Significance
Signaling by human epidermal growth factor receptor (HER) receptors relies on heteromeric interactions between four members of the family: EGF receptor, HER2, HER3, and HER4. These interactions remain remarkably poorly understood. Using super-resolution microscopy imaging and signaling assays, we demonstrate a rich scope of HER receptor organization patterns that are differentially influenced by ligands and coreceptor expression, resulting in unique phosphorylation signatures of HER receptors. We also show that there are fundamental differences in molecular mechanisms that govern HER receptor cross-activation, which do not always follow the canonical kinase dimerization mechanism. Our data underscore an emerging concept in the field that HER receptor signaling needs to be interpreted in the context of higher-order receptor oligomers, redefining the basic signaling unit relevant for receptor function.
Keywords: HER/ERBB receptors, receptor tyrosine kinase signaling, receptor clustering, STORM, EGFR activation
Abstract
Heteromeric interactions between the catalytically impaired human epidermal growth factor receptor (HER3/ERBB3) and its catalytically active homologs EGFR and HER2 are essential for their signaling. Different ligands can activate these receptor pairs but lead to divergent signaling outcomes through mechanisms that remain largely unknown. We used stochastic optical reconstruction microscopy (STORM) with pair-correlation analysis to show that EGF and neuregulin (NRG) can induce different extents of HER3 clustering that are dependent on the nature of the coexpressed HER receptor. We found that the presence of these clusters correlated with distinct patterns and mechanisms of receptor phosphorylation. NRG induction of HER3 phosphorylation depended on the formation of the asymmetric kinase dimer with EGFR in the absence of detectable higher-order oligomers. Upon EGF stimulation, HER3 paralleled previously observed EGFR behavior and formed large clusters within which HER3 was phosphorylated via a noncanonical mechanism. HER3 phosphorylation by HER2 in the presence of NRG proceeded through still another mechanism and involved the formation of clusters within which receptor phosphorylation depended on asymmetric kinase dimerization. Our results demonstrate that the higher-order organization of HER receptors is an essential feature of their ligand-induced behavior and plays an essential role in lateral cross-activation of the receptors. We also show that HER receptor ligands exert unique effects on signaling by modulating this behavior.
The human epidermal growth factor receptors (HERs/ErbBs) are essential regulators of development and adult homeostasis (1). All four of them, EGF receptor (EGFR; HER1), HER2 (ErbB2), HER3 (ErbB3), and HER4 (ErbB4), are at the focus of therapeutic efforts in a variety of human diseases. Most of what we know about their activation mechanism has been revealed by the studies on EGFR, which showed that ligand binding induces EGFR dimerization through a series of structurally well-defined interactions between the extracellular and intracellular receptor domains (2). These interactions result in the formation of an asymmetric kinase dimer in which one kinase (termed the “activator kinase”) is asymmetrically positioned to activate the second kinase (termed the “receiver kinase”) allosterically (3). The receiver kinase then is poised to phosphorylate the receptor tails, resulting in the recruitment of downstream signaling molecules and signal propagation.
One of the characteristic features of the HER receptor family is a significant degree of heteromeric interactions in response to ligand binding through which the receptors activate a variety of signaling pathways (1). These interactions are particularly important for signaling by the orphan receptor HER2 and the catalytically impaired HER3, which do not signal on their own under normal conditions. Although all HER receptors are assumed to form heterodimers in which the kinase domains recapitulate the asymmetric kinase homodimer characterized for EGFR (3), the molecular details of the heteromerization are largely not understood. The protein interfaces involved in the asymmetric kinase domain interactions are highly conserved among all HER receptors and have been structurally shown to support the formation of another active HER receptor homodimer, HER4/HER4, as well as an EGFR/HER3 heterodimer (4, 5). Enzymatic studies on the isolated kinase domains of HER receptors have also provided convincing evidence that their catalytic activation in heterodimers is dependent on the asymmetric dimer interface (4–7). On the other hand, because of a lack of structures of liganded extracellular domain heterodimers, we do not know how ligand binding promotes heterotypic interactions between the extracellular portions of these receptors. Functional studies in cells have shown that binding of a ligand cognate to only one HER receptor is sufficient to induce cross-activation of other HER receptors (8–10). How these interactions are further fine-tuned by ligands with different specificity for the individual receptors is presently not completely understood.
Several studies have indicated that heterodimeric HER receptor complexes are functionally distinct, depending on which cognate ligand activates them. The example of pairing between EGFR, which specifically binds its own ligands such as EGF, and the catalytically impaired HER3, which binds its own ligand neuregulin (NRG), is particularly intriguing. Signaling crosstalk between EGFR and HER3 plays a predominant role in signaling in the adult liver and in melanomas (11, 12) and in underlying poor response to therapies that directly target HER2 (13). The EGFR/HER3 pair can be activated either through EGF or NRG, but although the same active enzyme (EGFR) is engaged in both cases, the resulting pattern of receptor phosphorylation is significantly different when the EGFR/HER3 complex is induced by EGF or NRG stimulation (Fig. 1). EGF induces robust phosphorylation of both receptors, whereas NRG stimulation results in poor EGFR phosphorylation (Fig. 1) (10, 14). Based on the structural understanding of the EGFR/HER3 kinase interaction, these two kinases should interact in the same way in each case, with EGFR in the receiver position and HER3 serving as an allosteric activator. What then explains differences in phosphorylation outcomes?
Fig. 1.
Differential effects of NRG and EGF on the phosphorylation of coexpressed HER3 and EGFR. NR6 cells stably coexpressing mEos–HER3 and EGFR–GFP were serum starved for 6 h, stimulated with 10 nM EGF or 10 nM NRG for 2–60 min, and subsequently lysed. Western blot analysis of the lysates with the indicated antibodies reveals phosphorylation states of EGFR and HER3 as well as AKT and ERK. Antibodies used are listed in Table S1.
Although our mechanistic understanding of ligand-induced HER receptor activation has been developed primarily in the context of a dimer model, initial studies on the behavior of EGFR in response to EGF referred to higher-order oligomerization (15, 16). Subsequently, numerous studies have provided quantitative evidence that ligand-induced active EGFR complexes extend beyond dimers to larger oligomers. In some studies, such indications came from the experiments in which FRET was detected between two EGF ligand molecules; because of the structural constraints of their interaction, this transfer is most logically explained by receptor oligomerization (17–21). Other studies imaged fluorescently labeled EGFR itself using image correlation microscopy (ICS) (22, 23), fluorescence correlation spectroscopy (FCS) (24, 25), stochastic optical reconstruction microscopy (STORM) (26), FRET, fluorescence lifetime imaging microscopy (FLIM) (27, 28), spatial mapping of the receptor by immuno-electron microscopy (29, 30), or number and brightness analysis (31). This spectrum of experimental approaches all led to a unifying conclusion that EGFR is organized in larger clusters in response to ligand binding. More recent studies provide evidence that these higher-order multimers are needed to achieve the complete spectrum of EGFR phosphorylation (21, 25).
At present, it is uncertain whether in the HER family higher-order oligomerization is unique to EGFR or constitutes an essential component of signaling by all HER receptors. It is also unknown how this behavior of EGFR influences its interactions with other HER receptors and to what extent the formation of higher-order complexes is involved in HER receptor crosstalk initiated by different ligands. In this study we set out to understand the mechanism by which EGF and NRG stimulate different phosphorylation states of HER receptors. Using NR6 cells that do not express detectable levels of any HER receptors, we created stable cell lines expressing HER receptors and imaged these receptors in the range corresponding to physiological expression levels. We applied STORM analysis that combines conventional STORM superresolution imaging with a blinking correction algorithm. In STORM, the position of a single molecule can be determined with a precision of ∼20 nm (FWHM) over a wide range of receptor densities. As a result, this method provides an advantage over published single-molecule imaging analyses of HER receptors, which often are based on single-molecule tracking of a sparsely labeled HER receptor or its ligand and are confounded by contributions from unlabeled “dark” receptors that represent the endogenous receptor populations (17–22, 32, 33). Additionally, because many fluorophores commonly used for STORM blink erratically (34), we chose a fluorophore [a photo-activatable fluorescent protein (PAFP), mEos3.2, hereafter referred to as “mEos”] that enables correction of the blinking behavior in postimaging analysis. The correction eliminates the overcounting of receptors, which is inevitable when the quantification of receptor oligomerization is based on fluorescence intensity, as applied in previous STORM analysis of EGFR oligomerization (26). We then scored the blink-corrected molecular positions with a pair-correlation analysis, obtaining a reliable representation of the extent of receptor clustering based on the fluorescent image.
We show that when EGFR and HER3 are coexpressed, EGF induces robust phosphorylation of both receptors, but NRG stimulation leads only to HER3 phosphorylation. Using STORM with blink-correction analysis, we show that in response to the two different ligands, EGF and NRG, EGFR and HER3 populate distinct oligomeric states, with EGF stimulation resulting in higher-order clustering of both receptors driven by EGFR oligomerization. Moreover, we show that although phosphorylation of HER3 in response to NRG is a consequence of direct asymmetric dimer formation between EGFR and HER3, the asymmetric interaction between these two receptors is dispensable for their phosphorylation in response to EGF. Our data indicate that recruitment of HER3 into EGF-dependent EGFR clusters results in its phosphorylation through lateral propagation and is independent of the formation of the canonical active complex between the EGFR and HER3 kinase domains. In contrast, HER3 interaction with HER2 in response to NRG binding also proceeds through cluster formation but remains dependent on the canonical asymmetric interactions within the HER2/HER3 kinase dimer. Our data thus unravel unique features of membrane organization of HER receptors and a surprising complexity of the mechanisms that support ligand-induced crosstalk in the HER receptor family.
Results
EGF and NRG Induce Different Phosphorylation Receptor States in the EGFR/HER3 Complexes.
To examine the differences in the mechanism for receptor activation when signaling by the EGFR/HER3 complex is induced by EGF or by NRG at normal physiological levels of receptor expression, we created stable NR6 cell lines expressing HER receptor constructs. NR6 cells do not express detectable levels of HER receptors, minimizing the interference from endogenous HER receptors (Fig. 6 for HER2 and Fig. S1A for EGFR and HER3). Stable cell lines were sorted by flow cytometry (Fig. S1B), and we subsequently used STORM to image selectively cells that express 20–60 receptors per square micrometer on average, corresponding to the average density of the receptor observed under normal physiological conditions (Fig. S1C) (35). The receptors were tagged with mEos to allow quantitative analysis of HER receptors by STORM in later analyses. We fused mEos between the N-terminal signal peptide and the beginning of the mature extracellular domain of HER receptors (Fig. 2A). The mEos fusion did not affect the normal function of HER receptors in response to ligand binding (Fig. S1D).
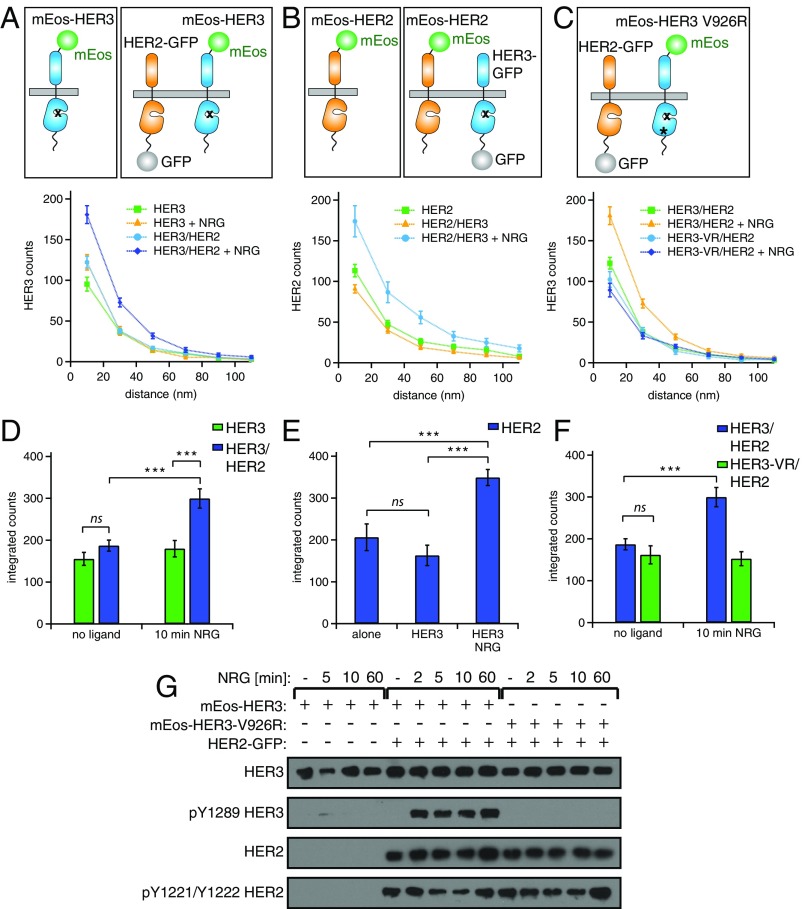
Fig. 6.
NRG induces clustering of HER3 in the presence of HER2 and its phosphorylation via the asymmetric kinase dimerization. (A–C, Upper) Cartoons depicting the different combinations of mEos–HER3, HER2–GFP, mEos–HER2, and mEos–HER3–V926R used in the experiments below. (Lower) Pairwise distance histograms calculated from the STORM images (as described in Fig. 2) for the following conditions. (A) Cells expressing mEos–HER3 alone or mEos–HER3 and HER2–GFP and stimulated for 10 min with 10 nM NRG. (B) Cells expressing mEos–HER2 alone or coexpressing HER3–GFP and stimulated for 10 min with 10 nM NRG. (C) Cells coexpressing mEos–HER3–V926R and HER2–GFP, and stimulated for 10 min with 10 nM NRG. (D–F) Integrated counts of pair-correlation data in A, B, and C, respectively, calculated as described in Fig. 2. In F, a relative comparison with the extents of clustering to the wild-type mEos–HER3 coexpressed with HER2–GFP is shown. ns, not significant. ***P < 0.001. (G) Western blot analysis of the lysates from NR6 cells expressing mEos–HER3 alone, mEos–HER3/HER2–GFP, or mEos–HER3–V926R/HER2–GFP cells were serum starved 6 h and were stimulated with NRG for 2–60 min.
Fig. S1.
Endogenous expression of HER receptors and functionality of tagged HER receptors in NR6 cells. (A) Western blot analysis of untransfected NR6 cells and NR6 cells stably expressing mEos–HER3, mEos–HER3/EGFR–GFP, or mEos–HER3/EGFR–V924R–GFP. Cells were grown for 24–36 h, serum starved for 6 h, stimulated for 10 min with 10 nM EGF (E) or 10 nM NRG (N), and then were lysed. Phosphorylation of HER3 was measured at Y1289, and phosphorylation of EGFR was measured at Y1068. (B) Cell-surface receptor levels were measured by flow cytometry analysis for mEos-tagged EGFR, HER2, and HER3 using an antibody capture assay (Quantum Simply Cellular beads; Bangs Laboratories, Inc.). NR6 cells expressing respective constructs and assay bead calibration populations were labeled with HER receptor allophycocyanin (APC)-conjugated antibodies. Expression levels were determined by interpolation against the antibody capture calibration curve. (C) The distribution of the number of receptors per cell that were accepted for STORM correlation analysis for the different receptors. The numbers were counted by STORM imaging and blink correction, which can be used to estimate the number of molecules per square micrometer. The surface area of the cell was estimated to be ∼800 square micrometers. (D) Western blot of HEK293 cellular lysates transfected with wild-type, mEos-, or GFP-tagged HER receptor constructs. To measure phosphorylation of the HER3 and HER2 tagged constructs, untagged HER2 and HER3 were coexpressed. Following 24 h transfection, cells were serum starved for 6 h, stimulated for 10 min with 10 nM EGF or 10 nM NRG, and then lysed. Phosphorylation of EGFR was measured at Y1068, phosphorylation of HER2 was measured at Y1221/Y1222, and phosphorylation of HER3 was measured at Y1289.
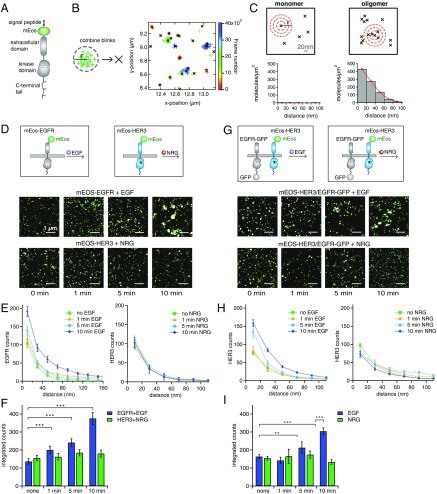
Fig. 2.
STORM imaging of EGFR and HER3 upon ligand stimulation. (A) Schematic representation of the mEos–HER receptor fusion. (B) Fluorescent blinks (colored crosses) from individual mEos-tagged receptors were combined based on a threshold radius (r) and time (color scale). The positions of all blinks were averaged to determine the blink-corrected position (denoted by a black x). (C) Molecular positions of a simulated monomer or oligomer. The number of molecules within each red shell (which has a thickness of the bin size, 20 nm) was counted and normalized for the area of each shell for each molecule in the image. Counts are summed in the pairwise distance histograms and show increasing height of bins at short distances with increasing size of oligomers because of the increase in the local density of molecules. Red curves reflect the bin height and are plotted instead of bins in all subsequent figures to allow simpler plotting of multiple overlapping histograms. (D, Upper) Cartoon schematic of the constructs and ligands used in the experiment. (Lower) Representative reconstructed STORM images of mEos–EGFR and mEos–HER3 organization at the plasma membrane of NR6 cells after stimulation with 10 nM EGF or 10 nM NRG for the indicated amount of time. Cells were imaged using TIRF illumination to detect receptors selectively at the plasma membrane. (E) STORM images of mEos–EGFR or mEos–HER3 under the indicated conditions were corrected for blinking as described in B; then the pairwise distance histograms were constructed from the blink-corrected molecular positions. For all experiments, the count values for each bin are the median of hundreds of nonoverlapping regions of interest (25 × 25 pixels) analyzed in total and were compiled from 10–12 cells; all error bars reflect the SE. Data were compiled from all experiments, including experiments performed on different days, to account for sample variability. Statistical significance for all plots was calculated using a two-sided Wilcoxon rank sum test for equal medians. (F) Sum of the counts (y-value, also termed “integrated counts” here) from the pairwise correlation histograms in E up to 80 nm (first four bins), representing the increase in total number of receptors above the average density within a radius of 80 nm. Error bars represent the sum of the four SEs of the bins that were summed. ***P < 0.001. (G) Cartoon schematic (Upper) and STORM images (Lower) of mEos–HER3 coexpressed with EGFR–GFP and stimulated with ligand (EGF or NRG) for the indicated times. (H) Pair-correlation analysis as described in E for STORM images of mEos–HER3 coexpressed with EGFR–GFP and stimulated with EGF or NRG. (I) Sums of counts calculated as in F for the above H pair-correlation histograms. **P < 0.01; ***P < 0.001.
To evaluate the extent of EGFR and HER3 activation upon stimulation with EGF or neuregulin 1β (NRG1β; referred to as “NRG” throughout the paper), we measured the phosphorylation status of key tyrosine residues in the C-terminal tails of EGFR and HER3 by Western blot analysis of the NR6 cell lines stably coexpressing tagged HER3 and EGFR receptors. As shown in Fig. 1, HER3 phosphorylation is comparable when cells cotransfected with HER3 and EGFR are stimulated with EGF or NRG. This HER3 phosphorylation translates to similar levels of serine-threonine kinase (AKT) phosphorylation, which is predominantly induced by phosphorylated HER3 (Fig. 1). In contrast, although phosphorylated EGFR is robustly detected in response to EGF, almost no signal is detected in response to NRG (Fig. 1). Consequently, phosphorylation of ERK, a main downstream target of EGFR, is also significantly lower in response to NRG than with EGF stimulation (Fig. 1).
The observed lack of EGFR phosphorylation in response to NRG is in agreement with previous observations (10, 14), but it is somewhat puzzling in the context of our understanding of the mechanism of HER receptor activation. According to this mechanism, EGFR and HER3 are expected to form an asymmetric kinase dimer upon ligand-induced heterodimerization. Because of severe impairment in catalytic activity, HER3 has been shown to act as the allosteric activator of EGFR when these two partners form an asymmetric dimer (6, 7). Because EGFR is the only active kinase in this complex, the resulting receptor phosphorylation is a result of its activity. Although phosphorylation of EGFR in cis in the active dimer has been shown to be less pronounced than in trans, it does occur (36). Why then cannot EGFR autophosphorylate itself when stimulated with NRG in the presence of HER3?
STORM-Based Measurement of Receptor Clustering.
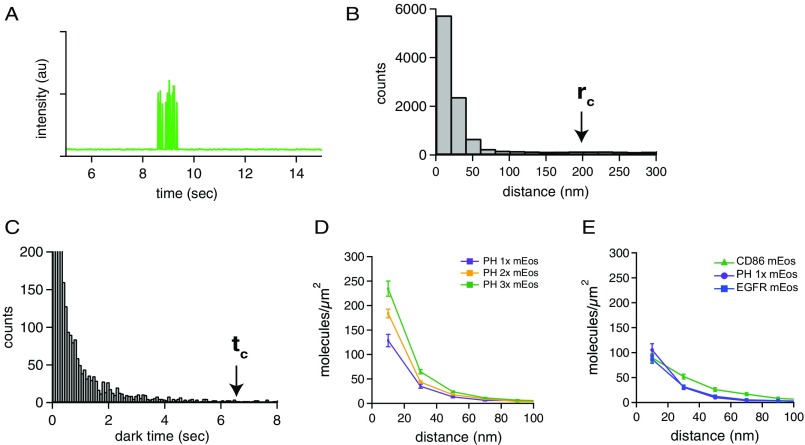
To understand the underlying mechanism for the difference in the response of the coexpressed EGFR and HER3 receptors to stimulation with their cognate ligands, we examined how ligand addition affects their spatial organization at the plasma membrane using our pair-correlation analysis of blink-corrected STORM images. The use of mEos as a fluorescent label of HER receptors lies at the core of our STORM analysis. Unlike organic fluorophores such as Alexa 647, whose blinking behavior can be erratic and thus subject to overcounting, some PAFPs, such as mEos, exhibit a brief burst of blinks and then photobleach and do not return to a bright state (Fig. S2A for a representative fluorescence time trace). The blinking behavior of mEos then can be deconvoluted by applying a blink-correction algorithm to discern whether fluorescent bursts originated from a single molecule or from multiple molecules within the spatial resolution (20 nm) (34). In this method, bursts of fluorescence (blinking) originating from a single molecule can be combined by applying thresholds in both the temporal and spatial dimension of the fluorescence localizations (Fig. 2B). Both thresholds were determined experimentally: the temporal threshold from the dark-state lifetime distribution, and the spatial threshold from the pair-correlation histogram of the raw data (Fig. S2 B and C). The pair-correlation histogram is the distribution of distances between any two pairs of localizations; this function is flat if all molecules are distributed randomly but exhibits a peak with increased local density of molecules around another molecule (Fig. 2C). The amplitude of the peak increases if the local density increases, and the width of the peak increases with cluster size (Fig. 2C). After combining the blinks originating from single molecules, we constructed a blink-corrected pair-correlation histogram, which we used as a quantitative readout of receptor clustering.
Fig. S2.
Threshold determination for blink correction and PH calibration. (A) An example of a fluorescence time trace of a single mEos-tagged receptor photoactivated with 405-nm light and imaged with 561-nm excitation light. The fluorophore turns on, blinks several times, and then turns off (photo bleaches) permanently. (B) Pairwise distance histogram of raw (not blink-corrected) STORM localizations of a presumed monomeric receptor (PH-1×). The cutoff radius (rc) used for the subsequent blink corrections was set at the 99th percentile of this histogram, around 200 nm. (C) Histogram of the dark times of a sparse, monomeric sample of mEos-tagged PH-1×. Dark times were determined by applying the blink correction to the STORM data, using a very large cutoff time, and then determining the 99th percentile of these dark times to set a threshold for the blink correction of all data. The cutoff time (tc) was determined to be around 6.4 s (400 frames). (D) One, two, or three copies of mEos were fused to the PH domain, a membrane-localized and presumed monomeric protein. These constructs were expressed in NR6 cells and imaged by STORM, and the blink-corrected pairwise distance histograms were calculated. Although the presumed monomeric curve is not flat, as was expected, progressive increases in peak amplitude were observed for the doubly and triply tagged proteins. This basal peak in the pairwise distance histogram could be caused by clustering of the PH domain proteins in the plasma membrane of mammalian cells, by membrane inhomogeneity, or by rare long-lived dark states of the chromophore. Therefore only relative changes in the pairwise distance histogram were used for all analysis. (E) Pairwise distance histogram for mEos-tagged CD86 and mEos-tagged PH-1× constructs plotted on the same graph as mEos–EGFR. All constructs were stably expressed in NR6 cell lines.
When Expressed Alone, EGFR, but Not HER3, Clusters in Response to Binding of the Cognate Ligand.
Using STORM, we first looked at how the organization of EGFR and HER3 at the plasma membrane changes in response to stimulation with their respective selective ligands, EGF and NRG, when these receptors are expressed alone. NR6 cells stably expressing mEos-tagged EGFR or HER3 were serum starved, stimulated with their respective ligands for varied periods of time, and subsequently fixed and imaged by STORM using total internal reflection fluorescence (TIRF) illumination to image the receptors selectively at the plasma membrane. Upon stimulation with EGF, we observed that EGFR molecules start to cluster as early as 1 min after EGF stimulation, forming increasingly larger clusters at later time points (Fig. 2 D–F). Using a counting radius of 50 nm (chosen because it is approximately equal to the FWHM spatial resolution of the raw STORM image), we created histograms of the number of receptors per cluster and estimated that these larger clusters contain ∼5–20 receptor molecules on average (Fig. S3A). This number is an estimate, and the exact number can depend on several parameters and aspects of the counting method; therefore we use it only as a general comparison between different receptor clusters. In contrast to EGFR, no measurable change in clustering could be detected for the catalytically impaired HER3 in the presence of NRG (Fig. 2 D–F).
Fig. S3.
Distributions of the number of receptors per cluster for EGFR and HER3. (A) Cells expressing mEos–EGFR were stimulated for 10 min with 10 nM EGF and were imaged and analyzed by STORM. The blink-corrected data were used to determine the number of receptors within a 50-nm radius of any other receptor. This radius was chosen because it is approximately equal to the FWHM of the localization precision of the raw STORM data; i.e., it does not indicate that clusters are 50 nm in size but only that they can be determined with 50-nm FWHM resolution. The number of receptors within these clusters with a 50-nm radius were counted and binned over the range indicated in the x-axis labels. (B) The analysis in A was applied to cells coexpressing mEos–HER3 and EGFR–GFP, which were stimulated with 10 nM EGF for 10 min.
Notably, the pair-correlation histograms describing EGFR and HER3 organization under the serum-starved conditions are not flat, suggesting that these receptors could exist in oligomeric states larger than monomeric in the basal condition (Fig. 2E). To understand the origin of this basal-level peak in the pair-correlation, we used a calibration system previously applied to count the number of molecules in clusters in yeast cells (34). In this method, different oligomeric states are modeled by fusion of an increasing number of mEos molecules to the pleckstrin homology (PH) domain of AKT, which is monomeric and localizes to the plasma membrane (34). We analyzed blink-corrected pair-correlation functions of 1×, 2×, and 3× repeats of mEos fused to the PH domain in NR6 cells. In contrast to the analysis performed in yeast, the correlation function of the monomeric PH-1×–mEos fusion in mammalian cells was not flat, even at relatively low (20 molecules/μm2) expression level (Fig. S2D).
As an additional control, we also analyzed the membrane organization of a single-pass transmembrane protein, CD86 [previously demonstrated to be monomeric (37)], tagged at the C terminus with mEos, and observed a similar peak in the pair-correlation function (Fig. S2E). Although there could be several explanations for this peak observed for both PH-1×–mEos and CD86–mEos, which did not appear in the counting study in yeast (34), it is possible that this behavior reflects intrinsic heterogeneity of the mammalian cell membrane environment, such as the presence of cholesterol-rich lipid microdomains or regions of the membrane that are not completely flat. These irregularities would be expected to prevent uniform distribution of membrane proteins. We therefore have refrained from interpreting the basal state of HER receptor clustering at the plasma membrane and instead focused on relative changes in HER receptor organization at the membrane in response to ligand stimulation.
EGF Induces HER3 Clustering in the Presence of EGFR.
We next used STORM to investigate the effect of EGF and NRG stimulation on HER3 and EGFR when they are coexpressed. Because PAFPs that can be paired with mEos for two-color STORM have substantially lower brightness, we focused on single-color experiments in which one receptor was tagged with mEos and another with the monomeric enhanced version of GFP. GFP labeling allowed the selection of cells that coexpress both receptors but did not interfere with the detection of mEos. We first looked at mEos–HER3 coexpressed with EGFR–GFP. Remarkably, HER3 organization was significantly different when cells were stimulated with NRG rather than EGF. In the presence of its own ligand, NRG, and EGFR coexpression, the extent of HER3 clustering was similar to that observed for HER3 alone upon NRG stimulation (Fig. 2 G and H). We reasoned that this behavior is compatible with NRG-induced HER3/EGFR heterodimers, which in our assay would not be detected as a change in the HER3 oligomerization state. In contrast, EGF induced significant clustering of HER3 at the membrane upon EGFR coexpression (Fig. 2 G and H), but not in the absence of EGFR (Fig. S4A). This effect could be seen as early as 5 min after EGF stimulation and was reminiscent of the behavior of EGFR in response to EGF when EGFR was expressed alone (Fig. 2D). In the complementary set of experiments, when mEos–EGFR was coexpressed with HER3–GFP, we observed that coexpression of HER3 did not affect EGFR behavior in a measurable way. EGFR still clustered in response to EGF but not in response to NRG (Fig. S4B). EGF-induced HER3 clusters were smaller in receptor number than EGFR clusters, averaging 5– 12 molecules in a cluster (Fig. S3B), as determined using the estimate described above for EGF-induced EGFR clusters.
Fig. S4.
Clustering of EGFR with HER3 with EGF but not with NRG. (A) mEos–HER3 was expressed alone (blue bars) or was coexpressed with EGFR–GFP (green bars) in NR6 cells and was stimulated with 10 nM EGF or 10 nM NRG for 10 min. Only EGF caused receptor clustering and did so only if HER3 was coexpressed with EGFR. (B) In contrast to A, here mEos was fused to EGFR, and GFP was fused to HER3. Cells coexpressing mEos–EGFR/HER3–GFP were stimulated with 10 nM NRG or 10 nM EGF for 10 min and were imaged by STORM.
These data show that although both EGF and NRG are capable of inducing HER3 phosphorylation in an EGFR-dependent manner, the underlying organization of HER3 and EGFR complexes at the membrane differs quite markedly in response to each ligand. EGFR and HER3 form smaller oligomers, likely dimers, in the presence of NRG and form higher-order clusters in the presence of EGF. This difference coincides with the pattern of EGFR phosphorylation. As with clustering, we observe EGFR phosphorylation only in the presence of EGF, not with NRG, suggesting that clustering of EGFR might be necessary for its efficient phosphorylation. Because we observed HER3 clustering only when HER3 was coexpressed with EGFR and stimulated with EGF, we hypothesized that HER3 clustering might be a direct consequence of EGF-induced EGFR clustering.
EGFR Clustering Is Necessary for EGF-Dependent HER3 Clustering.
To test our hypothesis that HER3 clustering is a consequence of EGFR clustering, we first looked at the functional requirements within EGFR that contribute to this behavior. We predicted that an intact kinase is essential for EGFR clustering because EGFR activation was previously linked to the formation of ligand-induced higher-order oligomers (25, 29, 32, 38). We introduced a mutation (V924R) that falls within the asymmetric dimer interface between the kinase domains and renders EGFR incapable of activation (3). We imaged cells expressing the mEos–EGFR–V924R mutant alone by STORM. The V924R mutation completely abrogated EGFR clustering in response to ligand binding across all EGF stimulation time points (Fig. 3 A and D), suggesting that the catalytic activity of EGFR is essential for its clustering. To ascertain that the inhibitory effect of the V924R mutation on EGFR clustering is caused by the inhibition of catalytic activity and not by the disruption of receptor interactions that rely on the interface centered around V924, we inhibited EGFR activity through an alternative strategy using the small-molecule kinase inhibitor gefitinib. Gefitinib binding is compatible with kinase asymmetric dimerization but blocks catalysis (39). The addition of EGF to cells in the presence of gefitinib also prevented the formation of higher-order EGFR clusters but, interestingly, increased the basal level of clustering (Fig. 3 B and E). We reasoned that this increased basal level might be caused by the stabilization of a preformed lower-order oligomeric state of EGFR, such as a dimer, as has been described previously for other type I EGFR kinase inhibitors (40). Cumulatively, our results show that EGFR clustering depends on its ability to form a catalytically active complex.
Fig. 3.
Clustering of EGFR and HER3 upon EGF is dependent on EGFR kinase activity. Pairwise distance histograms calculated as in Fig. 2 for the following conditions: (A) Cells expressing the mutant mEos–EGFR–V924R were stimulated with 10 nM EGF for the indicated time and were imaged and analyzed by STORM. (B) Cells expressing mEos–EGFR were treated with gefitinib (10 μM) during the entire period of serum starvation (6 h) and then were stimulated with 10 nM EGF. (C) Cells expressing both mEos–HER3 and EGFR–V924R–GFP were stimulated with 10 nM EGF or 10 nM NRG for 10 min and then were imaged and analyzed by STORM. (D–F) Integrated histograms were calculated from the pair-correlation data as described in Fig. 2 for the mEos–EGFR–V924R mutant and mEos–EGFR with gefitinib. Comparisons with compiled wild-type mEos–EGFR are shown in D and E, and the comparison with wild-type EGFR–GFP coexpressed with mEos–HER3 is shown in F. **P < 0.01; ***P < 0.001.
We then used the EGFR–V924R mutant to test if HER3 clustering in response to EGF depends on EGFR clustering. When mEos–HER3 was coexpressed with the EGFR–V924R–GFP, we could no longer detect HER3 clustering in response to EGF (Fig. 3 C and F). These results indicate that HER3 reorganization at the plasma membrane in response to EGF is linked directly to ligand-induced changes in the behavior of its interaction partner, EGFR.
HER3 Phosphorylation by EGFR Depends on the Ability of EGFR to Homo-Oligomerize.
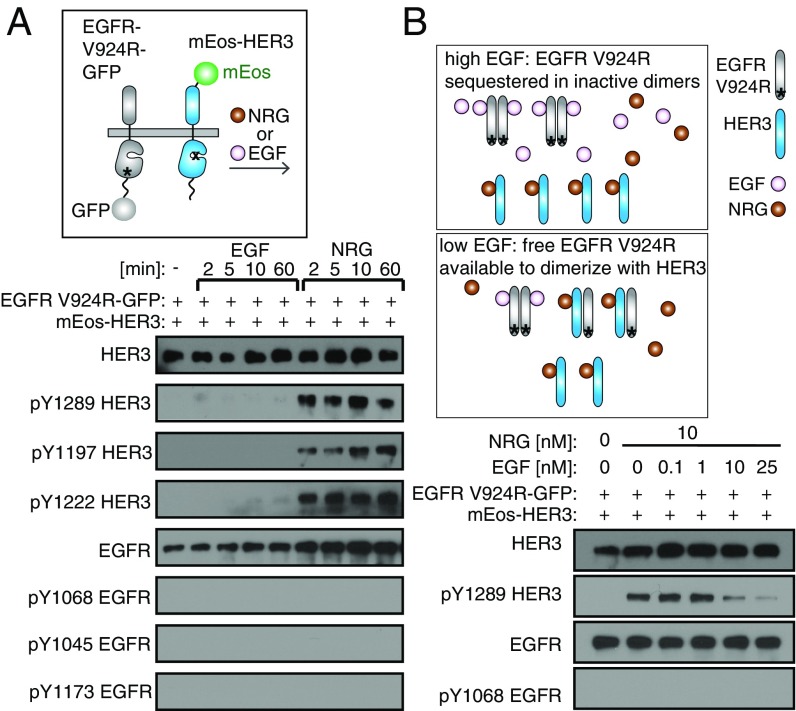
We then examined whether blocking EGFR homo-oligomerization while retaining its ability to interact with HER3 will influence the extent of HER3 phosphorylation induced by EGF. To this end, we used the EGFR–V924R mutant. Although this mutation blocks the ability of EGFR to self-activate by inhibiting its allosteric activator function, it preserves its function as a receiver kinase. The catalytic function of the EGFR–V924R mutant therefore can be activated by partnering with a receptor with an intact allosteric activator function, such as HER3 (3, 6).
In cells coexpressing EGFR–V924R with HER3, EGFR phosphorylation could no longer be detected in response to EGF (Fig. 4A). These data indicated that the EGFR phosphorylation detected upon EGF stimulation is primarily a result of EGFR activation through self-association and that HER3 cannot act as an allosteric activator receptor for the EGFR kinase under these circumstances. Moreover, we observed that in these cells, HER3 is no longer phosphorylated in response to EGF (Fig. 4A). This lack of phosphorylation does not result from the inability of the EGFR–V924R mutant to phosphorylate HER3, because, upon stimulation with NRG, HER3 was phosphorylated efficiently by EGFR–V924R (Fig. 4A). Our data therefore suggest that, upon stimulation with EGF, HER3 is phosphorylated by EGFR in a manner that is dependent on the ability of EGFR to form self-activating oligomers.
Fig. 4.
Homo-oligomerization of EGFR is required for HER3 phosphorylation in response to EGF. (A, Upper) Cartoon depicting EGFR–V924R–GFP and mEos–HER3 receptors. (Lower) Western blot analysis of the lysates from NR6 cells stably expressing mEos–HER3 and EGFR–V924R–GFP. The lysates were collected after stimulation with 10 nM EGF or 10 nM NRG for the indicated periods of time. (B, Upper) The cartoons provide a schematic representation of predicted interaction preferences between EGFR–V924R and HER3 under different ratios of EGF and NRG. (Lower) Western blot analysis of the lysates from NR6 cells stably expressing mEos–HER3 and EGFR–V924R–GFP. The lysates were collected after stimulation with 10 nM NRG and increasing concentrations of EGF (0–25 nM) for 10 min. Phosphorylation was monitored at Y1289 for HER3 and at Y1068 for EGFR with site-specific antibodies.
In the Presence of EGFR and HER3, EGF Preferentially Drives EGFR Homo-Oligomerization.
Our data indicate that, in response to NRG, HER3 phosphorylation proceeds via the formation of a canonical EGFR/HER3 heterodimer in which HER3 allosterically activates the EGFR kinase, but such heterodimers might not form in the presence of EGF. Rather, in the presence of EGF, liganded EGFR is much more likely to interact with another EGFR. Hence, if both EGF and NRG are present, EGFR/HER3 heterodimers will form only under nonsaturating levels of EGF when EGF-free EGFR molecules are available to interact with HER3 in the NRG-stabilized EGFR/HER3 heterodimers (Fig. 4B). To test this idea, we stimulated cells coexpressing HER3 and EGFR–V924R with a saturating concentration of NRG (10 nM) and a range of EGF concentrations. We predicted that at low concentrations of EGF an available pool of ligand-free EGFR molecules would be available to form NRG-induced HER3/EGFR–V924R heterodimers and support HER3 phosphorylation. Increasing concentrations of EGF should result in sequestering EGFR–V924R in dimers, which are catalytically inactive because of the inability of EGFR–V924R to form an asymmetric dimer, and therefore HER3 phosphorylation could no longer be supported. Indeed, in the absence or at low concentrations of EGF, NRG induced efficient HER3 phosphorylation by EGFR–V924R, reflecting these two receptors’ ability to heterodimerize efficiently through the asymmetric kinase dimer interface. In agreement with our prediction, HER3 phosphorylation decreased progressively with increasing EGF concentration, indicating a shift in the distribution of NRG-induced EGFR–V924R/HER3 heterodimers to inactive EGFR–V924R homo-oligomers (Fig. 4B).
HER3 Phosphorylation in Response to EGF Proceeds Through a Noncanonical Mechanism.
If EGF preferentially drives self-association of EGFR through asymmetric kinase dimerization (Fig. 4B), it would suggest that the chances of forming EGFR/HER3 asymmetric kinase dimers in response to EGF are minimal. Nevertheless, we observe that HER3 is robustly phosphorylated by the wild-type EGFR in response to EGF (Fig. 1), indicating that its phosphorylation might proceed through a mechanism that does not rely on the canonical asymmetric dimer interface formed between EGFR and HER3. We introduced a V926R mutation in HER3 that is equivalent to the V924R mutation in EGFR and that disrupts the allosteric activator interface between HER3 and EGFR (6). We observed that, in the presence of EGF, the HER3–V926R mutant is still robustly phosphorylated on a wide spectrum of phosphorylation sites (Fig. 5A). In the presence of NRG, however, introduction of the V926R mutation in HER3 completely blocks phosphorylation by EGFR (Fig. 5A). These findings demonstrate that EGF-induced crosstalk between EGFR and HER3 proceeds through a noncanonical mechanism whereby EGFR and HER3 do not rely on the formation of a direct asymmetric dimer. NRG induces intrinsically different interactions between HER3 and EGFR and engages the receptors through asymmetric heterodimerization of the kinase domains that relies on HER3’s function as an allosteric activator.
Fig. 5.
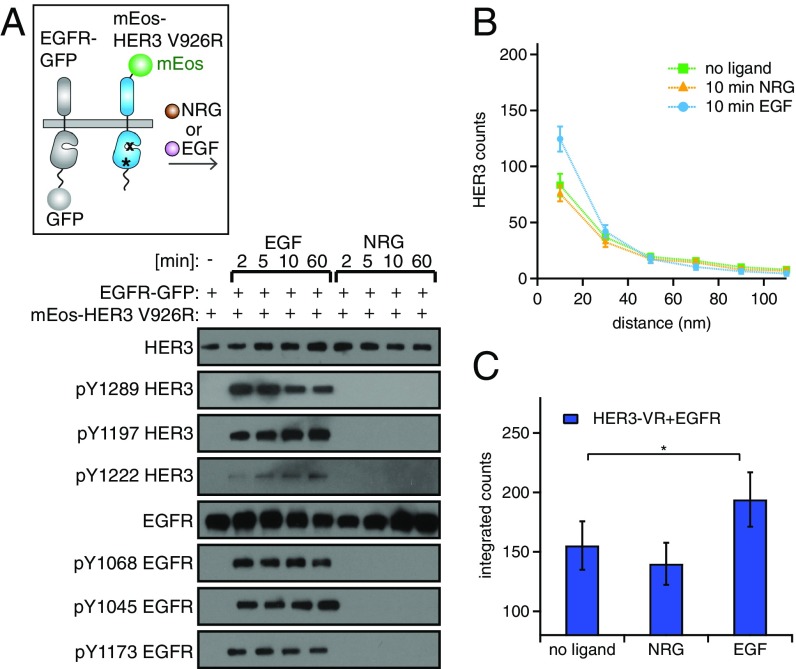
In the EGF-induced clusters, HER3 is phosphorylated by EGFR independently of asymmetric kinase dimerization. (A, Upper) Cartoon depicting the EGFR–GFP and mEos–HER3–V926R receptors. (Lower) Western blot analysis of the lysates from NR6 cells stably expressing mEos–HER3–V926R and EGFR–GFP. The lysates were collected after stimulation with 10 nM EGF or 10 nM NRG for the indicated periods of time. (B and C) Pairwise distance histograms (B) and integrated counts from those histograms calculated from the STORM images (C) (as described in Fig. 2) for cells coexpressing the mutant mEos–HER3–V926R and EGFR–GFP and stimulated with 10 nM EGF or 10 nM NRG for 10 min. *P < 0.05.
Although phosphorylation of HER3 in response to EGF does not proceed through the canonical dimerization mechanism, it is strictly dependent on EGFR’s ability to self-activate in EGF-induced homo-oligomers that we observed by STORM (Figs. 2 D–F and 4). Our imaging analysis of HER3 coexpressed with EGFR indicates that EGF also mobilizes HER3 to cluster, a behavior that is not observed with the addition of NRG (Fig. 2 G–I). We therefore assessed whether the HER3–V926R mutant is recruited to clusters; such recruitment could explain why it still retains the ability to be phosphorylated by EGFR in response to EGF, reflecting the behavior of wild-type HER3. In agreement with this assumption, we observed that HER3–V926R is indeed recruited to clusters efficiently (Fig. 5 B and C), mirroring the behavior of wild-type HER3. The ability of HER3–V926R to cluster also demonstrates that, like its phosphorylation, HER3 clustering in response to EGF is independent of the asymmetric dimerization interface localized in its kinase domain.
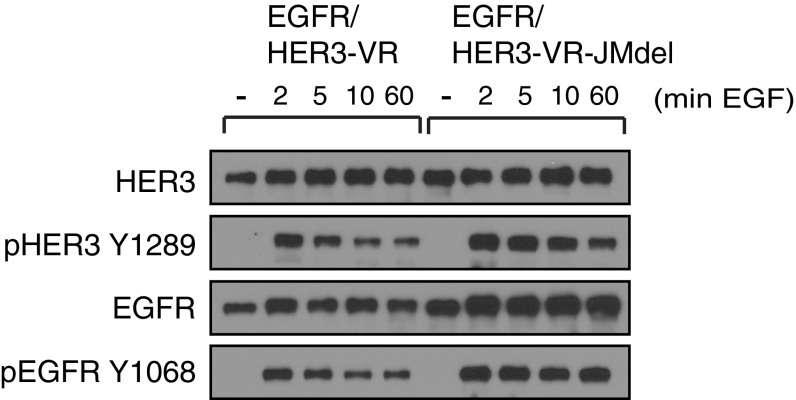
We subsequently tested if another conserved portion of the intracellular region of HER receptors, the juxtamembrane segment, could be responsible for mediating the interaction between HER3 and EGFR in the EGF-induced clusters and enable HER3 receptor tail phosphorylation. In active HER receptor asymmetric dimers, the juxtamembrane segment of the receiver kinase interacts with a binding site on the kinase domain of the activator kinase (41). In this asymmetric arrangement, the receiver kinase potentially could bind the juxtamembrane segment of a third receptor, enabling lateral signaling (Fig. S5). We deleted the juxtamembrane segment in HER3 to see if this deletion would disrupt its phosphorylation by EGFR in response to EGF. However, HER3 missing the juxtamembrane segment was still phosphorylated efficiently by EGFR upon stimulation with EGF (Fig. S5). This finding suggests that HER3 phosphorylation in the EGF-induced clusters follows a different mechanism that could involve a different region of the receptor or might be a result of a high local concentration of the enzyme (EGFR) and substrate (HER3) in the clusters.
Fig. S5.
The juxtamembrane region of HER3 is not a critical interface in lateral signaling. Western blot analysis of NR6 cells stably coexpressing EGFR–GFP and either mEos–HER3–V926R or mEos–HER3–V926R–JMdel, which is a juxtamembrane deletion mutant (residues 669–674, corresponding to the sequence EPLDPSE). Cells were grown for 24–36 h, serum starved for 6 h, then stimulated with 10 nM EGF, and subsequently lysed.
HER2 Clusters Together with HER3 in Response to NRG, but Their Signaling Relies on Asymmetric Kinase Dimerization.
In light of the fundamental differences with which ligands control EGFR and HER3 receptor organization, we examined how HER3 interacts with another dimerization partner, HER2. Like HER3, HER2 is an obligate heterodimerization partner, because it lacks its own ligands. Therefore, HER2 and HER3 can interact only in response to HER3-specific ligands. In such complexes, HER2 is assumed to act as the receiver kinase and to phosphorylate HER3.
We applied STORM to analyze the behavior of mEos–HER3 under conditions of HER2–GFP coexpression in the NR6 cells and stimulation with NRG. In contrast to our results in the analysis of the behavior of the HER3/EGFR complex upon NRG stimulation, membrane organization of HER3 changed significantly in response to NRG when HER2 was coexpressed (Fig. 6A). In the presence of HER2, HER3 displayed increasing levels of clustering upon NRG stimulation, to an extent similar to the HER3 clustering observed in the presence of EGF and EGFR coexpression (Figs. 2 H and I and 6 A and D). To test if HER3 clustering is accompanied by changes in membrane organization of HER2, we measured the behavior of mEos–HER2 in the presence of HER3 expression. mEos–HER2 also clustered significantly in response to NRG, suggesting that HER2 and HER3 interact upon ligand binding, forming higher-order oligomers at the plasma membrane (Fig. 6 B and E).
To investigate the nature of HER3 clusters associated with HER2 coexpression, we then probed whether they form independently of the asymmetric kinase dimerization interface, as we observed with EGF-induced HER3 clusters in cells coexpressing EGFR (Fig. 5B). Introduction of the V926R mutation in HER3 entirely abrogated its clustering upon NRG stimulation when HER2 was coexpressed (Fig. 6 C and F). This result suggests that these clusters are intrinsically different from the clusters that HER3 forms in the presence of EGFR and EGF. Consequently, breaking the allosteric activator interface by the V926R mutation blocked NRG-dependent HER3 phosphorylation by HER2 (Fig. 6G). Although in our assays we observed NRG-independent HER2 phosphorylation for all HER2 stable cell lines we have generated, HER2 did not engage with HER3 in active complexes in the absence of NRG, as evidenced by the lack of detectable HER3 phosphorylation under these conditions (Fig. 6G). Taken together, these results show that HER2/HER3 signaling proceeds through another unique route that involves higher-order interactions at the plasma membrane but is dependent on the formation of the canonical asymmetric kinase dimer interface.
Discussion
Oligomerization of receptor tyrosine kinases, traditionally considered as dimerization, constitutes a basic regulatory principle behind their activation by extracellular ligands (42). Our observations solidify the notion that EGFR signaling is controlled through the formation of oligomeric complexes that extend beyond a dimer. In agreement with previous studies, we observed the formation of receptor clusters that increase in size over time upon EGF stimulation. Our data show that the clusters reach the approximate size of ∼5–20 receptors upon 10 min of EGF stimulation, corroborating a number associated with spontaneously formed activated EGFR clusters on the cell surface of the human A431 carcinoma cells and perhaps suggesting a self-organizing principle within these clusters (28). We also observe that EGFR clustering is dependent on its kinase activity, as previously reported (28, 29), and we show that self-activation through the formation of an asymmetric kinase dimer is key to EGFR oligomerization. However, our data indicate that not all activating stimuli have the same effect on EGFR organization at the membrane, because when EGFR is recruited to a complex with another HER receptor, HER3, and activated via a HER3 cognate ligand, it does not cluster.
In contrast to EGFR, studies on the regulation of HER3 oligomerization in response to ligands have thus far yielded conflicting results. Cell-based approaches using indirect measurements, such as chemical crosslinking, nucleic acid aptamers, and bulk fluorescence complementation, provided evidence that HER3 oligomerizes before NRG binding, but upon binding the ligand it dissociates into monomeric receptors that are poised to heterodimerize with other HER receptors (43–45). More direct approaches, such as immunoelectron microscopy and quantum dot single-particle tracking analyses, have challenged this model and point to significant HER3 homodimerization and even clustering in response to NRG (30, 46). In contrast, no HER3 oligomers have been observed during biochemical studies of the isolated extracellular domain of HER3 (47, 48) or through studies of chimeric full-length receptors (49).
Our findings underscore the importance of considering HER3 behavior in the context of its molecular environment, which we were able to control by using a cell line that does not endogenously express HER receptors. We observed that, when expressed alone, HER3 does not form extensive homo-oligomers that are dissociated or formed upon ligand stimulation. However, when HER3 is coexpressed with EGFR or HER2, it engages in interactions that result in its clustering at the plasma membrane upon ligand binding. Intriguingly, this behavior is greatly influenced by the combination of the receptor type and the type of stimulating ligand. When coexpressed with EGFR, HER3 undergoes significant clustering only in the presence of EGF but not in the presence of NRG. When HER3 is coexpressed with HER2, NRG induces significant clustering of HER3, corroborating previous studies in which measurement of NRG-dependent HER3 clustering was conducted in cells with measurable levels of HER2 expression (30, 46).
Most importantly, our results have important implications for understanding the molecular mechanisms that underlie the cross-communication between HER3 and other HER receptors. In a seemingly reciprocal mode of interaction, such as activation of the HER3/EGFR complex by EGF or NRG, this receptor pair leads to divergent patterns of receptor phosphorylation depending on the ligand (Fig. 1). Our results show that these different patterns reflect changes in receptor organization induced by different ligands. We show that the lack of EGFR phosphorylation in response to NRG does not reflect its lack of engagement with HER3, because HER3 is robustly phosphorylated in an EGFR-dependent manner. Rather, EGFR cannot be phosphorylated because NRG is unable to induce homomeric interactions in EGFR. It is not completely clear why EGFR needs to homo-oligomerize to become phosphorylated efficiently. A preference for the phosphorylation of receptor tails in trans in the EGFR receptor dimer was reported previously (36) and could explain why EGFR phosphorylation is not favorable in an EGFR/HER3 heterodimer in which EGFR would need to phosphorylate its own tail (in cis). Another possibility is that EGFR autophosphorylation is simply not a very efficient process, and higher-order clustering of this receptor induced by its own ligands, such as EGF, increases the rate of autophosphorylation. In an analogous manner, NRG-dependent clustering of HER2 that we observe is likely necessary for its efficient phosphorylation as suggested before by the studies using aptamers that are predicted to selectively block these higher-order interactions (50).
Asymmetric kinase domain dimerization, with HER3 restricted to the allosteric activator position because of impaired catalytic activity, has been a benchmark for understanding how HER receptors activate in heteromeric complexes (3, 6). Here we uncover significant differences in how HER3 forms signaling complexes with EGFR and HER2 in response to different ligands (Fig. 7). In the presence of its own ligand, NRG, HER3 engages with its dimerization partners EGFR and HER2 by taking the function of an allosteric activator kinase, adhering to the asymmetric kinase dimerization mechanism. However, when EGFR homomeric complexes form preferentially in the presence of EGF, HER3 does not form asymmetric kinase dimers with EGFR. Although this phenomenon has been noted before (51), the underlying mechanism was unknown. We show that, upon EGF stimulation, HER3 follows the behavior of its interaction partner, EGFR, and forms clusters. We propose that this behavior facilitates HER3 phosphorylation by promoting activating interactions between EGFR and HER3 in which they engage in the kinase/substrate mode rather than kinase/activator mode. Although at present we do not know the molecular mechanism of HER3 clustering in response to EGF-induced EGFR activation, EGFR clustering was shown to be concurrent with the generation and notable rearrangement of anionic lipids in the plasma membrane (26, 29). EGFR clustering also was shown to be promoted by depletion of cholesterol, suggesting that the membrane environment in which EGFR clusters is unlikely to represent lipid rafts (24). These data indicate that biophysical changes in the membrane may create an environment conducive to the clustering of EGFR interaction partners with which EGFR otherwise would form only weak interactions. In contrast to EGF, NRG fails to induce clustering of HER3 and EGFR, but, intriguingly, it does promote clustering between HER2 and HER3. The importance of the asymmetric kinase dimerization in this process suggests that this clustering is intrinsically distinct from the clustering of HER3 and EGFR that we observe upon EGF treatment.
Fig. 7.
Summary of the underlying mechanistic differences in signaling by HER3-containing heteromeric complexes. (Left) Upon stimulation with EGF, EGFR clusters and self-activates through asymmetric kinase dimerization. Under these conditions, HER3 also clusters and is phosphorylated by EGFR without engaging with it through the asymmetric kinase dimer interface. (Right) NRG does not lead to clustering of either EGFR or HER3 when they are coexpressed and drives EGFR kinase activation through asymmetric kinase heterodimerization with HER3. In these dimers, HER3 plays a role of the allosteric activator. When HER2 and HER3 are coexpressed, they both cluster in response to NRG, and their signaling is dependent on HER3’s function as an allosteric activator of HER2.
In summary, our work brings mechanistic insights into the previously documented differences in signaling outcomes induced by stimulation with EGF and NRG. Our results show that, despite highly conserved structural features of HER receptors, not every ligand induces a receptor heterocomplex that is consistent with the asymmetric kinase dimerization mechanism. In these cases, higher-order oligomerization and cross-activation through lateral interactions in a kinase/substrate-like mode seem to come into play. As a result, there is a qualitative difference in the phosphorylation states of the engaged receptors, providing insight into how cells interpret signals delivered by different HER ligands. It is important to note that relative oligomerization states of HER receptors in response to ligands are most likely influenced by both their stoichiometry and relative expression levels. For example, in the presence of HER3, HER2, and EGFR, NRG might be more likely to favor the formation of the HER2/HER3 dimers rather than EGFR/HER3 dimers, because the former have been shown to form preferentially (52). Perturbing the level of expression of any individual receptors and the relative level of expression among HER family members could provide important information about the hierarchy of receptor interactions.
Our data also emphasize that HER receptor signaling needs to be interpreted in the context of higher-order oligomeric structures, in a manner that could be somewhat parallel to ephrin (Eph) receptors. In Eph receptor signaling, first step of ligand-mediated receptor autophosphorylation usually is not sufficient to generate functional responses, and the Eph receptors must form multimeric complexes to activate downstream signaling robustly (53). Moreover, the size of the Eph clusters has been correlated with the strength and type of the signaling response (53–55). Likewise, our data show that constriction of EGFR receptor within a heterodimer in which its kinase domain is efficiently activated is still not sufficient for its autophosphorylation, and EGFR undergoes robust phosphorylation only when allowed to cluster. It is tempting to speculate that regulation through control of the size of the signaling unit could be also operative for other receptor tyrosine kinases to control their net phosphorylation and functional outcomes of their activation. Finally, our findings have important implications for the treatment of human diseases in which HER3 signaling plays an important role. HER3 contributes to drug resistance to EGFR- or HER2-targeted therapeutics (56) and recently was discovered to carry a spectrum of mutations in human cancers (57). The most recognized function of this catalytically impaired receptor is serving as an allosteric activator of other HER receptors. Our data show that, under some conditions, HER3 can contribute to signaling independently of this function, emphasizing the need for the careful design of therapeutic strategies targeting HER3.
Materials and Methods
Plasmids and Cell Lines.
Murine stem cell virus (MSCV) plasmids and Plat-e cells were a generous gift from the Wells laboratory at the University of California, San Francisco (UCSF), and NR6 cells were a generous gift from the Moasser laboratory at UCSF. mEos3.2 was fused between the human EGFR, HER2, or HER3 signal peptide domains, residues 1–24 (EGFR), 1–22 (HER2), 1–19 (HER3), and the remainder of the sequence using a homemade Gibson assembly reagent into retrovirus-capable vectors (MSCV). Similarly, for mEos-tagged PH constructs, one, two, or three copies of mEos3.2 were fused at the N terminus of the PH domain of phospholipase C delta 1 (PLCΔ); for the CD86 construct, mEos3.2 was fused at the C terminus of the gene. Virus was produced by transient transfection of the plasmid into Plat-e cells using FuGENE (Promega), and virus-containing supernatant was collected after 2 d. Supernatant was filtered (0.22-μm pore diameter) and was added immediately to NR6 cells plated on the previous day; antibiotic (250 µg/mL hygromycin or 2 µg/mL puromycin) was added 24 h after viral infection. Cells were cultured in Gibco DMEM supplemented with 10% FBS, penicillin/streptomycin, sodium pyruvate, and nonessential amino acids. For cells expressing two different HER receptors, one receptor was tagged with monomeric EGFP at the C terminus after the terminal residue of the HER receptor (e.g., EGFR–GFP), and the other receptor was tagged with mEos at the N terminus after the signal peptide (e.g., mEos–HER3). The second construct also was introduced by stable transfection (as above) to a cell line already expressing the mEos-fused receptor, and coexpressing cells were selected by adding both hygromycin (250 µg/mL) and puromycin (2 µg/mL) to the medium.
Sample Preparation.
Eight-well chambered coverglass slides (Lab-Tek) were washed with 1 M KOH for 10 min and then were washed and coated with polylysine (0.01%) for 30 min. Stable cell lines expressing the proteins of interest were deposited on the washed glass slide and allowed to adhere for 36 h. Cells were serum starved for 6 h and then were stimulated for specified amounts of time with 10 nM EGF or NRG (saturating concentration) at 37 °C. Cells were fixed with 4% formaldehyde for 10 min at 20 °C, washed, and kept in PBS at 4 °C for up to 1 wk in the dark before imaging.
Microscopy.
Fixed cells were imaged using a home-built STORM inverted microscope consisting of 405-nm, 488-nm, 561-nm, and 647-nm lasers. The lasers were coaligned and reflected by a quad-band dichroic mirror through a 10× Olympus objective (NA 1.4), and fluorescence was collected from the same objective with an Andor EMCCD camera. Images were collected and processed with a home-written software program. Maximum laser power used during STORM measured before the objective was 0.03 mW for 405 nm and 65 mW for 561 nm. Cells expressing only one receptor fused to mEos were carefully located at low green intensity using a wide-field setting (20 Hz) to minimize bleaching of mEos. The cells then were illuminated with a high intensity (65 mW) 561-nm laser at 60 Hz, with an activation frame (405 nm) every 10 frames. By increasing the power on the 405-nm laser (zero to 0.03 mW) during imaging, mEos was photoconverted from a green to a red state and could be observed as single fluorophores. The activation frames were programmed to occur simultaneously with a brightfield image, which was used in postprocessing to correct for drift. Approximately 10,000–60,000 images were collected, until all mEos fluorophores were photobleached, while the 405-nm power was kept low enough to activate mEos only sparsely (about 5–30 fluorophores per frame) to avoid improper counting in the analysis. For cells expressing two receptors (e.g., mEos–HER3/EGFR–GFP), the GFP was much brighter than mEos, and cells could be located easily with low intensity in the green channel. Cells then were subjected to the STORM imaging, and if fluorophores appeared in the red channel, it was clear that cells coexpressed both receptors. In some cases, cells were located that did not contain mEos but did contain GFP; these cells were disregarded. Generally, cells were imaged only if they were within the range of expression that was appropriate for the analysis, 20–60 molecules/µm2, which is the value of the blink-corrected pair-correlation function at long distances (>1 µm). At lower expression levels the signal to background was insufficient, and at higher expression levels the movies became exceedingly long (more than 20 min), at which point autofluorescence caused by the 405-nm laser illumination could become problematic. The details of STORM image analysis is described in SI Materials and Methods.
Signaling Assays.
NR6 cells stably expressing HER receptors were plated at a density of 120,000 cells per 10-cm round plate and were allowed to adhere for 36 h. Cells were serum starved and stimulated with 10 nM (or as specified for the titration) ligand for specified amounts of time at 37 °C. Cells then were set on ice and lysed and collected in Tris (50 mM)/NaCl (150 mM) buffer containing 1% Triton X-100, 1 mM Na3VO4, 1 mM EDTA, 1 mM NaF, and one-fourth of a cOmplete Mini, EDTA-free protease inhibitor tablet (Roche) per 10 mL buffer. The lysates were spun down to remove larger organelles, and the supernatant was assayed for protein concentration using a BCA kit (Thermo Fisher). Samples were run on SDS/PAGE, transferred to a PVDF membrane, and blotted for with antibodies (Table S1 for a list of antibodies). After secondary incubation with HRP-linked antibody, ECL prime Western blotting detection agent (GE Healthcare) was added to the membrane, and membranes were imaged on a developer.
Table S1.
Antibodies used in this study
| Receptor | Phosphorylation site | Source |
| EGFR | Santa Cruz Biotechnology sc-03 | |
| pEGFR | Y1068 | Cell Signaling 2234 |
| Y1045 | Cell Signaling 2237 | |
| Y1173 | Cell Signaling 4407 | |
| HER3 | Cell Signaling XP D22C5 | |
| pHER3 | Y1289 | Cell Signaling 4791 |
| Y1197 | Cell Signaling 4561 | |
| Y1221/1222 | Cell Signaling 4784 | |
| HER2 | Santa Cruz Biotechnology sc-284 | |
| pHER2 | Y1221/1222 | Cell Signaling 2249 |
| AKT | Cell Signaling 2920 | |
| pAKT | S473 | Cell Signaling 4060 |
| ERK1/2 (MAPK) | p44/42 | Cell Signaling 4696 |
| pERK | T202/Y204 | Cell Signaling 4370 |
SI Materials and Methods
Analysis of STORM images was performed as follows. Localizations from the single molecules were processed using a home-written program. The point-spread function originating from the molecule was fit to an elliptical Gaussian and was accepted only within an appropriate range of size and shape to prevent the inclusion of dust or mobile or irregular molecules. The set of all localizations then was corrected for blinking as in ref. 33. A temporal threshold for blinking was set as the 99th percentile of the dark state lifetime distribution (Fig. S2C), in other words, the time below which 99% of the dark state times are included. A spatial threshold for blinking was determined by examining the pair-correlation function before blink correction of the PH–mEos3.2(1×) construct expressed in NR6 cells (Fig. S2B) and observing the minimum distance at which the correlation curve becomes flat. Physically, this distance represents the maximal radius at which blinking from the same molecule could occur, which is limited by the cumulative spatial resolution over the course of the blinking occurrences from any one molecule. Thresholds of 200 nm for the radius and 400 frames (6.67 s) were set, and any localizations occurring with these thresholds were combined and marked as a single molecule. These blink-corrected molecular positions then were used to calculate a corrected pair-correlation histogram, which calculates all the pairwise distances between the molecules per area. This histogram is further corrected for average density by subtracting the average baseline value (between 500–1,000 nm) from the function. Any differences in the peak height and width among different conditions then can be compared directly, even though different samples may contain slightly different densities of molecules. Comparing the peak size for a range of regions of interest of the same sample demonstrated that there was no strong correlation of peak size based solely on the density of receptors within the range chosen.
Acknowledgments
We thank Z. Gartner, J. Kung, N. Michael, and T. M. Thaker for critical reading of the manuscript and insightful discussions, S. Liang for many valuable insights; and R. McGorty and V. Pessino from the Huang laboratory for valuable assistance with STORM imaging and helpful discussions. This work was supported by American Cancer Society Postdoctoral Fellowship 124801-PF-13-365-01-TBE (to B.v.L.), National Institute of General Medical Sciences Grant R01 GM109176 (to N.J.), and National Institutes of Health New Innovator Award (DP2 OD008479 (to B.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617994114/-/DCSupplemental.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA, Schlessinger J, Ferguson KM. The EGFR family: Not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2014;6(4):a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Qiu C, et al. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16(3):460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Littlefield P, et al. Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations. Sci Signal. 2014;7(354):ra114. doi: 10.1126/scisignal.2005786. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 6.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci USA. 2009;106(51):21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monsey J, Shen W, Schlesinger P, Bose R. Her4 and Her2/neu tyrosine kinase domains dimerize and activate in a reconstituted in vitro system. J Biol Chem. 2010;285(10):7035–7044. doi: 10.1074/jbc.M109.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olayioye MA, et al. ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol Cell Biol. 1998;18(9):5042–5051. doi: 10.1128/mcb.18.9.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riese DJ, 2nd, van Raaij TM, Plowman GD, Andrews GC, Stern DF. The cellular response to neuregulins is governed by complex interactions of the erbB receptor family. Mol Cell Biol. 1995;15(10):5770–5776. doi: 10.1128/mcb.15.10.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinkas-Kramarski R, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15(10):2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 11.Ueno Y, et al. Heregulin-induced activation of ErbB3 by EGFR tyrosine kinase activity promotes tumor growth and metastasis in melanoma cells. Int J Cancer. 2008;123(2):340–347. doi: 10.1002/ijc.23465. [DOI] [PubMed] [Google Scholar]
- 12.Carver RS, Stevenson MC, Scheving LA, Russell WE. Diverse expression of ErbB receptor proteins during rat liver development and regeneration. Gastroenterology. 2002;123(6):2017–2027. doi: 10.1053/gast.2002.37060. [DOI] [PubMed] [Google Scholar]
- 13.Choi SH, Mendrola JM, Lemmon MA. EGF-independent activation of cell-surface EGF receptors harboring mutations found in gefitinib-sensitive lung cancer. Oncogene. 2007;26(11):1567–1576. doi: 10.1038/sj.onc.1209957. [DOI] [PubMed] [Google Scholar]
- 14.Crovello CS, Lai C, Cantley LC, Carraway KL., 3rd Differential signaling by the epidermal growth factor-like growth factors neuregulin-1 and neuregulin-2. J Biol Chem. 1998;273(41):26954–26961. doi: 10.1074/jbc.273.41.26954. [DOI] [PubMed] [Google Scholar]
- 15.Yarden Y, Schlessinger J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 1987;26(5):1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- 16.Yarden Y, Schlessinger J. Self-phosphorylation of epidermal growth factor receptor: Evidence for a model of intermolecular allosteric activation. Biochemistry. 1987;26(5):1434–1442. doi: 10.1021/bi00379a034. [DOI] [PubMed] [Google Scholar]
- 17.Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2(3):168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 18.Whitson KB, Beechem JM, Beth AH, Staros JV. Preparation and characterization of Alexa Fluor 594-labeled epidermal growth factor for fluorescence resonance energy transfer studies: Application to the epidermal growth factor receptor. Anal Biochem. 2004;324(2):227–236. doi: 10.1016/j.ab.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 19.Gadella TW, Jr, Jovin TM. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J Cell Biol. 1995;129(6):1543–1558. doi: 10.1083/jcb.129.6.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichinose J, Murata M, Yanagida T, Sako Y. EGF signalling amplification induced by dynamic clustering of EGFR. Biochem Biophys Res Commun. 2004;324(3):1143–1149. doi: 10.1016/j.bbrc.2004.09.173. [DOI] [PubMed] [Google Scholar]
- 21.Needham SR, et al. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat Commun. 2016;7:13307. doi: 10.1038/ncomms13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clayton AH, et al. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem. 2005;280(34):30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- 23.Kozer N, et al. Exploring higher-order EGFR oligomerisation and phosphorylation--a combined experimental and theoretical approach. Mol Biosyst. 2013;9(7):1849–1863. doi: 10.1039/c3mb70073a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saffarian S, Li Y, Elson EL, Pike LJ. Oligomerization of the EGF receptor investigated by live cell fluorescence intensity distribution analysis. Biophys J. 2007;93(3):1021–1031. doi: 10.1529/biophysj.107.105494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, et al. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife. 2016;5:5. doi: 10.7554/eLife.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Res. 2014;24(8):959–976. doi: 10.1038/cr.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clayton AH, Orchard SG, Nice EC, Posner RG, Burgess AW. Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors. 2008;26(6):316–324. doi: 10.1080/08977190802442187. [DOI] [PubMed] [Google Scholar]
- 28.Clayton AH, Tavarnesi ML, Johns TG. Unligated epidermal growth factor receptor forms higher order oligomers within microclusters on A431 cells that are sensitive to tyrosine kinase inhibitor binding. Biochemistry. 2007;46(15):4589–4597. doi: 10.1021/bi700002b. [DOI] [PubMed] [Google Scholar]
- 29.Ariotti N, et al. Epidermal growth factor receptor activation remodels the plasma membrane lipid environment to induce nanocluster formation. Mol Cell Biol. 2010;30(15):3795–3804. doi: 10.1128/MCB.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S, et al. Mapping ErbB receptors on breast cancer cell membranes during signal transduction. J Cell Sci. 2007;120(Pt 16):2763–2773. doi: 10.1242/jcs.007658. [DOI] [PubMed] [Google Scholar]
- 31.Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc Natl Acad Sci USA. 2010;107(38):16524–16529. doi: 10.1073/pnas.1002642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung I, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464(7289):783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 33.Low-Nam ST, et al. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat Struct Mol Biol. 2011;18(11):1244–1249. doi: 10.1038/nsmb.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puchner EM, Walter JM, Kasper R, Huang B, Lim WA. Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. Proc Natl Acad Sci USA. 2013;110(40):16015–16020. doi: 10.1073/pnas.1309676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawamoto T, et al. Quantitative assay of epidermal growth factor receptor in human squamous cell carcinomas of the oral region by an avidin-biotin method. Jpn J Cancer Res. 1991;82(4):403–410. doi: 10.1111/j.1349-7006.1991.tb01863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacs E, et al. Analysis of the role of the C-terminal tail in the regulation of the epidermal growth factor receptor. Mol Cell Biol. 2015;35(17):3083–3102. doi: 10.1128/MCB.00248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fricke F, Beaudouin J, Eils R, Heilemann M. One, two or three? Probing the stoichiometry of membrane proteins by single-molecule localization microscopy. Sci Rep. 2015;5:14072. doi: 10.1038/srep14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abulrob A, et al. Nanoscale imaging of epidermal growth factor receptor clustering: Effects of inhibitors. J Biol Chem. 2010;285(5):3145–3156. doi: 10.1074/jbc.M109.073338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11(3):217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gan HK, et al. The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor AG1478 increases the formation of inactive untethered EGFR dimers. Implications for combination therapy with monoclonal antibody 806. J Biol Chem. 2007;282(5):2840–2850. doi: 10.1074/jbc.M605136200. [DOI] [PubMed] [Google Scholar]
- 41.Jura N, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137(7):1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macdonald-Obermann JL, Adak S, Landgraf R, Piwnica-Worms D, Pike LJ. Dynamic analysis of the epidermal growth factor (EGF) receptor-ErbB2-ErbB3 protein network by luciferase fragment complementation imaging. J Biol Chem. 2013;288(42):30773–30784. doi: 10.1074/jbc.M113.489534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park E, Baron R, Landgraf R. Higher-order association states of cellular ERBB3 probed with photo-cross-linkable aptamers. Biochemistry. 2008;47(46):11992–12005. doi: 10.1021/bi8004208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kani K, Warren CM, Kaddis CS, Loo JA, Landgraf R. Oligomers of ERBB3 have two distinct interfaces that differ in their sensitivity to disruption by heregulin. J Biol Chem. 2005;280(9):8238–8247. doi: 10.1074/jbc.M410944200. [DOI] [PubMed] [Google Scholar]
- 46.Steinkamp MP, et al. erbB3 is an active tyrosine kinase capable of homo- and heterointeractions. Mol Cell Biol. 2014;34(6):965–977. doi: 10.1128/MCB.01605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297(5585):1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 48.Ferguson KM, Darling PJ, Mohan MJ, Macatee TL, Lemmon MA. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. EMBO J. 2000;19(17):4632–4643. doi: 10.1093/emboj/19.17.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger MB, Mendrola JM, Lemmon MA. ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett. 2004;569(1-3):332–336. doi: 10.1016/j.febslet.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q, Park E, Kani K, Landgraf R. Functional isolation of activated and unilaterally phosphorylated heterodimers of ERBB2 and ERBB3 as scaffolds in ligand-dependent signaling. Proc Natl Acad Sci USA. 2012;109(33):13237–13242. doi: 10.1073/pnas.1200105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kancha RK, von Bubnoff N, Duyster J. Asymmetric kinase dimer formation is crucial for the activation of oncogenic EGFRvIII but not for ERBB3 phosphorylation. Cell Commun Signal. 2013;11:39. doi: 10.1186/1478-811X-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzahar E, et al. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16(10):5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis S, et al. Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science. 1994;266(5186):816–819. doi: 10.1126/science.7973638. [DOI] [PubMed] [Google Scholar]
- 54.Egea J, et al. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron. 2005;47(4):515–528. doi: 10.1016/j.neuron.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 55.Salaita K, et al. Restriction of receptor movement alters cellular response: Physical force sensing by EphA2. Science. 2010;327(5971):1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sergina NV, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445(7126):437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaiswal BS, et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell. 2013;23(5):603–617. doi: 10.1016/j.ccr.2013.04.012. [DOI] [PubMed] [Google Scholar]