets in the Molecular Signatures Database: Hallmark Gene Sets, Canonical Pathways, Chemical and Genetic Perturbations, and Oncogenic Signatures. We reported the top 20 results from each gene set.
TCGA LUAD Differential Expression Analysis.
The TCGA human lung adenocarcinoma (LUAD) dataset was downloaded via Firehose from the Broad Institute Genome Analysis Center (gdac.broadinstitute.org), which comprises patients’ mRNA expression data (RSEM value, i.e., normalized RNA-seq by Expectation- Maximization). We applied t.test to compare the gene expression of tumor and normal samples and plotted the boxplot of the key genes using R (version 3.2.2).
Cell Culture, Treatments, and Viral Infection.
KP-KrasG12D/+ (established from mouse lung tumors with a genetic background of KrasG12D/+;p53−/−), KP-Kras−/− (CRISPR-based Kras knockout cell clones), and A549 (KRASG12S or sgKRAS) cells were cultured with DMEM with 10% (vol/vol) FBS, supplemented with penicillin–streptomycin and glutamine in a 5% CO2 incubator. 293fs cells were used to package lentivirus containing sgRNA and Cas9. KP and A549 cells were infected with packaged lentivirus twice and selected with puromycin. Kras−/− cells were infected with WZL-HRAS-V12 or WZL-Hygro retrovirus packaged using amphotropic phoenix cells. For single-clone isolation, we seeded 1,000 cells onto 100-mm dish and cultured cells for 1 wk. The single-cell clones were picked manually using pipette tips under the microscope and subsequently transferred to a 12-well plate. Each of the individual clones was further grown and analyzed for by immunoblot for protein detection or genomic DNA isolation for sequencing. For pooled cells, the surviving puromycin-resistant cells were collected and subjected to an immunoblot analysis for protein detection and PCR-based deep sequencing to identify gene editing. Dnmt1 and Ezh2 knockdown was performed using respective shRNAs (Tables S1 and S2). Chemical inhibitors, 5Aza (Sigma), SAHA (Selleckchem), and TSA (Sigma) were used at indicated concentrations.
Fluorescence-Activated Cell Sorting.
To detect membrane-bound mouse Fas or human FAS receptor, KP or A549 cells were trypsinized, washed, and incubated with FITC- or PE-conjugated Fas (BD 554258) or FAS antibody (ebioscience; 11-0959-42) for 1 h at 4 °C. After incubation, cells were washed, resuspended, and analyzed by fluorescence-activated cell sorting (FACS). To detect Annexin V-positive apoptotic cells, KP, A549, or DLD1 cells were incubated with indicated activating antibodies against mouse Fas (Jo2; 500 ng/mL; BD 554254; 24 h) or human FAS (EOS9.1; 250 ng/mL; ebioscience; 16-0958-81; 10 h) plus cycloheximide (CHX) (2.5 μg/mL) to induce apoptosis. Isotype control antibody was used in negative controls. After incubation, the total cells (both floating and adherent) were collected, washed, and incubated with FITC-conjugated Annexin V and propidium iodide (PI). Stained cells were then analyzed by FACS to quantify the percentage of apoptotic cells.
MeDIP Assay.
Methylated DNA immunoprecipitation assays were performed on Fas promoter in KrasG12D/+ and Kras−/− cells. Genomic DNA was first fragmented by sonication, methylated DNA was immunoprecipitated using anti-5 methyl-cytosine (5mC) antibody (Abcam), and 5mC levels were analyzed using Fas promoter primer sets with qPCR (Tables S1 and S2). Ten percent of input DNA was used as control.
Surveyor Assay.
Genomic DNA was purified from cell lines using Quick Extraction Buffer. For the surveyor nuclease assay, PCR products were purified with a QIAquick Gel Extraction Kit and treated with the Surveyor nuclease kit (Transgenomic). Digested DNA was electrophoresed on a 4–20% Novex TBE Gel (Life Technologies) and stained with ethidium bromide.
Xenograft Assay.
For xenograft experiments, one million KrasG12D/+ or Kras−/− cells were s.c. injected into NCrnu/nu mice (Taconic) and monitored for tumors. All animal study protocols were approved by the University of Massachusetts Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank T. Jacks, P. Zamore, W. Hahn, E. Sontheimer, V. Ambros, N. Wajapeyee, P. Sharp, M. Moore, M. Muzumdar, H. Yin, D. Anderson, and C. Mello for their insightful comments. We thank D. Conte and S. Deibler for critically editing this manuscript. We thank Y. Liu and E. Kittler at the University of Massachusetts core facilities for histology and deep-sequencing support. This work was supported by grants from NIH (R00CA169512 and DP2HL137167), the Worcester Foundation, the American Cancer Society (129056-RSG-16-093), and the Lung Cancer Research Foundation (to W.X.). Y.L. was supported by the China Scholarship Council (Grant 201506260151) and the Thousand Talent Plan funding (to Z.W.) from the Chinese government. M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The deep-sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information BioProject database (accession no. PRJNA356881).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620861114/-/DCSupplemental.
References
- 1.Heist RS, Engelman JA. SnapShot: Non-small cell lung cancer. Cancer Cell. 2012;21(3):448.e2. doi: 10.1016/j.ccr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351(6273):604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patricelli MP, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6(3):316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 5.Athuluri-Divakar SK, et al. A small molecule RAS-mimetic disrupts RAS association with effector proteins to block signaling. Cell. 2016;165(3):643–655. doi: 10.1016/j.cell.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormick F. The potential of targeting Ras proteins in lung cancer. Expert Opin Ther Targets. 2015;19(4):451–454. doi: 10.1517/14728222.2014.1000304. [DOI] [PubMed] [Google Scholar]
- 7.McCormick F. KRAS as a therapeutic target. Clin Cancer Res. 2015;21(8):1797–1801. doi: 10.1158/1078-0432.CCR-14-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin L, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400(6743):468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 10.Fisher GH, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15(24):3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jechlinger M, Podsypanina K, Varmus H. Regulation of transgenes in three-dimensional cultures of primary mouse mammary cells demonstrates oncogene dependence and identifies cells that survive deinduction. Genes Dev. 2009;23(14):1677–1688. doi: 10.1101/gad.1801809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4(7):1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuveson DA, et al. Endogenous oncogenic K-ras G12D stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5(4):375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 14.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapoor A, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158(1):185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158(1):171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson L, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11(19):2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 19.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514(7522):380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516(7531):428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mou H, Kennedy Z, Anderson DG, Yin H, Xue W. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med. 2015;7(1):53. doi: 10.1186/s13073-015-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15(7):387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenton RG, Hixon JA, Wright PW, Brooks AD, Sayers TJ. Inhibition of Fas (CD95) expression and Fas-mediated apoptosis by oncogenic Ras. Cancer Res. 1998;58(15):3391–3400. [PubMed] [Google Scholar]
- 25.Peli J, et al. Oncogenic Ras inhibits Fas ligand-mediated apoptosis by downregulating the expression of Fas. EMBO J. 1999;18(7):1824–1831. doi: 10.1093/emboj/18.7.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wajapeyee N, Malonia SK, Palakurthy RK, Green MR. Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors. Genes Dev. 2013;27(20):2221–2226. doi: 10.1101/gad.227413.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449(7165):1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peter ME, et al. The role of CD95 and CD95 ligand in cancer. Cell Death Differ. 2015;22(5):885–886. doi: 10.1038/cdd.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villa-Morales M, Fernández-Piqueras J. Targeting the Fas/FasL signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(1):85–101. doi: 10.1517/14728222.2011.628937. [DOI] [PubMed] [Google Scholar]
- 30.Feldser DM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue W, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci USA. 2014;111(34):E3553–E3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue W, et al. Response and resistance to NF-κB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov. 2011;1(3):236–247. doi: 10.1158/2159-8290.CD-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue W, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22(11):1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai J, et al. FAT4 functions as a tumour suppressor in gastric cancer by modulating Wnt/β-catenin signalling. Br J Cancer. 2015;113(12):1720–1729. doi: 10.1038/bjc.2015.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemann MT, et al. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci USA. 2004;101(25):9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 40.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113(6):703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 41.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260(5104):85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 43.Gore SD, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66(12):6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 44.Sarkar S, et al. Histone deacetylase inhibitors reverse CpG methylation by regulating DNMT1 through ERK signaling. Anticancer Res. 2011;31(9):2723–2732. [PubMed] [Google Scholar]
- 45.Maecker HL, Yun Z, Maecker HT, Giaccia AJ. Epigenetic changes in tumor Fas levels determine immune escape and response to therapy. Cancer Cell. 2002;2(2):139–148. doi: 10.1016/s1535-6108(02)00095-8. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, et al. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol. 2015;16(1):111. doi: 10.1186/s13059-015-0680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.