Significance
The work describes the first nuclear localization signal (NLS) peptide that not only promotes nuclear shuttling of a DNA damage response (DDR) protein but mediates a direct interaction with a deubiquitylase for enhanced stability. Its identification suggests that NLS peptides, aside from their canonical function in nuclear import, may have acquired additional properties. The study also reports on an important role of the ubiquitin-specific protease 7 (USP7)–ring finger protein 169 (RNF169) axis in driving DNA repair and poly (ADP-ribose) polymerase inhibition resistance. Several lines of evidence indicate that USP7 deubiquitylates and enforces RNF169-dependent DDRs. Together, these data highlight a critically important role of the USP7–RNF169 axis in genome stability maintenance.
Keywords: USP7, RNF169, DNA damage, DNA repair, deubiquitylation
Abstract
Loading of p53-binding protein 1 (53BP1) and receptor-associated protein 80 (RAP80) at DNA double-strand breaks (DSBs) drives cell cycle checkpoint activation but is counterproductive to high-fidelity DNA repair. ring finger protein 169 (RNF169) maintains the balance by limiting the deposition of DNA damage mediator proteins at the damaged chromatin. We report here that this attribute is accomplished, in part, by a predicted nuclear localization signal (NLS) that not only shuttles RNF169 into the nucleus but also promotes its stability by mediating a direct interaction with the ubiquitin-specific protease USP7. Guided by the crystal structure of USP7 in complex with the RNF169 NLS, we uncoupled USP7 binding from its nuclear import function and showed that perturbing the USP7–RNF169 complex destabilized RNF169, compromised high-fidelity DSB repair, and hypersensitized cells to poly (ADP-ribose) polymerase inhibition. Finally, expression of USP7 and RNF169 positively correlated in breast cancer specimens. Collectively, our findings uncover an NLS-mediated bipartite mechanism that supports the nuclear function of a DSB response protein.
Cells respond to DNA double-strand breaks (DSBs) by mounting a series of signal transduction events that culminate in cell arrest and DNA repair (1). Signal transduction entails the coordinated assembly of a cohort of DNA damage mediator proteins, including p53-binding protein 1 (53BP1) and receptor-associated protein 80 (RAP80), at the damaged chromatin, which, in turn, enforces checkpoint control and cell tolerance to DNA damage (2, 3). Paradoxically, although DSB loading of 53BP1 and RAP80 underlies robust activation of DNA damage responses (DDRs), they operate at the expense of high-fidelity DNA repair, because their productive accumulation at DSB-flanking chromatin blocks DNA end resection and suppresses RAD51-dependent homologous recombination (HR) repair (4–11). Exactly how cells achieve a balance between robust DSB signal transduction and HR repair remains to be defined, but several lines of evidence converge on the idea that limiting the extent of DSB ubiquitylation may selectively promote HR repair (12–15), highlighting the need for cell-intrinsic mechanisms to restrain chromatin responses arising from DSBs (16).
Intriguingly, one such cell-intrinsic strategy involves ring finger protein 169 (RNF169), a paralog of the RIDDLE (radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties) syndrome protein RNF168 (17, 18). In stark contrast to RNF168, which plays positive roles in amplifying ubiquitin-dependent DSB signals (18, 19), RNF169 is endowed with antagonistic properties in the DSB signal transduction pathway. Indeed, RNF169 competes for RNF168-catalyzed ubiquitin adducts and displaces 53BP1 and RAP80 from DSBs (14, 20, 21). Despite its putative role as an integral component of the DSB signal transduction cascade, how RNF169 is regulated mechanistically and how it contributes to the multipartite self-restraining mechanisms in DNA damage surveillance and repair processes remain to be established.
The ubiquitin-specific protease USP7 orchestrates diverse cellular and pathophysiological processes that range from immune responses to DNA damage tolerance. Although USP7 was canonically known for its role in regulating the MDM2/p53 pathway (22), the observation that p53 nullizygosity did not rescue the lethality of USP7 knockout (KO) animals suggests that USP7 also plays important functions outside the MDM2/p53 context (23). As a DNA damage-regulated deubiquitylating (DUB) activity (24), USP7 is reported to confer cell resistance to genotoxic stress by targeting a panel of DDR proteins, including Rad18 (25), RNF168 (26), and Claspin (27). Moreover, in line with antiapoptotic functions, USP7 is overexpressed in breast and prostate cancers (28, 29), and genetic ablation or chemical inhibition of USP7 DUB activity induced cell killing in vitro and tumor regression in vivo (30–34), underscoring the potential development of USP7 inhibitors as anticancer therapeutics.
Results
USP7 Is a Bona Fide Interacting Partner of RNF169.
As part of our effort in probing the self-restraining mechanisms of DSB signal transduction, we analyzed the RNF169 interactome. Although RNF169 limits excessive loading of the DNA damage-mediator proteins 53BP1 and RAP80 at DSBs (14, 20, 21), exactly how its activity is regulated is not known. To this end, we affinity-purified RNF169 protein complexes from 293T cells engineered to express S protein-Flag-Streptavidin–binding peptide (SFB)-tagged RNF169 stably. Following a two-step tandem affinity purification scheme (35) we subjected the RNF169 precipitates to mass spectrometric-based protein identification. A number of putative RNF169-interacting factors emerged, including the ubiquitin-specific protease USP7 and the Down syndrome kinase DYRK1A (Fig. 1A). We confirmed the RNF169–USP7 interaction using a coimmunoprecipitation (co-IP) approach involving ectopically expressed proteins (Fig. 1B) as well as those proteins expressed at endogenous levels (Fig. 1C). Our co-IP results also validated a number of other putative RNF169 interactions, including DYRK1A (Fig. 1B). To understand the regulatory basis for the USP7–RNF169 interaction better, we further mapped the USP7-binding domain on the RNF169 polypeptide. In addition to the previously characterized RNF169 mutants (20), we generated a series of RNF169 deletion mutants (Fig. 1D) and analyzed their ability to interact with USP7 (Fig. 1E). Our domain-mapping exercise uncovered that RNF169∆C6, which lacks amino acids 609–654, did not interact with USP7. Using bacterially expressed and purified proteins, we further showed that RNF169 interacts directly with USP7 in a manner that required amino acids 609–654 (Fig. 1F). Together, we concluded that USP7 is a bona fide RNF169-interacting factor.
Fig. 1.
RNF169 interacts directly with USP7. (A) Affinity purification of RNF169 protein complexes identified a list of RNF169-interacting proteins. Names and numbers of distinct peptides from top hits are shown. (B) U2OS RNF169 SFB cells were induced or not induced with doxycycline (Dox) for 24 h. Cell lysates were subjected to IP with Streptavidin beads, and RNF169-associated immunoprecipitates were analyzed by Western blotting analysis using indicated antibodies. DYRK1A and CHK1 were used as positive and negative controls, respectively. (C) Cell lysates derived from HeLa cells were incubated with Protein A agarose beads conjugated with anti-RNF169 antibody or control. Immunoprecipitates were detected with anti-RNF169 or anti-USP7 antibodies. (D) Schematic illustration of full-length (FL) and deletion mutants of RNF169 used in this study. MIU, motif interacting with ubiquitin; RING, Really Interesting New Gene. (E) HEK293T cells were transiently transfected with plasmids encoding Myc-USP7 and HA-tagged RNF169 wild type and mutants (as depicted in D). Cell lysates were incubated with anti-HA affinity matrix, and immunoprecipitates were blotted with anti-HA and anti-Myc antibodies. (F) RNF169 interacts directly with USP7. Purified GST-USP7 fusion proteins were incubated with purified MBP-RNF169 immobilized on amylose resin. (G and H) HEK293T cells were transfected with plasmids encoding SFB-tagged USP7 FL or mutants. Cell lysates were prepared for pull-down experiments against purified MBP-RNF169. A schematic illustration of USP7 constructs is shown. CD, catalytic domain; HA, human influenza hemagglutinin; IP, immunoprecipitation; TRAF, TNF receptor-associated factor.
RNF169 Docks on USP7 Ubiquitin-Like Domains.
USP7 plays pleiotropic roles to support cell proliferation and organismal development. USP7 interactions are mediated primarily through its N-terminal TNF receptor-associated factor or its C-terminal ubiquitin-like (UBL) domains. To dissect the USP7–RNF169 interaction further, we expressed USP7 protein fragments and analyzed their interaction with purified maltose-binding protein (MBP)–RNF169 using a pull-down approach (Fig. 1G). Results indicated that the USP7–RNF169 interaction is mediated by the UBLs. Resembling the binding interfaces between USP7 and other UBL domain-interacting factors, fine-mapping of USP7 UBLs revealed that UBL1–3 are most important in supporting its interaction with RNF169 (Fig. 1H).
Structural Basis of the USP7–RNF169 Interaction.
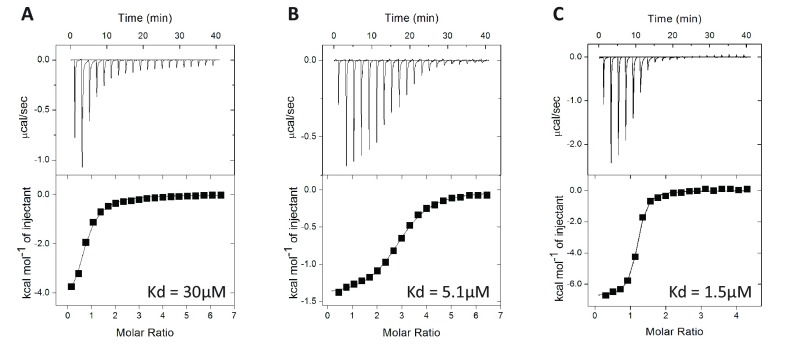
The USP7 UBL domains interacted with RNF169. Given that the UBLs also mediate USP7 interactions with other cellular (e.g., GMPS, DNMT1) and viral (ICP0) factors (36–38), we looked for possible conserved sequence determinants on the RNF169 C6 region (i.e., 609–654 aa), according to the KxxxK motif described previously (39). Similar to the KxxxK motif previously identified among USP7 UBL domain-binding proteins, visual inspection of the RNF169 C6 sequence also revealed a KxxxK motif (Fig. 2A). To test directly whether the KxxxK motif underlies the RNF169–USP7 interaction, a series of RNF169 peptides of varying lengths, all of which encompass the KxxxK motif, were synthesized. These RNF169 peptides included peptides of 27-aa length (617SLRRGRKRHCKTKHLEQNGSLKKLRQT643), 17-aa length (617SLRRGRKRHCKTKHLEQ633), 14-aa length (619RRGRKRHCKTKHLE632), 13-aa length (620RGRKRHCKTKHLE632), 12-aa length (620RGRKRHCKTKHL631), and 10-aa length (621GRKRHCKTKH630). Using isothermal titration calorimetry (ITC), we examined the interaction of these RNF169 peptides with purified USP7 UBL1–3 domains and identified the 13-aa mer as the optimal peptide that displayed low micromolar affinity for the USP7 UBL domains (Fig. 2B and Fig. S1). We also synthesized peptides that correspond to the GMPS and ICP0 KxxxK motif. ITC results indicated that under our experimental conditions, the RNF169 13-aa peptide exhibited comparable, if not higher, affinity for the USP7 UBL domains (Fig. 2B and Fig. S1).
Fig. 2.
Crystal structure of USP7 UBL1–3-RNF169620–632 peptide. (A) Alignment of KxxxK motif from RNF169 and other USP7 UBL domain-binding proteins. (B) Comparison of the dissociation constants of GMPS, ICP0, and RNF169 peptides with bacterially purified USP7 UBL1–3. (C, Left) Electrostatic surface representation of the USP7 UBL1–3 domain complexed with RNF169620–632 peptide. Acidic regions are shown in red, and basic regions are shown in blue. (C, Right) Ribbon representation of the complex of RNF169620–632 peptide and the USP7 UBL1–3 domain, with UBL1, UBL2, and UBL3 colored blue, gold, and gray, respectively. The RNF169 peptide is shown in stick representation; the missing loop (residues 668–673) connecting the UBL1 and UBL2 domains is represented using a dotted line. (D) Binding interface between RNF169620–632 peptide and USP7. Key hydrogen bonds bridging the interactions are highlighted by brown dashed lines. (E) Two acidic pockets (A and B) on the surface of UBL1–2 are optimized for the recognition of the lysine residues (K623 and K627) of RNF169. The “sliced” side view is plotted to show engagement of the side chains of the lysine residues in the binding pockets.
Fig. S1.
ITC curves for USP7 UBL1–3 over GMPS316–328 (A), ICP0615–627 (B), and RNF169620–632 (C) (Fig. 2B).
To explore the pattern of mutual recognition between USP7 and RNF169 further, the 13-aa RNF169 peptide was used to cocrystalize with USP7 UBL1–3 domains, and the crystal structure [Protein Data Bank (PDB) ID code 5GG4] was determined at 3.1-Å resolution with Rwork and Rfree refined to 21% and 26%, respectively (Fig. 2C and Table S1). Analysis of our structure revealed that the RNF169 peptide binds primarily to the negative charged surface formed by UBL1–2 domains (residues D758 to D764), and that these interactions are mainly mediated by hydrogen bonds and electrostatic attractions (Fig. 2D). At the binding interface, two crucial lysine residues (K623 and K627) of the RNF169 peptide were anchored onto two adjacent acidic pockets on the surface of the UBL1–2 domains, which constituted USP7 residues M637, K681, D684, and D762 and R628, E759, M761, and D764, respectively. Within these two pockets, the ε-amino groups of K623 and K627 of RNF169 interacted directly with the side chains of USP7 D762 and D764 via salt bridges, making crucial contributions to the interaction (Fig. 2E). Moreover, the backbone amino group of RNF169 T628 is also observed to form an important hydrogen bond with the side-chain carboxylate of USP7 E759. In addition, the guanidine group of RNF169 R624 makes contact with the side chain of USP7 D653, and the ε-amino group of RNF169 K629 interacts with the side chain of USP7 D758. Together, these hydrogen bonds may also help to stabilize the RNF169–USP7 interaction further.
Table S1.
Data collection and refinement statistics
| Data collection statistics | USP7–RNF169 |
| PBD ID code | 5GG4 |
| Data collection | |
| Wavelength, Å | 0.9776 |
| Space group | C121 |
| Cell dimensions | |
| a, b, c; Å | 162.195, 99.965, 124.299 |
| α, β, γ; ° | 90, 95.537, 90 |
| Resolution range, Å | 46.96–3.11 (3.221–3.11) |
| Rmerge, % | 11.5 (67.9) |
| I/σI | 10 (3.9) |
| Completeness, % | 99.9 (100) |
| Redundancy | 3.8 (3.6) |
| Wilson B-factor, Å2 | 83.1 |
| Refinement | |
| No. of reflections (overall) | 33,903 |
| No. of reflections (test set) | 1,783 |
| Rwork/Rfree, % | 22.6/26.8 |
| No. of atoms | |
| Proteins/ligands | 8,271/361 |
| B-factors, Å2 | |
| Protein/ligands | 83.52/83.41 |
| rmsd | |
| Bond length, Å | 0.005 |
| Bond angles, ° | 1.051 |
| Ramachandran plot, % | |
| Favored/allowed/outlier, % | 91.2/6.6/2.2 |
Values in parentheses are for the highest resolution shell.
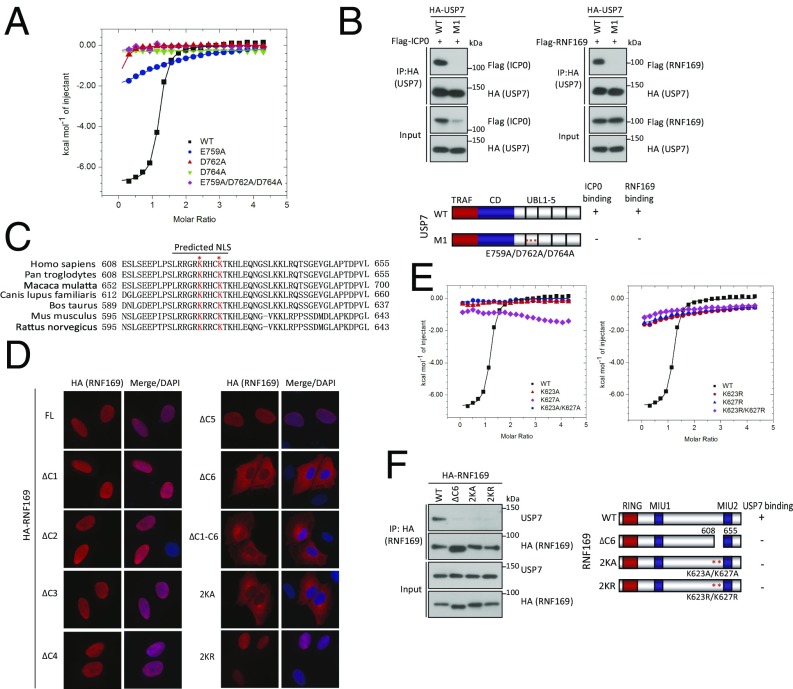
To extend our observations derived from the crystal structure, we designed a series of point mutations to target key residues at the USP7–RNF169 interface and evaluated their contributions to the complex formation. Evidently, alanine mutants of E759, D762, and D764 on the USP7 UBL1–3, alone or in combination, abolished interaction with the RNF169 13-aa peptide (Fig. 3A). Likewise, and consistent with a similar mode of interaction among USP7 UBL domain-interacting proteins, the USP7 mutant carrying alanine substitutions on E759, D762, and D764 (M1) did not interact with either ICP0 or RNF169 in co-IP experiments (Fig. 3B).
Fig. 3.
Critical residues of the USP7- and RNF169-binding interface. (A) Mutational analysis of the interaction between RNF169620–632 peptide and USP7 UBL1–3 mutants by ITC experiments. Mutations include the USP7 single-point mutants E759A, D762A, and D764A and the triple mutant E759A/D762A/D764A (M1). (B) HEK293T cells were cotransfected with HA-tagged USP7 (wild type and M1 mutant) and Flag-ICP0 or Flag-RNF169. Cell lysates were incubated with anti-HA agarose beads, and immunoprecipitates were analyzed using anti-HA and anti-Flag antibodies. (C) Sequence alignment of the USP7-binding region on RNF169 (608–655 aa) of different species. Key lysine residues are shown in red. (D) U2OS cells were transfected with HA-RNF169 (wild type and mutants) and their subcellular localization was examined by indirect immunofluorescence staining using anti-HA antibodies. Nuclei were visualized by DAPI staining. (Magnification: 60×.) (E) Mutational analysis of the interaction between USP7 UBL1–3 and the RNF169620–632 peptide by ITC experiments. Mutations include RNF169 K623A, K627A, and K623A/K627A (Left) and K623R, K627R, and K623R/K627R (Right). (F) Cell lysates derived from HEK293T cells expressing indicated HA-tagged RNF169 proteins were incubated with anti-HA agarose beads. Immunoprecipitates were separated by SDS/PAGE, and Western blotting experiments were performed using anti-HA and anti-USP7 antibodies.
Dual Functions of the RNF169 KxxxK Motif.
The USP7-binding domain on the RNF169 polypeptide (i.e., KxxxK motif) resided within a stretch of basic amino acids. This fact prompted us to speculate whether this stretch may be where the RNF169 nuclear localization signal (NLS) lies (40). Indeed, bioinformatics prediction of putative NLS on the RNF169 polypeptide indicated that it falls within the C6 region (Fig. 3C). We thus experimentally mapped the RNF169 NLS using our RNF169 deletion mutants (Fig. 1D) by examining the subcellular localization of these RNF169 deletion mutants, including the USP7-binding defective RNF169∆C6. Consistent with the idea that the RNF169 NLS overlaps with the USP7-binding KxxxK motif, the RNF169∆C6 mutant was cytoplasmic (Fig. 3D). Likewise, alanine substitutions of RNF169 K623 and K627 (K623A/K627A; hereafter referred to as 2KA) perturbed both its nuclear retention (Fig. 3D) and its ability to interact with USP7 (Fig. 3 E and F). Because such a RNF169 mutant (2KA) would preclude us from testing the significance of the RNF169–USP7 interaction, we were compelled to generate a separation-of-function RNF169 mutant that can be shuttled into the nucleus but remains USP7-binding defective. To this end, we noted that RNF169 K623 and K627 fitted perfectly into their respective binding pockets on USP7 (Fig. 2E and Fig. S2; details are provided in Discussion), raising the possibility that bulky arginine substitutions on K623 and K627 (hereafter referred to as 2KR) may be sufficient to impair the RNF169–USP7 interaction. We also compared and contrasted the binding interface between USP7 UBL1–3 and RNF169 with other USP7 UBL-binding proteins, including UHRF1 and ICP0 (Fig. S3). Because arginine substitutions would retain basic charges on the NLS, we envisaged that RNF169 2KR may be imported into the nucleus. In line with our hypothesis, RNF169 2KR was expressed in the cell nucleus (Fig. 3D) but was not able to interact with USP7 (Fig. 3 E and F).
Fig. S2.
“Sliced” side view of the binding pockets reveals engagement of the side chains of the lysine residues (A and C) or the arginine residues (B and D) in the binding pockets.
Fig. S3.
Superpositions of USP7 UBL1–3 domains of the RNF169–USP7 complex (PDB ID code 5GG4) with the domains of the ICP0–USP7 (A, PDB ID code 4WPH) and UHRF1–USP7 (B, PDB ID code 5C6D) complexes. RNF169, ICP0, and UHRF1 peptides are shown in green, red, and yellow, respectively. The orientations of the structures in the enlarged panels are slightly adjusted to illustrate the side-chain engagement of the lysine residues better.
USP7 Deubiquitylates and Stabilizes RNF169.
USP7 is endowed with DUB activity, and it has been reported to stabilize a number of DDR factors. To examine whether USP7 may promote RNF169 stability, we performed Western blotting analyses on cell lysates derived from USP7-depleted cells. We targeted USP7 using two independent siRNAs and immunoblotted for RNF169 using our previously characterized anti-RNF169 antibodies (20). Notably, genetic ablation of USP7 led to marked reduction in RNF169 protein levels (Fig. 4A), lending credence to the idea that USP7 may stabilize RNF169. We also chemically inhibited USP7 DUB activity using the small molecules P22077 (32) and P5091 (31). Consistent with previous reports where USP7 inhibition stabilized p53, P22077 and P5091 pretreatment resulted in marked elevation in p53 levels (Fig. 4B), especially in nonirradiated cells. More importantly, chemical inhibition of USP7 correlated with substantial reduction of RNF169 (Fig. 4B), suggesting that USP7, in a DUB-dependent manner, stabilizes RNF169.
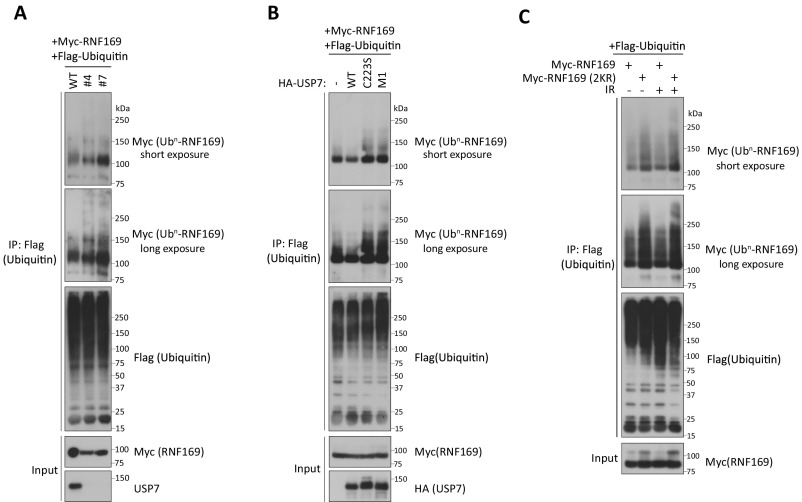
Fig. 4.
USP7 deubiquitylates and stabilizes RNF169. (A) HeLa cells pretreated with indicated siRNAs were harvested, and cell lysates were subjected to Western blotting analysis with indicated antibodies. *Nonspecific bands. (B) HeLa cells were treated with DMSO, P22077 (20 μM), or P5091 (15 μM) for 2 h before ionizing radiation (IR) treatment (10 Gy). Cells were harvested 4 h after IR treatment, and lysates were immunoblotted with indicated antibodies. (C) Lysates derived from USP7 KO HeLa cells (clones 4 and 7) and control HeLa cells were analyzed by Western blotting experiments using indicated antibodies. (D) USP7 KO HeLa cells and their parental cells were treated with 50 μg/mL cycloheximide (CHX), and cells were harvested at indicated time points for processing. Western blotting experiments were performed using indicated antibodies to evaluate the expression of RNF169. Quantification is shown, and data represent mean ± SEM from three independent experiments (*P < 0.01; **P < 0.001). WT, wild type. (E) HeLa cells and their USP7 KO derivatives were treated with MG132 (10 μM) for 4 h. Cell lysates were prepared, and IP experiments were performed under denaturing conditions using Protein A beads conjugated with anti-RNF169 antibodies. Western blotting experiments were performed using indicated antibodies. (F) Strep-Flag-USP7 proteins were incubated at 37 °C for 10 min with immobilized Flag-ubiquitylated proteins, including Myc-RNF169. Thereafter, reaction products were boiled in Laemmli buffer, separated by SDS/PAGE, and processed for Western blotting using anti-Myc antibodies to determine the ubiquitylation status of Myc-RNF169. (G) Cell lysates derived from control or USP7 KO HeLa cells and their reconstituted counterparts (WT and mutants) were subjected to Western blotting analysis. Relative RNF169 expression from three experiments is plotted (***P < 0.001 vs. HeLa). ns, not significant. (H) USP7 KO HeLa cells were cotransfected with Flag-RNF169, HA-tagged USP7 (WT and mutants), or HA vector. Cells were treated with 10 μM MG132 for 4 h and were subsequently processed for IP experiments using anti-Flag beads under denaturing conditions. Western blotting analyses were performed using indicated antibodies. (I) RNF169 KO HeLa cells reconstituted with WT RNF169 or its mutants (∆C6 or 2KR) were subjected to 50 μg/mL CHX, and cells were harvested at indicated time points. Proteins were separated by SDS/PAGE and were immunoblotted with indicated antibodies. Quantification is shown and represents data of three independent experiments (**P < 0.01 vs. WT). (J) RNF169 KO HeLa cells stably expressing Flag-tagged RNF169 (WT and 2KR) were treated with or without IR before the ubiquitylation status of RNF169 proteins was analyzed. Procedures are essentially as described in H. (K) USP7 deubiquitylated RNF169, but not its 2KR mutant, in vitro. The experiment was performed essentially as in F. (L) Representative histological photomicrographs demonstrating direct correlation between nuclear RNF169 and nuclear USP7 expression in breast cancer. Statistics analyses revealed a significant correlation (P < 0.05) between immunohistochemical expression of RNF169 and USP7 (Materials and Methods). (Magnification: 10× in L; scale bars, 20 μM in Insets.)
To extend these observations and to test the functional significance of USP7 DUB activity and its binding to RNF169 in DDRs more efficiently, we generated USP7 KO cells using the CRISPR/CAS9 methodology. We designed two guide (g)RNA sequences to target the USP7 genetic locus and screened for USP7 KO cells by Western blotting analyses (Materials and Methods). In line with results derived from RNA interference (RNAi) studies, USP7-inactivated cells expressed RNF169 at lower levels compared with their parental cells (Fig. 4C). The fact that a reduction in RNF169 protein levels was observed in independent clones derived from two separate USP7-targeting events argues against the possibility of off-target effects. To corroborate the idea that USP7 promotes RNF169 stability, we also performed the cycloheximide assay to monitor RNF169 turnover. Because the steady-state protein level of RNF169 is reduced in USP7 KO cells (Fig. 4C), we adjusted protein loading such that RNF169 protein level is similar among wild-type and KO cells at time 0 for better comparison. Consistently, translation inhibition led to appreciable RNF169 turnover and was markedly increased in USP7 KO cells (Fig. 4D). Because USP7 plays established roles as a deubiquitinase, we further tested whether USP7 may deubiquitylate RNF169. We pretreated USP7 proficient cells and KO derivatives with proteasome inhibitor MG132, immunoprecipitated RNF169 under denaturing conditions, and examined the ubiquitylation status of RNF169. In support of a role of USP7 in DUB and stabilizing RNF169, ubiquitylated species of RNF169 were substantially increased in USP7 KO cells (Fig. 4E and Fig. S4A). To test directly if USP7 deubiquitylates RNF169, we purified USP7 and RNF169 proteins. We first purified USP7 proteins from 239T cells overexpressing either wild-type USP7 or its DUB-inactive mutant C223S (Fig. 4F, Top). We also obtained ubiquitylated RNF169 protein preparations from 293T cells that ectopically expressed ubiquitin and Myc-tagged RNF169. Using these preparations, we found that coincubating USP7, but not its catalytic inactive mutant (C223S), with ubiquitylated RNF169 protein preparations led to robust RNF169 deubiquitylation (Fig. 4F).
Fig. S4.
(A) HeLa cells [wild type (WT)] and their USP7-deficient derivatives (clones 4 and 7) were cotransfected with Flag-ubiquitin and Myc-RNF169 expression constructs. Twenty-four hours after transfection, cells were treated with MG132 (10 μM for 4 h) and were subsequently processed for IP experiments under denaturing conditions (buffer containing 1% SDS) using anti-Flag agarose beads. Western blotting analyses were performed using indicated antibodies. (B) USP7 KO HeLa cells (clone 4) were cotransfected with Flag-ubiquitin, Myc-RNF169, and HA-tagged USP7 (WT and mutants) or an empty HA vector. Cells were processed, and the ubiquitylated status of RNF169 was analyzed as described in A. (C) RNF169 KO HeLa cells were cotransfected with Flag-ubiquitin and Myc-RNF169 or Myc-RNF169 (2KR), and were treated with or without ionizing radiation (IR) before being processed to determine the ubiquitylation status of RNF169 essentially as described for A.
To evaluate whether the RNF169-stabilizing effect requires USP7 DUB activity or its interaction with RNF169 in vivo, we reconstituted USP7 KO cells (clone 4) with USP7 alleles that encode wild-type USP7, its DUB-inactive C223S mutant, or its RNF169-binding defective M1 mutant. By immunoblotting for endogenous RNF169 at steady states, we found that reintroduction of USP7, but not its DUB or M1 mutants, restored RNF169 expression to a level that resembles the level in isogenic parental HeLa cells (Fig. 4G). Furthermore, by analyzing the ubiquitylation status of Flag epitope-tagged RNF169, we found that RNF169 proteins were more readily ubiquitylated in USP7 C223S and M1 mutant-expressing cells (Fig. 4H and Fig. S4B), supporting the idea that USP7 deubiquitylates RNF169 in a manner that requires its DUB activity and its ability to interact with RNF169.
To test the importance of the USP7–RNF169 complex formation further in conferring RNF169 stability, we performed a cycloheximide experiment to determine the stability of RNF169 and its USP7-binding defective mutant in a RNF169-null background. In agreement with an important role of USP7 binding in conferring RNF169 protein stability, the RNF169∆C6 and the 2KR mutant, both of which were USP7-binding defective (Fig. 3F), displayed significantly faster turnover rates compared with wild-type RNF169 (Fig. 4I). These observations are in line with the higher level of ubiquitylated species of RNF169 2KR compared with its wild-type counterpart (Fig. 4J and Fig. S4C). Finally, compromising the USP7–RNF169 interaction severely hampered USP7-dependent RNF169 deubiquitylation in vitro (Fig. 4K). Together, these results are entirely consistent with the notion that RNF169 protein stability is enhanced via its interaction with USP7, and that the stabilizing effect can be seen in both unperturbed and irradiated cells. Together, we concluded that USP7, via its interaction with RNF169, deubiquitylates and promotes RNF169 stability.
Nuclear RNF169 Expression Shows a Direct Correlation with Nuclear USP7 in Breast Cancer.
USP7 is frequently amplified in breast cancers (cBioPortal; Fig. S5). To lend credence to the direct relationship between USP7 and RNF169 in vivo, immunohistochemical expression of USP7 and RNF169 was studied on a panel of breast cancer samples (141 cases) with a high proportion of invasive ductal carcinoma (118 cases). Details of the profiles of breast cancer cases, including tumor stages and tumor grades, are described in Table S2. To this end, we evaluated nuclear staining intensities of USP7 and RNF169 (Fig. 4L), and performed Pearson and χ2 analyses. The nuclear expression of USP7 and RNF169 was scored semiquantitatively according to their intensity and percentage of stained tumor cells. The median values were used as a cutoff between high and low nuclear expression. Importantly, statistical analyses revealed a significant positive correlation between nuclear USP7 and nuclear RNF169 protein levels (P = 0.034; Fig. 4L and Tables S3 and S4), providing support for an in vivo role of USP7 in regulation of RNF169 protein stability.
Fig. S5.
Cross-cancer alteration summary for USP7. Cutoff at altered samples = 5% (data from cBioPortal). adeno, adenocarcinoma; Broad, Broad Institute; BRCCRC, British Columbia Cancer Research Center; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; DLBCL, diffuse large C-cell lymphoma; FHCRC, Fred Hutchinson Cancer Research Center; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; MPNST, malignant peripheral nerve sheath tumor; MSKCC, Memorial Sloan Kettering Cancer Center; NEPC, neuroendocrine prostate cancer; PCNSL, primary central nervous system lymphoma; pub, publication; TCGA, The Cancer Genome Atlas.
Table S2.
Profile of breast cancer cases in TMA
| Description | No. of cases, % |
| Patients | 141 |
| Year range of patients | 1993–2003 (10 y) |
| Medium age of first diagnosis, y | 56 (24–85) |
| Mean of survival months | 116.3261 (4–254) |
| Tumor stage | |
| I | 16 (11.35%) |
| II | 25 (17.73%) |
| III | 6 (4.26%) |
| IV | 5 (3.55%) |
| Missing | 89 (63.12%) |
| Tumor grade | |
| 1 | 20 (14.18%) |
| 2 | 30 (21.28%) |
| 3 | 76 (53.90%) |
| Missing | 15 (10.64%) |
| Lymph node metastasis | |
| Positive | 67 (47.52%) |
| Negative | 60 (42.55%) |
| Missing | 14 (9.93%) |
| Estrogen receptor | |
| Positive | 73 (51.77%) |
| Negative | 25 (17.73%) |
| Missing | 43 (30.50%) |
| Progesterone receptor | |
| Positive | 50 (35.46%) |
| Negative | 38 (26.95%) |
| Missing | 53 (37.59%) |
Table S3.
χ2 correlation between RNF169 and USP7 expression in breast cancer
| Parameters | USP7 expression | Total | |
| Low | High | ||
| RNF169 expression | |||
| Low | 28 | 15 | 43 |
| High | 23 | 30 | 53 |
| Total | 51 | 45 | 96 |
The nuclear expression of USP7 and RNF169 was scored semiquantitatively according to their intensity and percentage of stained tumor cells. The median values were used as a cutoff between high and low expression. The χ2 correlation between RNF169 and USP7 expression showing significant correlation, P = 0.034.
Table S4.
Pearson correlation between RNF169 and USP7 expression in breast cancer
| Parameters | RNF169 expression | USP7 expression |
| RNF169 expression | ||
| Pearson correlation | 1 | 0.216* |
| Sig. (two-tailed) | 0.034 | |
| N | 108 | 96 |
| USP7 expression | ||
| Pearson correlation | 0.216* | 1 |
| Sig. (two-tailed) | 0.034 | |
| N | 96 | 104 |
The nuclear expression of USP7 and RNF169 was scored semiquantitatively according to their intensity and percentage of stained tumor cells. The median values were used as a cutoff between high and low nuclear expression. The Pearson correlation between RNF169 and USP7 nuclear expression showing significant (Sig.) positive correlation is P = 0.034.
Correlation is significant at the 0.05 level (two-tailed).
USP7 Promotes RNF169 Loading at DSBs.
RNF169 limits excessive accumulation of DNA damage mediator proteins 53BP1 and RAP80 at DSBs by competing for RNF168-catalyzed ubiquitin adducts (14, 20, 21). Given that RNF169 readily accumulates at ionizing radiation-induced foci (IRIF), we first examined whether USP7 may be important in mobilizing RNF169 in ionizing radiation-treated cells. We speculated that USP7 may enforce RNF169 functions at DSBs by enhancing its stability. To this end, we first silenced USP7 using siRNAs and then examined RNF169 IRIF in U2OS cells with stable expression of Flag-tagged RNF169. Indirect immunofluorescence staining experiments revealed that USP7 knockdown compromised Flag-tagged RNF169 IRIF, at least in part, by reducing RNF169 protein levels (Fig. 5 A–C). Similar effects were observed when experiments were conducted to examine IRIF of endogenous RNF169 (Fig. 5 D–F). Furthermore, pretreatment of cells with USP7 inhibitor P22077 or P5901 resulted in a marked reduction in RNF169 protein and accumulation at DSBs (Fig. 5 G–I), indicating that USP7 DUB activity is important in driving RNF169 accumulation at DSBs.
Fig. 5.
USP7 promotes RNF169 loading onto DSBs. (A) U2OS cells stably expressing Flag-RNF169 were transfected with small interfering control (siCTR) or USP7-targeting siRNAs (siUSP7-1 or siUSP7-2). Forty-eight hours after transfection, cells were subjected to IR treatment (10 Gy) and were subsequently processed for immunostaining experiments using anti-Flag and anti–γ-H2AX antibodies. (B) Quantification is shown for results derived from three independent experiments. Data represent mean ± SEM (**P < 0.01 vs. siCTR). (C) Western blotting analysis was performed to assess RNAi-mediated knockdown efficiency in A. (D–F) RNF169 IRIF were assessed in HeLa cells following essentially the same procedures described in A–C (**P < 0.01 vs. siCTR). (G–I) U2OS cells stably expressing Flag-RNF169 were treated with DMSO, P22077 (20 μM), or P5091 (15 μM). Cells received IR treatment (10 Gy) and were processed for immunostaining or Western blotting experiments 4 h afterward. Quantification of results in G is shown, and the results are derived from three independent experiments (***P < 0.001). Western blotting analyses were performed using standard procedures with indicated antibodies. (J) Parental HeLa cells and USP7 KO and their derivatives (WT, C223S, M1) were subjected to IR treatment (10 Gy). Cells were preextracted before paraformaldehyde fixation 6 h afterward, and were processed for immunostaining experiments using anti-RNF169 and anti–γ-H2AX antibodies. (K) Quantification of results derived from three independent experiments is shown, and depicts the percentage of cells with RNF169 IRIF (***P < 0.001). ns, not significant. (Magnification: 60×.)
To examine more definitively the requirement of USP7 in supporting RNF169 docking at DSBs, we also reconstituted USP7 KO HeLa cells with wild-type USP7 to exclude off-target effects (Fig. 4G) and showed that reconstitution with HA-tagged USP7 restored RNF169 IRIF (Fig. 5 J and K). This observation was in sharp contrast to USP7 KO cells that were engineered to express the USP7 DUB mutant (C223S) or the RNF169-binding defective mutant (M1) (Fig. 5 J and K). Together, we concluded that USP7 promotes RNF169 stability and IRIF in manners that required its DUB activity.
The USP7–RNF169 Axis Promotes HR Repair and Confers PARP Inhibition Resistance.
RNF169 promotes high-fidelity DNA repair, possibly by restraining the ubiquitin-mediated DSB signaling events (14). Because USP7 promotes RNF169 stability, we first tested whether down-regulation of USP7 may also compromise DSB repair using a gene conversion-based DSB repair reporter (Fig. 6A). Following two rounds of siRNA transfection, cells were electroporated with an Isce1 expression construct to induce a DSB, and the percentage of cells positive for GFP was subsequently monitored by flow cytometric analysis. In resemblance to RNF169 inactivation, cells depleted of USP7 impaired DSB repair (Fig. 6 B and C). To examine the importance of USP7 and its ability to interact with RNF169 in supporting DSB repair, we reconstituted RNF169 KO cells with wild-type RNF169 or its mutant alleles (Fig. 6D). Consistent with the idea that USP7 enforces RNF169-dependent DDRs, reintroduction of wild-type RNF169, but not its USP7-binding defective mutant 2KR, enhanced high-fidelity DSB repair (Fig. 6D). Given that HR deficiency confers cell hypersensitivity to poly (ADP-ribose) polymerase inhibition (PARPi), we performed clonogenic survival assays to determine the roles of the USP7–RNF169 axis in response to Olaparib. Although reconstituting RNF169 KO cells with wild-type RNF169 protected cells from PARPi, cells expressing the USP7-binding defective 2KR mutant did not alleviate PARP hypersensitivity (Fig. 6 E and F). Given the putative role of USP7 in stabilizing RNF169, we reasoned that if the RNF169 2KR mutant was overexpressed, that overexpression might compensate for its relatively higher turnover rate. To this end, we transfected RNF169 KO HeLa cells with increasing molar amounts of RNF169 2KR plasmids and assayed for HR repair using the plasmid-based direct repeat (DR)-GFP reporter (Fig. 6 G and H). Consistent with the idea that USP7 enforces RNF169-dependent DNA repair by enhancing its stability, overexpression of RNF169 2KR promoted HR repair to the same level in cells expressing wild-type RNF169 (Fig. 6 G and H). We also reconstituted USP7 KO cells with wild-type USP7, its DUB-inactive C223S mutant, and the RNF169-binding defective mutant (M1) to evaluate the contribution of the USP7–RNF169 interaction in HR repair. Whereas reintroduction of wild-type USP7 led to marked elevation in HR repair events, cells expressing the DUB mutant were indistinguishable from its KO counterparts, indicating that USP7 promotes HR repair in a manner that requires its DUB activity (Fig. 6 I and J). Importantly, expression of the RNF169-binding defective USP7 mutant failed to promote HR repair to levels supported by wild-type USP7 (Fig. 6 I and J), supporting the notion that the USP7–RNF169 interaction contributes, at least in part, to USP7-dependent HR repair. Together, these data reveal a role of USP7 in promoting both RNF169-dependent and RNF169-independent DDR functions.
Fig. 6.
USP7-RNF169 axis promotes HR repair. (A) Schematic of gene conversion-based HR reporter is shown. GFP positivity reflects DSB repair via the HR pathway. (B) DR-U2OS cells pretreated with indicated siRNAs were transfected with plasmid encoding the I-Sce1 endonuclease. Flow cytometric analysis of the GFP-positive cell population was performed 48 h posttransfection. Data represent mean ± SEM from three independent experiments (**P < 0.01; ***P < 0.001). (C) Western blotting experiment was performed to assess RNAi-mediated knockdown efficiency in cells used in B. (D) RNF169 KO HeLa cells stably expressing vector only or RNF169 (WT and mutant) were electroporated with plasmids encoding I-Sce1 and DR-GFP. Cells were harvested and subjected to flow cytometric analysis 48 h postelectroporation. Data represent mean ± SEM from three independent experiments (*P < 0.05). (E) HeLa RNF169 KO cells stably expressing vector only and RNF169 (WT and mutants) were treated with indicated doses of the PARPi Olaparib. Cells were allowed to grow for 10 d before they were stained with Coomassie Blue. Data represent mean ± SEM from three independent experiments (**P < 0.01; ***P < 0.001). (F) Western blotting analysis of the protein level of RNF169 in cells used in D and E. (G and H) RNF169 KO HeLa cells were mock-transfected or transiently transfected with expression constructs encoding either HA-tagged RNF169 (WT) or its 2KR mutant in addition to the plasmid-based DR-GFP reporter and an Isce1 plasmid to evaluate HR repair efficiency. HA-RNF169 2KR plasmid was transfected at increasing molar ratios to cells transfected with WT RNF169 (1:1, 2:1, and 3:1 ratios). Cells were subjected to flow cytometric analysis (G) or Western blotting analysis (H) using indicated antibodies. Results represent mean ± SEM from three independent experiments (*P < 0.05). (I and J) USP7 KO HeLa cells reconstituted with empty vector (HA), WT USP7 (WT), its DUB mutant (C223S), or the RNF169-binding defective mutant (M1) were subjected to the gene conversion assay to evaluate HR repair efficiency. (I) Results represent mean ± SEM from three independent experiments (*P < 0.05; ***P < 0.001). (J) Western blotting analysis was performed to assess HA-USP7 and RNF169 expression. HA, human influenza hemagglutinin.
Discussion
To promote cell survival and organismal development, cells have put in place self-limiting mechanisms to prevent DDRs from exceeding physiological thresholds. Indeed, unrestrained DDRs can lead to undesirable DNA rearrangements, prolonged cell arrest, or premature cell commitment to apoptosis (16). As such, the identification of Ring finger protein RNF169, which antagonizes ubiquitin-mediated DSB signals (14, 20, 21), represents an exciting opportunity to dissect how cells limit chromatin responses at DSBs. Notably, despite its implicated roles as a negative regulator of DSB responses, exactly how RNF169 is regulated is not known. In this study we document an unprecedented role of an NLS that supports the nuclear function of RNF169. We show that the RNF169 NLS serves not only to target the Ring finger protein into the nucleus but also enforces its occupancy at DSBs by mediating its interaction with USP7, a deubiquitylase that protects it from proteolytic degradation. Intriguingly, this dual utility of NLS seems to be unique to RNF169, because USP7-binding KxxxK motif and NLS do not overlap in other USP7 UBL-binding proteins. Accordingly, genetic ablation or chemical inhibition of USP7 destabilizes RNF169, compromises high-fidelity DSB repair, and hypersensitizes cells to PARPi. Together, we propose that the USP7–RNF169 axis is integral to the DSB signal transduction cascade and allows fine-tuning of chromatin responses in the DSB microenvironment (Fig. 7).
Fig. 7.
Proposed working model. In the presence of USP7, RNF169 is stabilized and productively assembles at DSBs to facilitate HR-driven DNA repair. In the absence of USP7, RNF169 undergoes active degradation, resulting in attenuated accumulation at DSBs and impaired HR repair.
There has been a surge of interest in elucidating the complex structures of the USP7 UBL domain with its interacting proteins (39, 41–44). Among them, two crystal structures detailing USP7 UBL1–3 domains with peptides derived from the proteins ICP0 and UHRF1 were recently reported (39, 42). In these structures, the two lysine residues from ICP0 and UHRF1 peptides were anchored on the acidic patch across the surface of the UBL1 and UBL2 domains via hydrogen bonds and electrostatic attractions, undertaking major tasks for their mutual recognition. In the complex structure of the USP7–ICP0 complex, K620 and K624 from ICP0 insert their respective side chains into two binding pockets situated ∼12 Å apart. One pocket (pocket A) is mainly constituted by USP7 residues M637, K681, D684, and D762, whereas the other one (pocket B) is composed of residues R628, E759, M761, and D764. On the contrary, in the complex structure of USP7–UHRF1, K659 buries its side chain into the pocket B when K657 also interacts with the peripheral region of pocket B, suggesting that the two lysine residues share the same binding pocket, leaving pocket A unoccupied. In our structure, RNF169 binds the USP7 UBL domains in a very similar mode as ICP0, occupying both pockets A and B using K627 and K623, respectively. It is very interesting that although the three peptides derived from ICP0, UHRF1, and RNF169 all bear the conserved KxxxK motif (except that UHRF1 has an extra K in the middle), they bind UBL domains in two distinctive ways. In addition, a number of lysine and arginine residues are found in the vicinity of the USP7-binding KxxxK motif. Intriguingly, even though both K and R maintain the same charge and a long side chain, arginine has not been observed to make contacts with the two above-mentioned binding pockets, suggesting that these two pockets might only be large enough to accommodate the side chains of lysine residues. Consistent with this speculation, simple replacement of the side chain of lysine residues in our structure with the side chain of arginine residues revealed steric hindrance of the binding pockets (Fig. S2), which is further corroborated by our ITC results examining the binding between USP7 and RNF169 K623R, K627R, and 2KR mutants (Fig. 3E). Taken together, we propose that USP7 UBL domains use the acidic pocket(s) to localize the KxxxK motif of its interacting partners and that pocket B turns out to be more crucial in the binding selectivity, considering it is the only lysine-occupied pocket in all three available crystal structures. We propose that the specific binding patterns may vary depending on the position of the lysines and different residue composition surrounding the KxxxK motif.
RNF169 serves structural roles at DSBs as it competes for and physically occupies RNF168-catalyzed ubiquitin adducts (14, 20, 21). Given the dynamic remodeling nature of the DSB-flanking chromatin, one would envisage that RNF169 may be subjected to stringent regulation. In line with this idea, we found that RNF169 turnover is actively driven by the ubiquitin/proteasome system. In an attempt to identify the ubiquitylating activity, we examined cells depleted of candidate E3 ubiquitin ligases, including RNF168 (20), but were unable to detect changes in RNF169 stability (Fig. S6A). Similarly, the observation that RNF169 and its RING-inactivated mutant displayed similar half-lives also argues against autoubiquitylation as the primary driver of RNF169 degradation (Fig. S6 B and C). Although further work will be required to appreciate the homeostatic balance of RNF169 fully, our current findings firmly establish the ubiquitin-specific protease USP7 as a key deubiquitinase that promotes RNF169 stability and highlight how USP7, via RNF169, promotes high-fidelity DSB repair and cell resistance to genotoxic stress.
Fig. S6.
(A) U2OS or HeLa cells were treated with nontargeting [small interfering control (siControl)] or RNF168-specific (siRNF168) siRNAs. Cell lysates were separated by SDS/PAGE and were subjected to Western blotting analyses using indicated antibodies. The asterisk denotes nonspecific bands. (B) RNF169 KO HeLa cells reconstituted with WT RNF169 or its Really Interesting New Gene (RING) domain deletion mutant (ΔRING) were subjected to 50 μg/mL cycloheximide (CHX). Cells were harvested at the indicated time point for processing. Proteins were separated by SDS/PAGE and were processed for immunoblotting analyses using indicated antibodies. (C) Quantification is shown and represents data (mean ± SEM) derived from three independent experiments.
Deubiquitylases have emerged as having key regulatory activities with diverse roles (45), including tumor suppressor functions (46). Among the dozens of deubiquitinases, USP7 is arguably one of the most studied deubiquitylases; and in the DDR arena, it serves a multitude of genome integrity protective roles by stabilizing a cohort of DDR factors, including Rad18 (25), CHK1 (47), DNA polymerase eta (48), XPC (49), and Claspin (27). Our findings here not only add RNF169 to the expanding repertoire of USP7 functions; in addition, the fact that the USP7–RNF169 axis supports high-fidelity DNA repair raises the possibility that misexpression of USP7 may compromise genome stability. Although USP7 has been associated with a number of human cancers, it would be of significant interest to examine how perturbing the USP7–RNF169 axis may also contribute to other genome instability-associated human diseases.
Prompted by the recent characterization of small molecules that specifically target USP7 (30–33), and their potentials as anticancer therapeutics, a thorough understanding of the pharmacological effects of USP7 inhibition is pivotal for clinical evaluation of these promising small molecules. Our uncovering of an early role of USP7 in the DDR represents an exciting direction to pursue in defining the biological roles of the deubiquitinases further. Intriguingly, our cell survival assays indicated that perturbing the USP7–RNF169 complex formation rendered cells hypersensitive to Olaparib and suggest that inhibitors that specifically inactivate the USP7–RNF169 interaction may prove useful, alone or in combination with PARPi, in selective killing of HR-deficient human cancers. Because all current USP7 inhibitors target its DUB activity, our findings provide a good rationale for drug USP7 UBL interactions when treating human cancers.
Materials and Methods
Cell Cultures.
U2OS, 293T, and HeLa cells were originally from the American Type Culture Collection and were cultured in DMEM supplemented with 10% (vol/vol) FBS at 37 °C in 5% CO2. For generation of cell lines with doxycycline-inducible expression of RNF169, the RNF169 cDNA was cloned in frame 3′ of the SFB sequence. U2OS cells were infected with lentiviral particles carrying RNF169-SFB expression constructs and were subsequently selected by 2 mg/mL G418. Expression of RNF169-SFB was induced by supplementing cell culture media with doxycycline at 1 μg/mL for 12 h. Generation of cell lines with constitutive expression of RNF169 (wild type and mutants) and USP7 (wild type and mutants) was performed as previously described (50).
Generation of RNF169 and USP7 KO Cells.
RNF169 KO cells were generated using transcription activator-like effector nuclease (TALEN) technology. The target sequence was 5′-TGCTGGGCGAGTGCGCCCGCcgcagccaacccgagCGCTGCCGCCCGCGCCGGGA-3′. USP7 KO cells were generated using the CRISPR-Cas9 method as previously described (51). Briefly, cells were cotransfected with vectors that encode the modified target-specific gRNA, human Cas9 (hCas9), and GFP at a molar ratio of 4:5:1. Cells were sorted for GFP positivity 48 h after transfection, and were individually seeded into 96-well plates. Positive clones were confirmed by Western blotting analysis. Guide RNA sequences designed to target USP7 were 5′ ATTTCGCACAAAACACGGA 3′ (USP7-gRNA1) and 5′ AGAAGCACACAGGCTACGT 3′ (USP7-gRNA2). USP7 KO 4 and 7 were derived from USP7-gRNA1 and USP7-gRNA2, respectively.
Plasmids and siRNAs.
The Myc-USP7 plasmid was a gift from Lori Frappier (The University of Toronto, Toronto, ON, Canada). For all other epitope-tagged RNF169 and USP7 expression constructs (wild type and mutants), cDNAs were subcloned into pDONR201 using Gateway technology (Invitrogen) and were subsequently transferred to Gateway-compatible destination vectors for bacterial or mammalian expression studies. All constructs were verified by sequencing. The gRNA and hCas9 cloning vectors were gifts from George Church (Addgene plasmids 41824 and 41815, respectively). The pDR-GFP was a gift from Maria Jasin (Addgene plasmid 26475). For RNAi-mediated depletion experiments, cells were transfected twice with either nontargeting control or target siRNAs (Dharmacon) using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. Sequences for the siRNAs designed to target USP7 were 5′-ACCCUUGGACAAUAUUCCUdTdT-3′ (siUSP7 1) and 5′-AGUCGUUCAGUCGUCGUAUUU-3′ (siUSP7 2). Sequences of RNF169-targeting siRNAs were previously described (20).
Antibodies and Chemicals.
Antibodies against γ-H2AX, 53BP1, RNF168, and RNF169 were described previously (20). Rabbit polyclonal antibodies against DYRK1A were raised in a GST-tagged DYRK1A N-terminal fragment (corresponding to amino acid resides 1–499) and were purified using a column conjugated with MBP-tagged DYRK1A N terminus proteins. Details of other antibodies and their sources are as follows: anti-CHK1 (SC8408; Santa Cruz Biotechnology), FK2 (04263; Millipore), anti-PCNA (PC10; Santa Cruz Biotechnology), anti-Actin (A5441; Sigma), anti-Ubiquitin (05944; Millipore), anti-Flag (M2, F3165; Sigma), anti-Myc (sc-40; Santa Cruz Biotechnology), anti-HA (MMS-101R; Covance), anti-USP7 (A300-033A; Bethyl Laboratories), and anti-p53 (sc126; Santa Cruz Biotechnology). Chemicals and their sources are as follows: MG132 (C2211; Sigma), deubiquitinase inhibitor 1,10-phenanthroline monohydrate (P9375; Sigma), and USP7 inhibitors P22077 (S7113; Selleckchem) and P5091 (S7132; Selleckchem).
IP and Western Blotting.
For whole-cell extracts, cells were lysed with NETN buffer [20 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, 0.5% Nonidet P-40, and 1 mM EDTA] supplemented with BitNuclease (Biotool) on ice for 15 min. Cell lysates were boiled with SDS loading buffer. Proteins were separated by SDS/PAGE, transferred to PVDF membranes, and blotted with indicated antibodies. For co-IP experiments, cells were lysed with NETN buffer for 15 min on ice, followed by centrifugation at 18,407 × g (Eppendorf Centrifuge, Hamburg, Germany, 5424R, 24-place Aerosol-tight fixed-angle rotor) for 10 min at 4 °C. Supernatants were incubated with either Streptavidin beads (GE Healthcare) or anti–HA-conjugated agarose beads (Biolegend) for 4 h at 4 °C with rotation. Beads were subsequently washed three times with NETN buffer and boiled with SDS loading buffer.
In Vivo Ubiquitination Assay.
HeLa cells stably expressing HA-Flag epitope–tagged RNF169 were treated with 10 μM MG132 for 4 h before harvesting. Cells were lysed with denaturing buffer [20 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 0.5% Nonidet P-40, 0.5% deoxycholate, 0.5% SDS, and 1 mM EDTA] supplemented with the DUB inhibitor 1,10-phenanthroline monohydrate on ice for 10 min, followed by boiling at 95 °C for 5 min. The cell lysates were cooled on ice for another 5 min before incubating with anti-Flag (M2) beads for 4 h at 4 °C. Beads were washed four times with denaturing buffer and boiled with SDS loading buffer. To detect endogenous RNF169 ubiquitination, cell lysates were incubated with anti-RNF169 antibody, together with Protein A agarose beads, at 4 °C overnight. Reciprocal IP experiments by immunoprecipitating Flag-ubiquitin were performed essentially the same as above, except that cells were lysed in denaturing buffer containing 1% SDS.
Protein Purification.
MBP-tagged RNF169 (wild type and mutant) and GST-tagged USP7 fusion proteins were expressed in Escherichia coli BL21 cells induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37 °C for 6 h. For MBP-tagged protein, cells were harvested and resuspended in MBP-binding buffer (20 mM Tris⋅HCl, 200 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, and 10 mM beta-mercaptoethanol), followed by sonication for 30 s, twice. The extracts were centrifuged at 16,200 × g (Hitachi Centrifuge, Tokyo, CR21GII, R20A2) for 30 min, and the supernatants were collected and incubated with amylose resins at 4 °C overnight. Bound proteins were washed three times with MBP-binding buffer and resuspended for pull-down assay. For GST-tagged protein, cells were resuspended in NETN buffer, followed by sonication and centrifugation. The supernatant was incubated with glutathione Sepharose beads at 4 °C overnight. After washing with NETN buffer three times, the bound protein was eluted with 50 mM Tris (pH 8.8) containing 20 mM reduced glutathione.
MBP Pull-Down Assay.
For pull-down assays, MBP-RNF169 resins were incubated with cell lysates derived from 293T cells expressing the indicated proteins for 4 h at 4 °C. Resins were washed four times with NETN buffer and boiled with SDS loading buffer. Protein complexes were subjected to Coomassie Blue staining or Western blotting analyses with indicated antibodies. For pull-down experiments involving bacterially expressed and purified proteins, resins coupled with full-length or truncated MBP-RNF169 were incubated with GST-USP7 fusion protein for 4 h at 4 °C, followed by the same steps described above.
Cycloheximide Chase Experiment.
Cells were seeded onto 60-mm dishes with cell culture media supplemented with 50 μg/mL cycloheximide. Cells were harvested at indicated time points, and lysates were subjected to Western blotting experiments to analyze protein expression at indicated time points. Image J software (NIH) was used to quantify the relative expression level compared with control treatment.
Immunofluorescence Staining.
Procedures for indirect immunofluorescence experiments were as previously described (50). Briefly, cells grown as monolayers on coverslips were exposed to ionizing radiation (10 Gy). Cells were washed with PBS and fixed with 3% (wt/vol) paraformaldehyde for 15 min at room temperature. Subsequently, cells were permeabilized in 0.5% Triton solution for 30 s. To detect endogenous RNF169 IRIF, cells were preextracted with Triton solution for 30 s, followed before fixing with paraformaldehyde. Cells were stained by sequential incubation of primary antibodies and secondary fluorophore-conjugated antibodies for 45 min, respectively. Images were acquired using an Olympus BX51 fluorescence microscope.
Homologous Recombination Assay.
For experiments conducted with U20S DR-GFP cells, cells were electroporated with the I-Sce1 expression construct pCBASce (5 μg) after two rounds of siRNA transfection, and were collected 48 h afterward for flow cytometric analysis. To compare the HR rates in RNF169- or USP7-null HeLa cells, KO cells and their reconstitution counterparts were cotransfected with pCBASce (5 μg) and DR-GFP (5 μg), and they were harvested 48 h posttransfection for flow cytometric analyses.
Clonogenic Survival Assay.
Cells were seeded on 60-mm dishes at a density of 1,500 cells per dish at day 0. After 24 h of cell seeding, cells were treated with Olaparib at indicated doses or its solvent DMSO as a control. Cells were then allowed to grow for 10 d, and were fixed with Coomassie Blue solution. The plates were photographed, and colony counts were quantified using QuantityOne software.
In Vitro Ubiquitylation Assay.
Streptavidin-binding peptide–Flag (Strep-Flag)-tagged USP7 proteins were purified from 293T cells transiently transfected with expression constructs of USP7 wild type or its DUB mutant C223S. Strep-Flag-USP7 proteins were immunoprecipitated by incubating cell lysates with Streptavidin beads with gentle agitation at 4 °C, and were subsequently eluted with 1 mg/mL biotin. Ubiquitylated RNF169 protein preparations were obtained from 293T cells that ectopically expressed Flag-ubiquitin and Myc-RNF169 (or its 2KR mutant). Flag-tagged ubiquitylated proteins, including Myc-RNF169, were immunoprecipitated under denaturing conditions [20 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 0.5% Nonidet P-40, 0.5% deoxycholate, 1% SDS, and 1 mM EDTA supplemented with the DUB inhibitor 1,10-phenanthroline monohydrate]. Immobilized Flag-ubiquitylated proteins were washed once with denaturing buffer and then with NETN buffer. To evaluate whether USP7 deubiquitylates RNF169 in vitro, purified Strep-Flag-USP7 or its DUB mutant was coincubated with Flag-ubiquitylated proteins at 37 °C for 10 min. Thereafter protein samples were boiled in Laemmli buffer, separated by SDS/PAGE, and processed for Western blotting using anti-Myc antibodies.
Statistical Analysis.
Data are represented as mean ± SEM of at least three independent experiments, and the Student’s t test (two-tailed) was used to compare two groups for independent samples. P < 0.05 was considered as statistically significant.
Crystallization and Structure Determination.
For the structural determination and ITC experiments, DNA-encoding UBL1–3 domain (561–891 aa) was subcloned into pET28a-modified plasmid (Novagan), which contains an 8× His-sumo tag and a ULP1 cleavage site at the N terminus. The plasmids were subsequently transformed into BL21 (Gold) cell strains. The cells were initially grown at 37 °C, when cell density reached OD600 at 1.0, induced by IPTG, with a final concentration of 0.2 mM, and growth was continued at 16 °C for 24 h. Cells were collected and suspended with nickel-nitrilotriacetic acid (Ni-NTA)–binding buffer [20 mM Tris⋅HCl and 1 M NaCl (pH 8.0)]. After sonic lysis, the lysate was cleared by centrifugation and purified through the Ni-NTA column, and the eluent was mixed with ULP1 cleavage enzyme at 10 °C overnight. The protein samples were further purified by MonoQ and Superdex 200 pg (16/60), and then transferred into storage buffer [20 mM Tris⋅HCl and 200 mM NaCl (pH 8.0)] and concentrated to 1 mM.
UBL1–3 (1 mM) protein samples and RNF169 13-aa peptide (620RGRKRHCKTKHLE632) were mixed at a 1:2 ratio and incubated on ice for 1 h. Initial crystals were grown in hanging drops at 20 °C, which contain 0.1 μL of sample and 0.1 μL of buffer, and equilibrated against 100 μL of reservoir solution containing 0.2 M sodium chloride, 8% (wt/vol) PEG 8000, and 0.1 M sodium cacodylate (pH 6.0). The crystals for data collection were refined in solution containing 0.2 M sodium chloride, 6% (wt/vol) PEG 8000, and 0.1 M sodium cacodylate (pH 5.8). For data collection, crystals were transferred into cryoprotectant supplemented with 30% (vol/vol) glycerol and flash-frozen in liquid nitrogen. The X-ray data were collected at beamline BL18U1 of the Shanghai Synchrotron Radiation Facility. The initial data were indexed, integrated, and scaled by the HKL3000 package. The phase was solved by molecular replacement using Phenix, corresponding to PDB ID code 2YLM as the search model. The model build and refinement were carried out by Coot and Phenix.
ITC Measurements.
The wild-type and mutant USP7 UBL1–3 domains were purified without removing the 8× His-sumo tag and stored in storage buffer. Protein concentration was determined based on their respective UV absorption at 280 nm. A MicroCal iTC200 system (GE Healthcare) was used to conduct the ITC measurements. The ITC experiments involved 20 injections of 2 μL of peptide into 200 μL of protein with a spacing time of 120 s. All ITC measurements were carried out at 20 °C. The resultant ITC curves were processed with ORIGEN 7.0 (MicroCal) software using a one-site fitting model.
Tissue Microarray.
One hundred forty-one cases of breast cancer diagnosed between the years 1993 and 2003 with clinical follow-up data were retrieved from the records of the Department of Pathology, Queen Mary Hospital of Hong Kong, with approval by the Institutional Review Board of The University of Hong Kong. The patient profile is summarized in Table S2. Histological sections of all cases were reviewed by a pathologist; representative paraffin tumor blocks were chosen as donor blocks for each case, and the selected areas were marked for construction of tissue microarray (TMA) blocks. The total number of cases that could be assessed and scored was 108 for RNF169 expression and 104 for USP7 expression.
Immunohistochemistry.
The TMA sections were deparaffinized and rehydrated by incubation with xylene and decreasing concentrations of ethanol. Citrate buffer (0.01 M, pH 6.0) was used for antigen retrieval. The slides were immersed into 3% H2O2/methanol for 10 min at room temperature to quench endogenous peroxidase. After rinsing in 0.05% Tween in PBS (PBST) twice, 200 μL of primary antibody was added to each section and incubated at 4 °C overnight. The primary antibodies used were rabbit polyclonal anti-RNF169 antibody diluted at 1:50 (20) (Fig. S7) and rabbit polyclonal anti-USP7 antibody diluted at 1:500 (Bethyl Laboratories, Inc.). The slides were then washed in PBST, and four to five drops of DAKO polymer were applied on each section and incubated at room temperature for 30 min. After washing, chromogen DAB/substrate reagent was added onto the slides and the slides were incubated for a further 3 min. Finally, the slides were dehydrated with increasing concentrations of ethanol, followed by clearing in xylene, and then mounted.
Fig. S7.
HeLa cells pretreated with control siRNAs (siControl) or RNF169-targeting siRNAs (siRNA169) were lysed with NETN buffer supplemented with Bitnuclease. Cell lysates were processed for Western blotting analysis using indicated antibodies.
An Aperio ScanScope system (Aperio Technology) was used to visualize and assess for RNF169 and USP7 expression. The immunostained TMA slides were scanned by ScanScope scanners. The high-resolution tissue section images captured by ScanScope were automatically stored in the Spectrum Database. Individual stained TMA spots were assessed on the computer screen with the use of Aperio’s image viewer, ImageScope.
Tissue sections were scored according to their intensity and the percentage of stained tumor cells in the whole core. Staining was detectable in both the cytoplasm and the nucleus for RNF169 and USP7, but because it is the nuclear expression of RNF169 and USP7 that is functional and relevant to this work, only evaluation of nuclear staining was performed. The intensity of nuclear staining was scored as follows: 1 = weak, 2 = moderate, and 3 = strong. The percentage of cells positively stained was scored as follows: 1 ≤ 25%, 2 ≤ 50%, 3 ≤ 75%, and 4 > 75%. For each case, a final score was obtained by multiplying the score of intensity and the score of percentage, with 12 being the maximum final score. To avoid subjectivity in evaluation, scoring was done by two independent individuals.
Because nuclear RNF169 staining showed significant variation in the intensity of nuclear staining within each tissue core, nuclear scoring was assessed using the H score system, which involves the summation of an individual (intensity × percentage cell) score for each intensity level. Nuclear USP7, on the other hand, showed fairly uniform intensity of nuclear staining within each tissue core; thus, scoring for each intensity level would be redundant and the predominant intensity score could be used for assessment. Statistical analysis was performed using the SPSS program, version 20. The correlation between RNF169 and USP7 expression was analyzed by bivariate Pearson correlation analysis, as well as by χ2 analysis. The median value of RNF169 and USP7 was used as a cutoff between high and low levels of expression, respectively. P values of less than or equal to 0.05 were considered statistically significant.
Acknowledgments
We thank Drs. Lori Frappier and Maria Jasin for sharing invaluable reagents, and acknowledge technical support from Dr. Jing Guo and the Faculty Core Facility (The University of Hong Kong) with imaging and flow cytometric analyses. We thank the staff from the BL18U1 beamline of the National Center for Protein Science Shanghai at the Shanghai Synchrotron Radiation Facility for assistance during data collection. This work is supported by the Research Grants Council Hong Kong (Projects 17108314 and 17102516), by the Outstanding Young Research Award (The University of Hong Kong) (to M.S.Y.H.), by National Natural Science Foundation of China Projects 31270782 and 91540103 (to Q.G.), and by the Committee on Research and Conference Grants (The University of Hong Kong) Project 201309176148 (to U.-S.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinate and structure factor for the USP7-RNF169 complex structure have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5GG4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616602114/-/DCSupplemental.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huen MS, Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci. 2010;35(2):101–108. doi: 10.1016/j.tibs.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwertman P, Bekker-Jensen S, Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol. 2016;17(6):379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- 4.Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem. 2011;286(15):13669–13680. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y, et al. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011;25(7):685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42(3):319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callen E, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153(6):1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escribano-Díaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49(5):872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49(5):858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339(6120):700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato K, et al. Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol Cell. 2014;53(4):617–630. doi: 10.1016/j.molcel.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 13.Sy SM, et al. Critical roles of ring finger protein RNF8 in replication stress responses. J Biol Chem. 2011;286(25):22355–22361. doi: 10.1074/jbc.M111.232041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulsen M, Lukas C, Lukas J, Bekker-Jensen S, Mailand N. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J Cell Biol. 2012;197(2):189–199. doi: 10.1083/jcb.201109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Typas D, et al. The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80. Nucleic Acids Res. 2016;44(6):2976. doi: 10.1093/nar/gkv1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altmeyer M, Lukas J. To spread or not to spread--chromatin modifications in response to DNA damage. Curr Opin Genet Dev. 2013;23(2):156–165. doi: 10.1016/j.gde.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Stewart GS, et al. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci USA. 2007;104(43):16910–16915. doi: 10.1073/pnas.0708408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136(3):420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 19.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Feng W, Jiang J, Deng Y, Huen MS. Ring finger protein RNF169 antagonizes the ubiquitin-dependent signaling cascade at sites of DNA damage. J Biol Chem. 2012;287(33):27715–27722. doi: 10.1074/jbc.M112.373530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panier S, et al. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol Cell. 2012;47(3):383–395. doi: 10.1016/j.molcel.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 22.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17(1):86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kon N, et al. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29(9):1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khoronenkova SV, et al. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell. 2012;45(6):801–813. doi: 10.1016/j.molcel.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zlatanou A, et al. USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene. 2016;35(8):965–976. doi: 10.1038/onc.2015.149. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Q, Sharma N, He J, Wani G, Wani AA. USP7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing RNF168. Cell Cycle. 2015;14(9):1413–1425. doi: 10.1080/15384101.2015.1007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J Cell Biol. 2009;184(1):13–19. doi: 10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, et al. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J Clin Invest. 2016;126(6):2205–2220. doi: 10.1172/JCI85747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song MS, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455(7214):813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reverdy C, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19(4):467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Chauhan D, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altun M, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18(11):1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Colland F, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8(8):2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 34.Das DS, et al. A novel hypoxia-selective epigenetic agent RRx-001 triggers apoptosis and overcomes drug resistance in multiple myeloma cells. Leukemia. 2016;30(11):2187–2197. doi: 10.1038/leu.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huen MS, et al. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell. 2010;37(6):854–864. doi: 10.1016/j.molcel.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Knaap JA, et al. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17(5):695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Holowaty MN, et al. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278(32):29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- 38.Du Z, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3(146):ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfoh R, et al. Crystal structure of USP7 ubiquitin-like domains with an ICP0 peptide reveals a novel mechanism used by viral and cellular proteins to target USP7. PLoS Pathog. 2015;11(6):e1004950. doi: 10.1371/journal.ppat.1004950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lange A, et al. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J Biol Chem. 2007;282(8):5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim RQ, van Dijk WJ, Sixma TK. Structure of USP7 catalytic domain and three Ubl-domains reveals a connector α-helix with regulatory role. J Struct Biol. 2016;195(1):11–18. doi: 10.1016/j.jsb.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Zhang ZM, et al. An allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1. Cell Reports. 2015;12(9):1400–1406. doi: 10.1016/j.celrep.2015.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng J, et al. Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nat Commun. 2015;6:7023. doi: 10.1038/ncomms8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faesen AC, et al. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011;44(1):147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 45.Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 46.Fraile JM, Quesada V, Rodríguez D, Freije JM, López-Otín C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene. 2012;31(19):2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 47.Alonso-de Vega I, Martín Y, Smits VA. USP7 controls Chk1 protein stability by direct deubiquitination. Cell Cycle. 2014;13(24):3921–3926. doi: 10.4161/15384101.2014.973324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian J, et al. USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene. 2015;34(36):4791–4796. doi: 10.1038/onc.2014.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He J, et al. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J Biol Chem. 2014;289(39):27278–27289. doi: 10.1074/jbc.M114.589812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Y, Feng W, Sy SM, Huen MS. ATM-dependent phosphorylation of the Fanconi anemia protein PALB2 promotes the DNA damage response. J Biol Chem. 2015;290(46):27545–27556. doi: 10.1074/jbc.M115.672626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]