Significance
Arginine vasopressin receptor 1a (Avpr1a) and oxytocin receptor (Oxtr) are evolutionarily conserved loci that affect socio-reproductive behavior in many animals. That these loci affect fitness and exhibit substantial genetic variation in wild populations raises questions about the processes that maintain genetic variation at these loci. We show that the length of microsatellites located in the 5′ regulatory regions of Avpr1a and Oxtr are associated with reproductive success and gene expression in the brain. Crucially, balancing selection through sexually antagonistic fitness effects and density-related social influences is capable of maintaining microsatellite length polymorphisms at both genes. The action of sex and population density operating at two loci indicates that balancing selection may maintain diversity at many other behavioral loci.
Keywords: Avpr1a, Oxtr, sexual conflict, density-dependent selection, Myodes glareolus
Abstract
Most variation in behavior has a genetic basis, but the processes determining the level of diversity at behavioral loci are largely unknown for natural populations. Expression of arginine vasopressin receptor 1a (Avpr1a) and oxytocin receptor (Oxtr) in specific regions of the brain regulates diverse social and reproductive behaviors in mammals, including humans. That these genes have important fitness consequences and that natural populations contain extensive diversity at these loci implies the action of balancing selection. In Myodes glareolus, Avpr1a and Oxtr each contain a polymorphic microsatellite locus located in their 5′ regulatory region (the regulatory region-associated microsatellite, RRAM) that likely regulates gene expression. To test the hypothesis that balancing selection maintains diversity at behavioral loci, we released artificially bred females and males with different RRAM allele lengths into field enclosures that differed in population density. The length of Avpr1a and Oxtr RRAMs was associated with reproductive success, but population density and the sex interacted to determine the optimal genotype. In general, longer Avpr1a RRAMs were more beneficial for males, and shorter RRAMs were more beneficial for females; the opposite was true for Oxtr RRAMs. Moreover, Avpr1a RRAM allele length is correlated with the reproductive success of the sexes during different phases of reproduction; for males, RRAM length correlated with the numbers of newborn offspring, but for females selection was evident on the number of weaned offspring. This report of density-dependence and sexual antagonism acting on loci within the arginine vasopressin–oxytocin pathway explains how genetic diversity at Avpr1a and Oxtr could be maintained in natural populations.
Most variation in behavior has a substantial genetic basis. Identifying loci that underpin the expression of behavior is central to our understanding of the evolution and adaptive significance of behavioral diversity (1, 2). Although many studies have found an association between genotype and behavior (2–4), few have quantified the eco-evolutionary dynamics of these genetic polymorphisms. A corollary of the diversity of behaviors exhibited in wild populations is the action of balancing selection (3, 5), a general term for mechanisms that promote fitness of alternate genotypes, including density-dependent selection (1), negative frequency-dependent selection (6), heterozygote advantage (7), and sexual antagonism (8, 9). Density- and frequency-dependent selection, for example, can maintain polymorphisms at the foraging gene in laboratory populations of Drosophila melanogaster (1, 10). However, the lack of evidence for the conditions that drive balancing selection on behavioral loci in natural settings creates a challenge to behavioral genetics in understanding the dynamics of behavioral loci in real-world scenarios. Genes within the arginine vasopressin–oxytocin pathway present a classic opportunity to meet this challenge; its constituent loci have been subject to extensive study because they exert major effects on animal behavior (5, 11, 12).
The neurotransmitters vasopressin and oxytocin are evolutionarily conserved, with the vasopressin–oxytocin pathway regulating social and reproductive behaviors in many mammals including humans (5, 11, 13, 14). The behaviors associated with vasopressin and oxytocin are often mediated by the density of their receptors, notably arginine vasopressin receptor 1a (V1aR) and oxytocin receptor (OTR), in specific regions of the brain (5, 11–13). The genetic basis of the variation in V1aR density and its concomitant effect on behavior has been studied comprehensively in microtine voles (5, 15–17). In the prairie vole Microtus ochrogaster, arginine vasopressin receptor 1a (Avpr1a) expression and V1aR density in specific regions of the brain correlate with allele length at a regulatory region-associated microsatellite (RRAM) located in the 5′ regulatory region of the Avpr1a gene (15, 16), and longer Avpr1a RRAM alleles are associated with greater partner preference and male parental care in the laboratory (15). This intraspecific pattern of an association between Avpr1a RRAM allele length and V1aR expression in the brain and/or socio-reproductive behavior extends to other mammals. In chimpanzees, genetic diversity at one Avpr1a RRAM locus is associated with sociality (18). In humans, allele length at the Avpr1a RRAM locus RS3 is correlated with gene expression in the hippocampus (19) and with male pair bonding (14), altruism (19), and maternal behavior (20), whereas allele length at a second Avpr1a RRAM locus (RS1) correlates with autism and promoter activity (21). An association between RRAM allele length and transcriptional activity is not unique to Avpr1a and has been shown in other genes and in diverse taxa (22, 23).
In contrast to Avpr1a, no genetic polymorphism in the 5′ regulatory region of the oxytocin receptor (Oxtr) that associates with variation in OTR density in the brain has been identified. Nonetheless, the region ∼1–5 kbp upstream of Oxtr is important for the regulation of this gene’s expression (24, 25), and other studies have found an association between behavior and SNPs within an intron or in the 3′ UTR of Oxtr (26, 27). Moreover, there is appreciable variation in OTR density within and among rodents (12, 28). Variation of OTR density in the nucleus accumbens is associated with partner preference and maternal care in prairie vole females, such that individuals with higher OTR density show more alloparental care than individuals with lower OTR densities (29). In short, many studies have provided convincing evidence that variation in Oxtr expression has a prominent role in regulating social and sexual behavior in many animals (30, 31).
Polymorphisms at the Avpr1a RRAM are associated with the reproductive success of rodents in the laboratory (32) and in some (5, 33, 34), but not all (17), field experiments. There have been few attempts to quantify the fitness consequences of polymorphisms at the Oxtr regulatory region, or indeed the natural levels of genetic diversity, at this locus. Nonetheless, the extensive variation in OTR density in the brains of male prairie voles presumably impacts fitness, because the distribution of OTR density in the brain predicts male mating success (12). Directional selection is expected to erode fitness-associated genetic diversity toward an optimum value (35), but wild rodent populations contain extensive standing genetic variation, at least at Avpr1a. For example, wild prairie vole populations have more than 15 alleles at the Avpr1a RRAM locus (15, 17, 33, 34), and Okhovat et al. (5) identified an excess of intermediate-frequency alleles at nucleotide sites within Avpr1a as compared with putative neutral loci. A combination of putative fitness effects and extensive genetic and phenotypic diversity at Avpr1a and Oxtr are compelling evidence for the action of balancing selection. However, experimental manipulations that explicitly test this prediction are lacking (5).
In line with a well-established gene–brain–behavior model in rodents (5, 11, 15, 16) and in primates (19, 36), the bank vole Myodes glareolus presents a good model to study selection operating on polymorphisms in the regulatory regions of Avpr1a and Oxtr (SI Materials and Methods). Here, we quantify the roles of sex and population density in determining the fitness of bank voles with different genotypes at Avpr1a and Oxtr. Assessing the role of sex follows the predominantly sexually divergent roles adopted by loci within the vasopressin–oxytocin pathway: variation in V1aR density in the brain typically is associated with the expression of behaviors in males that include spatial memory, mating behavior, offspring care, and aggressiveness (5, 15, 37), whereas variation in OTR density in the brain is associated more with female behaviors, such as maternal aggression, mother–infant bonding, and same-sex social interactions (30, 38, 39). Quantifying whether there is an interaction between genotype and population density is an extension of the ecology of many species, notably the prairie vole (40) and the bank vole (41), whose populations naturally experience periodic fluctuations in density that alter the extent of intraspecific competition, e.g., for food and breeding territories (41). Bank voles experience a decrease in reproductive success and survival probability as population density increases, and thus environmental heterogeneity can favor alternate genotypes (42). To determine whether balancing selection can maintain high standing genetic variation, we first used artificial selection to create sufficient numbers of bank voles with distinct genotypes at RRAM loci for both Avpr1a and Oxtr. Next, we allowed animals to compete naturally for territories and mates, and then we quantified the fitness components of different genotypes.
SI Materials and Methods
Sequencing the Coding Sequence and 5′ Regulatory Region of Avpr1a and Oxtr in the Bank Vole.
Sequences of the coding and the 5′ regulatory regions of the Avpr1a and Oxtr loci from the bank vole genome were obtained by primer walking. Initial primers for PCR were designed against the conserved regions of these genes using sequence data for these loci in GenBank derived from other rodent species (prairie vole, M. ochrogaster, accession numbers Avpr1a: GU954384.1, Oxtr: NW_004949099.1; montane vole, Microtus montanus, accession number Avpr1a: GU954414.1; house mouse, M. musculus, accession numbers Avpr1a: NM_016847.2, Oxtr: NM_001081147.1; Norway rat, Rattus norvegicus, accession numbers Avpr1a: CM000237.2, Oxtr: CH473957.1). Subsequent pairs of primers for PCR and sequencing were designed against the specific bank vole sequence derived from previous sequencing. Sequences were obtained by Sanger sequencing using Big Dye chemistry v.3.1 and capillary electrophoresis on an ABI3100 (Applied Biosystems).
Genotyping of Avpr1a and Oxtr RRAM in the Bank Vole.
Using Primer3, we designed primers that amplified the Avpr1a and Oxtr RRAMs: Avpr1a_RRAM_forward, 5′-AGC TCC TAG TTT AAA AGC CC-3′ and Avpr1a_RRAM_reverse 5′-GAA CCA GTG AGG ATG ACA GG-3′, and Oxtr_RRAM forward 5′-AAG ATT TCT CTC AGG GTT GGT G-3′ and Oxtr_RRAM_reverse 5′-CTC TCA GAG ATG TAG GAA CCT TG-3′.
DNA was extracted from tissue samples using the Qiagen DNeasy Tissue kit and a Kingfisher magnetic particle processor (Thermo Fisher Scientific). Alleles were amplified in 10-µL PCRs that contained 1.5 µL DNA (5–50 ng in total), 0.05 U DreamTaq (Fermentas), 1× DreamTaq Buffer (Fermentas), 2 mM each dNTP, 0.3 µM reverse primer, and 0.3 µM labeled forward primer (the Avpr1a primer was labeled with VIC dye, and the Oxtr primer was labeled with NED dye). Thermocycling conditions were 95 °C for 3 min and then 35 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s. PCR products were pooled with a LIZ600 size standard, separated by capillary electrophoresis on an ABI3100, and sized using GeneMapper v.3.7 (Applied Biosystems).
No evidence of any mutation events in the germ line that could have been shown as a mismatch in Avpr1a or Oxtr microsatellite lengths between the generations could be found. This result is in line with a study by Donaldson and Young (15), who did not find any mutations in the Avpr1a RRAM when they compared the founder population and individuals in the seventh generation.
Evolutionary Divergence of the Avpr1a and Oxtr Sequences in the Bank Vole and Other Rodents.
We aligned the coding sequences of Avpr1a in bank vole, prairie vole, montane vole, house mouse, and Norway rat and Oxtr coding sequences in bank vole, prairie vole, house mouse, and Norway rat using MUSCLE. Sequence alignments were imported into MEGA5, which was used to calculate Kimura two-parameter distances between both loci for all pairs of species; this procedure provided a total of 1,258 and 1,138 nucleotide positions in the Avpr1a and Oxtr datasets, respectively. The coding sequences at Avpr1a and Oxtr are conserved within microtine voles, with bank voles showing less than 3% divergence with prairie and montane voles (Table S5). There was ∼12% (Avpr1a) and 8% (Oxtr) sequence divergence between bank voles and house mouse and Norway rat (Table S5).
Table S5.
Pairwise comparison of the number of base substitutions per site at Avpr1a and Oxtr coding sequences among rodent species, based on the Kimura two-parameter model (5)
| Locus | Species | ||||
| Bank vole | Prairie vole | Montane vole | House mouse | ||
| Avpr1a | Prairie vole | 0.0235 | |||
| Montane vole | 0.0210 | 0.0121 | |||
| House mouse | 0.1200 | 0.1220 | 0.1240 | ||
| Norway rat | 0.1204 | 0.1212 | 0.1252 | 0.0963 | |
| Oxtr | Prairie vole | 0.0297 | |||
| House mouse | 0.0823 | 0.0804 | |||
| Norway rat | 0.0823 | 0.0804 | 0.0589 | ||
There is less than 3% divergence in the coding regions of Avpr1a and Oxtr in the bank vole and the prairie vole.
Breeding for Avpr1a and Oxtr Genotypes.
Wild-caught animals used for the construction of selection lines were captured in 2010 and 2011 from 20 trapping locations in central Finland (62°37′N, 26°20′E). Each location contained four Ugglan Special multiple-capture live traps (Grahnab) located at the corners of a 15 × 15 m square. The trapping habitat was coniferous forest, predominantly composed of Norway spruce (Picea abies), Scots pine (Pinus sylvestris), and various shrubs (e.g., Calluna sp., Vaccinium spp.). There were no differences in the average length of Avpr1a or Oxtr RRAM alleles between the years 2010 and 2011 (mean ± SE for Avpr1a: 497 bp ± 0.6 bp and 497 bp ± 0.6 bp, for Oxtr: 288 bp ± 0.5 bp and 287 bp ± 0.7 bp for years 2010 and 2011, respectively). Use of study animals followed the ethical guidelines for animal research in Finland. Animals were housed in the Experimental Animal Unit, University of Jyväskylä, Finland in standard Makrolon Type III cages (43 × 26 × 15 cm) with sawdust and hay for bedding, with food (Labfor 36; Lactamin AB) and water ad libitum, at 22 °C and on a 16-h/8-h light/dark photoperiod. All animals were implanted with electronic identification microchips (Trovan Unique) using antiseptic techniques. Except where stated, a small sample of ear tissue was taken as a source of DNA for genotyping.
We paired wild-caught individuals with known Avpr1a and Oxtr genotypes and then genotyped their offspring to create the appropriate pairs for further breeding. We kept a pedigree to ensure that all animals in these breeding pairs were unrelated. The level of relatedness among the founding stock of wild-caught animals is unknown, but the relatedness of these animals is likely to be low, because bank vole populations in central Finland are large. For example, the estimated density of 7–30 bank voles/ha (59) implies a population of of 70,000–300,000 animals in the area of our trapping locations. Of course, young bank voles caught in the same traps may be kin, but such animals were kept separate for breeding. Breeding pairs were placed in separate cages, and the males were removed after 14 d of cohabitation. Females were monitored daily for births. Pups were sexed within 24 h of birth, and the tip of tail was sampled as a source of DNA. Twenty-day-old offspring were weaned by separating mother and offspring in different cages, and different sexes of the offspring were separated when 40 d old.
Interaction Between Avpr1a and Oxtr Genes.
Avpr1a and Oxtr are independent loci. Avpr1a and Oxtr are on separate chromosomes in other genomes (e.g., in M. musculus, Avpr1a resides in chromosome 10 and Oxtr in chromosome 6), and therefore there was no evidence of linkage disequilibrium between bank vole Oxtr and Avpr1a RRAM genotypes in the sample of wild-caught individuals (Pearson correlation P = 0.920, n = 325, linkage disequilibrium test P = 1.00, SE = 0, n = 325; Genepop version 4.2).
Because short and long genotypes are rare at both loci, the background Oxtr genotype in the animals bred for length variation in Avpr1a is at the center of the allele frequency distribution; i.e., all animals possess an Oxtr of medium length or a length between medium and short (i.e., between 284.7 and 288.5 bp); and the reverse is true for the animals selected for length variation in Oxtr (Avpr1a length between 493.5 bp and 495.1 bp).
Gene Expression Associated with Avpr1a and Oxtr RRAM.
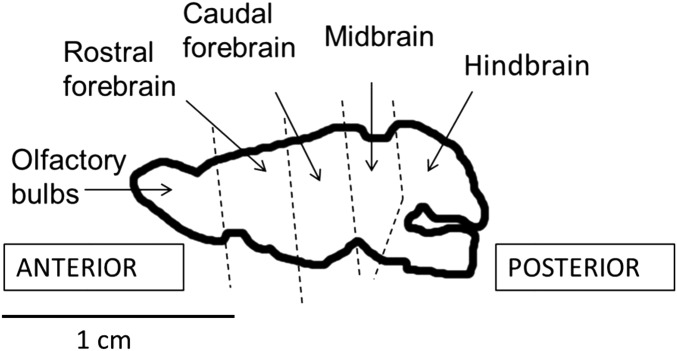
To assess whether alleles at Avpr1a and Oxtr RRAM loci were associated with the level of gene expression in bank vole brains, we dissected brains from 60 animals (30 Avpr1a and 30 Oxtr individuals); the number of individuals of each sex and the genotypes [i.e., short (SS), medium (MM), and long (LL)] were the same in each group. Brains were divided into five specific regions: olfactory bulbs, rostral forebrain, caudal forebrain, midbrain, and hindbrain, (Fig. S4). The samples were preserved in RNAlater (Qiagen) immediately after dissection and then were stored at −80 °C until analysis (∼3 mo later). RNA was extracted using a TRIzol/RNeasy kit followed with a clean-up step for purification of RNA using a Pure Link RNA Mini Kit (Ambion) or a Pure Link RNA Micro Kit (Ambion) for the small olfactory bulbs. RNA quantity was determined by fluorescence-based quantitation assay using a Qubit fluorometer (Invitrogen).
Fig. S4.
The lateral view of bank vole brain regions chosen for gene-expression analyses of Avpr1a and Oxtr: olfactory bulbs, rostral forebrain, caudal forebrain, midbrain, and hindbrain.
Variation in gene expression in the different genotypes was determined by quantitative RT-PCR in 10-μL final reaction volumes that contained ∼75 ng RNA, 5 μL power SYBR Green master mix (Applied Biosystems), 0.08 μL RT enzyme mix (Applied Biosystems), 1.5 μM forward primer, and 0.25 μM reverse primer (or 2 μM β-actin primer mix to amplify the β-actin housekeeping gene) (Ambion). Forward and reverse Avpr1a primers were 5′-GCC TAC GTG ACC TGG ATG AC-3′ and 5′-CGC CAG ATG TCG TAG CAG AT-3′, respectively; forward and reverse Oxtr primers were 5′-GTC ACA TGG ATC ACG CTT GC-3′ and 5′-CGT CTT GAG TCC CAG GTT CT-3′, respectively. We amplified the β-actin housekeeping gene using primers originally designed for mouse but which are conserved in M. glareolus: forward primer 5′-TGC GTG ACA TCA AAG AGA AG-3′ and reverse primer 5′-GAT GCC ACA GGA TTC CAT A-3′. Thermal cycling conditions on a 7500 Fast Real-Time PCR System (Applied Biosystems) were 30 min at 48 °C, followed by 10 min at 95 °C and then 40 cycles of 15 s at 95 °C and 1 min at 60 °C. A melting-curve analysis was performed to check the specificity of PCR product. Efficiencies of quantitative PCR reactions (β-actin = 104%, Avpr1a = 114%, Oxtr = 88%) were calculated against serial dilutions of cDNA using 0.8–20, 37.5–200, and 6.5–200 ng of template per reaction for β-actin, Avpr1a, and Oxtr, respectively. The amount of Avpr1a or Oxtr cDNA, normalized to the amount of the reference gene β-actin cDNA, was calculated by the comparative cycle threshold method using 7500 Software for 7500 Fast Real-Time PCR v.2.0.6 (Applied Biosystems).
Field Experiments.
The Avpr1a experiment was repeated over 2 y (2012 and 2013), and the Oxtr experiment was replicated twice during the year 2013. Each experimental enclosure (40 × 50 m) was surrounded by a galvanized sheet metal fence that was 1.2 m high and embedded 0.5 m into the ground to prevent escape by burrowing. The study individuals relied on natural food resources in the enclosures. Although the fences prevent bank voles from escaping, they do not prevent possible predation by birds and mammals and thus they reflect natural conditions. Each enclosure contained 20 trapping sites (Ugglan Special multiple-capture live traps; Grahnab) that were laid out on a regular 10-m grid and baited during trappings with sunflower seeds and potato.
The Avpr1a experiment consisted of a total of 40 females and 40 males in eight low-density enclosures and 50 females and 50 males in five high-density enclosures. The low-population-density treatment contained five females and five males per enclosure, and the high-population-density treatment contained 10 females and 10 males per enclosure. Each genotype (i.e., SS, SM, MM, LM, or LL) was equally represented in each enclosure so that there was one male and one female of each genotype in the low-density enclosures, and there were two males and two females of each genotype in the high-density enclosures. The Oxtr experiment consisted of a total of 27 females and 27 males in nine low-density enclosures and 42 females and 42 males in seven high-density enclosures. For Oxtr, the low-population-density treatment contained three females and three males per enclosure; the high-population-density treatment contained six females and six males per enclosure. Again, each of three genotypes (SS, MM, or LL) was equally represented in each enclosure. The animals were released at low- and high-population densities and were assigned to different treatments so that animals’ weight did not differ significantly in the different population densities (generalized linear model, P > 0.1 for both Avpr1a and Oxtr experiments).
In nature, bank vole populations experience periodic fluctuations in density so vole numbers can vary by as much as 90% in crash years and by as much as 30% in peak years (60). Thus trying to establish low and high population densities that reflect natural populations is both difficult and irrelevant. We used similar population densities in our earlier studies (5, 43) and have shown that they confer conditions for sufficient male–male and female–female competition. We did not release opposite-sex siblings into the same enclosures. In this study the individuals participated in one reproductive bout in which they could produce one litter. Females were released into the enclosures first to allow them to compete for and establish territories, and males were released 4 d later to allow breeding. After 18 d, all individuals were live-trapped and taken to the laboratory; the trapability of bank voles is high (>80%), and trapping was continued until no new individuals were caught after three consecutive trapping sessions. The proportions of individuals that survived until trapping were as follows: In the low-density-population experiments, 76% of individuals in the Avpr1a experiment and 81% of individuals in the Oxtr experiment survived. In the high-density-population experiments, 59% of the individuals in Avpr1a experiment and 71% of the individuals in Oxtr experiment survived. The level of predation in the enclosures cannot be determined, because no individual-based tracking system was available, and it was not known if an individual died from a disease, predation, or some other factor.
In the laboratory, females were monitored daily for pregnancies and births. The pups were sexed within 24 h after birth, and tissue samples taken from the tip of tail to allow a microsatellite-based paternity analysis, as discussed see below. The number of newborn offspring was not affected by the number of days the mothers stayed in the laboratory before giving birth (GLMM, Avpr1a: P = 0.211 for allele length, P = 0.001 for density, P = 0.713 for time in the laboratory; Oxtr: P = 0.839 for allele length, P = 0.297 for density, P = 0.099 for time in the laboratory).
Within 3 d after birth, the mothers and their pups (in their breeding cage) were returned to the mother’s original enclosure and were released near the trap in which the mother had been caught. The cages were left open so the mothers could safely carry pups back to their nests. When the pups were recruited to the population (∼30 d old), all individuals were trapped out of the enclosures. Thus, our variables of reproductive success (see Materials and Methods, Statistical Analyses) combine data on both fecundity and survival selection.
In the Avpr1a experiment, 62.5% and 30% of males and 80% and 40% of females reproduced in low- and high-density enclosures, respectively. In the Oxtr experiment, 63% and 45.2% of males and 81.5% and 61.9% of females reproduced in low- and high-density enclosures, respectively.
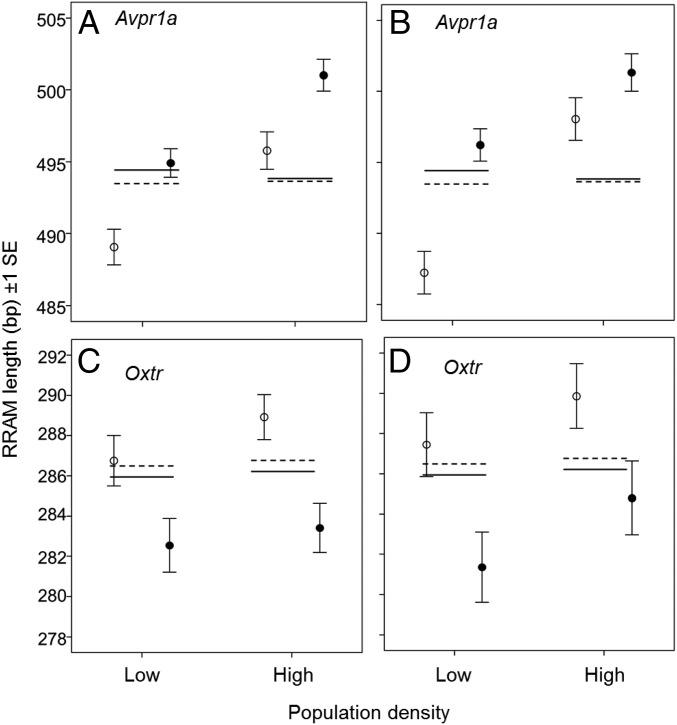
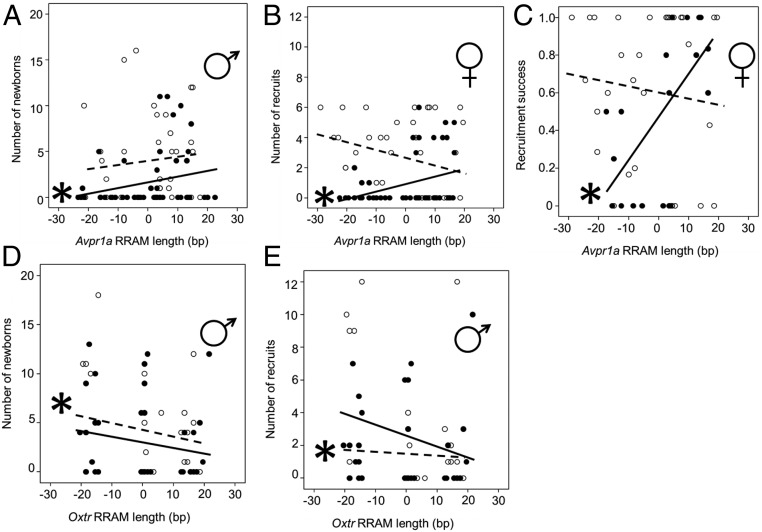
In Fig. 2, the data points comprise paternally and maternally derived alleles at the Avpr1a RRAM (A and B) and Oxtr RRAM (C and D) loci. For the Avpr1a experiment, there were 159 newborn and 112 recruited bank vole offspring in the low-population-density treatment and 82 newborn and 54 recruited offspring in the high-population-density treatment (Table S6). In the Oxtr experiment, there were 111 newborn and 73 recruited offspring in the low-population-density treatment and 132 newborn and 65 recruited offspring in the high-population-density treatment (Table S6). In Fig. 3, each data point represents one adult individual. The sample sizes for the Avpr1a experiment were n = 40 and 50 males at low and high density, respectively (A), n = 40 and 50 females at low and high density, respectively (B), and 32 and 20 females at low and high density, respectively (C). The sample sizes for the Oxtr experiment were n = 27 and 42 males at low and high density, respectively (D) and (E) n = 27 and 42 males at low and high density, respectively (E).
Fig. 2.
Sex-specific and density-dependent selection of Avpr1a and Oxtr loci in the field. Mean lengths (bp ± 1 SE) of the paternally (black circles) and maternally (white circles) derived alleles at Avpr1a (A and B) and Oxtr (C and D) RRAM loci in bank vole offspring produced at high and low population densities. Newborn offspring are shown in A and C; recruited offspring are shown in B and D. Reproductively successful males had significantly longer Avpr1a alleles than females, whereas the opposite pattern was observed for Oxtr allele lengths. Furthermore, increasing population density selected for longer Avpr1a alleles in both sexes. The mean Avpr1a (A and B) and Oxtr (C and D) RRAM allele lengths in the parental generation are shown as solid (males) and dashed (females) lines.
Table S6.
The total number of newborn offspring of each sex at the low and high population density in the Avpr1a and Oxtr experiments
| Sex | Low density | High density | ||
| Avpr1a | Oxtr | Avpr1a | Oxtr | |
| Newborn males | 86 | 54 | 50 | 55 |
| Newborn females | 73 | 57 | 32 | 77 |
Fig. 3.
Effect of sex and population density (solid trend lines and black circles indicate high density; white circles and dashed trend lines indicate low density) upon variation in fitness components of different RRAM genotypes at Avpr1a (A–C) and Oxtr (D and E). (A) Longer Avpr1a RRAM alleles are associated with more newborn offspring in males at high population density. (B and C) Longer Avpr1a RRAM alleles are associated with more recruited offspring (B) and greater recruitment success (the ratio of the number of recruited offspring to the number of newborn offspring) (C) in females. (D and E) For Oxtr, shorter RRAM alleles were associated with greater numbers of newborn (D) and recruited (E) offspring in males at low population density. Allele lengths are shown as population-centered values of the mean length for each sex. Symbols indicate the sex that is included in the plot; only data with a significant interaction between allele length and density are shown (Table 1 and Table S3). Significant effects of allele length in densitywise GLMM analyses are indicated by asterisks.
Paternity Analysis.
Offspring maternity is known, because pups were born in the laboratory. Offspring paternity was determined by genotyping all animals at six microsatellite loci (MSCg10A11, 6-FAM dye; MSCg13G2, 6-FAM dye; MSCg15F7, VIC dye; MSCg16E2, NED dye; MSCg17E9, PET dye; and MSCg6G11; NED dye) using published PCR conditions (61) and fluorophores added to the 5′ ends of the forward primer (Applied Biosystems). All genotypes were resolved by capillary electrophoresis on an ABI3100 alongside a LIZ600 (Applied Biosystems) size standard. Paternities were assigned with 95% statistical confidence using Cervus v. 3.0 using the simulation and procedure of “most likely candidate with known mother” with logarithm of the odds (LOD) scores.
Results
Genetic Diversity and Gene Expression.
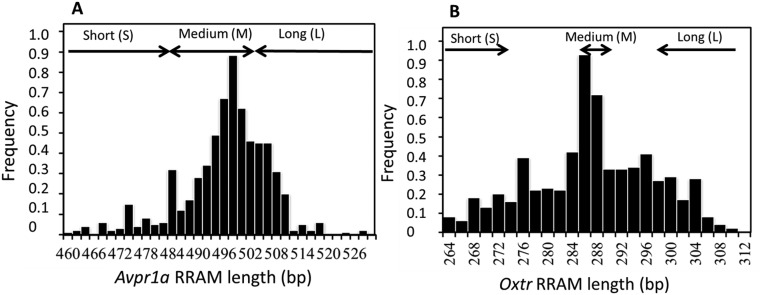
Both Avpr1a and Oxtr RRAM loci exhibit high levels of genetic variation in natural bank vole populations. After genotyping 325 individuals, we observed 31 alleles at the Avpr1a RRAM that varied between 460 and 528 bp in length and that had a qualitatively normal distribution around the most frequent alleles, which were between 496 bp and 502 bp long (Fig. S1A). At the Oxtr RRAM, we uncovered 24 alleles that varied between 264 and 310 bp in length, with the most frequent alleles being between 286 and 290 bp in length (Fig. S1B).
Fig. S1.
Frequency distribution of alleles at the Avpr1a (A) and Oxtr (B) RRAM loci based on the genotypes of 325 wild-caught bank voles, M. glareolus, from central Finland. Also shown are the arbitrary classifications of allele length as short, medium, or long alleles that were used to balance the numbers of contrasting genotypes in each experimental enclosure.
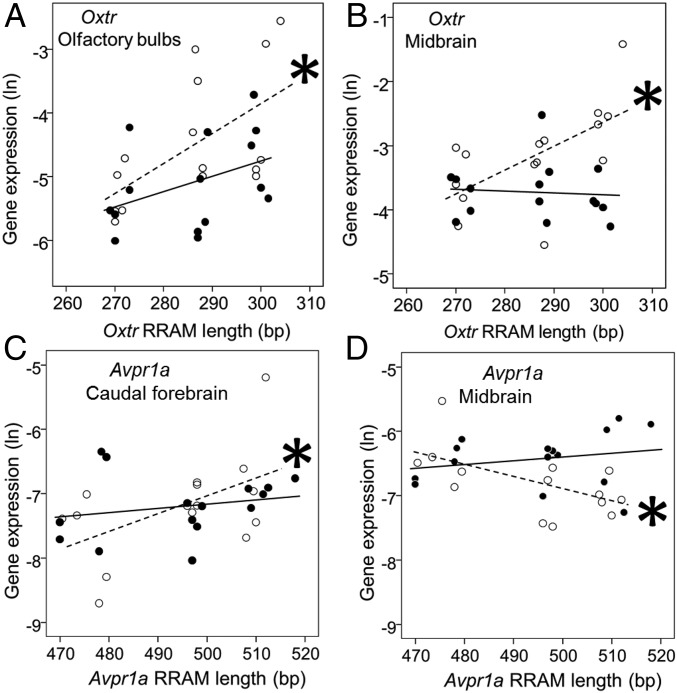
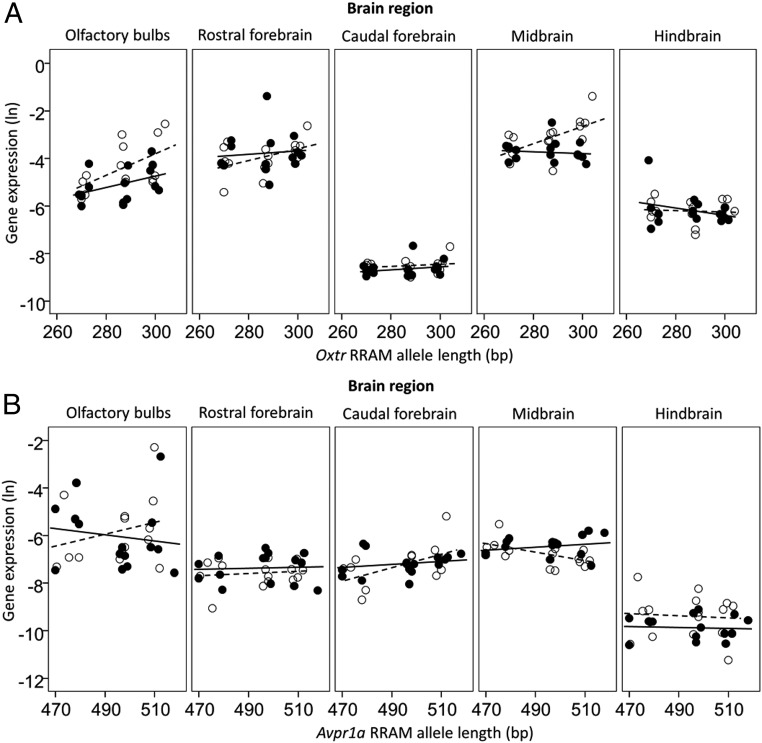
In Oxtr RRAM loci, gene expression was sex-specific in different regions of the brain (P = 0.019) (Fig. 1, Fig. S2, and Tables S1 and S2), and the association between Oxtr RRAM allele lengths and gene expression interacted with brain regions (P = 0.004) (Fig. 1, Fig. S2, and Tables S1 and S2). Longer Oxtr RRAM alleles were associated with increased gene expression in the olfactory bulbs (Fig. 1A) and in the midbrain of females (Fig. 1B). At Avpr1a RRAM loci, expression differed in brain regions in females (P < 0.001) (Fig. 1, Fig. S2, and Tables S1 and S2): Longer Avpr1a RRAM alleles were associated with increased gene expression in the caudal forebrain (Fig. 1C) and decreased gene expression in the midbrain (Fig. 1D).
Fig. 1.
Significant associations between gene expression (ln-transformed) of Oxtr (A and B) and Avpr1a (C and D) and RRAM allele lengths in the olfactory bulbs (A), the midbrain (B and D), and the caudal forebrain (C) of the bank vole. White circles and dashed lines represent females, and black circles and solid lines represent males. Fig. S2 and Tables S1 and S2 for full details of statistical tests of gene expression in all brain regions and Fig. S4 for illustration of brain regions. Significant sex-specific associations according to linear models (Table S2) are indicated by asterisks.
Fig. S2.
Associations between gene expression (ln-transformed) of Oxtr (A) and Avpr1a (B) and the length of the Oxtr and Avpr1a RRAM alleles in the olfactory bulbs, rostral forebrain, caudal forebrain, midbrain, and hindbrain of the bank vole. White circles and dashed lines indicate females, and black circles and solid lines indicate males. See Tables S1 and S2 for full details of statistical tests of gene expression in all brain regions.
Table S1.
Linear models to quantify effects of Avpr1a and Oxtr RRAM allele length, brain region (hindbrain taken as a reference), and sex on gene expression (ln-transformed) of Avpr1a and Oxtr in M. glareolus
| Gene, effect | Locus | Estimate | df | F | P |
| Oxtr | |||||
| Allele length | −0.009 | 1,135 | 4.699 | 0.032 | |
| Brain region | 4,135 | 3.007 | 0.021 | ||
| Olfactory bulbs | −11.719 | 0.001 | |||
| Rostral forebrain | −4.641 | 0.166 | |||
| Caudal forebrain | −6.449 | 0.055 | |||
| Midbrain | −5.257 | 0.117 | |||
| Sex | −0.230 | 1,135 | 2.510 | 0.115 | |
| Brain region × allele length | 4,135 | 4.075 | 0.004 | ||
| Olfactory bulb × allele length | 0.045 | 0.022 | |||
| Rostral forebrain × allele length | 0.025 | 0.036 | |||
| Caudal forebrain × allele length | 0.014 | 0.230 | |||
| Midbrain × allele length | 0.027 | 0.021 | |||
| Brain region × sex | 4,135 | 3.046 | 0.019 | ||
| Olfactory bulb × sex | 0.634 | 0.025 | |||
| Rostral forebrain × sex | −0.113 | 0.686 | |||
| Caudal forebrain × sex | 0.144 | 0.607 | |||
| Midbrain × sex | 0.599 | 0.034 | |||
| Avpr1a | |||||
| Allele length | 0.003 | 1,143 | 0.421 | 0.518 | |
| Brain region | 4,143 | 82.507 | <0.001 | ||
| Olfactory bulbs | 3.682 | <0.001 | |||
| Rostral forebrain | 2.147 | <0.001 | |||
| Caudal forebrain | 2.416 | <0.001 | |||
| Midbrain | 3.013 | <0.001 | |||
| Sex | 0.020 | 1,143 | 0.020 | 0.888 |
The estimate, F-statistic (F), and degrees of freedom (df) from the models are reported for each effect. Interactions between effects are shown with “x.” Significant (P < 0.05) effects are highlighted in bold.
Table S2.
Linear models to quantify the effects of Oxtr and Avpr1a RRAM allele lengths on the expression (ln-transformed) of Avpr1a and Oxtr in five different brain regions of M. glareolus
| Locus, brain region | Sex | Allele length | Allele length × sex | |||||||||
| Estimate | df | F | P | Estimate | df | F | P | Estimate | df | F | P | |
| Oxtr | ||||||||||||
| Olfactory bulbs | −2.580 | 1,26 | 0.775 | 0.387 | 0.010 | 1,26 | 9.006 | 0.006 | 0.010 | 1,26 | 0.945 | 0.340 |
| Rostral forebrain | −2.267 | 1,26 | 0.599 | 0.446 | 0.003 | 1,26 | 1.681 | 0.206 | 0.008 | 1,26 | 0.569 | 0.457 |
| Caudal forebrain | 0.059 | 1,26 | 0.002 | 0.961 | 0.002 | 1,26 | 1.074 | 0.310 | 0.000 | 1,26 | 0.000 | 0.996 |
| Midbrain | −4.737 | 1,26 | 5.326 | 0.029 | −0.001 | 1,26 | 4.346 | 0.047 | 0.017 | 1,26 | 5.914 | 0.022 |
| Hindbrain | −1.422 | 1,26 | 0.417 | 0.524 | −0.006 | 1,26 | 1.095 | 0.305 | 0.005 | 1,26 | 0.412 | 0.527 |
| Avpr1a | ||||||||||||
| Olfactory bulbs | −7.949 | 1,26 | 1.099 | 0.304 | −0.006 | 1,26 | 0.107 | 0.747 | 0.016 | 1,26 | 1.126 | 0.298 |
| Rostral forebrain | −0.671 | 1,26 | 0.044 | 0.836 | 0.001 | 1,26 | 0.174 | 0.680 | 0.001 | 1,26 | 0.032 | 0.860 |
| Caudal forebrain | −4.469 | 1,26 | 1.955 | 0.174 | 0.003 | 1,26 | 5.231 | 0.031 | 0.009 | 1,26 | 1.960 | 0.173 |
| Midbrain | 5.222 | 1,26 | 5.319 | 0.029 | 0.003 | 1,26 | 1.589 | 0.219 | −0.011 | 1,26 | 5.632 | 0.025 |
| Hindbrain | 1.121 | 1,26 | 0.077 | 0.783 | 0.000 | 1,26 | 0.092 | 0.763 | −0.002 | 1,26 | 0.053 | 0.620 |
The estimate, F-statistic (F), and degrees of freedom (df) from the models are reported for each main effect and interaction. Allele length × sex refers to the interaction between allele length and sex. Significant (P < 0.05) effects are highlighted in bold.
Effect of Avpr1a and Oxtr RRAM Genotype upon Fitness.
We released more than 300 mature bank voles (Avpr1a, n = 180; Oxtr, n = 138) with different Avpr1a and Oxtr RRAM genotypes into experimental field populations (Avpr1a, n = 13; Oxtr, n = 16) that contained an equal number of voles of each genotype. These animals were allowed to compete and reproduce at high and low population densities (SI Materials and Methods, Field Experiments). We observed that RRAM allele length at both loci had a significant effect on reproductive success that was contingent on both sex and population density (Figs. 2 and 3, Table 1, and Table S3).
Table 1.
GLMMs to quantify effects of sex, population density, and Avpr1a and Oxtr RRAM allele lengths on three different components of reproductive success: number of newborn offspring, number of recruited (weaned) offspring, and recruitment success in the bank vole M. glareolus
| Fitness component, locus–sex combination | Allele length | Population density | Allele length × population density | |||
| Estimate | P | Estimate | P | Estimate | P | |
| Number of newborn offspring | ||||||
| Avpr1a male | −0.007 | 0.422 | −0.419 | 0.093 | 0.050 | 0.011 |
| Avpr1a female | 0.000 | 0.950 | −0.257 | 0.070 | 0.011 | 0.370 |
| Oxtr male | −0.024 | <0.001 | −0.011 | 0.936 | 0.023 | 0.011 |
| Oxtr female | −0.001 | 0.850 | −0.062 | 0.630 | 0.001 | 0.900 |
| Number of recruited offspring | ||||||
| Avpr1a male | −0.003 | 0.800 | −0.518 | 0.160 | 0.046 | 0.130 |
| Avpr1a female | 0.007 | 0.325 | −0.776 | 0.006 | 0.053 | 0.009 |
| Oxtr male | −0.037 | <0.001 | −0.240 | 0.463 | 0.045 | 0.002 |
| Oxtr female | 0.002 | 0.840 | −0.119 | 0.520 | 0.002 | 0.900 |
| Recruitment success | ||||||
| Avpr1a male | 0.035 | 0.051 | 0.131 | 0.853 | ||
| Avpr1a female | 0.127 | 0.002 | 0.669 | 0.289 | −0.146 | <0.001 |
| Oxtr male | 0.006 | 0.354 | 0.431 | 0.481 | −0.036 | 0.169 |
| Oxtr female | 0.007 | 0.869 | 0.516 | 0.442 | −0.011 | 0.629 |
Allele length × population density refers to the interaction between allele length and population density. Significant (P < 0.05) effects are highlighted in bold.
Table S3.
GLMMs to quantify the effects of sex, population density, and Avpr1a and Oxtr RRAM allele lengths on three components of reproductive success: number of newborn offspring, number of recruited (weaned) offspring, and recruitment success in M. glareolus
| Fitness component, locus-sex combination | Allele length | Population density | Allele length × population density | ||||||||||||
| Estimate | df | F | Z | P | Estimate | df | F | Z | P | Estimate | df | F | Z | P | |
| Number of newborn offspring | |||||||||||||||
| Avpr1a male | −0.007 | −0.80 | 0.422 | −0.419 | −1.68 | 0.093 | 0.050 | 2.54 | 0.011 | ||||||
| Avpr1a female | 0.000 | 0.06 | 0.950 | −0.257 | −1.81 | 0.070 | 0.011 | 0.89 | 0.370 | ||||||
| Oxtr male | −0.024 | −3.59 | <0.001 | −0.011 | −0.08 | 0.936 | 0.023 | 2.53 | 0.011 | ||||||
| Oxtr female | −0.001 | −0.18 | 0.850 | −0.062 | −0.48 | 0.630 | 0.001 | 0.12 | 0.900 | ||||||
| Number of recruited offspring | |||||||||||||||
| Avpr1a male | −0.003 | −0.26 | 0.800 | −0.518 | −1.42 | 0.160 | 0.046 | 1.50 | 0.130 | ||||||
| Avpr1a female | 0.007 | 0.98 | 0.325 | −0.776 | −2.76 | 0.006 | 0.053 | 2.62 | 0.009 | ||||||
| Oxtr male | −0.037 | −3.30 | <0.001 | −0.240 | −0.73 | 0.463 | 0.045 | 3.16 | 0.002 | ||||||
| Oxtr female | 0.002 | 0.20 | 0.840 | −0.119 | −0.64 | 0.520 | 0.002 | 0.13 | 0.900 | ||||||
| Recruitment success | |||||||||||||||
| Avpr1a male | 0.035 | 1, 37 | 4.08 | 0.051 | 0.131 | 1, 37 | 0.04 | 0.085 | |||||||
| Avpr1a female | 0.127 | 1, 48 | 10.39 | 0.002 | 0.669 | 1, 48 | 1.15 | 0.289 | −0.146 | 1, 48 | 18.66 | <0.001 | |||
| Oxtr male | 0.006 | 1, 32 | 0.88 | 0.354 | 0.431 | 1, 32 | 0.51 | 0.481 | −0.036 | 1, 32 | 1.98 | 0.169 | |||
| Oxtr female | 0.007 | 1, 44 | 0.03 | 0.869 | 0.516 | 1, 44 | 0.60 | 0.442 | −0.011 | 1, 44 | 0.24 | 0.629 | |||
Allele length × population density refers to the interaction between allele length and population density. The estimate, Z-statistic (Z), F-statistic (F), and degrees of freedom (df) from the models are reported for each main effect and interaction. Significant (P < 0.05) effects are highlighted in bold.
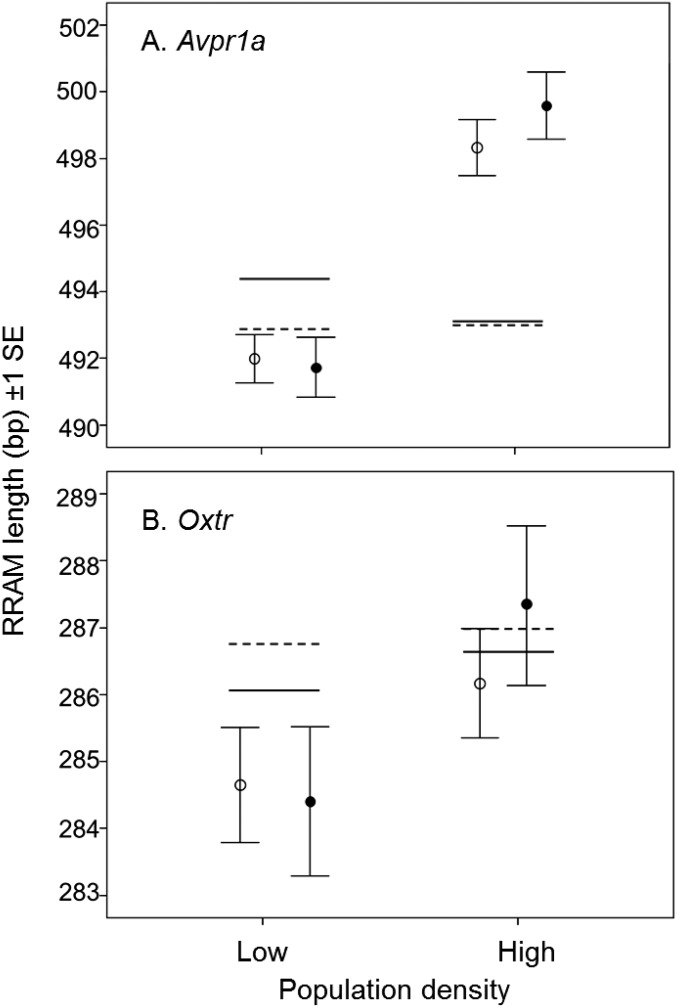
Females with longer Avpr1a RRAM alleles produced significantly more offspring that survived to recruitment (weaned offspring) at high population density (P = 0.009 allele length × density) (Table 1 and Table S3); more precisely, an ∼20 bp increase in Avpr1a RRAM allele length corresponds to an additional recruitment of one offspring per female at high population density. Male reproductive success also showed an interaction of population density × Avpr1a RRAM allele length, but with the main effect operating on the number of sired offspring (newborn animals) (P = 0.011) (Table 1 and Table S3), not the number of recruited offspring (P = 0.130) (Table 1 and Table S3). At high density, an ∼30 bp increase in Avpr1a RRAM allele length in males corresponded to the production of one more newborn offspring, but Avpr1a genotype had no apparent fitness effect at low density. Oxtr had a significant impact only on male reproductive success at low population density; males with shorter alleles sired more offspring (P < 0.001 for allele length; P = 0.011 for allele length × density) (Fig. 3D, Table 1, and Table S3) and achieved more recruited offspring (P < 0.001 for allele length; P = 0.002 for allele length × density) (Fig. 3E, Table 1, and Table S3). In effect, reducing a male Oxtr genotype by about 20 bp corresponds to an increase in fitness of one additional recruited offspring at the low population density.
Additional evidence for sex-specific optima was apparent by quantifying the lengths of the maternal and paternal alleles in the offspring (Fig. 2). There was a trend to produce offspring with longer RRAM alleles at a high population density (Fig. S3), although this density effect was significant only for Avpr1a (Table S4). At both high and low population densities, offspring inherited significantly longer Avpr1a alleles from males than from females (P < 0.001 for origin of allele) (Fig. 2 A and B and Table S4). At Oxtr, we found the opposite pattern, with offspring inheriting significantly longer RRAM alleles from their mothers than from their fathers (P = 0.004 for origin of allele) (Fig. 2 C and D and Table S4). These results indicate sex-specific fitness optima for both Avpr1a and Oxtr alleles.
Fig. S3.
Mean lengths (bp ± 1 SE) of the two copies (maternal and paternal) of the Avpr1a (A) and Oxtr (B) RRAM in offspring produced at high and low population densities. Newborn offspring are indicated by white circles, and recruited offspring are indicated by black circles. The mean Avpr1a and Oxtr RRAM allele lengths of the parental generation are shown as solid lines for males and as dashed lines for females.
Table S4.
GLMMs to assess the effect of Avpr1a and Oxtr microsatellite RRAM allele lengths (dependent variable) in paternally and maternally derived alleles (origin of allele) for newborn and recruited offspring in relation to population density in M. glareolus
| Locus, effect | Newborn offspring | Recruited offspring | ||||||
| Estimate | t | df | P | Estimate | t | df | P | |
| Avpr1a | ||||||||
| Origin of allele | 5.931 | 4.520 | 239 | <0.001 | 9.144 | 6.105 | 163 | <0.001 |
| Population density | 6.309 | 2.587 | 11 | 0.025 | 10.281 | 3.483 | 11 | 0.005 |
| Origin of allele × density | −0.516 | −0.230 | 239 | 0.819 | −5.644 | −2.156 | 163 | 0.033 |
| Oxtr | ||||||||
| Origin of allele | −4.216 | −2.889 | 241 | 0.004 | −6.096 | −3.177 | 135 | 0.002 |
| Population density | 1.558 | 0.456 | 31 | 0.652 | 0.914 | 0.224 | 14 | 0.826 |
| Origin of allele × density | −1.299 | −0.656 | 241 | 0.512 | 1.033 | 0.368 | 135 | 0.713 |
Origin of allele and population density are main effects; origin of allele × density refers to the interaction between allele length and population density. Significant (P < 0.05) effects are highlighted in bold. The estimate, t-statistic (t), and degrees of freedom (df) from the models are reported for each main effect and interaction.
Discussion
Genes within the arginine vasopressin–oxytocin pathway provide some of the best-studied models of the link from gene to brain to socio-sexual behavior (5, 11–14), but the mechanisms that can maintain high phenotypic and genetic variation in these loci are not known (5). Our field experiments show how RRAM genotypes at both Avpr1a and Oxtr affect reproductive success, in agreement with some work on the Avpr1a RRAM in the prairie vole (33, 34), and provide insight into the dynamics of the Oxtr locus. The major advance in understanding the eco-evolutionary dynamics of the arginine vasopressin–oxytocin pathway is that both loci have sex- and population density-specific fitness optima. Genetic diversity at these loci thus has adaptive relevance in natural settings and is likely maintained by balancing selection.
That sex and population density interact to vary the fitness optima for alleles at Avpr1a and Oxtr RRAM loci provides plausible mechanisms for the maintenance of genetic diversity at these loci (33, 34). Apparent functional divergence between sexes can maintain polymorphisms by generating different optimal trait values between the sexes via sexual antagonism (8, 9). A taxonomically widespread influence of sexually antagonistic alleles is supported by empirical studies on quantitative traits (e.g., testosterone level, body size) (6, 43) and at specific loci (44–46). Some authors have argued that sexual antagonism alone may be insufficient to account for most natural patterns of genetic diversity (6) but that instead some interaction with changes in social environment, such as fluctuation in population density, is required (6, 42).
Changes in population density (40, 41) can impact components of fitness through intraspecific competition (e.g., for food, mates, and territories). Competitive interactions for resources are often resolved by an individual’s level of aggression, a behavior regulated by Avpr1a and Oxtr (13, 47). Interestingly, male prairie voles with divergent Avpr1a genotypes enjoy similar overall fitness that is achieved via different mechanisms, being associated with either an apparent capability to monopolize a female partner or increased extra-pair fertilization (5). Okhovat et al. (5) suggested that population density could dictate the strength and direction of selection acting on divergent Avpr1a genotypes, with population density cycles thus maintaining genetic diversity. Therefore it is relevant that we observed an interaction between population density and Avpr1a RRAM allele length in bank voles, in contrast to a field study on prairie voles in which males with shorter Avpr1a RRAM alleles enjoyed greater reproductive success irrespective of density treatment (33), likely a response to the greater competition at high population density. More generally, high population density selects for longer alleles at both loci and in both sexes of the bank vole (Fig. 2). By analogy, these results imply selection for increased gene expression (15, 17, 19, 21), raising the possibility that the optimum female Avpr1a genotype at high population density represents a shift toward the male optimum genotype, and the male allelic optimum for Oxtr at high population density represents a shift toward the female optimum (Fig. 2). Conversely, there is the possibility of sex-specific gene expression associated with genotype (e.g., Avpr1a in the midbrain) (Fig. 1D) and for still further fine-scale variation in V1aR and OTR receptor density in the brain (12, 15). Indeed, no association between genotype and behavior was identified in female prairie voles at Avpr1a (15). Processes such as the activation of hormone receptors can drive sex-specific gene expression; for example, estrogen receptor mediates the transcriptional activity of many genes, including the expression of Oxtr (48). Nonetheless, examining several processes in tandem demonstrates how intralocus sexual conflict can be dynamic through an interrelationship with the social environment (6, 42). Interactions between different mechanisms of balancing selection have a fundamental role in maintaining diversity.
Intraspecific interactions determine reproductive success, but the severity and timing of competition often differ between sexes (6, 35, 41, 49, 50). Quantifying the numbers of offspring from birth to recruitment revealed sex-specific timing of selection acting on Avpr1a. Bank vole males do not perform parental care, and male reproductive success is determined primarily by intrasexual competition for mating opportunities when males establish a dominance hierarchy (41). Female bank voles compete intrasexually for breeding territories and then protect and care for their young (41). Consistent with this sexual dichotomy in natal care, Avpr1a RRAM allele length affects the outcome of competition for mating opportunities in males (i.e., newborn offspring), whereas for females the critical period of selection on the Avpr1a genotype occurs during maternal care (i.e., weaning). Avpr1a genotypes associate with aggressive behavior (13), but we do not know if this association is the mechanism by which Avpr1a genotype affects intrasexual competition in bank voles. For example, female choice may determine male reproductive success, because laboratory studies indicate that female prairie voles prefer to mate with males that have longer Avpr1a alleles (32). In females, our data imply a role for the expression of behaviors associated with the protection of offspring, e.g., from infanticide by intruders (51), and/or mother–offspring social dynamics rather than intrasexual competition for territories. In bank voles, the Avpr1a and Oxtr genotype impacts the outcome of sexual interactions in nature, but with the timing and mechanisms differing between sexes indicating pleiotropy.
Variation in Oxtr expression is associated typically with maternal affiliative behavior (30, 38) and aggression (39). Despite sex-specific fitness optima for Oxtr, the nonsignificant association between female Oxtr RRAM allele length and reproductive success indicates that other factors, e.g., environmental plasticity or epistatic interactions of gene networks, have a greater role in driving female reproductive success in the wild. Nonetheless, our data indicate that Avpr1a has a prominent role underlying female reproductive success, counter to laboratory studies on prairie voles (15) but consistent with work on other rodents (39) and in humans (20). Conversely, the significant role of Oxtr in determining male bank vole reproductive success adds support to evidence that OTR density in the brain predicts mating tactics and reproductive success in male prairie voles (12). Indeed, greater oxytocin induces partner-preference formation (30). However, in bank voles, longer Oxtr alleles appear costly for males, especially at low population density; this difference presumably reflects the promiscuous and monogamous mating systems of bank voles and prairie voles, respectively. In short, we find no evidence that this gene has stereotyped sex-limited fitness effects.
That genetic diversity at both RRAM loci affects fitness shows how polymorphisms in microsatellite allele length can represent important functional genetic variation. This finding is consistent with evidence that microsatellites often are associated with gene-regulatory elements (52). Changes in microsatellite allele length may alter the position of regulatory DNA motifs, such as transcription factor-binding sites. Positional changes in regulatory motifs can alter transcriptional activity (23, 53) and represent one mechanism by which changes in microsatellite length can affect gene expression. Microsatellite allele length is associated with the level of expression of many genes (4, 23), including Avpr1a in several vertebrates (17, 21, 54). Of course, regulation of gene expression extends to genomic features beyond the action of microsatellite allele length, even at Avpr1a. For example, Turner et al. (55) did not find a correlation between RRAM allele length and V1aR brain expression in an interspecific comparison of deer mice (Peromyscus) species. More specifically, a detailed functional genetic analysis of Avpr1a in the prairie vole uncovered how SNPs and methylation of CpG sites affect gene expression of V1aR in the brain and associated male socio-sexual behaviors (5). Multifaceted control of gene expression, both genetic and epigenetic, may explain the failure to establish a link between Avpr1a RRAM length and behavior, such as fidelity, in some field studies on prairie voles (17, 33, 56) and the variation in gene expression among similar RRAM genotypes (Fig. 1 and Fig. S2). Indeed, the failure to maintain a significant association between candidate genetic polymorphisms and behavior across different populations highlights the substantial challenge in quantifying evolutionary dynamics at behavioral loci in wild populations (57).
Uncovering the mechanism(s) by which the Oxtr and Avpr1a RRAMs might affect gene expression in the bank vole and identifying the influence of other potential modifiers of gene expression require detailed functional genomic analyses. Nonetheless, an analogous influence of sex and population density on the two genes Oxtr and Avpr1a is convincing support of a direct influence of allele length. What is most relevant for our understanding of the eco-evolutionary dynamics of Avpr1a and Oxtr in natural bank vole populations is that, although these loci having duplicated and then adopted more specialized roles in mammals over roughly 100 Mya (58), both density-dependent selection and sexual antagonism act on both loci.
Materials and Methods
Model Species.
The bank vole, M. glareolus, is a small rodent that inhabits forests and fields in the Palearctic; its distribution extends from Europe into western Siberia (41). Female bank voles are philopatric and defend their breeding territories; males are more dispersive and do not make provision for their young; both sexes mate multiply (41).
Avpr1a and Oxtr.
M. glareolus contains microsatellite loci in the 5′ regulatory region (i.e., RRAM) of both Avpr1a and Oxtr (SI Materials and Methods, Sequencing the Coding Sequence and 5′ Regulatory Region of Avpr1a and Oxtr in the Bank Vole and Table S5). At Avpr1a, the RRAM consists of (CA) and (GA) dinucleotide motifs and is located ∼920 bp upstream of Avpr1a exon 1. The Avpr1a RRAM appears conserved in many rodents; e.g., a RRAM that also is rich in (CA) and/or (GA) motifs is located some 903, 963, 965, and 980 nt upstream of Avpr1a exon 1 in the prairie vole (15, 16), mouse, Norway rat, and in eight species of deer mice (55), respectively. The Oxtr RRAM in the bank vole comprises a mixture of predominantly (CT)n/(GA)n dinucleotide motifs that are located immediately (∼10 bp) upstream of the oxytocin receptor transcript variant X1 and 1,448 bp upstream of the oxytocin receptor transcription start site in Mus musculus.
To quantify natural levels of polymorphisms in the Avpr1a and Oxtr RRAM loci, we caught 325 wild bank voles from central Finland from 20 trapping locations that were scattered over an area of ∼100 km2. All animals were genotyped using the primers and PCR conditions described in SI Materials and Methods, Genotyping of Avpr1a and Oxtr RRAM in the Bank Vole. The use of the animals followed the principles of Directive 2010/63/EU (License no. ESAVI/3834/04.10.03/2011) as well as all the institutional guidelines for animal research in Finland.
Selective Breeding of Animals with Distinct Avpr1a and Oxtr Genotypes.
Individuals with short (i.e., ≤484 bp and ≤274 bp in Avpr1a and Oxtr, respectively) and long (i.e., ≥504 bp and ≥298 bp in Avpr1a and Oxtr, respectively) alleles at the Avpr1a and Oxtr RRAM were rare in natural populations (Fig. S1). We therefore used selective breeding to produce sufficient unrelated animals with short and long alleles (as well as animals with medium-length alleles) at both loci (SI Materials and Methods, Breeding for Avpr1a and Oxtr Genotypes for details). This procedure allowed us to balance each field enclosure with contrasting genotypes, i.e., animals with short (S) alleles (Avpr1a: 460–484 bp; Oxtr: 264–274 bp), medium (M) (Avpr1a: 486–504 bp Oxtr: 286–290), or long (L) alleles (Avpr1a: 504–528 bp; Oxtr: 298–310 bp) as well as individuals with a combination of S and M (SM) alleles or L and M (LM) alleles.
Effect of Avpr1a and Oxtr RRAM Genotypes upon Reproductive Success.
We determined the relative effects of Avpr1a and Oxtr RRAM genotype, sex, and population density on reproductive success under seminatural conditions in outdoor enclosures at the Konnevesi Research Station, University of Jyväskylä (62°37′ N, 26°20′ E) (SI Materials and Methods, Field Experiments for details). To manipulate the degree of breeding selection among individuals, we established higher- and lower-population-density treatments. Animals of opposite sex with a common ancestor in the selective breeding pedigree were not released into the same enclosure to avoid possible inbreeding-avoidance effects. For Avpr1a, the lower-population-density treatment (n = 8 populations) contained five females and five males per enclosure, and the higher-population-density treatment (n = 5 populations) contained 10 females and 10 males per enclosure; each genotype (i.e., SS, SM, MM, LM, or LL) was equally represented in each enclosure, so that there were one male and one female of each genotype at the lower density and two males and two females of each genotype at the higher density. For Oxtr, the lower-population-density treatment (n = 9 populations) contained three females and three males per enclosure, and the higher-population-density treatment (n = 7 populations) contained six females and six males per enclosure; again each of three genotypes (SS, MM, and LL) was equally represented in each enclosure. The number of individuals differed in the Avpr1a and Oxtr experiments because of constraints in producing enough heterogeneous (SM or ML) animals for the Oxtr populations.
Animals were allowed to move, establish territories, and reproduce. After 16 d we began to trap animals on a regular trapping grid to identify breeding females. All trapped animals were measured in the laboratory, where the pregnant females were maintained and monitored until they gave birth; females and pups were returned to the enclosures within 3 d after birth (SI Materials and Methods, Field Experiments). We determined the parentage of all pups (Avpr1a, n = 241; Oxtr, n = 243; see details in Table S6) at birth using microsatellite genotyping (SI Materials and Methods, Genotyping of Avpr1a and Oxtr RRAM in the Bank Vole) and followed their survival to recruitment. Thus, our variables of reproductive success (Statistical Analyses) combine data for both breeding and fecundity selection as well as survival selection.
Statistical Analyses.
We used generalized linear mixed models (GLMMs) to analyze the effect of Avpr1a and Oxtr RRAM genotype on (i) the number of newborn offspring, (ii) the number of recruited offspring, and (iii) the recruitment success (the ratio of the number of recruited offspring to the number of newborn offspring) (Table 1 and Table S3). Sexes were examined separately. The GLMMs quantified whether the numbers of newborn or recruited offspring (dependent variables) could be predicted by the independent variables of allele length (centered value of the mean length of Avpr1a or Oxtr RRAM alleles), population density (high or low), and their interaction. Variation between years and enclosures in the Avpr1a experiment and replicates and enclosures in the Oxtr experiment were accounted for by including them as random factors. Numbers of newborn and recruited offspring were examined using a zero-inflated negative binomial model (ZINB) with a Poisson distribution, using glmmadmb in R v. 3.1.1 (R Development Core Team 2014). Recruitment success was examined using GLMM (events-trials, binomial distribution, and logit link function) in SPSS (IBM SPSS Statistics 22). The difference in the length of maternally and paternally derived Avpr1a and Oxtr RRAM alleles (Table S4) was analyzed using population density and origin of allele (maternal or paternal RRAM allele) and their interactions as independent variables. Offspring ID nested within litter and experimental enclosure was included as a random effect. We used linear models with R v.3.1.1 (R Development Core Team 2014) to analyze the effects of allele length, sex, brain region, and their interactions on the expression of Avpr1a and Oxtr (Table S1). Avpr1a and Oxtr expression also was analyzed separately for each brain region (Table S2).
Ethical Approval.
Use of study animals followed the ethical guidelines for animal research in Finland.
Acknowledgments
We thank the staff of the Experimental Animal Unit and Konnevesi Research Station, University of Jyväskylä, and S. Huttunen, S. Kyröläinen, P. Lehmann, M. Väätäinen, and T. Niittynen for logistical support; A. van t’Hof for cloning and sequencing; and E. Kallio, B. Crespi, the Crawford laboratory, three anonymous reviewers for insightful comments, and J. Valkonen for statistical advice. This work was supported by the Biological Interactions Doctoral Programme (E.L.); by Academy of Finland Grants 257340, 119200, 115961, and 140767 (to E.K.), 257729 (to M.M.), and 118603, 109165, 204284, and 268670 (to T.M.); and by the Center of Excellence in Evolutionary Research of the Academy of Finland.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
2Authors after the first author are listed in alphabetical order.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621228114/-/DCSupplemental.
References
- 1.Sokolowski MB, Pereira HS, Hughes K. Evolution of foraging behavior in Drosophila by density-dependent selection. Proc Natl Acad Sci USA. 1997;94(14):7373–7377. doi: 10.1073/pnas.94.14.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322(5903):896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendesky A, Bargmann CI. Genetic contributions to behavioural diversity at the gene-environment interface. Nat Rev Genet. 2011;12(12):809–820. doi: 10.1038/nrg3065. [DOI] [PubMed] [Google Scholar]
- 4.Fondon JW, 3rd, Hammock EA, Hannan AJ, King DG. Simple sequence repeats: Genetic modulators of brain function and behavior. Trends Neurosci. 2008;31(7):328–334. doi: 10.1016/j.tins.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Okhovat M, Berrio A, Wallace G, Ophir AG, Phelps SM. Sexual fidelity trade-offs promote regulatory variation in the prairie vole brain. Science. 2015;350(6266):1371–1374. doi: 10.1126/science.aac5791. [DOI] [PubMed] [Google Scholar]
- 6.Mokkonen M, et al. Negative frequency-dependent selection of sexually antagonistic alleles in Myodes glareolus. Science. 2011;334(6058):972–974. doi: 10.1126/science.1208708. [DOI] [PubMed] [Google Scholar]
- 7.Penn DJ, Damjanovich K, Potts WK. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc Natl Acad Sci USA. 2002;99(17):11260–11264. doi: 10.1073/pnas.162006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chippindale AK, Gibson JR, Rice WR. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc Natl Acad Sci USA. 2001;98(4):1671–1675. doi: 10.1073/pnas.041378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wedell N, Kvarnemo C, Tregenza T. Sexual conflict and life histories. Anim Behav. 2006;71(5):999–1011. [Google Scholar]
- 10.Fitzpatrick MJ, Feder E, Rowe L, Sokolowski MB. Maintaining a behaviour polymorphism by frequency-dependent selection on a single gene. Nature. 2007;447(7141):210–212. doi: 10.1038/nature05764. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson ZR, Young LJ. Oxytocin, vasopressin, and the neurogenetics of sociality. Science. 2008;322(5903):900–904. doi: 10.1126/science.1158668. [DOI] [PubMed] [Google Scholar]
- 12.Ophir AG, Gessel A, Zheng DJ, Phelps SM. Oxytocin receptor density is associated with male mating tactics and social monogamy. Horm Behav. 2012;61(3):445–453. doi: 10.1016/j.yhbeh.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldwell H, Young W., III . Oxytocin and vasopressin: Genetics and behavioral implications. In: Lim R, Lajtha A, editors. Handbook of Neurochemistry and Molecular Neurobiology. 3rd Ed. Springer; New York: 2006. pp. 573–607. [Google Scholar]
- 14.Walum H, et al. Genetic variation in the vasopressin receptor 1a gene (AVPR1A) associates with pair-bonding behavior in humans. Proc Natl Acad Sci USA. 2008;105(37):14153–14156. doi: 10.1073/pnas.0803081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammock EA, Young LJ. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308(5728):1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- 16.Donaldson ZR, Young LJ. The relative contribution of proximal 5′ flanking sequence and microsatellite variation on brain vasopressin 1a receptor (Avpr1a) gene expression and behavior. PLoS Genet. 2013;9(8):e1003729. doi: 10.1371/journal.pgen.1003729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ophir AG, Campbell P, Hanna K, Phelps SM. Field tests of cis-regulatory variation at the prairie vole avpr1a locus: Association with V1aR abundance but not sexual or social fidelity. Horm Behav. 2008;54(5):694–702. doi: 10.1016/j.yhbeh.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Staes N, et al. Chimpanzee sociability is associated with vasopressin (Avpr1a) but not oxytocin receptor gene (OXTR) variation. Horm Behav. 2015;75(1):84–90. doi: 10.1016/j.yhbeh.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Knafo A, et al. Individual differences in allocation of funds in the dictator game associated with length of the arginine vasopressin 1a receptor RS3 promoter region and correlation between RS3 length and hippocampal mRNA. Genes Brain Behav. 2008;7(3):266–275. doi: 10.1111/j.1601-183X.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- 20.Avinun R, Ebstein RP, Knafo A. Human maternal behaviour is associated with arginine vasopressin receptor 1A gene. Biol Lett. 2012;8(5):894–896. doi: 10.1098/rsbl.2012.0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tansey KE, et al. Functionality of promoter microsatellites of arginine vasopressin receptor 1A (AVPR1A): Implications for autism. Mol Autism. 2011;2(1):3. doi: 10.1186/2040-2392-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324(5931):1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: Structure, function, and evolution. Mol Biol Evol. 2004;21(6):991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- 24.Inoue T, et al. Structural organization of the human oxytocin receptor gene. J Biol Chem. 1994;269(51):32451–32456. [PubMed] [Google Scholar]
- 25.Young LJ, et al. The 5′ flanking region of the monogamous prairie vole oxytocin receptor gene directs tissue-specific expression in transgenic mice. Ann N Y Acad Sci. 1997;807(1):514–517. doi: 10.1111/j.1749-6632.1997.tb51955.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen FS, et al. Common oxytocin receptor gene (OXTR) polymorphism and social support interact to reduce stress in humans. Proc Natl Acad Sci USA. 2011;108(50):19937–19942. doi: 10.1073/pnas.1113079108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skuse DH, et al. Common polymorphism in the oxytocin receptor gene (OXTR) is associated with human social recognition skills. Proc Natl Acad Sci USA. 2014;111(5):1987–1992. doi: 10.1073/pnas.1302985111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross HE, Young LJ. Oxytocin and the neural mechanisms regulating social cognition and affiliative behavior. Front Neuroendocrinol. 2009;30(4):534–547. doi: 10.1016/j.yfrne.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olazábal DE, Young LJ. Species and individual differences in juvenile female alloparental care are associated with oxytocin receptor density in the striatum and the lateral septum. Horm Behav. 2006;49(5):681–687. doi: 10.1016/j.yhbeh.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Mokkonen M, Crespi BJ. Genomic conflicts and sexual antagonism in human health: Insights from oxytocin and testosterone. Evol Appl. 2015;8(4):307–325. doi: 10.1111/eva.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumsta R, Heinrichs M. Oxytocin, stress and social behavior: Neurogenetics of the human oxytocin system. Curr Opin Neurobiol. 2013;23(1):11–16. doi: 10.1016/j.conb.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Castelli FR, Kelley RA, Keane B, Solomon NG. Female prairie voles show social and sexual preferences for males with longer avpr1a microsatellite alleles. Anim Behav. 2011;82(5):1117–1126. [Google Scholar]
- 33.Solomon NG, et al. Polymorphism at the avpr1a locus in male prairie voles correlated with genetic but not social monogamy in field populations. Mol Ecol. 2009;18(22):4680–4695. doi: 10.1111/j.1365-294X.2009.04361.x. [DOI] [PubMed] [Google Scholar]
- 34.Harris MN, et al. The role of avpr1a microsatellite length on reproductive success of female Microtus ochrogaster. Behaviour. 2014;151(8):1185–1207. [Google Scholar]
- 35.Roff DA. Evolution of Life Histories: Theory and Analysis. Chapman & Hall; New York: 1992. pp. 1–548. [Google Scholar]
- 36.Anestis SF, et al. AVPR1A variation in chimpanzees (Pan troglodytes): Population differences and association with behavioral style. Int J Primatol. 2014;35(1):305–324. [Google Scholar]
- 37.Ophir AG, Wolff JO, Phelps SM. Variation in neural V1aR predicts sexual fidelity and space use among male prairie voles in semi-natural settings. Proc Natl Acad Sci USA. 2008;105(4):1249–1254. doi: 10.1073/pnas.0709116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rilling JK, Young LJ. The biology of mammalian parenting and its effect on offspring social development. Science. 2014;345(6198):771–776. doi: 10.1126/science.1252723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosch OJ, Neumann ID. Both oxytocin and vasopressin are mediators of maternal care and aggression in rodents: From central release to sites of action. Horm Behav. 2012;61(3):293–303. doi: 10.1016/j.yhbeh.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Getz LL, Hofmann JE, McGuire B, Dolan TW. Twenty-five years of population fluctuations of Microtus ochrogaster and M. pennsylvanicus in three habitats in east-central illinois. J Mammal. 2001;82(1):22–34. [Google Scholar]
- 41.Mills SC, Mokkonen M, Koskela E, Mappes T. Genotype-by-environment interactions and reliable signaling of male quality in bank voles. In: Hunt J, Hosken DJ, editors. Genotype-by-Environment Interactions and Sexual Selection. John Wiley & Sons, Ltd.; Chichester, UK: 2014. pp. 241–264. [Google Scholar]
- 42.Mappes T, et al. Frequency and density-dependent selection on life-history strategies--a field experiment. PLoS One. 2008;3(2):e1687. doi: 10.1371/journal.pone.0001687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox RM, Calsbeek R. Cryptic sex-ratio bias provides indirect genetic benefits despite sexual conflict. Science. 2010;328(5974):92–94. doi: 10.1126/science.1185550. [DOI] [PubMed] [Google Scholar]
- 44.Dean R, Perry JC, Pizzari T, Mank JE, Wigby S. Experimental evolution of a novel sexually antagonistic allele. PLoS Genet. 2012;8(8):e1002917. doi: 10.1371/journal.pgen.1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rostant WG, Kay C, Wedell N, Hosken DJ. Sexual conflict maintains variation at an insecticide resistance locus. BMC Biol. 2015;13(1):34. doi: 10.1186/s12915-015-0143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barson NJ, et al. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature. 2015;528(7582):405–408. doi: 10.1038/nature16062. [DOI] [PubMed] [Google Scholar]
- 47.Storm EE, Tecott LH. Social circuits: Peptidergic regulation of mammalian social behavior. Neuron. 2005;47(4):483–486. doi: 10.1016/j.neuron.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Ivell R, et al. The structure and regulation of the oxytocin receptor. Exp Physiol. 2001;86(2):289–296. doi: 10.1113/eph8602185. [DOI] [PubMed] [Google Scholar]
- 49.Johnston SE, et al. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature. 2013;502(7469):93–95. doi: 10.1038/nature12489. [DOI] [PubMed] [Google Scholar]
- 50.Mokkonen M, Koskela E, Mappes T, Mills SC. Evolutionary conflict between maternal and paternal interests: Integration with evolutionary endocrinology. Integr Comp Biol. 2016;56(2):146–158. doi: 10.1093/icb/icw053. [DOI] [PubMed] [Google Scholar]
- 51.Mappes T, et al. Advantage of rare infanticide strategies in an invasion experiment of behavioural polymorphism. Nat Commun. 2012;3(1):611. doi: 10.1038/ncomms1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawaya S, et al. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS One. 2013;8(2):e54710. doi: 10.1371/journal.pone.0054710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vardhanabhuti S, Wang J, Hannenhalli S. Position and distance specificity are important determinants of cis-regulatory motifs in addition to evolutionary conservation. Nucleic Acids Res. 2007;35(10):3203–3213. doi: 10.1093/nar/gkm201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim MM, et al. Enhanced partner preference in a promiscuous species by manipulating the expression of a single gene. Nature. 2004;429(6993):754–757. doi: 10.1038/nature02539. [DOI] [PubMed] [Google Scholar]
- 55.Turner LM, et al. Monogamy evolves through multiple mechanisms: Evidence from V1aR in deer mice. Mol Biol Evol. 2010;27(6):1269–1278. doi: 10.1093/molbev/msq013. [DOI] [PubMed] [Google Scholar]
- 56.McGraw LA, Davis JK, Thomas PJ, Young LJ, Thomas JW. NISC Comparative Sequencing Program BAC-based sequencing of behaviorally-relevant genes in the prairie vole. PLoS One. 2012;7(1):e29345. doi: 10.1371/journal.pone.0029345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korsten P, et al. Association between DRD4 gene polymorphism and personality variation in great tits: A test across four wild populations. Mol Ecol. 2010;19(4):832–843. doi: 10.1111/j.1365-294X.2009.04518.x. [DOI] [PubMed] [Google Scholar]
- 58.Yamashita K, Kitano T. Molecular evolution of the oxytocin-oxytocin receptor system in eutherians. Mol Phylogenet Evol. 2013;67(2):520–528. doi: 10.1016/j.ympev.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 59.Koivula M, Koskela E, Mappes T, Oksanen TA. Cost of reproduction in the wild: Manipulation of reproductive effort in the bank vole. Ecology. 2003;84(2):398–405. [Google Scholar]
- 60.Rikalainen K, Aspi J, Galarza JA, Koskela E, Mappes T. Maintenance of genetic diversity in cyclic populations-a longitudinal analysis in Myodes glareolus. Ecol Evol. 2012;2(7):1491–1502. doi: 10.1002/ece3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rikalainen K, Grapputo A, Knott E, Koskela E, Mappes T. A large panel of novel microsatellite markers for the bank vole (Myodes glareolus) Mol Ecol Resour. 2008;8(5):1164–1168. doi: 10.1111/j.1755-0998.2008.02228.x. [DOI] [PubMed] [Google Scholar]