Significance
Type 1 diabetes (T1D) is an increasing medical health problem caused by autoimmune T cells killing insulin-producing β cells in the islets of Langerhans. When the majority of β cells are destroyed, unless exogenous insulin is administered, ketoacidosis and death follow. However, providing exogenous insulin does not prevent the resultant complications of stroke, heart disease, visual impairment, or faulty wound healing, indicating the need to preserve β cells in the prediabetic stage to insure endogenous insulin production. We show that blockage of IFN-α signaling prior to clinical T1D disease by antibody or a sphingosine-1 receptor agonist prevents autoaggressive T cells from entering the islets and killing β cells. The result is aborting T1D by acting at the prediabetic stage.
Keywords: type I interferon, IFN-alpha, S1PR1, type 1 diabetes
Abstract
Blockade of IFN-α but not IFN-β signaling using either an antibody or a selective S1PR1 agonist, CYM-5442, prevented type 1 diabetes (T1D) in the mouse Rip-LCMV T1D model. First, treatment with antibody or CYM-5442 limited the migration of autoimmune “antiself” T cells to the external boundaries around the islets and prevented their entry into the islets so they could not be positioned to engage, kill, and thus remove insulin-producing β cells. Second, CYM-5442 induced an exhaustion signature in antiself T cells by up-regulating the negative immune regulator receptor genes Pdcd1, Lag3, Ctla4, Tigit, and Btla, thereby limiting their killing ability. By such means, insulin production was preserved and glucose regulation maintained, and a mechanism for S1PR1 immunomodulation described.
Type 1 diabetes (T1D) is an autoimmune disorder defined by infiltration of autoreactive lymphoid cells into the islets of Langerhans that destroy insulin-producing β cells (1). By the time of clinical diagnosis, T cells have destroyed 60–80% or more of total β cells, resulting in high blood glucose levels as a result of low insulin production. Prevention of ketoacidosis and death require lifelong delivery of exogenous insulin. However, daily insulin therapy is associated with increased prevalence of debilitating pathologies of cardiovascular, central and peripheral nervous, ophthalmic, and peripheral vascular systems among others.
A role for type I IFN in autoimmune disease was first reported by Notkins’ laboratory (2) and pancreata removed at necropsy from humans with T1D displayed significant increases in type I IFN (3, 4). Treatment of humans having hairy cell leukemia (5) or hepatitis C virus (6) with IFN-α was associated with induction or acceleration of the diabetogenic process, and recent longitudinal studies demonstrated that a IFN-I gene signature of individuals at risk for developing T1D preceded clinical onset (7, 8). Direct evidence for an association of IFN-I with T1D was shown by Stewart et al. (9) in transgenic (tg) mice and strengthened in studies with NOD mice (10, 11). Unanue and coworkers (10) found IFN-I transcriptional signatures within the islets preceded T-cell activation. McDevitt and coworkers (11) reported treatment of 2- to 3-wk-old NOD mice with antibody to IFNAR1 delayed the onset and decreased the incidence of T1D. Using the virus-induced Rip-LCMV T1D model, Zinkernagel and coworkers (12) demonstrated that genetic ablation of Ifnar could delay onset of T1D. However, the mechanism of action by type I IFN was unknown. Here, we report studies that define the species of type I IFN and mechanism involved causing T1D and therapeutic approaches to prevent diabetes by preserving β-cell function.
Results and Discussion
To uncover the pathological role(s) of IFN-I, a viral mouse model of T1D (13) was used in which several parameters mimic immunological and histopathological components of human T1D and the “self” antigen recognized by specific autoimmune T cells causing T1D was known. Such antigen-specific autoimmune T cells were isolatable, quantifiable, and transferable. This model was accomplished by placing the glycoprotein (GP) of lymphocytic choriomeningitis virus (LCMV) under the transcriptional control of the rat insulin promoter (RIP) in C57BL/6 mice (13). In this model (Rip-LCMV), β cells express the known MHC-I restricted CD8 CTL GP immunodominant epitopes for the LCMV GP (amino acids 33–41 and 276–286, respectively) and the MHC-II restricted CD4 T-cell epitope GP61–80. By expressing the GP transgene only within the islets, CD8+ T cells were found to be necessary for causing T1D, whereas CD4+ T cells were not (14). Rip-LCMV mice challenged i.p. with 2 × 105 pfu of LCMV developed T1D defined as blood glucose >250 mg/dL and a robust lymphoid cell infiltration into the islets with significant islet destruction within 12–21 d after viral infection (Fig. 1). A virus (LCMV-GPV) constructed having mutations in the glycoprotein (GP33–41: GP 38 F/L and GP276–286: GP282 G/D) failed to generate CTL and develop T1D (Fig. S1). Further, CTL clones generated against the GP33 and GP276 epitopes do not lyse targets expressing LCMV-GPV (15, 16). These findings indicated the specificity of the virus/CTL/Rip-LCMV model.
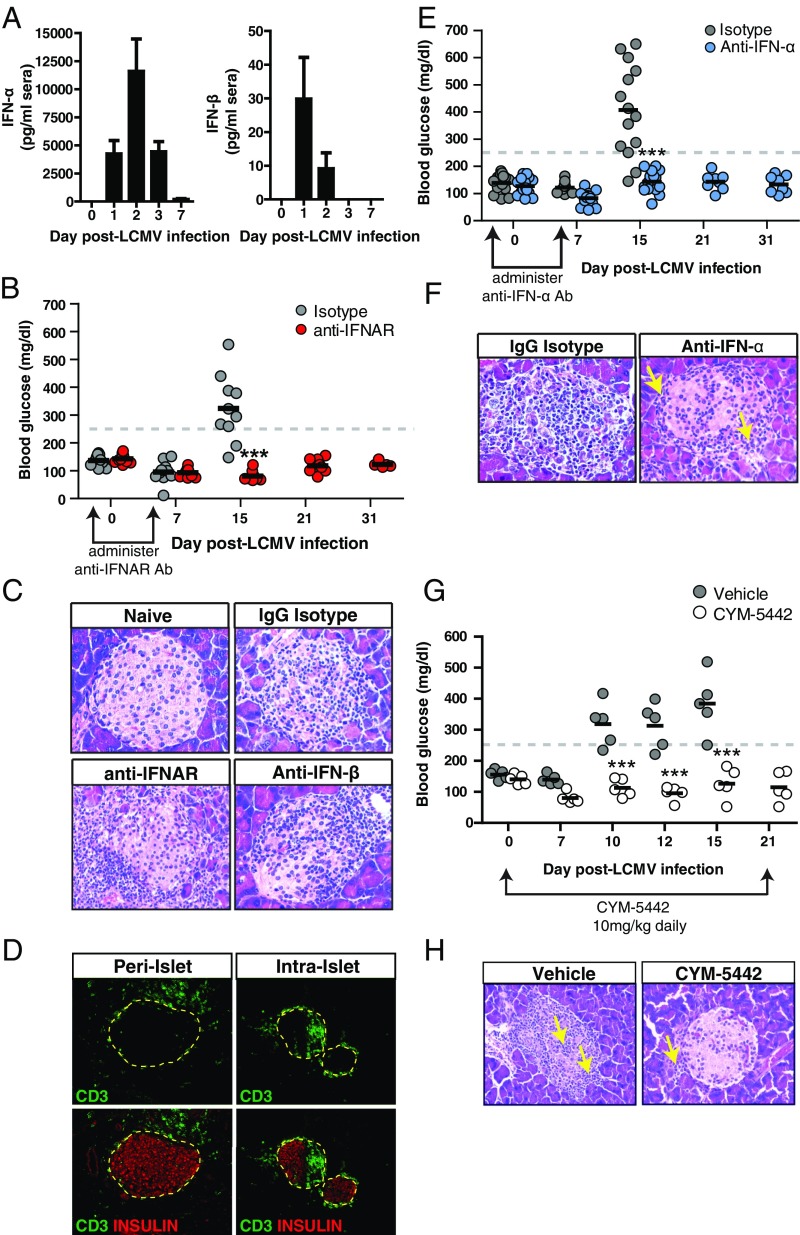
Fig. 1.
IFN-α signaling is required for development of T1D. Rip-LCMV mice were infected with 2 × 105 pfu LCMV i.p. and treated with either IgG isotype control antibody or antibody to IFNAR, to IFN-α or IFN-β at day −1 and day 5 p.i. (A) Serum levels of IFN-α and IFN-β were measured 0, 1, 2, 3, and 7 d after LCMV infection by ELISA. (B) Blood glucose levels over a 31-d postinfection period following IFNAR blockade. (C) Representative H&E-stained sections of pancreatic islets within Rip-LCMV mice at day 15 p.i. (D) Immunofluorescence (I.F.) analysis islets of peri-islet (anti-IFNAR) or intraislet (IgG isotype) accumulation of CD3+ T cells at day 15 p.i. (E) Blood glucose levels over a 31-d postinfection period following anti-IFNα blockade. (F) H&E analysis of islets from anti-IFNα–treated mice at day 15 p.i. (G) Blood glucose measurements were taken over a 21-d postinfection observation period during CYM-5442 treatment. (H) H&E analysis of islets showed presence of lymphoid cells outside of the islet (yellow arrow) of CYM-5442–treated mice at day 15 p.i. In contrast, vehicle control display massive lymphoid cell infiltration into the islet (yellow arrows). Serum IFN-I levels represent average from eight mice from two independent experiments. H&E and I.F. images are representative of at least three mice per group. ***P < 0.001, two-way ANOVA.
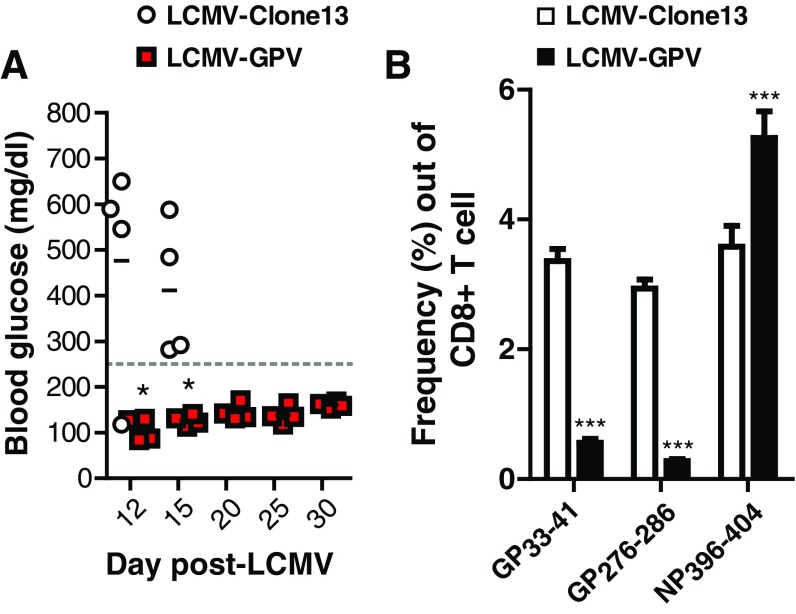
Fig. S1.
Rip-LCMV mice infected with LCMV-GPV fail to develop T1D. Rip-LCMV mice were infected i.p. with 2 × 105 LCMV-Clone13 or LCMV-GPV, and blood glucose levels was monitored. (A) Mice receiving LCMV-GPV did not develop T1D (BG < 250 mg/dL) during a 30-d observation period, whereas 4/4 mice receiving LCMV-Clone13 developed T1D (BG > 250 mg/dL) by day 15 p.i. (B) Splenocytes were isolated at day 7 p.i. and stained with anti-CD3, anti-CD8, and CD8+ T-cell–specific LCMV-GP33–41, LCMV-GP276–286, or LCMV-NP396–404 tetramer. Flow data represents three mice per group. One-way ANOVA analysis was used to assess differences between the groups. *P < 0.05; ***P < 0.001.
Intraperitoneal inoculation of Rip-LCMV mice with LCMV resulted in the expression of type I IFN in the sera in the first 24 h after infection (p.i.) (Fig. 1A). IFN-β peaked at 24 h p.i., whereas IFN-α peaked at 48 h p.i (Fig. 1A). Rip-LCMV mice were treated 1 d before infection and at day 5 p.i. with an antibody against IFN-alpha-beta receptor 1 (anti-IFNAR-Clone MAR1-5A3) (17). Fifteen days p.i., 8/10 isotype control mice (80%) developed diabetic blood glucose levels of >250 mg/dL which increased to levels 350–600 mg/dL in all (10/10) mice, necessitating euthanasia. In contrast, mice (10/10) treated with anti-IFNAR maintained normal blood glucose levels (<250 mg/dL, range 145–200 mg/dL) over an observation period of 31 d (Fig. 1B). Examination of islets within anti-IFNAR–treated mice revealed significant reduction in lymphoid cells infiltrating into the islets, but accumulation of lymphoid cells around the islets (Fig. 1C) at day 15 p.i. compared with IgG isotype-treated controls. Immunofluorescence analysis at day 21 p.i. of IFNAR antibody-treated mice confirmed that CD3+ T cells were predominantly localized to the peri-islet border as opposed to inside the islets (Fig. 1D). A statistically significant reduction (P < 0.01) in the islet pathology score was observed with anti-IFNAR–treated mice compared with IgG control-treated mice at day 15 p.i (Fig. S2).
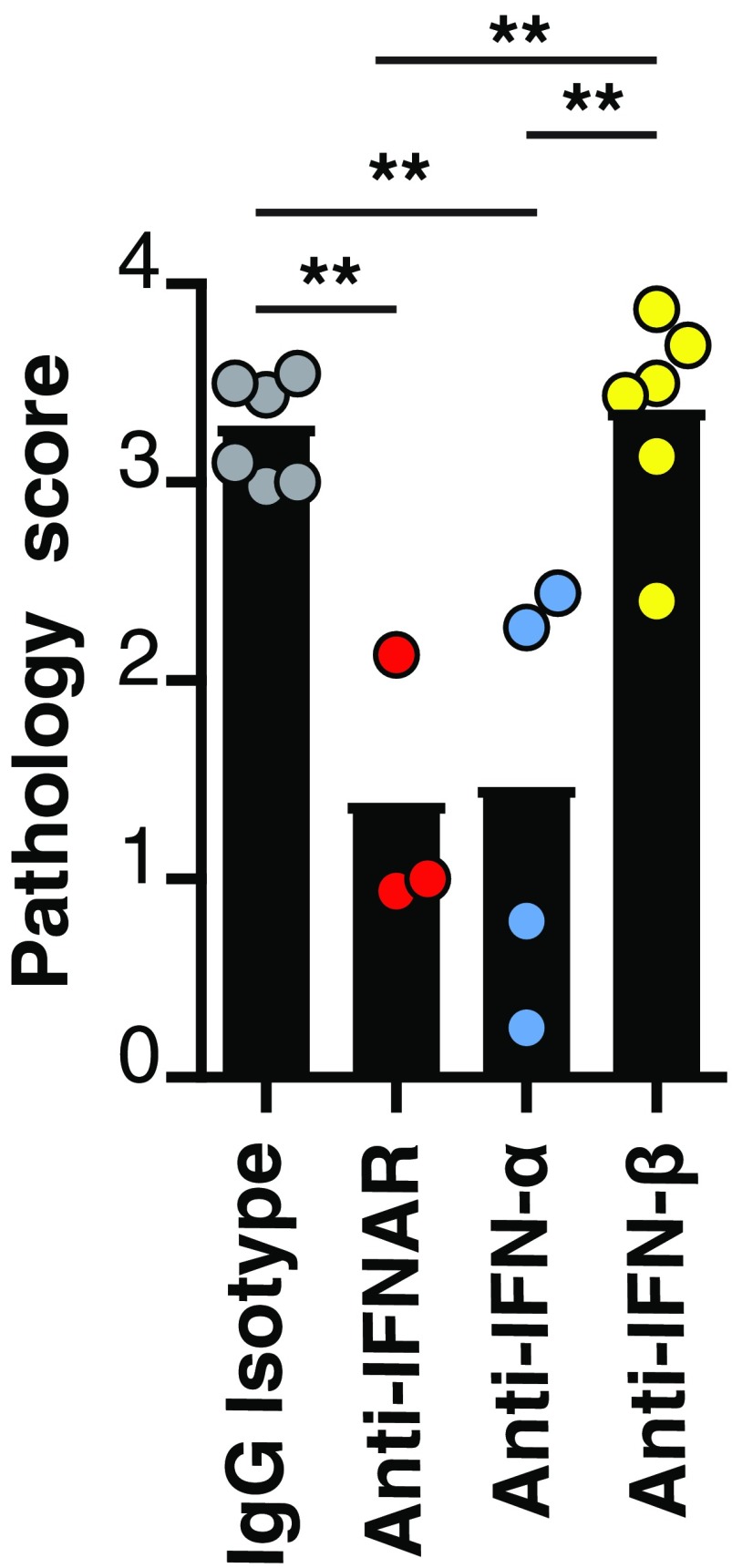
Fig. S2.
Blockade of IFN-α significantly reduces islet immunopathology in Rip-LCMV mice infected with virus. Rip-LCMV mice were treated with IgG isotype control antibody (1 mg; Clone PIP), antibody to IFNAR (1 mg; Clone: MAR1-5A3), antibody to IFN-α (1 mg; Clone: TIF-3C5), or antibody to IFN-β (0.25 mg; Clone: HDβ-4A7) 1 d before infection with LCMV and at 5 d p.i. Pancreata were removed at day 15 p.i., cut, and stained with H&E. A pathology score index was used (Materials and Methods) for at least 10 islets per mouse by using two independent observations. Each value represents the average pathology score out of all of the islets for each mouse. One-way ANOVA analysis with a Bonferroni post hoc test was used to assess differences between the groups. **P < 0.01.
IFN-α, but Not IFN-β, Is a Required Signal for Autoreactive T Cells to Enter the Islets.
We determined whether IFN-α or IFN-β was responsible for causing T1D. Within the mouse, type I IFN is divided into one IFN-β molecule and 14 IFN-α species of molecules. Antibody-mediated neutralization of IFN-β (clone HDβ-4A7) was associated with intraislet penetration of lymphoid cells (Fig. 1C) and resulted in a similar islet pathology score to control mice (Fig. S2). In contrast, blockade of IFN-α aborted lymphocyte trafficking into the islets and T1D. For these studies, we used a neutralizing antibody (clone TIF-3C5) that targets six murine IFN-α species (IFN-αA, IFN-1, IFN-4, IFN-5, IFN-11, and IFN-13) (18, 19). Anti–IFN-α prevented T1D over the 31 d after LCMV infection (Fig. 1E). Inspection of the pancreatic islets within anti-IFNα–treated mice revealed peri-islet cuffing of lymphoid cells (Fig. 1F, yellow arrows) with negligible lymphocytes inside the islets. Corresponding ameliorated pathology of the islets led to a significantly reduced (P < 0.01) pathology score compared with controls (Fig. S2).
Targeting Sphingosine-1-Phosphate Receptor 1 Signaling Is a Pharmacologically Relevant Approach to Abrogate Lymphoid Migration into the Islets to Preserve Insulin-Producing β Cells and Abort T1D.
Sphingosine-1-phosphate receptor 1 (S1PR1) agonists like CYM-5442 modulate lymphocyte trafficking and significantly alter IFN-α autoamplification acting primarily on pDCs to reduce IFN-α expression by promoting the turnover of IFNAR at the cell surface, resulting in reduced STAT1 phosphorylation and inhibition of type I IFN autoamplification (20). To determine whether modulation of the S1PR1 signaling would abort the development of T1D, Rip-LCMV tg mice infected with virus and receiving daily administration of CYM-5442 (10 mg/kg) showed normal blood glucose levels over a 21-d observation period compared with isotype control in which four out of five mice displayed diabetic levels (>250 mg/dL) by day 10 p.i. and all by day 15 p.i (P < 0.001) (Fig. 1G). Pancreata of the CYM-5442–treated group at day 15 p.i. revealed inflammatory T cells located around the outside of the islet but absence inside the islet (Fig. 1H).
To track GP-specific CD8+ T cells, we transferred 20,000 congenic Thy1.1+ CD8+ T cells (P14) from TCR tg mice that recognize the LCMV GP33–41 epitope (21) into Rip-LCMV mice 2 d before LCMV infection. Pancreata within anti-IFNAR–treated mice were devoid of infiltrating P14 T cells at day 15 p.i. In virus-infected Rip-LCMV mice treated with anti-IFN-α, P14 T cells were found localized at peri-islet walls and not within the islets at day 7 p.i (Fig. 2A). In contrast, mice treated with IgG isotype control displayed robust P14 T-cell infiltration into the islet interior as early as day 7 p.i (Fig. 2A).
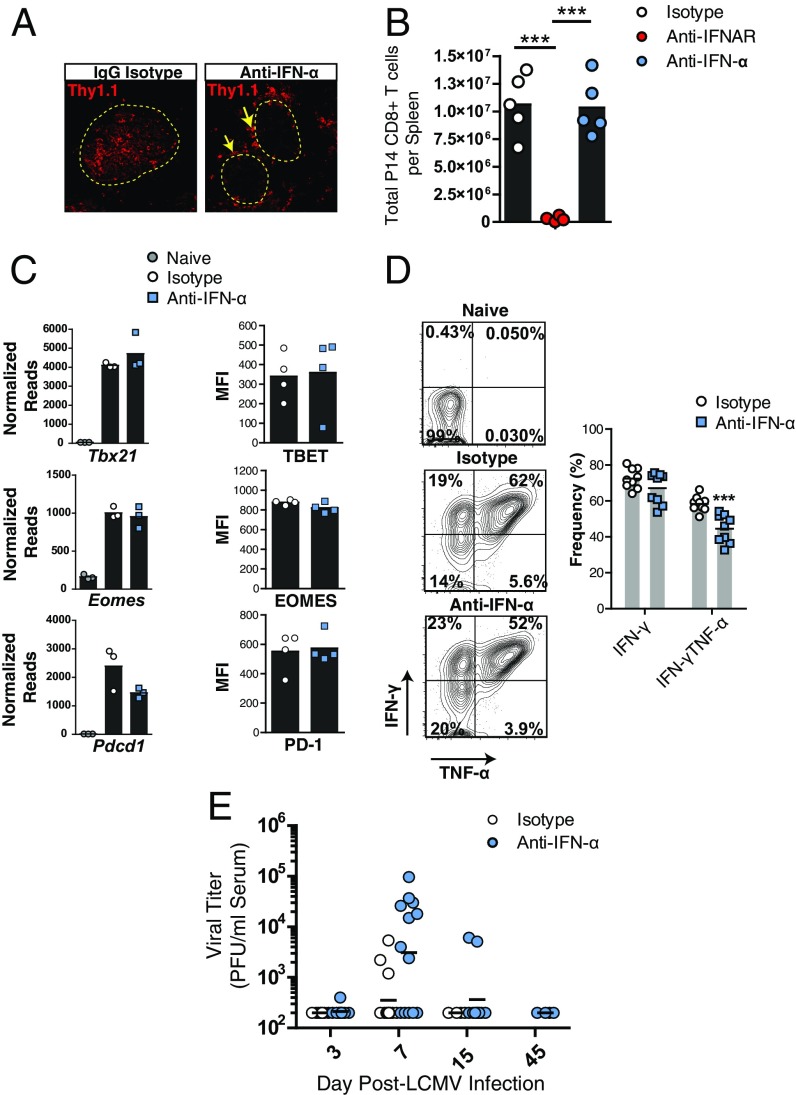
Fig. 2.
Blockade of IFN-α signaling prevents the migration of anti-self GP-specific T cells into the islets but does not limit their expansion or effector activity in the spleen. Twenty thousand P14 CD8+ T cells were adoptively transferred into Rip-LCMV mice 2 d before LCMV infection. (A) Immunofluorescence analysis of islets at day 7 p.i. revealed P14 T cells localized to the islet border (yellow arrows) in anti-IFNα–treated mice. (B) Numbers of P14 CD8+ T cells in the spleens at day 7 p.i. (C) RNA-seq read counts and protein expression of Tbx21 (TBET), Eomes (EOMES), and Pdcd1 (PD-1) on P14 T cells at day 7 p.i. in the spleen. (D) Intracellular IFN-γ and TNF-α was measured in P14 T cells following ex vivo peptide restimulation of splenocytes with the LCMV GP33–41 peptide at day 7 p.i. (E) Viral titers in the serum. Immunofluorescence analysis was representative of three pancreata per group. Ex vivo peptide restimulation was performed on two independent experiments with at least four mice per group. Flow cytometry data were generated from at least four mice per group. Viral titer analysis represents at least 11 mice per group. ***P < 0.001, one-way ANOVA.
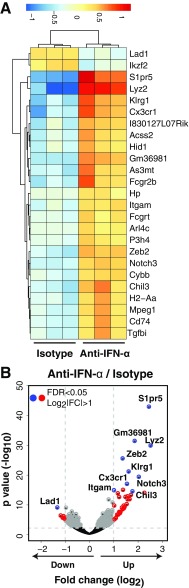
We did not detect differences in the number of P14 T cells in the spleens of IgG isotype-treated control mice compared with anti–IFN-α treated mice at day 7 p.i., whereas a dramatic reduction in P14 T cells was observed following anti-IFNAR treatment (Fig. 2B). Through RNA-sequencing (seq) analysis, we found several genes traditionally associated as phenotypic markers of effector CD8+ T-cell differentiation, e.g., Zeb2, Klrg1, Notch3, and the migratory receptors Cx3cr1 and S1pr5 were up-regulated within P14 cells isolated from the spleens following blockade of IFN-α signaling (Fig. S3). No differences were detected in mRNA or protein expression of Tbx21 (TBET), Eomes (EOMES) or the immune regulatory receptor Pdcd1 (PD-1) on anti-IFN-α–treated mice (Fig. 2C). P14 T cells isolated from the spleens of anti-IFN-α–treated mice showed a significant reduction in the joint expression of both IFN-γ and TNF-α but similar expression of IFN-γ following ex vivo restimulation with LCMV-GP33–41 peptide (Fig. 2D). Blockade of IFN-α resulted in similar viral titers in the serum compared with IgG isotype-treated control mice (Fig. 2E). Collectively, these results provide evidence that blockade of IFN-α with antibody use is not halting the expansion or functional activity of anti-self GP-specific T cells. The anti–IFN-α antibody blocks at least 6 of the 14 IFN-α species (18, 19) and suggests a species of IFN-α not targeted by the antibody to IFN-α used was sufficient to allow P14 T cells to expand in response to LCMV infection.
Fig. S3.
Blockade of IFN-α increases the expression of CD8+ T-cell effector genes within P14 T cells. P14 T cells were sorted by FACS from the spleens at day 7 p.i. and subject to RNA-seq analysis. (A) We identified 41 differentially expressed genes (FDR < 0.05; log2 |FC| > 1; of total 428 genes at FDR < 0.05) between IgG isotype control and anti-IFN-α–treated mice, with the top 25 most differentially expressed genes by P value shown in the heat map. (B) Volcano plot shows several genes up-regulated within anti-IFN-α–treated mice that are associated with CD8+ T-cell function.
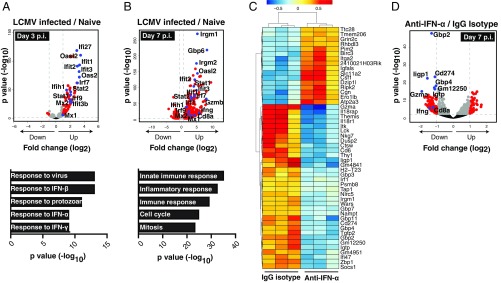
A Type I IFN Gene Signature Precedes CD8+ T-Cell Infiltration into the Pancreatic Islets Following Infection of Rip-LCMV Mice.
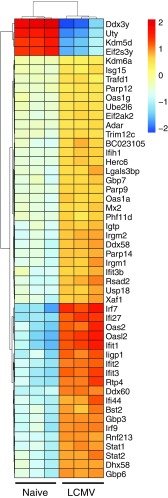
We investigated how blockade of IFN-α may impact islet-specific gene expression and control leukocyte migration into the islets. We found 204 differentially expressed genes (FDR-adjusted P < 0.05) within the islets of the IgG isotype control group, of which 75 had a log2|FC|>1 compared with naïve controls at day 3 p.i (Fig. S4). A significant enrichment for genes associated with early detection of RNA viruses including Ddx58 (RIG-I) and Ifih1 (MDA5), genes involved in the formation of the IFN-stimulated gene factor 3 (ISGF3) complex (Stat1, Stat2, Irf9) and several other IFN-stimulated genes (ISGs) including Irf7, Ifit-1, Ifit-2, Ifit-3, and Mx2 was noted (Fig. 3A). The IFN-I gene signature at day 3 p.i. was supported by gene ontology annotation that showed an enrichment for IFN-I–induced response pathways (Fig. 3A). Several IFN-I–induced genes remained significantly up-regulated day 7 p.i (Fig. 3B). Further, expression of T-cell–specific genes including Cd8, Gzmb, and Ifng was found at day 7 p.i., supporting the histological evidence of a robust GP-specific CD8+ T-cell presence within the islets of IgG isotype-treated control mice at this time-point (Fig. 2A).
Fig. S4.
IFN-I–induced genes are up-regulated within the islets of Rip-LCMV mice following infection with LCMV. Unsupervised hierarchal clustering of the top 50 most differentially expressed genes in the pancreatic islets of naïve and LCMV-infected Rip-LCMV mice at day 3 p.i. Male mice were used for naïve controls, whereas female mice were infected with LCMV. Ddx3y, Uty, Kdm5d, and Eif2s3y show reduced expression because they were not expressed in the female group.
Fig. 3.
IFN-I gene signature is observed within the pancreatic islets following infection with LCMV. RNA isolated from pancreatic islets of Rip-LCMV mice was subject to RNA-seq analysis. (A) Volcano plot highlights 75 differentially expressed genes (log2 |FC| > 1 and FDR < 0.05; of total 204 genes at FDR < 0.05) between naïve and LCMV-infected mice at day 3 p.i. with corresponding gene ontology enrichment analysis below. Labeled in blue are genes associated with IFN-I signaling. Ddx3y, Uty, Kdm5d, and Elf2s3y were excluded from the volcano plot because they are Y chromosome-linked genes not found in LCMV-infected female mice. (B) Volcano plot shows 1,825 differentially expressed genes (log2 |FC| > 1 and FDR < 0.05) between naïve and LCMV-infected mice at day 7 p.i. with corresponding gene ontology annotation below. (C) Heat map showing top 50 most differentially expressed genes between anti–IFN-α and IgG isotype control at day 7 p.i. Color scale is variance with respect to row means. (D) Volcano plot highlights 251 differentially genes (log2 |FC| > 1 and FDR < 0.05). Gene ontology annotation was acquired from genes sets comprising genes with log2 |FC| > 1 and FDR < 0.05. RNA-seq data were generated from islet RNA of three individual mice per group.
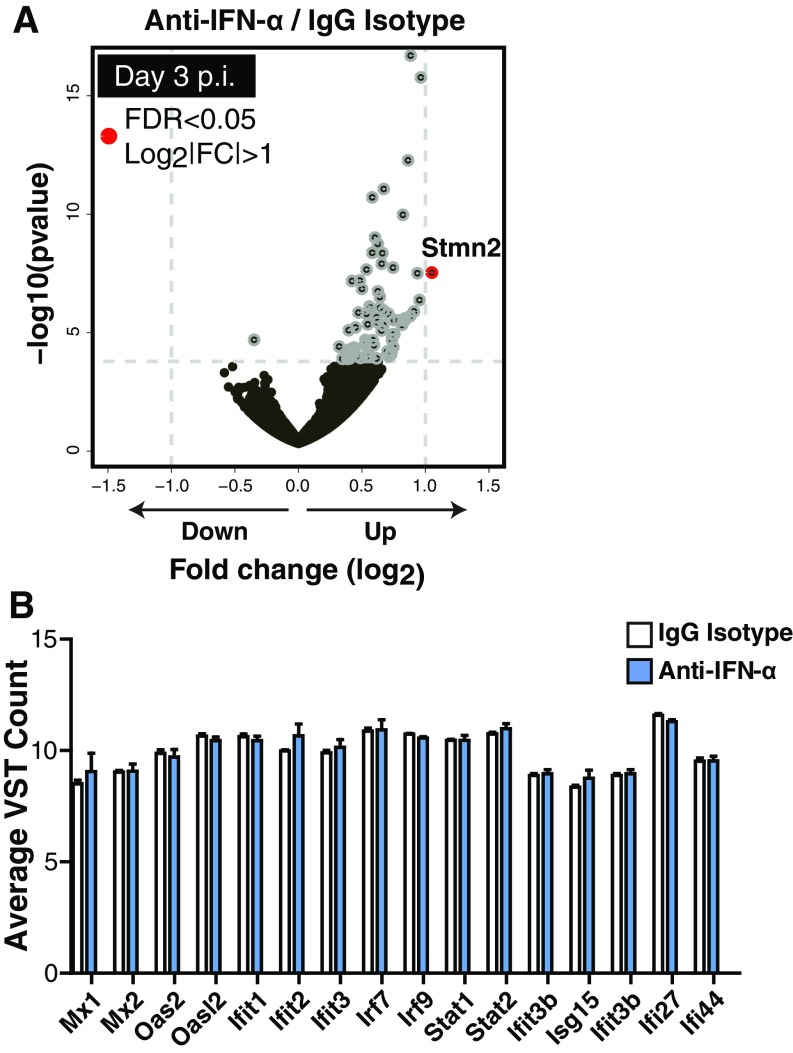
Significant differences in gene expression within the islets of mice treated with antibody to IFN-α compared with IgG isotype control were not detected at day 3 p.i. in any immune- or inflammation-related pathways (Fig. S5A), and levels of type I IFN targets genes were comparable between the two groups (Fig. S5B). However, an increase in IFN-β in the serum following anti–IFN-α treatment was found. The extra IFN-β may likely have enhanced IFN-I–induced gene expression in the islets, because IFN-β has a 20- to 30-fold higher affinity to IFNAR than IFN-α, and results in more potent signaling transduction through IFNAR. At day 7 p.i., a significant reduction in the expression of genes associated with CD8+ T cells including Gzma, Ifng, and CD8a occurred (Fig. 3 C and D).
Fig. S5.
Neutralization of IFN-α by antibody blockade did not induce transcriptional changes within the pancreatic islets of Rip-LCMV mice at day 3 p.i. Rip-LCMV mice were treated with anti–IFN-α or IgG isotype control and killed at day 3 p.i. for islet isolation. (A) Volcano plot indicates that Stmn2 was the only identified gene with a log |FC| > 1 and FDR < 0.05 (of total 74 genes at FDR < 0.05). (B) Variance stabilizing transformation (VST) adjusted read counts of IFN-I target genes between anti–IFN-α and IgG isotype control.
Increased Expression of Negative Immune Regulating Surface Molecules on Autoreactive T Cells Is Observed Following Treatment with the Selective S1PR1 Agonist, CYM-5442.
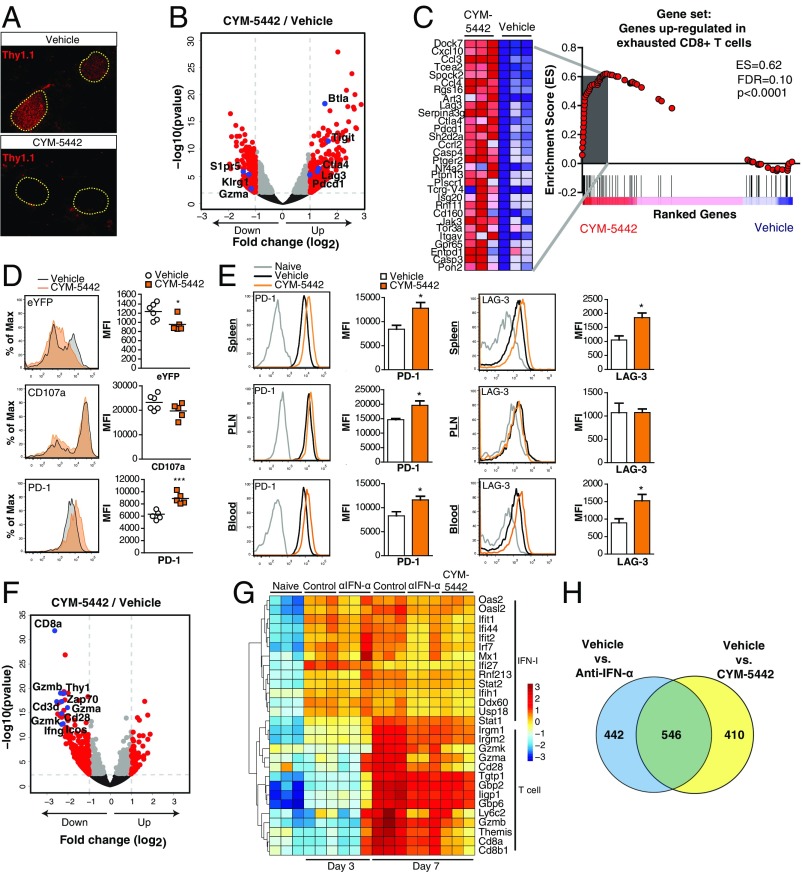
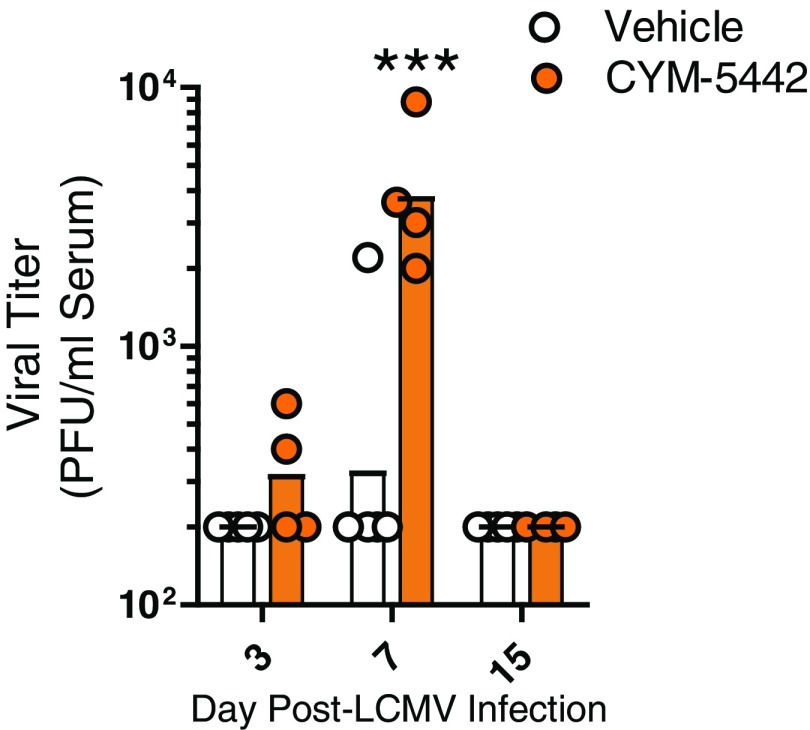
The phenotypic convergence of anti-IFN-α and S1PR1 agonist inhibition of T1D led us to hypothesize there might be shared checkpoints at the molecular and cellular level whereby targeting S1PR1 prevented T1D disease. Adoptive transfer of P14 T cells resulted in their migration to the islets but were restricted to geographical areas outside and surrounding the islets at day 7 p.i. in CYM-5442–treated mice (Fig. 4A). RNA-sequencing (seq) analysis on P14 T cells from the spleen showed that CYM-5442 treatment reduced mRNA expression of genes associated with activated CD8+ T-cell effectors including Klrg1, Gzma, and S1pr5 (Fig. 4B). Importantly, several genes that inhibit the immune response of T cells and cause T-cell exhaustion (22) were up-regulated including Pdcd1 (encoding PD-1), Lag3, Ctla4, Btla, and Tigit (Fig. 4B). The finding was supported by gene set enrichment analysis (GSEA) where we found that genes up-regulated in exhausted CD8 T cells isolated during chronic LCMV infection (22) were similarly up-regulated in P14 T cells from CYM-5442–treated mice (Fig. 4C). We next determined whether CYM-5442 treatment influenced GP-specific CD8 T-cell responses within the spleens of transgenic mice that express enhanced YFP (eYFP) from an IRES element placed after the stop codon of IFN-γ mRNA (23). Ex vivo peptide restimulation with LCMV GP33–41 peptide resulted in a significant (P < 0.05) reduction in median fluorescence intensity (MFI) of GP33–41 tetramer-positive CD8+ T cells expressing eYFP and, thus, IFN-γ in the CYM-5442 group (Fig. 4D). CYM-5442 treatment also resulted in a reduction in the MFI of the degranulation marker CD107a, whereas PD-1 expression was elevated (Fig. 4D). Examination of P14 T cells in Rip-LCMV mice treated with CYM-5442 revealed a significant (P < 0.05) increase in the surface expression of PD-1 on P14 T cells in the spleen, pancreatic lymph nodes, and blood, whereas LAG3 expression was significantly increased on P14 cells in the spleen and blood at day 7 p.i (Fig. 4E). CYM-5442 treatment resulted in a significant increase in viral titers in the serum compared with vehicle-treated mice at day 7 p.i., but fell below limits of detection by day 15 p.i (Fig. S6). Together, these results suggest that anti-self GP-specific T cells have reduced function in response to CYM-5442 treatment that results in increased viral replication.
Fig. 4.
S1PR1 agonist, CYM-5442, induced an exhaustion signature on LCMV-GP–specific CD8+ T cells and prevented their entry into the pancreatic islets. P14 T cells were sorted by FACS from the spleens of CYM-5442 or vehicle-treated Rip-LCMV mice and analyzed by RNA-seq. (A) Immunofluorescence analysis of Thy1.1+ P14 T cells in pancreatic tissue from CYM-5442 or vehicle-treated Rip-LCMV mice at day 7 p.i. (B) Volcano plot of genes differentially are expressed (log2 |FC| > 1 and FDR < 0.05) within the CYM-5442 group; T-cell activation regulating molecules are labeled in blue. (C) GSEA of an exhaustion gene signature in P14 T cells. (D) Analysis of eYFP, CD107a and PD-1 expression on GP33–41 specific CD8+ T cells following ex vivo LCMV-GP peptide restimulation. (E) Expression of PD-1 and LAG3 on P14 CD8+ T cells in the spleen, pancreatic lymph nodes (PLN), and blood at day 7 p.i. from vehicle or CYM-5442 treated Rip-LCMV mice. (F) Volcano plot shows selected expressed genes within the islets of CYM-5442 vs. vehicle-treated mice. (G) Heat map of selected IFN-I and CD8+ T-cell genes within the islets across all experimental groups. Color scale is variance with respect to row means. (H) Venn diagram comparing differentially expressed genes (FDR < 0.05) in the islets at day 7 p.i. shared between anti–IFN-α and CYM-5442–treated groups. RNA-seq data are from three mice per group. eYFP data are from one experiment with at least five mice per group. PD-1 and LAG3 expression on P14 T cells (CD3+CD8+CD90.1+) is from one experiment with at least three mice per group *P < 0.05; ***P < 0.001, Student’s t test.
Fig. S6.
CYM-5442 treatment increases viral replication within Rip-LCMV mice. Viral titers were measured within the serum of vehicle and CYM-5442–treated mice. ***P < 0.001 two-way ANOVA.
Comparable Gene Expression in the Pancreatic Islets Is Observed Following Antibody to IFN-α and CYM-5442 Treatment.
RNA-seq examination of islets from CYM-5442–treated mice at 7 p.i. mimicked the reduction in CD8+ T-cell–specific genes observed following IFN-α blockade as a significant reduction in CD8a, Thy1, Gzma, Gzmb, Cd3d, and Ifng occurred (Fig. 4F). The expression of selected IFN-I and CD8+ T-cell–related genes in the islets of all treatment groups at day 3 and 7 p.i. was examined (Fig. 4G). At day 3 p.i., control mice showed up-regulation of IFN-I genes that was comparable to anti–IFN-α treatment. At day 7 p.i., IFN-I–induced genes remained elevated in control mice whereas a reduction was observed in the IFN-I gene signature following anti–IFN-α or CYM-5442 treatment. CD8+ T-cell related genes were not up-regulated at day 3 p.i., but were detected at day 7 p.i. Reduced expression of the CD8+ T-cell genes was observed in anti–IFN-α and CYM-5442–treated mice at day 7 p.i (Fig. 4G). Further, we found 546 (39%) differentially expressed genes (FDR < 0.05) shared between the anti–IFN-α and CYM-5442 groups compared with isotype control (Fig. 4H). Thus, S1PR1 agonist prevented T1D by suppressing IFN-α signaling (20, 24), restricting the migration of anti-self T cells to geographic areas outside the islets and likely by contributing to T-cell exhaustion through up-regulation of negative immune regulators like PD-1 and LAG3.
In conclusion, we present four findings. First, IFN-I signaling, specifically IFN-α but not IFN-β, is essential for causing T1D. Second, a selective S1PR1 agonist CYM-5442 prevented progression from prediabetes to T1D. Third, we provided evidence that type I IFN-α, and not IFN-β, was essential for entry of autoreactive T cells into the islets. Of interest is the report by Meyer et al. (25) on humans with autoimmune T1D due to mutations in autoimmune regulator (AIRE). A subset of patients with AIRE deficiency that generated self-reactive neutralizing antibodies specific for type I IFN-α1, IFN-α2, IFN-α5, IFN-α8, and IFN-α14 were protected from T1D whereas, in contrast, those not possessing anti–IFN-α antibodies developed T1D. Thus, a prominent role for IFN-α signaling in T1D is now found in both animal models of T1D and a subset of humans with AIRE deficiency. Importantly, CYM-5442 blocks not all but ∼80–90% of type I IFN, thereby maintaining sufficient IFN-α to generate CTLs and antibodies to combat viral infections (20, 24, 26), while preventing T1D. Fourth, CYM-5442 therapy enhanced the expression of negative immune regulating surface molecules on autoreactive T cells, further limiting the ability of any autoimmune T-cell that might enter the islets from killing β cells.
The selective and potent S1PR1 agonist CYM-5442 (27, 28) is a predecessor compound to the clinical agonist ozanimod (29), now completing phase 3 clinical trials for multiple sclerosis (30) and ulcerative colitis (31). S1PR1 agonists are clinically effective immunomodulators because of multistep interdiction of immunopathology. S1PR1 is expressed on lymphocytes, endothelium, and plasmacytoid dendritic cells (32). Receptor modulation inhibits lymphocyte recirculation and egress from thymus and secondary lymphoid organs, while also suppressing cytokine amplification and immunopathology, without impeding the development of antiviral immunity and effective immunological memory for the control of pathogens (33–35).
Direct effects are shown here on lymphocyte migration to pancreatic islets coupled with IFN-α modulation and up-regulation of negative immune regulators including PD-1 and LAG-3. This multistep interdiction of autoimmunity by S1PR1 agonist suggests that similar therapy might be considered for humans during the prediabetic period when autoantibodies to GAD65 and insulin are present and blood glucose levels are normal. We are testing this possibility by using S1PR1 agonist therapy in a second model of Rip-LCMV–induced T1D where the viral transgene is placed in the thymus and the islets. By this manipulation, high avidity-specific GP CD8+ T cells are eliminated, resulting in a prediabetic period of months before onset of T1D (14).
Materials and Methods
Seven- to eight-week-old mice, all H-2b C57BL/6, were used. Mice were bred and maintained in pathogen-free conditions in the animal facility at The Scripps Research Institute (TSRI). All mouse procedures were in accordance with TSRI Animal Research Committee guidelines. Generation and characterization of Rip-LCMV and P14 Tgs have been reported (13, 14, 36). LCMV-Clone13 was grown and quantitated as published (13, 14, 16, 18). LCMV-GPV variants were made and used as described (15, 16, 36). Stock parental and GPV virus both titered 4–5 × 107 pfu/mL. Mice were injected with 2 × 105 pfu LCMV i.p (13, 14, 36). Rip-LCMV mice were treated with either IgG isotype control antibody (1 mg; Clone PIP; Leinco Technologies), antibody to IFNAR (1 mg; Clone: MAR1-5A3; Leinco Technologies), antibody to IFN-α (1 mg; Clone: TIF-3C5; Leinco Technologies) or antibody to IFN-β (0.25 mg; Clone: HDβ-4A7; Leinco Technologies) 1 d before infection with LCMV and at 5 d p.i. CYM-5442 was resuspended in sterile water at 0.5 mg/mL and injected i.p. daily at 10 mg/kg. Blood glucose (mg/dL) was measured on blood extracted from the retro-orbital plexus by using a TRUEresult blood glucose meter (Trividia Health). Following ribosomal depletion of P14 T-cell RNA samples or pancreatic RNA samples, libraries were constructed and sequenced on an Illumina NextSeq500 by using 75-bp single reads. The mm10 reference genome, build GRCm38 v79, was downloaded from Ensembl, and reads mapped to it by using STAR v2.5.2a with the gene counting feature with the following settings:–runThreadN 5–readFilesCommand zcat–outSAMtype None–quantMode GeneCounts. Unstranded read gene counts were parsed in R and differential expression computed by using DESeq2 using a cutoff of adjusted P value <0.05. Adobe Illustrator was used for data visualization. Gene ontology analysis of biological processes was performed by using Genecodis (genecodis.cnb.csic.es/).
SI Materials and Methods
Histology and Immunofluorescence Analysis.
Pancreata were removed, fixed in zinc formalin (10%), cut on a microtome, and stained with hematoxylin and eosin (H&E). An islet “pathology score” index was calculated by measuring the severity of cellular infiltration for each islet based on a 0–4 scale (0 = no peri-islet or intraislet localization of lymphoid cells, 1 = peri-islet inflammation with no intraislet lymphoid cells; 2 = intraislet inflammation < 25% of total islet; 3 = 25–50% intraislet inflammation; 4 = 50–100% intraislet inflammation). At least 10 islets were scored per mouse. For immunofluorescence (IF), pancreata were snap frozen in OCT by using an ethanol/dry ice slurry and tissue was cut at 10 μM on a cryomicrotome. Slides were dessicated for 1 h at room temperature and stored at −80 °C. For antibody staining, slides were placed in 4% paraformaldehyde (10 min), washed in PBS, and blocked with 5% normal goat serum. Primary antibody stains were carried out in PBS supplemented with 1% BSA and 0.3% Triton X-100 (for intracellular antigens) by using the following: FITC-anti-CD90.1 (1:200, Biolegend), Rabbit anti-FITC (1:200, Thermo). Fluorescently conjugated alexafluor (Thermofisher) secondary antibodies were diluted at 1:1,000 and mounted with DAPI fluoromount-G (SouthernBiotech).
P14 T-Cell Isolation for RNA-Seq.
To perform RNA-seq analysis on P14 CD8+ T cells within Rip-LCMV-GP Tg mice, 20,000 Thy1.1+ CD8+ P14 T cells were adoptively transferred into Tg mice 2 d before infection with LCMV-Clone13. At day 7 p.i., spleens were removed and single-cell suspension prepared. P14 CD8+ T cells were purified from total splenocyte preparations before FACS by a CD8+ T-cell negative selection kit (Stem Cells), were stained with mouse anti-mouse Thy1.1 (Clone OX-7; Biolegend) and rat anti-mouse CD8a PE (Clone 53.6.7; BD Pharmingen) for 30 min on ice. Cells were then sorted on a MoFlo Astrios Cell Sorter (Beckman Coulter). RNA from 1 × 106 sorted P14 CD8+ T cells was isolated via an RNAqueous RNA purification kit (Ambion). RNA integrity was measured via Agilent Bioanalyzer, and samples with RNA integrity numbers >9 were used.
GSEA.
GSEA (37) was used to test whether specific transcriptional signatures were enriched in P14 T cells isolated from Rip-LCMV mice treated with CYM-5442. In brief, GSEA-ranked genes between phenotypic subgroups based on their differential gene expression. A calculated enrichment score based on how frequent genes at the top or bottom of the differentially ranked gene list appeared in a list of genes up-regulated in exhausted CD8+ T cells during chronic LCMV infection (22). A normalized enrichment score, FDR-corrected q value, and P value was calculated by following 10,000 random permutations of the two groups. GSEA was used to test the null hypothesis that the transcriptional signature observed in P14 T cells isolated from CYM-5442 or vehicle-treated mice was not significantly enriched for the specified gene set. GSEA was used to test the null hypothesis that the transcriptional signature observed in P14 T cells isolated from CYM-5442 or vehicle-treated mice was not significantly enriched for the following genes: Tnfsf6, Pbx3, Gp49b, Cd244, Ccl3, Eomes, Casp3, Plscr1, Kdt1, Ctla4, Pdcd1, Ier5, Rgs16, A430109m19rik, Tnfrsf9, Penk1, Coch, Ptpn13, Tcrg-v4, Nr4a2, Cd160, Ptger4, Ccl4, Wbp5, Gpr56, 1110067d22rik, Entpd1, Sh2d2a, 42982, Isg20, Trim47, Serpina3g, Casp4, 9130009c22rik, C79248, Lag3, Nr4a2, Nftac1, Car2, C330007p06rik, Gpd2, 2700084l22rik, Rnf11, Capzb, Tubb2, Bub1, Jak3, 9130410m22rik, Cd9, 1810054d07rik, Rcn, 2010100o12rik, Sybl1, Etf1, Cpa3, Cd7, Art3, 1810035l17rik, Atf1, Prkwnk1, Mtv43, Cit, Ccrl2, Adfp, D8ertd531e, Tcea2, Myh4, Tnfrsf1a, Spp1, S100a13, Pon2, Ai181996, G1p2, Tank, Shkbp1, 2510004l0rik, D15ertd781e, Icsbp1, Bc024955, Gdf3, Itgav, 1110006i15rik, Cpsf2, Klk6, Cpt2, Lman2, Tor3a, Crygb, Gpr65, Mki67, Tcrb-v13, Nptxr, Snrpb2, Ndfip1, Ptger2, Zfp91, Spock2, 5730469m10rik, Cxcl10, Gcin, Trim25, Wbscr5, Mox2, Dock7, Pawr, and Chl1.
Pancreatic Islet Isolation.
Pancreata obtained from Rip-LCMV-GP mice were placed in 5 mL of cold RPMI 1640. To dissociate tissue, 3 mL of collagenase P (1.2 U/mL) was injected directly into the pancreas by using a 30 G needle and digested at 37 °C following addition of 7 mL of warm RPMI 1640. After 30 min, collagenase P solution was removed and 10 mL of ice-cold RPMI 1640 added. Tubes were shaken vigorously for 1 min to break apart tissue, which were passed through a wide-mesh sieve to remove nondissociated debris. Following several washing steps, RPMI 1640 media was removed and 10 mL of prewarmed Ficoll-Paque Plus (GE Healthcare) was added. A 5-mL RPMI 1640 overlay was added to Ficoll, and cells were spun at 931 × g for 13 min at room temperature to separate islets from exocrine cells. After the spin, islets were isolated at the Ficoll/RPMI 1640 interface by using a plastic Pasteur pipet, spun in cold RPMI 1640 media at 233 × g for 5 min, and added onto an inverted 70 μM filter to further remove nonislet debris and placed into a Petri dish with RPMI 1640 and 10% FBS. At least 50 purified islets were handpicked with a pipette per pancreas before RNA extraction.
Flow Cytometry.
Spleens were digested by using a mixture of collogenase/DNase before homogenation on a 100 μM filter by using a butt-end of a syringe. RBCs were lysed for 3 min in 1× RBC lysis buffer. Rat anti-mouse CD4 (Biolegend), CD8 (Biolegend), IFN-γ (BD Pharmingen), TNF-α (Biolegend), CD107a (Biolegend) were used at a 1:100 dilution.
Ex Vivo Peptide Restimulation Assay and CD107a Degranulation Assay.
Splenocytes were seeded onto round-bottom 96-well plates at 1 × 106 cells per well in complete media (10% FBS, l-glutamine, penstrep, NEAA, Sodium Pyruvate, Hepes, BME) supplemented with LCMV-specific peptides (GP33–41 and GP276–286). After a 1-h incubation, Brefeldin A (Sigma) was added at a 1:500 dilution and cells incubated for 5 h at 37 °C. Surface antigens were stained for 30 min on ice, cells fixed and permeabilized by using Cytofix/Cytoperm (BD Pharmingen), and stained for intracellular proteins including IFN-γ and TNF-α. For the CD107a degranulation assay, splenocytes were seeded at 1 × 106 cells per well in Complete media supplemented with Golgistop (1:1,500). Anti-CD107a (Biolegend) was added at 1:100 at the beginning of the assay, cells incubated for 6 h at 37 °C, and then stained for surface antigens.
Acknowledgments
We thank Beatrice Cubitt for technical assistance. This work was supported by NIH Grant AI009484 (to M.B.A.O.), NIH T32 Training Grants 5T32AI007354-2 and 5T32AI007244-33 (B.S.M.), the Jeanette Bertea Hennings Foundation (B.S.M.), and the Skaggs-Oxford Scholarship (to J.Z.). This is publication 29431 from the Department of Immunology and Microbial Science, The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE96541).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700878114/-/DCSupplemental.
References
- 1.Herold KC, Vignali DA, Cooke A, Bluestone JA. Type 1 diabetes: Translating mechanistic observations into effective clinical outcomes. Nat Rev Immunol. 2013;13:243–256. doi: 10.1038/nri3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooks JJ, et al. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 3.Huang X, et al. Interferon expression in the pancreases of patients with type I diabetes. Diabetes. 1995;44:658–664. doi: 10.2337/diab.44.6.658. [DOI] [PubMed] [Google Scholar]
- 4.Foulis AK, Farquharson MA, Meager A. Immunoreactive alpha-interferon in insulin-secreting beta cells in type 1 diabetes mellitus. Lancet. 1987;2:1423–1427. doi: 10.1016/s0140-6736(87)91128-7. [DOI] [PubMed] [Google Scholar]
- 5.Guerci AP, et al. Onset of insulin-dependent diabetes mellitus after interferon-alfa therapy for hairy cell leukaemia. Lancet. 1994;343:1167–1168. doi: 10.1016/s0140-6736(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 6.Fabris P, et al. Type 1 diabetes mellitus in patients with chronic hepatitis C before and after interferon therapy. Aliment Pharmacol Ther. 2003;18:549–558. doi: 10.1046/j.1365-2036.2003.01681.x. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira RC, et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes. 2014;63:2538–2550. doi: 10.2337/db13-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallionpää H, et al. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes. 2014;63:2402–2414. doi: 10.2337/db13-1775. [DOI] [PubMed] [Google Scholar]
- 9.Stewart TA, et al. Induction of type I diabetes by interferon-alpha in transgenic mice. Science. 1993;260:1942–1946. doi: 10.1126/science.8100367. [DOI] [PubMed] [Google Scholar]
- 10.Carrero JA, Calderon B, Towfic F, Artyomov MN, Unanue ER. Defining the transcriptional and cellular landscape of type 1 diabetes in the NOD mouse. PLoS One. 2013;8:e59701. doi: 10.1371/journal.pone.0059701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, et al. Interferon-alpha initiates type 1 diabetes in nonobese diabetic mice. Proc Natl Acad Sci USA. 2008;105:12439–12444. doi: 10.1073/pnas.0806439105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang KS, et al. Toll-like receptor engagement converts T-cell autoreactivity into overt autoimmune disease. Nat Med. 2005;11:138–145. doi: 10.1038/nm1176. [DOI] [PubMed] [Google Scholar]
- 13.Oldstone MB, Nerenberg M, Southern P, Price J, Lewicki H. Virus infection triggers insulin-dependent diabetes mellitus in a transgenic model: Role of anti-self (virus) immune response. Cell. 1991;65:319–331. doi: 10.1016/0092-8674(91)90165-u. [DOI] [PubMed] [Google Scholar]
- 14.von Herrath MG, Dockter J, Oldstone MB. How virus induces a rapid or slow onset insulin-dependent diabetes mellitus in a transgenic model. Immunity. 1994;1:231–242. doi: 10.1016/1074-7613(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 15.Emonet SE, Urata S, de la Torre JC. Arenavirus reverse genetics: New approaches for the investigation of arenavirus biology and development of antiviral strategies. Virology. 2011;411:416–425. doi: 10.1016/j.virol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewicki H, et al. CTL escape viral variants. I. Generation and molecular characterization. Virology. 1995;210:29–40. doi: 10.1006/viro.1995.1314. [DOI] [PubMed] [Google Scholar]
- 17.Sheehan KC, et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 18.Ng CT, et al. Blockade of interferon Beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe. 2015;17:653–661. doi: 10.1016/j.chom.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheehan KC, Lazear HM, Diamond MS, Schreiber RD. Selective blockade of interferon-α and -β reveals their non-redundant functions in a mouse model of West Nile Virus infection. PLoS One. 2015;10:e0128636. doi: 10.1371/journal.pone.0128636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teijaro JR, et al. S1PR1-mediated IFNAR1 degradation modulates plasmacytoid dendritic cell interferon-α autoamplification. Proc Natl Acad Sci USA. 2016;113:1351–1356. doi: 10.1073/pnas.1525356113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pircher H, Bürki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 22.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teijaro JR, et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146:980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer S, et al. APECED patient collaborative AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell. 2016;166:582–595. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teijaro JR, Walsh KB, Rice S, Rosen H, Oldstone MB. Mapping the innate signaling cascade essential for cytokine storm during influenza virus infection. Proc Natl Acad Sci USA. 2014;111:3799–3804. doi: 10.1073/pnas.1400593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Cabrera PJ, et al. Full pharmacological efficacy of a novel S1P1 agonist that does not require S1P-like headgroup interactions. Mol Pharmacol. 2008;74:1308–1318. doi: 10.1124/mol.108.049783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Cabrera PJ, et al. S1P(1) receptor modulation with cyclical recovery from lymphopenia ameliorates mouse model of multiple sclerosis. Mol Pharmacol. 2012;81:166–174. doi: 10.1124/mol.111.076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott FL, et al. Ozanimod (RPC1063) is a potent sphingosine-1-phosphate receptor-1 (S1P1 ) and receptor-5 (S1P5 ) agonist with autoimmune disease-modifying activity. Br J Pharmacol. 2016;173:1778–1792. doi: 10.1111/bph.13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen JA, et al. RADIANCE Study Group Safety and efficacy of the selective sphingosine 1-phosphate receptor modulator ozanimod in relapsing multiple sclerosis (RADIANCE): A randomised, placebo-controlled, phase 2 trial. Lancet Neurol. 2016;15:373–381. doi: 10.1016/S1474-4422(16)00018-1. [DOI] [PubMed] [Google Scholar]
- 31. Celgene (2016) Oral ozanimod showed histologic improvements in patients with ulcerative colitis in the phase 2 TOUCHSTONE trial. BusinessWire.
- 32.Rosen H, Stevens RC, Hanson M, Roberts E, Oldstone MB. Sphingosine-1-phosphate and its receptors: Structure, signaling, and influence. Annu Rev Biochem. 2013;82:637–662. doi: 10.1146/annurev-biochem-062411-130916. [DOI] [PubMed] [Google Scholar]
- 33.Teijaro JR, et al. Protection of ferrets from pulmonary injury due to H1N1 2009 influenza virus infection: Immunopathology tractable by sphingosine-1-phosphate 1 receptor agonist therapy. Virology. 2014;452-453:152–157. doi: 10.1016/j.virol.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh KB, et al. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proc Natl Acad Sci USA. 2011;108:12018–12023. doi: 10.1073/pnas.1107024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandala S, et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 36.Oldstone MB, Edelmann KH, McGavern DB, Cruite JT, Welch MJ. Molecular anatomy and number of antigen specific CD8 T cells required to cause type 1 diabetes. PLoS Pathog. 2012;8:e1003044. doi: 10.1371/journal.ppat.1003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]