Significance
Regulatory networks change during evolution. A protein that controls many genes in one species may control a different set of genes in another. We examined how mRNA networks evolve, focusing on the PUF (Pumilio and FBF) family of RNA-binding proteins. These govern stability and translation of hundreds of mRNAs and enable coordinate regulation of discrete biological outcomes. To understand how RNA networks evolve, we used knowledge of the RNA specificity of each PUF protein to predict its mRNA targets and directly identified mRNAs bound to each protein in divergent fungi via biochemical methods. We find networks controlled by one protein switch during evolution to be controlled by another and that proteins with different specificities can share, gain, or lose batteries of mRNAs.
Keywords: RNA regulation, PUF proteins, evolution, 3′UTR elements
Abstract
Alterations in regulatory networks contribute to evolutionary change. Transcriptional networks are reconfigured by changes in the binding specificity of transcription factors and their cognate sites. The evolution of RNA–protein regulatory networks is far less understood. The PUF (Pumilio and FBF) family of RNA regulatory proteins controls the translation, stability, and movements of hundreds of mRNAs in a single species. We probe the evolution of PUF–RNA networks by direct identification of the mRNAs bound to PUF proteins in budding and filamentous fungi and by computational analyses of orthologous RNAs from 62 fungal species. Our findings reveal that PUF proteins gain and lose mRNAs with related and emergent biological functions during evolution. We demonstrate at least two independent rewiring events for PUF3 orthologs, independent but convergent evolution of PUF4/5 binding specificity and the rewiring of the PUF4/5 regulons in different fungal lineages. These findings demonstrate plasticity in RNA regulatory networks and suggest ways in which their rewiring occurs.
Coordinated regulation of genes and mRNAs pervades biology. Cells simplify the challenge by coregulating groups of functionally related genes via the same regulatory proteins. In this way, cells establish “networks” consisting of a site-specific regulatory protein, the downstream targets it controls, and the recognition sequences in DNA or RNA that link each target to its regulator. The discrimination of specific sequences in DNA and RNA is a cornerstone of biological regulation in living systems, as these binding sites determine which genes will be controlled.
Transcriptional networks display a remarkable capacity to evolve, as evidenced by comparative genomic studies. Networks can change at the individual gene level, in which specific transcriptional targets are gained or lost via changes in transcription factor binding sites within genomic DNA (1–4). Over extended periods of evolutionary time, the entire set of targets of a regulatory protein can change, until this regulator effectively controls a distinct set of genes. Alternatively, coregulated genes can fall under the control of different regulators. In this case, each individual target acquires DNA sites that are recognized by the alternate regulator. Ribosomal protein genes in fungi are a striking example: Their coregulation has been maintained for over a billion years of fungal evolution despite regulation by completely different systems in different species (5–7). This type of wholesale rewiring requires concerted evolution across the suite of targets. Although the mechanisms through which transcriptional networks evolve are unclear, it has been proposed to involve a period of “redundant” regulation, during which the same group of targets is temporarily controlled by two different regulatory systems, followed by loss of the ancestral connections (6, 8, 9). Rewiring often correlates with duplication and divergence of the responsible regulator (10–13).
The evolution of RNA regulatory networks is less well understood. A single RNA-binding protein (RBP) often controls groups of RNAs with common biological functions (14–17) and so governs their stability, translation, and localization. RBPs in the same protein family exhibit similar though not identical RNA-binding specificities (18, 19). Differences in family members generate divergence in their regulatory networks. 3′UTRs, like introns, are particularly favorable targets for network divergence, as they are relatively unconstrained in length and sequence. Alternative polyadenylation events, in which a single pre-mRNA species can give rise to two or more mRNAs that either contain or lack regulatory sites, provide a mechanism of network divergence unique to mRNAs (20–22). Other molecular mechanisms for the evolution of binding elements and mRNA–protein regulatory networks exist but are largely opaque.
Here we focus on the evolution of RNA–protein networks. We concentrate on the PUF (Pumilio and FBF) family of RBPs, because they are exemplary regulators of cytoplasmic mRNAs and provide a strong foundation based on genetics, biochemistry, and structural biology. PUF proteins generally bind single-stranded RNA elements in the 3′UTR to trigger mRNA repression, activation, or decay and can influence localization as well (23–27). PUF family members are present in all eukaryotes and maintain stem cells and promote memory formation in metazoa (28, 29). RNA targets for the canonical Saccharomyces cerevisiae PUF proteins have been identified by immunoprecipitation methods, including RIP-chip, HITS-CLIP, and RNA tagging (14, 30–32). Each PUF protein binds a group of functionally related mRNAs, forming distinct networks that are each enriched for specific cellular processes. For example, of the 476 tagged RNAs bound to S. cerevisiae Puf3, 191 encode proteins that contribute to mitochondrial organization (32).
PUF proteins bind different RNAs due to variations in a conserved protein scaffold. The proteins consist of eight largely helical repeats (PUF repeats). Three amino acids within each repeat can potentially contact a single RNA base (23, 33). The sequence of the 5′-end of most PUF binding elements is UGUR (where R is U or C), and the 3′-end is AU or UA. Differences in RNA binding specificity among PUF proteins are due to changes in the curvature of a conserved, largely helical PUF protein scaffold, which imposes different limitations on the RNAs that can bind with high affinity (30). RNA nucleotides in the binding element contort to accommodate the protein scaffold (30, 34, 35). For example, in S. cerevisiae, Puf3 has high specificity for an 8-nucleotide (nt) PUF element, Puf4 instead recognizes a related 9-nt element, and Puf5 binds most tightly to 9-nt and 10-nt elements (14, 30, 34, 36). Each of their binding elements begins with UGUR and ends with UA. Their specificities diverge through the presence or absence of internal bases in the RNA binding element. The identities of these bases generally are unimportant.
Elegant computational studies of mRNAs across fungal species support a model in which the Puf3 regulon, which controls nuclear-encoded mitochondrial transcripts in S. cerevisiae, was rewired during evolution (37). Bioinformatics revealed that many of these mRNAs possessed 9-nt elements in filamentous fungi and 8-nt elements in S. cerevisiae (37, 38). However, PUF–RNA networks have not been identified experimentally in any fungal species besides S. cerevisiae.
To probe the evolution of RNA regulatory networks at a molecular level, we directly identified RNAs bound to multiple PUF proteins in budding and filamentous fungi. Our studies revealed multiple independent rewiring events in the Puf3, Puf4, and Puf5 networks during fungal evolution. We show that the binding specificities of PUF orthologs from the filamentous fungi Neurospora crassa and Aspergillus nidulans are similar to one another as well as to their S. cerevisiae counterparts. We find strong evidence for at least two independent rewiring events for Puf3 orthologs and demonstrate the independent but convergent evolution of Puf4/5 binding specificities and the rewiring of the Puf4/5 networks in different fungal lineages. Our findings demonstrate the plasticity of RNA regulatory networks, how these networks emerge, and ways in which they have been rewired.
Results
Pattern of PUF Binding Elements Suggests Rewiring.
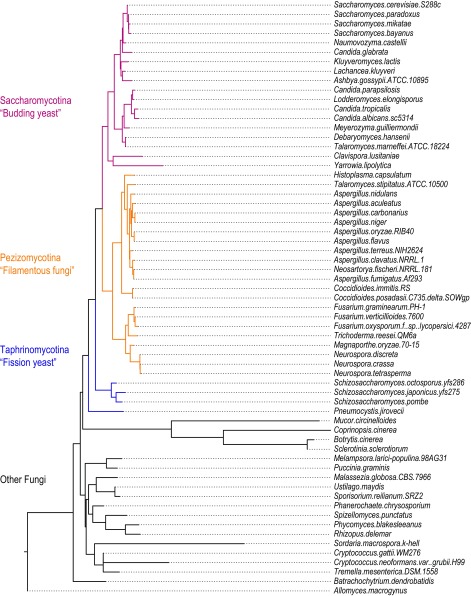
We identified matches to the known binding elements of S. cerevisiae PUF proteins in the 3′UTRs of orthologous RNAs in 62 species of fungi to investigate the evolution of binding sites across orthologous gene sets (Fig. 1A; full Phylogenetic tree in Fig. S1). The phylum represents at least 400 million years of evolution and includes species with strong foundations in genomics and functional information (39). We focused on species from two subphyla of Phylum Ascomycota: Saccharomycotina (budding yeasts) and Pezizomycotina (filamentous fungi) (40) (Fig. 1A). We considered in our analysis only mRNAs with a clear ortholog in at least 50% of species—that is, mRNAs that encode orthologus proteins—as defined previously (41, 42).
Fig. 1.
Conservation of putative PUF binding elements in Asycomocota fungi. (A) Phylogenetic tree of selected fungi (see full tree in Fig. S1). Colored branches represent each subphylum. Pink, Saccharomycotina (budding yeasts); orange, Pezizomycontina (filamentous fungi); blue, Taphrinomycontina (fission yeast), black, other fungi. (B) Representation of 8-, 9-, and 10-nt PWM models used to parameterize the log-likelihood function. Height of base represents probability of a base at each position in the binding element. (C–E) k-means clustering of orthologous transcripts based on log-likelihood scores for putative PUF binding elements in 3′UTRs for species in A, as shown in the key. Each row plots the log-likelihood PWM-match scores for an orthologous 3′UTR, and each column represents a species (with the phylogenetic tree shown above). Clustering was done independently for each heat map. Only clusters with a GO term P value lower than E-9 were highlighted (except cluster 2). Shown are the log-likelihood scores based on the 8-nt binding elements (C) found in one or more of 4425 orthologous transcripts. (D) The 9-nt binding elements found in 4,898 transcripts. (E) The 10-nt binding elements found in 4,423 transcripts.
Fig. S1.
Phylogenic tree for 62 species of fungi. Maximum likelihood tree was constructed using ribosomal RNA sequences (58).
We identified matches to PUF binding elements in the 3′UTRs of orthologous mRNAs, comparing against sequence motifs that correspond to the major binding elements of the canonical S. cerevisiae PUF proteins (14). Because the S. cerevisiae PUF proteins bind different lengths of sequence elements, each bearing a UGU at one end and a UA or AU at the other (see the Introduction), we searched separately for matches to the 8-, 9-, and 10-nt PUF position–weight matrices (PWMs) defined by previous biochemical analysis of S. cerevisiae Puf3 (Sc_Puf3), Puf4 (Sc_Puf4), and Puf5 (Sc_Puf5) (14, 30, 34, 36). For each sequence in each 3′UTR, we determined the log-likelihood ratio of the PWM model compared with a background genomic mononucleotide model (Fig. 1B). We assigned to each 3′UTR the best log-likelihood ratio identified for each PWM. For each PWM comparison, we then clustered groups of orthologous genes based on the assigned log-likelihood score (Fig. 1 C–E; gene ontology (GO) enrichments for each cluster are found in Table S1).
Table S1.
GO enrichments for each putative PUF binding element cluster
| Cluster no. | GO term | P value |
| 8-nt putative binding element | ||
| 0 | None | |
| 1 | None | |
| 2 | None | |
| 3 | None | |
| 4 | None | |
| 5 | None | |
| 6 | Vesicle-mediated transport | 5.63E-05 |
| Golgi vesicle transport | 0.025405 | |
| 7 | None | |
| 8 | None | |
| 9 | Mitochondrion organization | 1.19E-178 |
| Mitochondrial translation | 1.22E-102 | |
| 10 | None | |
| 11 | Cytoplasmic translation | 1.22E-12 |
| 12 | None | |
| 9-nt putative binding element | ||
| Single-organism process | 0.000581 | |
| 1 | Transmembrane transport | 1.46E-15 |
| 2 | None | |
| 3 | None | |
| 4 | Cytoplasmic translation | 1.32E-33 |
| Ribosomal small subunit biogenesis | 5.48E-08 | |
| 5 | Viral life cycle | 9.73E-42 |
| Symbiosis, encompassing mutualism | 9.73E-42 | |
| Transposition, RNA-mediated | 2.23E-34 | |
| Transposition | 3.45E-33 | |
| 6 | None | |
| 7 | Meiotic cell cycle | 0.001215 |
| Sporulation | 0.008913 | |
| 8 | Regulation of pheromone-dependent | 0.008338 |
| Regulation of signal transduction | 0.008338 | |
| 9 | None | |
| 10 | None | |
| 11 | Ribosome biogenesis | 1.10E-35 |
| Ribonucleoprotein complex biogenesis | 1.08E-32 | |
| rRNA processing | 7.17E-24 | |
| 12 | None | |
| 10-nt putative binding element | ||
| 0 | Chromatin modification | 1.84E-11 |
| Chromatin organization | 3.85E-10 | |
| 1 | None | |
| 2 | None | |
| 3 | None | |
| 4 | Chromosome organization | 6.94E-04 |
| Chromatin organization | 1.12E-03 | |
| 5 | None | |
| 6 | None | |
| 7 | Cytoplasmic translation | 2.28E-06 |
| Anion transport | 3.61E-04 | |
| Nitrogen compound transport | 3.98E-04 | |
| 8 | Positive regulation of Rho GTPase activity | 2.58E-03 |
| 9 | None | |
| 10 | Mitochondrial translation | 2.95E-05 |
| 11 | None | |
| 12 | None |
Striking patterns of conservation and evolution of PUF binding-sequence enrichment emerged across groups of orthologous genes. However, we detected no clear clusters for which enrichment of a single PUF PWM was conserved in all fungi. Many clusters exhibit functional relatedness. A few appear to lack it [e.g., cluster 2 in the Eurotiomycetes (Aspergilli)] (Fig. 1C).
In Saccharomycotina, the 8-nt cluster 1 and the 9-nt cluster 5 have GO term enrichments for nuclear-encoded “mitochondrion” (P value 1.4 E-96) and “ribosome biogenesis” (P value 4.0E-21), respectively, as noted previously (14, 37, 38). Strikingly, the enrichment for similar-length PUF binding elements was not conserved across other fungi: The enrichment for 8-nt-long sequences in cluster 1 was only seen for budding yeasts, whereas enrichment for 9-nt elements in clusters 5 were observed for sublineages in filamentous fungi. Clusters of conservation restricted to the Pezizomycotina include 9-nt clusters 3 and 4 and 10-nt cluster 6 (Fig. 1 D and E). All three of these clusters were enriched for mitochondrial function, but notably, none was enriched for the 8-nt PUF sequences known to mediate their control in S. cerevisiae. Interestingly, 9-nt cluster 3 was enriched for genes for both mitochondrion (3.1E-51) and ribosomal protein (4.1E-9). (A complete list of GO enrichments for each cluster is in Table S1.)
Predicted Switch Between Budding and Filamentous Fungi.
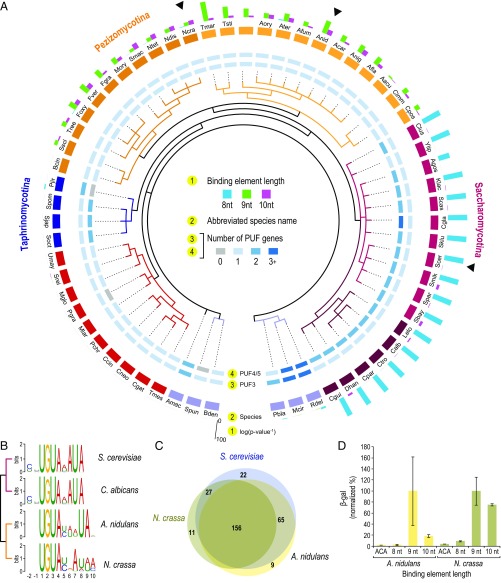
The 309 transcripts in cluster 1 (Fig. 1C) are highly enriched for 8-nt elements, consistent with their binding to Sc_PUF3 (14, 32). We refer to these transcripts as the “mitochondrial cluster.” In filamentous fungi, other clusters are enriched for mitochondria-related functions but contain transcripts with putative 9-nt or 10-nt binding elements (Fig. 1 D and E, clusters 3, 4, and 6). Our data are consistent with bioinformatic analyses that suggested a “rewiring” event for this group (37, 38). They further suggest widespread evolution of enriched sequences among transcripts in the mitochondrial cluster. We determined the enrichment of each length of putative binding element for transcripts in the mitochondrial cluster across 62 fungi (Fig. 2A, ring 1). All of the budding yeasts exhibit significant enrichment of 8-nt sequences in their 3′UTRs but not of 9-nt and 10-nt elements. Conversely, none of the filamentous fungi have significant enrichment for the 8-nt site. Instead, the filamentous fungi exhibit enrichment for putative binding elements 9 or 10 nt long (Fig. 2A, ring 1). No other clade showed a significant enrichment of putative binding elements in these orthologs. These results were supported by unbiased motif discovery in the groups’ 3′UTRs (43), which identified PWMs matching the 9- and 10-nt binding elements seen in A. nidulans and N. crassa transcripts orthologous to the mitochondrial cluster (Fig. 2B). Most of the filamentous fungi transcripts orthologous to the mitochondrial cluster have a PUF binding element, as is the case in S. cerevisiae (Fig. 2C). Thus, putative PUF binding elements in the mitochondrial cluster extend to filamentous fungi but have diverged so that they are of a different length, including 10-nt elements not previously detected.
Fig. 2.
Evolution of PUF–mitochondrial RNA network. (A) Enrichment of PUF binding elements in transcripts from the mitochondrial cluster across Ascomycota fungi. Abbreviations used for fungal species are provided in Fig. S5. Ring 1 represents binding element enrichment orthologs in each species. The inner ring is the phylogenic species tree. Black arrows mark the species used in C. (B) MEME-derived PWMs identified in 3′UTRs of mitochondrial cluster transcripts in each species. (C) Overlap of genes in mitochondrial cluster containing a putative PUF binding element for three species. (D) Relative luminescence values as a proxy for binding affinity for each PUF protein binding to each PUF site as assayed by yeast-three hybrid assays (48).
Fig. S5.
Abbreviations used for fungal species. Full species names were abbreviated as shown in Fig. 2.
Evolution of Putative Binding Elements Despite Conservation of PUF Binding Specificities.
Two models could explain the differences in binding site lengths: Transcripts in the mitochondrial cluster could be regulated by the same PUF ortholog whose binding specificity changed during evolution; alternatively, the sequence of the binding elements could have changed, while the protein specificity did not. To address this, we investigated the specificities of PUF proteins between species. All proteins containing a Pumilio domain for all 62 species were aligned, and then a protein tree was generated by maximum likelihood (Fig. S2) (44–46).
Fig. S2.
Phylogenic tree of PUF proteins. Maximum likelihood tree was constructed using all proteins predicted to contain a Pumilo binding domain (45).
Nearly all species have a single ortholog of Puf3 (Fig. 2A, ring 3). In contrast, Saccharomycotina and Schizosaccharomycetes have two paralogs that belong to a Puf4/Puf5 clade, whereas Pezizomycotina and other fungi have only one copy of the Puf4/5 protein (Fig. 2A, ring 4). This suggests there was one ancestral gene, which we refer to as PUF4/5, that was duplicated in the Saccharomycotina (37) and then again in the Schizosaccharomycetes. This is particularly intriguing because S. cerevisiae Puf4 and S. cerevisiae Puf5 have different binding specificities, suggesting that their binding preferences diverged after duplication (14, 30).
To distinguish between changes in protein specificity or binding site sequence, we assayed the RNA binding specificities of A. nidulans PUF4/5 (An_PUF4/5; AN10071) and N. crassa PUF-4/5 (Nc_PUF4/5; NCU16560.1) using a series of RNAs with binding elements ranging in length from 8 to 10 nt, using the three-hybrid system (47). In this assay, levels of LacZ expression are directly related to binding affinity (48). A. nidulans PUF4/5 bound best to sequences matching the 9-nt PUF binding element, similar to the known binding preference of Sc_Puf4, but also bound weakly to a 10-nt element (Fig. 2D). In contrast, N. crassa PUF-4/5 (Nc_PUF-4/5) bound well to both 9- and 10-nt-long binding elements (Fig. 2D). The broader specificity of Nc_PUF-4/5 echoes the expanded binding specificity of Sc_Puf5 (30). Together, these data suggest that the ancestral PUF4/5 protein could bind both 9-nt and 10-nt elements and that the known specificities for the S. cerevisiae proteins emerged after the PUF4/5 duplication (Discussion).
The binding specificities of the filamentous fungi PUF4/5 orthologs and the enrichment for putative 9- and 10-nt elements in the orthologs of the mitochondrial cluster suggest that there was a “regulator switch,” in which A. nidulans and N. crassa underwent a change in the protein that mediates control of orthologous mitochondrial RNAs. In this hypothesis, mRNAs that possess 8-nt elements in S. cerevisiae, which are recognized by Sc_Puf3, instead possess 9- or 10-nt elements in Neurospora and Aspergillus and are likely bound by PUF4/5 in those species.
In Vivo Targets of S. cerevisiae Puf3 and N. crassa PUF4/5 Demonstrate a Regulatory Switch.
To test the regulator switch hypothesis, we identified mRNAs bound in vivo to Sc_Puf3, N. crassa PUF3 (Nc_PUF3), and Nc_PUF4/5. We cross-linked RNA–protein complexes in vivo with UV irradiation of intact cells, immunopurified these protein–RNA complexes, and identified the RNAs by high throughput sequencing (“HITS-CLIP”) (49). We recently analyzed Sc_Puf5 using the same methods and included those data in the meta-analysis (30). The S. cerevisiae PUF genes were TAP-tagged and the N. crassa PUF genes were FLAG-tagged; all genes were integrated by homologous recombination at the endogenous chromosomal locus under control of the natural promoter. The three biological replicates for each protein displayed excellent reproducibility (Fig. 3 A–C).
Fig. 3.
HITS-CLIP data. (A–C) Reproducibility of biological replicates. Height of CLIP peaks were log-transformed and then plotted on each axis. Data were colored based on the precipitated protein (Sc_PUF3, blue; Nc_PUF3, red; Nc_PUF4/5, green). Spearman’s correlation coefficients (ρ), associated P values (P), and Pearson’s correlation coefficient (r) are indicated in A (ρ = 0.69, 0.84, 0.64; P = 0, 0, 0; r = 0.77, 0.96, 0.81; n = 500), B (ρ = 0.64, 0.85, 0.76; P = 0, 0, 0; r = 0.99, 0.99, 0.99; n = 835), and C (ρ = 0.85, 0.92, 0.79; P = 0, 0, 0; r = 0.71, 0.97, 0.67; n = 746). (D–G) Examples of binding peaks (read depth) for COX17, a canonical target of Sc_PUF3. YLL009C (COX17), NCU0058 (al-2), and NCU02530 (cox17) are depicted, with ORFs annotated in blue and the likely binding elements displayed beneath. (H) Overlapping CLIP targets for each protein. Only genes with orthologs in S. cerevisiae and N. crassa are included. RPM, reads per million mapped reads.
We identified 500, 835, and 746 significant CLIP peaks for Sc_Puf3, Nc_Puf3, and Nc_PUF4/5, respectively. These arise from 7.6, 8.4, and 14.3 million uniquely assignable reads (for complete mapping statistics, Table S2). Data quality in HITS-CLIP is often assessed by the presence of binding elements in peaks of read enrichment (50–52). In our data, 90%, 68%, and 99% of Sc_Puf3, Nc_PUF3, and Nc_PUF4/5 binding peaks, respectively, contain one or more sequences matching the PWMs derived for each set of targets, suggesting our sequencing data identified genuine PUF protein targets.
Table S2.
Summary of high throughput sequencing reads
| Internal ID | Sample ID | Raw reads | Reads after dup removal | % remaining | Reads after clipping | Mapped, unique | % mapped | Notes |
| DW115 | Sc Puf3 A | 19,151,614 | 7,735,761 | 40 | 6,723,006 | 5,260,964 | 78 | |
| DW116 | Sc Puf3 B | 14,524,322 | 5,782,164 | 40 | 3,161,397 | 1,709,752 | 54 | |
| DW125 | Sc Puf3 C | 7,626,399 | 3,099,312 | 41 | 1,557,163 | 634,971 | 41 | |
| DW100_102 | Nc Puf3 A | 1,179,860 | 456,082 | 39 | 402,543 | 354,391 | 88 | * |
| DW111 | Nc Puf3 B | 14,899,440 | 11,974,763 | 80 | 7,856,464 | 5,162,488 | 66 | |
| DW112 | Nc Puf3 C | 12,072,745 | 9,770,367 | 81 | 5,285,092 | 2,843,762 | 54 | |
| DW101_103 | Nc Puf5 A | 1,355,794 | 1,096,425 | 81 | 983,281 | 877,507 | 89 | * |
| DW113 | Nc Puf5 B | 18,748,631 | 14,990,087 | 80 | 12,394,324 | 10,541,429 | 85 | |
| DW114 | Nc Puf5 C | 11,393,103 | 9,959,443 | 87 | 5,274,058 | 2,918,173 | 55 |
Libraries were sequenced on Illumina MiSeq.
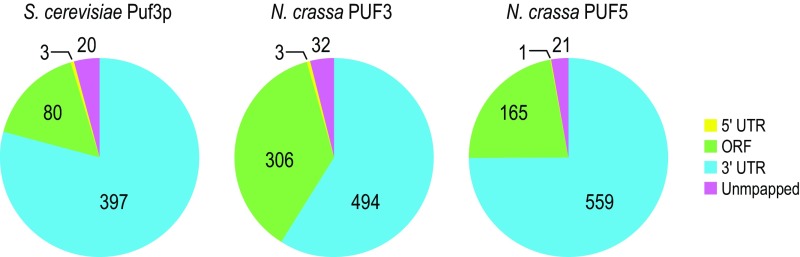
The peaks, predominantly found in the 3′UTRs of mRNAs, represent 465, 803, and 686 transcripts bound by Sc_Puf3, Nc_PUF3, and Nc_PUF4/5, respectively (Fig. S3 and Dataset S1). Representative CLIP peaks are shown in Fig. 3 D–G for COX17 orthologs, a well-characterized target of Sc_Puf3 (Fig. 3D) that is not efficiently bound by Sc_Puf5 (Fig. 3E). Strikingly, the Neurospora COX17 ortholog was bound by Nc_PUF5 but not Nc_PUF3 (Fig. 3 F and G). In all cases, binding peaks were centered over the known binding elements in the S. cerevisiae COX17 mRNA or the two predicted elements in the N. crassa ortholog. This trend was seen for the profiles of the five loci with the largest binding signal in the N. crassa and S. cerevisiae Puf3 and PUF4/5 proteins (Fig. S4 A–C).
Fig. S3.
Distribution of peaks in RNAs. Shown are mRNA regions where peaks are found for Sc_PUF3, Nc_PUF3, and Nc_PUF4/5. Sc_PUF5 from previous study was included for comparison (30).
Fig. S4.
Example HITS-CLIP peaks. Shown are five example HITS-CLIP peaks from each PUF protein. (A) Sc_PUF3, (B) Nc_PUF3, and (C) Nc_PUF4/5. Each line represents a separate biological replicate. Likely PUF binding element found in each peak is below the gene cartoon.
To assess whether targets were shared between Sc_Puf3 and N. crassa PUFs, we determined their overlap across those species (Fig. 3H). The overlap in transcripts bound by Sc_Puf3 and Nc_PUF4/5 was much more significant (6.1E-41, hypergeometric distribution test) than the overlap between Sc_Puf3 and Nc_PUF3 (5.0E-3, hypergeometric distribution test) (Fig. 3H): Orthologs of 42% of Sc_Puf3 targets were bound by Nc_PUF-4/5, whereas only 17% were bound by Nc_PUF3. We conclude that Sc_Puf3 and Nc_PUF4/5 proteins bind a similar set of RNAs, even more than do the two PUF3 orthologs.
PUF Binding Specificities Across Species.
To determine the in vivo sequence preference of each PUF protein, we identified PWMs based on overrepresented sequences in targets from the HITS-CLIP dataset (43). The binding preferences of the N. crassa PUFs were strikingly similar to those of their orthologous proteins in S. cerevisiae (Fig. 4 A and C). Sc_Puf3-bound sites contained sequences matching the known 8-nt binding element comprised of a 5′ UGU and a 3′ AUA (Fig. 4A). In addition to the core PUF binding element, ∼50% of the sequences associated with Sc_Puf3 contain a cytosine at the minus 2 position, which enhances binding in vitro and biological activity in vivo (32, 36). Nc_PUF3-bound regions contained 8-nt sequences similar to the known Sc_Puf3 binding element, but the PWM lacked the –2C enrichment seen in S. cerevisiae (Fig. 4B) (14, 32, 36). The PWM identified for targets of Sc_Puf5 contains the canonical 5′ UGUA with apparent degeneracy in the 3′-end; we previously showed that the single matrix includes discrete 8–12-nt-long binding elements that differ in spacer length (30). When training a single PWM on Nc_PUF4/5 targets, we identify the same type of apparent degeneracy (Fig. 4D) that can be explained as a combination of 9-nt- and 10-nt-long elements (Fig. 4E). The PWMs derived from in vivo targets agree well with our bioinformatic predictions of sequence enrichment (Fig. 2B) and the binding specificities measured ex vivo (Fig. 2D). We conclude that the in vivo binding preferences of PUF3 orthologs is highly conserved and that the binding preference of the single Nc_PUF4/5 is similar to that of Sc_Puf5.
Fig. 4.
In vivo binding elements of PUF proteins. MEME-derived PWMs from CLIP data. (A) S. cerevisiae PUF3, (B) N. crassa PUF3, (C) S. cerevisiae PUF5 (30), (D) N. crassa PUF4/5. (D) The previously defined Par-CLIP of Homo sapiens PUF3 ortholog, PUM2, defined by PhyloGibbs (52, 59). (E) Deconvolution of Nc_PUF4/5 composite binding element (D) into binding elements of 9 nt and 10 nt in length. (F) Mitochondrion GO term enrichment for all mRNAs targets of each PUF protein. (G) Venn diagram depicting the overlap between PUF targets from the mitochondrial cluster. (H) Nc_PUF3 and Nc_PUF4/5 interaction peaks identify separate binding elements for each PUF protein. One biological replicate for Nc_PUF3 and Nc_PUF4/5 are plotted in red and brown, respectively. Likely PUF binding elements are highlighted. RPM, reads per million mapped reads.
Evolution of PUF Target Networks in N. crassa.
To determine the functional enrichment for the targets of each PUF protein, we performed GO analyses. As expected, the Sc_Puf3 targets exhibited a strong enrichment for nuclear-encoded mitochondrial proteins (P value 1E-103) and translation factors (P value 6.5E-37) (Fig. 4F and Dataset S2). The targets of Nc_PUF4/5 were dramatically enriched for those encoding nuclear-encoded mitochondrial proteins (P value 1E-25); Nc_PUF3 targets also were enriched for this annotation, although much more modestly (P value 1E-5) (Fig. 4F and Dataset S2). In agreement with these enrichments, we found that 50% (155/309) of orthologous transcripts in the mitochondrial cluster (Fig. 1C) were bound by Sc_Puf3 in S. cerevisiae, whereas 44% (137/309) were bound by Nc_PUF4/5 and 13% (39/309) by Nc_PUF3 in N. crassa (Fig. 4G). Interestingly, of the 39 targets of Nc_PUF3 in the mitochondrial cluster, 71% were also bound by Nc_PUF4/5 (Fig. 4G). These transcripts often contained both 8-nt- and 9/10-nt-long putative PUF binding elements. Two examples of cotargets of Nc_PUF-3 and Nc_PUF-4/5 are transcripts CBP3 and NCU07386, which harbor both binding elements, one under each respective peak (Fig. 4H). Thus, a subset of mitochondrial-related transcripts are bound by both PUF4/5 and PUF3 through their respective recognition elements. Nc_PUF4/5 targets were enriched for other functional annotations beyond mitochondrial transcripts. The group of targets was also enriched for those encoding cytosolic ribosomal proteins (P value 3E-16), which is intriguing, as previously identified Sc_Puf4 targets are enriched for transcripts encoding ribosome biogenesis proteins (14) (Discussion).
Evidence for Multiple Rewiring Events and the Emergence of New Networks.
Our data suggest that orthologous transcripts have evolved different lengths of PUF binding elements in their 3′UTRs and hence are regulated by different PUF proteins. To further investigate this, we assembled a combined set of all orthologous transcripts bound by Sc_Puf3, Nc_PUF3, and Nc_PUF4/5, or Sc_Puf4 (identified in a previous study by a different method) (14). We clustered this combined set of transcripts based on putative PUF binding elements identified in 3′UTRs of each species’ orthologs across 62 fungal species (Fig. 1A). This analysis captured differential enrichment of putative PUF elements in the orthologous mRNAs, all of which are targets identified in our CLIP studies.
Several striking patterns emerged that implicate additional PUF regulatory rewiring and the emergence of networks. As reported above, transcripts encoding mitochondrial proteins (Fig. 5, cluster A) are enriched for 8-nt elements in S. cerevisiae and, to some extent in N. crassa, but are strongly enriched for 9-nt and 10-nt elements in filamentous fungi. Interestingly, many transcripts linked to mitochondrial envelope and organization (Fig. 5, cluster B) were specifically depleted of PUF binding elements in the Neurospora clade. In contrast, cluster C (Fig. 5) was bound by Nc_PUF3 and enriched for the 8-nt PUF element—strikingly, this group was enriched for transcripts encoding hydrolases and cellulose-binding proteins, functions that may be important for the optimal growth of this filamentous fungus. This cluster may represent the emergence of a new network in that species. Cluster D reveals ribosomal protein mRNAs with 9-nt elements in Neurospora but not in S. cerevisiae. Cluster E reveals mRNAs with 9-nt elements in S. cerevisiae (and recognized by its Puf4 and Puf5 proteins), which again may imply the emergence of a new network during evolution. Finally, we observe an enrichment of PUF targets in Neurospora that are responsive to light (Nc_PUF3: 233, P value 1.7E-13, Nc_PUF4/5: 257, P value 1.1E-22; hypergeometric distribution test). Thus, this regulatory system is also impacted by PUF regulation.
Fig. 5.
Evolution of PUF elements in PUF-bound transcripts. (A–E) k-means clusters. Orthologs of transcripts bound by Sc_PUF3, Sc_PUF4 (14), Nc_PUF3, or Nc_PUF4/5 were aligned and clustered based on the log-likelihood match of 3′UTR sequences to the 8-nt, 9-nt, and 10-nt PUF PWMs (Materials and Methods). Gray indicates a lack of ortholog in that species. Targets bound by each protein are annotated by colored bars to the left of the figure. Enriched GO terms for each cluster are shown to the right (S. cerevisiae in black and N. crassa in tan).
To visualize the overlaps, we depicted the connection between every orthologous target and its cognate binding protein(s) (Fig. 6). Each target of the four proteins (Sc_Puf3, Sc_Puf5, Nc_PUF3, or Nc_PUF4/5) is represented as a node (small circle). Many of the transcripts with mitochondrial functions are bound by Sc_Puf3 and Nc_PUF4/5 in the respective species, whereas others are uniquely associated with one protein or another (Fig. 6A). The fact that many connections are unique, but with targets that encode proteins in the same biological process, implies that PUF regulation of this biological function is conserved, whereas the specific mRNAs that are regulated diverged (Fig. 6 B–D).
Fig. 6.
Functional relatedness of all mRNAs bound by PUF3 and PUF4/5 proteins of S. cerevisiae and N. crassa. All orthologous targets of PUF proteins defined by CLIP are plotted as nodes (gray balls). Edges (lines) link the PUF to the RNAs it binds. Targets are colored based on their annotated function: (A) mitochondria (purple), (B) ribosome (red), (C) hydrolase (black), and (D) light responsive (blue).
The Emergence of New Sites Underlies Rewiring.
Two evolutionary routes could produce an apparent switch from 8-nt PUF3 regulation to 9/10-nt PUF4/5 regulation. An element of one length could change into a different length simply by gain or loss of nucleotides within the element; such changes are particularly facile, as the identity of bases in the central region of the site is unimportant in binding (see the Introduction). Alternatively, a new binding element could arise elsewhere in the same 3′UTR before the original motif degenerates. We find a preponderance of evidence for the latter model. Nc_PUF4/5 targets in the mitochondrial cluster are significantly enriched for transcripts with multiple binding elements per mRNA UTR (average of 2.04 elements per 3′ UTR) compared with all Nc_PUF4/5 targets (average of 1.67) (P value 6.5E-4, two-tailed t test). Furthermore, transcripts within a species can be bound by both proteins, as previously detailed for the cotargeting of CBP3 and NCU07386 by Nc_PUF-3 and Nc_PUF-4/5 (Fig. 4H). Although element-length switching likely exists, the presence of targets with two sites in a single 3′UTR suggests a transient intermediate may often exist in which both proteins act on the same mRNAs.
Discussion
Our work provides compelling experimental support for extensive rewiring of PUF-dependent regulatory networks in fungi and insights into how that occurred. Computational analyses prompted models of the rewiring of the nuclear-encoded mitochondrial network (5, 37, 38). Our biochemical data provide molecular evidence for such a regulator switch, reveal multiple rewiring events, and support fluiditiy in the emergence of new PUF–RNA networks.
The most parsimonious model for the evolution of the mitochondrial networks posits that mitochondrial transcripts (along with cytosolic ribosomal protein mRNAs and other transcripts) were regulated by both PUF3 and PUF4/5 in the common ancestor (Fig. 7). The relative prominence of PUF3 versus PUF4/5 control of mitochondrial transcripts evolved in the modern-day lineages, with PUF3 playing the predominant role in Sacharamycotina and PUF4/5 the major role in filamentous fungi. Orthologous PUF3 proteins have stringent specificity for 8-nt-long elements in virtually every organism tested to date, including mammalian PUM (14, 30, 52, 53). We show that, in contrast, Nc_PUF-4/5 and An_PUF4/5 can bind 9 nt, and even 10 nt in the case of Nc_PUF4/5 long binding elements (Fig. 2D), as does Sc_Puf5 (30). Together, this suggests that the ancestral mitochondrial transcripts were regulated by PUF3 and PUF4/5 bound to their 8-nt and 9/10-nt 3′ UTR elements, respectively.
Fig. 7.
Evolutionary model of PUF network rewiring. Colored boxes represent orthologous sets of mRNA targets. Solid arrows indicate PUF binding of the denoted binding element enriched in the target mRNA UTRs; dashed arrows represent weak binding or secondary modes of regulation (Discussion for details). Dashed colored boxes represent the acquisition or loss of transcripts from the denoted regulon. Mt, mitochondrial; Ribo, ribosomal.
Several events produced specialization of the PUF networks along the branch leading to budding yeasts. Our phylogenetic analysis indicates that PUF4/5 duplicated after the split of budding and filamentous fungi and then diverged in binding preference; broad binding specificity was likely ancestral (37). We propose that Sc_Puf5 retained the ancestral specificity whereas Sc_Puf4 specialized to bind a 9-nt element. These changes likely involved alterations in the topology of the PUF scaffold (30). As protein binding specificity evolved, so too did the elements of the downstream targets: Mitochondrial transcripts bound by Sc_Puf3 evolved toward 8-nt elements, whereas transcripts bound by Sc_Puf4 and Sc_Puf5 evolved 9-nt or maintained mixed-length PUF sequences, respectively. It is intriguing that Sc_Puf4 targets are enriched for ribosome-biogenesis transcripts, which are functionally related to the ribosomal protein transcripts bound by the orthologous Nc_PUF4/5 in N. crassa. We propose that the ancestral PUF4/5 protein regulated both mitochondrial and cytosolic ribosome-biogenesis functions but that this coregulation was lost in budding yeasts when the two networks specialized toward PUF3- and PUF4-dependent control, respectively. This may have facilitated a further decoupling of mitochondrial and cytosolic ribosomal protein gene expression in respiro-fermentative yeasts, as previously proposed to have occurred through upstream transcription factor rewiring (54, 55). The acquisition of sugar-responsive PUF3 regulation could have provided selective pressure to evolve and maintain the PUF3 network in respiro-fermentative yeasts. Sc_Puf3 is extensively phosphorylated when S. cerevisiae is depleted of glucose and switches from a repressor to an activator of mitochondrial mRNAs (24). Acquisition of regulated activation via phosphorylation may have committed the budding yeast lineage to using Puf3 to control mitochondrial RNAs.
A different story emerges for the evolutionary trajectory in the filamentous fungi. Our experimental data demonstrate PUF rewiring of the mitochondrial targets. Orthologs of Sc_Puf3-bound targets display strong enrichment for 9-nt or 10-nt sites in filamentous species. We show here that these orthologs of Sc_Puf3 targets are bound by Nc_PUF4/5 in N. crassa and to a lesser extent by Nc_PUF3. We find one set of transcripts that is strongly enriched for 9-nt PUF elements in the Neurospora species and, to a lesser extent, in the budding yeasts; however, orthologs in the Aspergilli are enriched primarily for 8-nt PUF elements (Fig. 5, cluster D, and Fig. 7, purple box). This observation suggests a second hand-off of targets potentially regulated by PUF3 in Aspergilli. Given the enrichment in this group for ribosome-related functions and for 9-nt elements in budding-yeast transcripts, we propose that a subset of these mRNAs are regulated by Puf4 in S. cerevisiae (Fig. 5, cluster D Sc_PUF4 binding profiles). This model suggests a complex interplay between three different PUF regulators in three branches of fungi and implicates a second independent rewiring of PUF3 regulation.
Our data also demonstrate an additional, independent evolution of PUF4/5 binding specificity in Aspergilli. Whereas the single PUF4/5 protein in N. crassa binds 9-nt and 10-nt sequences equally well, An_PUF4/5 strongly prefers the 9-nt-long element (Fig. 2D). Concordantly, the mitochondrial transcripts from Neurospora species are nearly equally enriched for 9-nt and 10-nt elements, whereas those from Aspergilli are significantly biased toward 9-nt elements (Fig. 2A, arrows). Together, these data indicate that Aspergilli have evolved both in terms of PUF4/5 specificity and in the 3′UTR sequences found in hundreds of its transcripts. It also confirms that there have been two independent origins of PUF4/5 binding preference: once after the split of Aspergilli and Neurospora species and again after PUF4/5 duplication in the Saccharomycotina lineage. It is plausible that the determinants for 9-nt versus 10-nt site preference in the PUF4/5 proteins may be relatively simple to change; modest changes in the curvature of the PUF scaffold may be sufficient (30, 35, 56). The related binding specificities of PUF proteins and the relaxed binding preferences of the ancestral PUF4/5 protein likely both contributed.
Regulatory redundancy is a recurring theme in transcription-factor network rewiring. Ancestral and newly derived regulators are proposed to cofunction until ancestral regulation is lost, completing the “handoff” of targets. In most examples, that redundancy is provided by the co-occurrence of distinct transcription factor binding sites (6, 8, 57). This model of multiple elements likely pertains to rewiring of the PUF regulons. For example, many mitochondrial transcripts bound by Nc_PUF3 are also bound by Nc_PUF4/5—in these cases, binding appears to occur through 8-nt or 9/10-nt elements, respectively. The co-occurrence of multiple elements may have facilitated evolution toward predominantly PUF3-dependent regulation in the budding yeasts versus primarily PUF4/5-dependent regulation in the filamentous fungi, simply through loss of one class of binding site. Once initiated, this situation could have amplified the selective pressure for other functionally related targets to acquire 8-nt or 9/10-nt elements in the respective lineages to maintain coregulation under the new regulatory system.
PUF proteins bind a set of related binding elements—this, combined with the relaxed specificity of PUF4/5, could provide an alternate model of redundant regulatory systems. PUF elements in the target transcripts could evolve by de novo creation of new binding sequences and loss of ancestral sites from elsewhere in the 3′UTRs. Alternatively, an existing PUF element could rapidly switch into a distinct-length PUF sequence through simpler changes. Insertion or deletion of bases into the PUF-element spacer region could instantly change PUF element length and thus change which PUF protein binds that element. Alternatively, addition of a simple UA dinucleotide downstream of an existing 8-nt element (e.g., UGUANNUA) could switch specificity from PUF3 to PUF4/5.
The recurrent and large-scale rewiring of PUF networks across fungal species may reflect the simplicity with which specificities can be changed via simple alterations of the PUF scaffold and its binding elements. It is intriguing that many of the rewiring events we report here are linked to groups of transcripts whose functional relationships are associated with species-specific differences in niche or environmental responsiveness. 3′UTR-based networks such as these may evolve particularly rapidly, given the rapid divergence of 3′UTRs in sequence and length. This may enable rewiring of transcript modules to different upstream regulatory systems that respond to novel signals as species evolve. It will be of interest to determine which features of PUF network evolution are idiosyncratic and which are shared with other families of mRNA regulatory proteins.
Materials and Methods
Detailed methods and descriptions of yeast three-hybrid assay, HITS-CLIP, N. crassa manipulations, and accession numbers can be found in SI Materials and Methods. Ascomycota fungi were chosen based on previously determined orthology (41). Phylogenetic tree shown in Fig. 1A and Fig. S1 was calculated using ribosomal RNA sequences (58), based on the maximum likelihood model executed in the PhyML program (45). Sequences of 300 bases downstream of the translation termination codon were obtained from organism-specific databases. Each 3′UTR sequence was probed for matches to PMWs representing PUF binding elements. The sequence with the high log-likelihood score was filtered to retain orthologous transcripts in which >50% of species had an ortholog. The remaining genes were then k-means clustered with k = 13. (We chose 13 clusters based on the “elbow” method, which considers the percentage of variance as a function of the number of clusters.) Additional descriptions of bioinformatic analyses are provided in SI Materials and Methods.
SI Materials and Methods
Bioinformatics.
Ascomycota fungi were chosen based on previously determined orthology (41). Phylogenetic tree shown in Fig. 1A and Fig. S1 was calculated using ribosomal RNA sequences (58), based on the maximum likelihood model executed in the PhyML program (45). Sequences of 300 bases downstream of the translation termination codon were obtained from organism-specific databases. Each 3′UTR sequence was probed for matches to PMWs representing PUF binding elements, using a custom perl script. For each n-mer of each 3′UTR sequence, a log-likelihood score was calculated comparing the likelihood of the PWM model (derived from RIP-chip data) (14) with a background model based on mononucleotide frequency taken from the genome. The sequence with the high log-likelihood score was reported by the script and assigned to the corresponding gene. The dataset was filtered to retain orthologous transcripts in which >50% of species had an ortholog. The remaining genes were then k-means clustered with k = 13. We chose 13 clusters based on the elbow method, which considers the percentage of variance as a function of the number of clusters. Clustering was visualized using Java Treeveiw (60). Functional enrichments were generated using DAVID, and Benjamini–Hochberg-corrected P values are reported (61, 62). Light responsive enrichments for PUF targets were calculated by hypergeometric distribution test [R, phyper (233, 2354, 8,458, 703, lower.tail = FALSE) – Nc_PUF3, phyper (257, 2,353, 8,459, 686, lower.tail = FALSE) – Nc_PUF4/5]. Unbiased motif analysis was done with MEME (options: -dna -mod zoops -nmotifs 3 -minw 6 -maxw 15) using 25 nt flanking the summit of each CLIP-peak (50 nt total) as the input sequences (43).
Yeast Three-Hybrid Assay.
The RNA-binding domains of A. nidulans PUF4 and N. crassa PUF5 were cloned into an activation-domain protein fusion plasmid, pGADT7 (63). Sequences for the RNA were cloned into the Hybrid RNA plasmid, p3HR2 (63). S. cerevisiae strain YBZ-1 1 [MATa, ura3-52, leu2-3, -112, his3-200, trp1-1, ade2, LYS2::(LexAop)-HIS3, URA3::(lexAop)-lacZ, and LexA-MS2 coat (N55K)] was transformed using standard lithium acetate procedures. Luminescence was assayed following the manufacturer’s instructions with a micorpalte reader (BioTech Synergy 4). Raw luminescence was normalized to OD660, and each biological replicate (n = 3) was averaged and SDs were calculated.
HITS-CLIP.
S. cerevisiae HITS-CLIP was preformed as in ref. 30, except that immunoprecipitated material was treated with RNAseR (Epicentre: RNR07250) following overnight ligation to a 3′ DNA adapter. PCR duplicates were removed using a random 5′ barcode introduced during adapter ligation. Unique sequencing reads were mapped to their respective genomes with bowtie2 (bowtie2 -D 20 -R 3 -N 1 -L 20 -i S,1,0.50) (64). CLIP peaks were defined using Piranha [Piranha -s -b 50 -a (defined empirically for each dataset) -v -p 0.01] (65). Only peaks present in all replicates were considered for further analysis.
N. crassa Manipulations.
N. crassa strains harboring a chromosomally integrated C-terminal FLAG tag at each PUF locus were generated as described in ref. 66. Conidia were used to inoculate liquid cultures (VM liquid supplemented with l-Histidine), which were harvested after 2 d of growth shaking at 30 °C onto filter paper, UV-crosslinked for 7 min (254-nm wavelength) on each side, and then frozen in liquid nitrogen. After cell disruption with mortar and pastel, lysate was cleared, RNase A treated as above, and immunoprecipitated with anti-FLAG M2 Magnetic Beads (Sigma) equilibrated according to the manufacturer’s protocol. The S. cerevisiae protocol was then followed beginning with the final IgG bead washing step.
Supplementary Material
Acknowledgments
We thank the M.W. and Kimble laboratories, and Elena Sorokin, in particular, for helpful discussions of this work. We also thank Laura Vanderploeg of the UW Biochemistry Media Lab for help with figures. This work was supported by NIH Grants R01 GM035690 and R01 GM093061 (to E.U.S.), F32 GM097821 (to A.D.K.), R01 GM083989 (to A.P.G.), and R01 GM50942 (to M.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617777114/-/DCSupplemental.
References
- 1.Doniger SW, Fay JC. Frequent gain and loss of functional transcription factor binding sites. PLOS Comput Biol. 2007;3(5):e99. doi: 10.1371/journal.pcbi.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wohlbach DJ, Thompson DA, Gasch AP, Regev A. From elements to modules: Regulatory evolution in Ascomycota fungi. Curr Opin Genet Dev. 2009;19(6):571–578. doi: 10.1016/j.gde.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villar D, Flicek P, Odom DT. Evolution of transcription factor binding in metazoans—Mechanisms and functional implications. Nat Rev Genet. 2014;15(4):221–233. doi: 10.1038/nrg3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whiteway M, Tebung WA, Choudhury BI, Rodríguez-Ortiz R. Metabolic regulation in model ascomycetes--Adjusting similar genomes to different lifestyles. Trends Genet. 2015;31(8):445–453. doi: 10.1016/j.tig.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Gasch AP, et al. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2(12):e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102(20):7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogues H, et al. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29(5):552–562. doi: 10.1016/j.molcel.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol. 2007;17(12):1007–1013. doi: 10.1016/j.cub.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443(7110):415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- 10.Reece-Hoyes JS, et al. Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol Cell. 2013;51(1):116–127. doi: 10.1016/j.molcel.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker CR, Hanson-Smith V, Johnson AD. Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science. 2013;342(6154):104–108. doi: 10.1126/science.1240810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conant GC, Wolfe KH. Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet. 2008;9(12):938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Zhang YE, Long M. New genes in Drosophila quickly become essential. Science. 2010;330(6011):1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2(3):E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keene JD. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet. 2007;8(7):533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 16.Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: A gatekeeper of inflammatory gene expression. Trends Biochem Sci. 2009;34(7):324–331. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 18.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell ZT, et al. Cooperativity in RNA-protein interactions: Global analysis of RNA binding specificity. Cell Reports. 2012;1(5):570–581. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayr C. Evolution and biological roles of alternative 3'UTRs. Trends Cell Biol. 2016;26(3):227–237. doi: 10.1016/j.tcb.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangone M, et al. The landscape of C. elegans 3'UTRs. Science. 2010;329(5990):432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3'UTRs. Nature. 2011;469(7328):97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18(3):150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 24.Lee C-D, Tu BP. Glucose-regulated phosphorylation of the PUF protein Puf3 regulates the translational fate of its bound mRNAs and association with RNA granules. Cell Reports. 2015;11(10):1638–1650. doi: 10.1016/j.celrep.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: Does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55(7):359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- 26.Quenault T, Lithgow T, Traven A. PUF proteins: Repression, activation and mRNA localization. Trends Cell Biol. 2011;21(2):104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA. 2011;2(4):471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
- 28.Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci USA. 2010;107(8):3936–3941. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13(4):286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 30.Wilinski D, et al. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat Commun. 2015;6:8213. doi: 10.1038/ncomms9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeberg MA, et al. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 2013;14(2):R13. doi: 10.1186/gb-2013-14-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lapointe CP, Wilinski D, Saunders HAJ, Wickens M. Protein-RNA networks revealed through covalent RNA marks. Nat Methods. 2015;12(12):1163–1170. doi: 10.1038/nmeth.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, McLachlan J, Zamore PD, Hall TMT. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110(4):501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 34.Miller MT, Higgin JJ, Hall TMT. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol. 2008;15(4):397–402. doi: 10.1038/nsmb.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12(11):945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- 36.Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci USA. 2009;106(48):20192–20197. doi: 10.1073/pnas.0812079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogan GJ, Brown PO, Herschlag D. Evolutionary conservation and diversification of Puf RNA binding proteins and their mRNA targets. PLoS Biol. 2015;13(11):e1002307. doi: 10.1371/journal.pbio.1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H, Guo X, Xu L, Gu Z. Rewiring of posttranscriptional RNA regulons: Puf4p in fungi as an example. Mol Biol Evol. 2012;29(9):2169–2176. doi: 10.1093/molbev/mss085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor JW, Berbee ML. Dating divergences in the Fungal Tree of Life: Review and new analyses. Mycologia. 2006;98(6):838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- 40.Sugiyama J, Hosaka K, Suh S-O. Early diverging Ascomycota: Phylogenetic divergence and related evolutionary enigmas. Mycologia. 2006;98(6):996–1005. doi: 10.3852/mycologia.98.6.996. [DOI] [PubMed] [Google Scholar]
- 41.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449(7158):54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 44.Mitchell A, et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015;43(Database issue):D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.SenGupta DJ, et al. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93(16):8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hook B, Bernstein D, Zhang B, Wickens M. RNA-protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA. 2005;11(2):227–233. doi: 10.1261/rna.7202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29(7):607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.König J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galgano A, et al. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3(9):e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ihmels J, et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309(5736):938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 55.Thompson DA, et al. Correction: Evolutionary principles of modular gene regulation in yeasts. eLife. 2013;2:e01114. doi: 10.7554/eLife.01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Opperman L, Wickens M, Hall TMT. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc Natl Acad Sci USA. 2009;106(48):20186–20191. doi: 10.1073/pnas.0812076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lavoie H, et al. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8(3):e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siddharthan R, Siggia ED, van Nimwegen E. PhyloGibbs: A Gibbs sampling motif finder that incorporates phylogeny. PLOS Comput Biol. 2005;1(7):e67. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saldanha AJ. Java Treeview--Extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 61.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 63.Koh YY, Wickens M. Determining the RNA specificity and targets of RNA-binding proteins using a three-hybrid system. Methods Enzymol. 2014;539:163–181. doi: 10.1016/B978-0-12-420120-0.00009-8. [DOI] [PubMed] [Google Scholar]
- 64.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uren PJ, et al. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28(23):3013–3020. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Honda S, Selker EU. Tools for fungal proteomics: Multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics. 2009;182(1):11–23. doi: 10.1534/genetics.108.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.