Abstract
Menin (MEN1) is a tumor-suppressor protein in neuroendocrine tissue. Therefore, we tested the novel hypothesis that menin regulates cholangiocarcinoma proliferation. Menin and miR-24 expression levels were measured in the following intrahepatic and extrahepatic cholangiocarcinoma (CCA) cell lines, Mz-ChA-1, TFK-1, SG231, CCLP, HuCCT-1, and HuH-28, as well as the nonmalignant human intrahepatic biliary line, H69. miR-24 miRNA and menin protein levels were manipulated in vitro in Mz-ChA-1 cell lines. Markers of proliferation and angiogenesis (Ki-67, vascular endothelial growth factors A/C, vascular endothelial growth factor receptors 2/3, angiopoietin 1/2, and angiopoietin receptors 1/2) were evaluated. Mz-ChA-1 cells were injected into the flanks of nude mice and treated with miR-24 inhibitor or inhibitor scramble. Menin expression was decreased in advanced CCA specimens, whereas miR-24 expression was increased in CCA. Menin overexpression decreased proliferation, angiogenesis, migration, and invasion. Inhibition of miR-24 increased menin protein expression while decreasing proliferation, angiogenesis, migration, and invasion. miR-24 was shown to negatively regulate menin expression by luciferase assay. Tumor burden and expression of proliferative and angiogenic markers was decreased in the miR-24 inhibited tumor group compared to controls. Interestingly, treated tumors were more fibrotic than the control group. miR-24–dependent expression of menin may be important in the regulation of nonmalignant and CCA proliferation and may be an additional therapeutic tool for managing CCA progression.
Cholangiocarcinoma (CCA) is a biliary epithelial adenocarcinoma associated with late diagnosis, poor long-term survival, and limited responsiveness to current therapies.1 The topographical range of biliary histology contributes to the heterogeneous presentation of CCA, although its classification as intrahepatic or extrahepatic remains to be anatomically based.2 Cholangiocytes proliferate in response to damage and various endothelial stressors. During cell proliferation, cholangiocytes adopt a neuroendocrine-like phenotype via autocrine/paracrine signaling by cytokines (IL-6), vascular endothelial growth factors (VEGFs), and neuropeptides.3, 4 Surgery is potentially curative for early disease, but few patients are surgical candidates and 5-year survival rates remain low.1, 2 Clearly, there is a need for advanced diagnostic strategies and improved targeted therapies for CCA.
Menin, encoded by the MEN1 (multiple endocrine neoplasia type I) tumor-suppressor gene, is a 610–amino acid, 67-kDa nuclear protein that is ubiquitously expressed in all tissues and evolutionarily conserved, but shares little sequence homology with other proteins.5 Although menin's function has not been comprehensively elucidated, several studies have suggested that it is a scaffold protein involved in diverse cell functions, including binding and regulating transcription factor activity,6 modifying histone proteins and chromatin structure,7, 8 and DNA repair.9, 10 Loss of heterozygosity at the MEN1 gene locus (11q13) inactivates or deletes menin, leading to tumorigenesis. Patients with MEN1 syndrome develop parathyroid neoplasms (95%), gastro-entero-pancreatic tract neuroendocrine tumors (40%), and pituitary adenomas (30%), as well as tumors in nonendocrine tissues, such as lipomas and cutaneous angiofibromas.11
Since menin's discovery as a tumor-suppressor protein in MEN1 syndrome, it has been shown to regulate cell proliferation in the lung, stomach, liver, breast, and prostate tissue12, 13, 14, 15, 16; however, menin's role in liver carcinogenesis has not been widely studied. One study has shown that menin expression is down-regulated in hepatocellular carcinoma (HCC), and that overexpressing menin in vitro decreased cell proliferation and gene expression of inflammatory cytokines.14 However, another study has shown that menin is up-regulated in HCC samples from patients with underlying cirrhosis and promotes HCC formation via interaction with mixed-lineage leukemia (MLL) histone methyltransferase complex and overexpression of homeobox A genes.17 Although menin appears to play a role in HCC formation, its role in CCA development and progression has not been studied.
Recent evidence demonstrates that menin and miR-24 form a negative regulatory feedback network to tightly control cell cycle and apoptotic genes.18, 19 miR-24 targets menin's 3′-untranslated region, resulting in decreased menin protein expression. Conversely, Vijayaraghavan et al18 showed that the menin-MLL protein complex is present upstream of miR-24 in both of its chromosome locations and that overexpression of menin increases miR-24 expression. miR-24 has been implicated as an oncogene in a host of other cancers, particularly in the gastrointestinal tract.20, 21, 22, 23
We propose that menin and miR-24 contribute to a regulatory negative-feedback loop that maintains cholangiocyte proliferation and that dysregulation leads to malignant growth. We have demonstrated the novel finding that menin is down-regulated in human CCA cell lines and advanced-stage human CCA samples and modulation of menin expression alters cholangiocarcinoma proliferation. We have also shown that miR-24 expression is up-regulated in human CCA cell lines and human CCA samples and modulation of miR-24 expression alters cholangiocarcinoma proliferation through changes in menin expression.
Materials and Methods
Materials
Reagents were purchased from Sigma (St. Louis, MO), unless otherwise indicated. Human Men1 siRNA and control vectors were purchased from Santa Cruz Biotechnology (Dallas, TX) and transfected with Human MEN1 siRNA (sc-35922) (Santa Cruz Biotechnology). Human pCMV6-Entry MEN1 cDNA and control vector were purchased from Origene (Rockville, MD) and transfected with Lipofectamine 2000 Transfection reagent (ThermoFisher Scientific, Grand Island, NY). miRIDIAN microRNA Human hsa-miR-24-1 5p mimic (C-300495-07-0005), hairpin inhibitor (IH-300495-08-0005), and negative controls were purchased from GE Dharmacon (Lafayette, CO) and were transfected with Lipofectamine RNAiMAX Transfection reagent (ThermoFisher Scientific). The RNeasy Mini Kit for RNA purification and all of the following primers were purchased from Qiagen (Valencia, CA): Menin (MEN1, NM_000244), glyceraldehyde-3-phosphate dehydrogenase (NM_002046), MKI67 (NM_001145966), VEGF-A (NM_001025366), VEGF-C (NM_005429), VEGF receptor (VEGFR)-2 (NM_VEGFR-2), VEGFR-3 (NM_002020), angiopoietin 1 (ANG-1, NM_001146), angiopoietin 2 (ANG-2, NM_001118887), angiopoietin receptor 1 (TIE-1, NM_001253357), and angiopoietin receptor 2 (TIE-2, NM_000459).
Antibodies used were as follows: hsa-miR-24-1 (002440 TaqMan; ThermoFisher Scientific), U6 snRNA (004394 TaqMan), Menin (A300-105A; Bethyl Laboratories, Montgomery, TX), Ck-19 (Ab15463; Abcam, Cambridge, MA), Ki-67 (Ab15580; Abcam), p-extracellular signal regulated kinase (ab50011; Abcam), and CD31 (89C2; Cell Signaling Technology, Danvers, MA).
In Vitro Studies
Cell Lines
We studied six human CCA cell lines: Mz-ChA-1, TFK-1, SG231, CCLP-1, HuCC-T1, and HuH-28. The human intrahepatic CCA cell lines CCLP-1,24 HuCC-T1,25 and SG23126 were a gift of Dr. Anthony J. Demetris (University of Pittsburgh, Pittsburgh, PA). The human extrahepatic CCA line, Mz-ChA-1,27 was a gift from Dr. Greg Fitz (UT Southwestern Medical Center, Dallas, TX). The human intrahepatic biliary cell line, HuH-28,28 and the human extrahepatic biliary TFK-1 cells29 were obtained from the Cancer Cell Repository (Tohoku University, Sendai, Japan); the cell lines were maintained as described.30 The human immortalized, nonmalignant, cholangiocyte cell line, H69, was obtained from Dr. Gregory J. Gores (Mayo Clinic, Rochester, MN).31
Expression of Menin in Nonmalignant and CCA Cells and cDNAs from Control and CCA Patients
Real-time PCR analysis32 of H69 and all CCA cell lines was evaluated using 1 μg of RNA. Glyceraldehyde-3-phosphate dehydrogenase was used as a control primer along with the SYBR Green real-time PCR kit (SABiosciences, Frederick, MD). A ΔΔCT analysis was performed using H69 as the control sample.33 Data are expressed as relative mRNA levels ± SEM (n = 3).
We evaluated the expression of MEN1 mRNA in cDNAs obtained from three control patients and three CCA patients (Table 1); these samples were obtained from Dr. Pietro Invernizzi (Humanitas Research Hospital, Rozzano, Italy) under a protocol number approved by the Ethics Committee of the Humanitas Research Hospital; the protocol was also reviewed by the Veterans' Administration Institutional Review Board (Temple, TX) and Research & Development Committee. The use of human tissue was also approved by the Texas A&M Health Science Center College of Medicine Institutional Review Board (College Station, TX). Real-time PCR was performed in triplicate.
Table 1.
Human CCA Biopsy Samples
| Human CCA biopsy sample∗ | Grade | Age, years | Diagnosis |
|---|---|---|---|
| 2 | 1 | 73 | Intrahepatic cholangiocarcinoma. Degree: G1. Histopathological staging: pT1. |
| 8 | 2 | 66 | Intrahepatic cholangiocarcinoma. Grade: G2. Growth pattern: solid. Histopathological staging: pT2a. |
| 6 | 3 | 67 | Cancer of the bile ducts: type, histological tubule-papillary; degree, histological G3. Histopathological staging TNM v7: pT1 N1. |
CCA, cholangiocarcinoma; TNM v7, Tumor Node Metastasis, 7th edition.
Sex was not available.
For immunoblotting analysis,34 protein was obtained and quantified from whole cell lysates from nonmalignant and CCA cell lines, and 40 μg was loaded into each well. Blots were blocked overnight and then stained with menin antibody at a 1:2000 dilution and β-actin antibody at a 1:5000 dilution. Data are expressed as fold change (means ± SEM, n = 3) of the relative expression after normalization with β-actin (housekeeping).
Measurement of menin expression was also performed using flow cytometry, as described.34 Briefly, H69 and Mz-ChA-1 cells (the CCA lines implanted into athymic mice) were harvested from culture plates using TrypLE solution (ThermoFisher Scientific), and washed with appropriate dilution of fixation buffer (eBioscience, San Diego, CA). Cells were resuspended at a minimum of 5 × 105 cells/mL in 1× permeabilization buffer (eBioscience, Inc., San Diego, CA) and incubated for 15 minutes at room temperature with menin antibody at a dilution of 1:100. Then, Alexa Fluor 488 conjugated secondary antibody was added to suspension at a dilution of 1:50 and cells were incubated for 15 minutes at room temperature in the dark. Cells incubated without antibody or with only Alexa Fluor 488 conjugated secondary antibody were used as negative controls. Cells were analyzed using FACSCalibur (Becton Dickinson, San Jose, CA), with CellQuest Pro software version 5.2 (BD Bioscience, San Jose, CA). At least 10,000 events in the light scatter (side scatter/forward scatter) were acquired. The expression of menin was identified and gated on FL1-A/Count plots. The relative quantity of the selected proteins (mean selected protein fluorescence intensity) is expressed as mean FL1-A (samples)/mean FL1-A (secondary antibodies only) (n = 3).
Modulation of Menin Expression, Proliferation, and Angiogenesis in Vitro
Mz-ChA-1 cells were targeted for the transient knockdown of menin expression using a human MEN1 siRNA (Santa Cruz Biotechnology) (sc-35922) along with siRNA Transfection Reagent (Santa Cruz Biotechnology) (sc-29528), according to the vendor's instructions. Mz-ChA-1 cells were targeted for menin overexpression using the pCMV6-Entry expression vector containing MEN1 cDNA was purchased from Origene32 and was transfected with the Lipofectamine 2000 Transfection reagent (ThermoFisher Scientific), according to protocol. Transfected cell lines were allowed to grow for 24 to 48 hours before harvesting.
The menin-containing plasmid was amplified using ElectroMAX DH5α Competent Cells (Qiagen) and Invitrogen imMedia agar and liquid media and isolated using Qiagen mini kit, according to vendors' instructions. Plasmid yields were quantified using ND-1000 NanoDrop (ThermoFisher Scientific). The pCMV6-Entry vector without the Men1 insert was used as a control.
Transfection efficacy was assessed by measuring menin expression by real-time quantitative PCR and flow cytometry.33, 35 Cell proliferation was measured by Ki-67 real-time PCR expression, migration via wound healing,36 invasion via Boyden chamber, and angiogenesis via VEGF-A, VEGF-C, VEGFR-2, VEGFR-3, ANG-1, ANG-2, TIE-1, and TIE-2 real-time PCR expression.
Wound Healing Assay
Mz-ChA-1 cells were plated onto 6-well plates, as described above, and transfected 24 hours later. Twenty-four hours after transfection, cells were tryspinized and reseeded onto a new 6-well plate with twice the number of cells. Twenty-four hours later, a wound was initiated in the cell monolayer using a small pipette tip and a 19-inch gauge needle.36 Phase contrast images were taken at 0, 3, 6, 24, and 48 hours with a Nikon Eclipse TS100 microscope, and NIS-elements D version 4.00.07 software was used to measure the width of the wound (Nikon, Melville, NY). Data are expressed as fold-change of relative widths compared with 0-hour measurements of their respective transfected cell type ± SEM (n = 6). Statistical significance was evaluated between control and transfected cell types at respective time points.
Boyden Chamber Migration Assay
Migration of Mz-ChA-1 cells was evaluated using QCM ECMatrix Cell Invasion Assay (Millipore, Billerica, MA), according to manufacturer's protocol. Fluorescence was read at 480/520 nm using a VersaMax microplate reader (Molecular Devices, Sunnyvale, CA). Data are expressed as fold-change of transfected cell lines relative to control cell lines ± SEM (n = 7).
Proliferation Assay
Cell proliferation was measured by the CellTiter 96 Aqueous nonradioactive cell proliferation assay (Promega, Madison, WI).32 Absorbance was measured at 490 nm on a microplate spectrophotometer (VersaMax; Molecular Devices). Data were expressed as the degree of change of treated cells compared with vehicle-treated controls (n = 7). A proliferation assay was used to assess the effects of altered MEN1 and miR-24 expression on CCA cell growth.
Expression of miR-24 in Nonmalignant and CCA Cells and miRNA-Sequencing Data from Control and CCA Patients
miRNA was isolated from samples using the Ambion mirVana miRNA isolation kit (ThermoFisher Scientific), per protocol.34 cDNA libraries were generated using TaqMan microRNA RT kit, per protocol (ThermoFisher Scientific) and TaqMan MicroRNA Assay hsa-miR-24 primers (ThermoFisher Scientific). TaqMan Universal PCR Master mix was used for real-time PCR, and U6 snRNAs were used for endogenous control (ThermoFisher Scientific). A ΔΔCT analysis was performed using normal H69 as controls. Data are expressed as fold-change of relative microRNA levels ± SEM (n = 3).
miRNA-sequencing data were obtained from nine human CCA patients from the The Cancer Genome Atlas Research Network (http://cancergenome.nih.gov, last accessed April 7, 2016) via the Genomic Data Commons Data Portal The Cancer Genome Atlas–CHOL. Briefly, the manifest files as well as the miRNA expression quantification files were downloaded for all 36 CCA patient samples contained in the The Cancer Genome Atlas database. File manipulation was performed with Python, Python Software Foundation, Python Language Reference version 2.7. (http://www.python.org, last accessed April 7, 2016). Example coding for this process can be found at https://github.com/ehrl1ch/RNA-Seq (last accessed April 7, 2016). miR-24 expression is displayed as normalized miRNA transcript count for each tumor sample compared with its matched human control. A t-test was performed to assess statistical significance.
Luciferase Construct
Experiments were performed to test whether miR-24 directly interacts with menin to change its expression. Luciferase constructs were obtained from Dr. Judy S. Crabtree (Louisiana State University, Baton Rouge, LA). These constructs consisted of a 1600-bp fragment of human MEN1 3′-untranslated region cloned into a pmirGLO vector (pmirGLO-MEN1). The plasmid was amplified and isolated according to the protocol described above. A total of 5 × 105 Mz-ChA-1 cells per assay were cotransfected using techniques described above with pmirGLO-MEN1 at 0.5 μg/well along with mir-24-1 mimic or hairpin inhibitor in addition to negative control at the concentration of 5 pmol/0.5 mL medium. After 24 hours, luciferase levels were measured using Dual-Glo Stop & Glo, per vendor's instructions (Promega), imaged using ThermoScientific Varioskan Lux and analyzed using ThermoScientific Skanit software version 4.1 (ThermoFisher Scientific). Data are expressed as fold-change of firefly/renilla luminescence ± SEM (n = 3).
Modulation of miR-24 in Vitro
We next performed experiments to determine whether miR-24 changes in Mz-ChA-1 cells lead to changes in menin expression and subsequently CCA proliferation. Mz-ChA-1 cells were targeted for miR24 knockdown using miR-24 hairpin inhibitor (GE Dharmacon; IH-300495-08-0005) using the Lipofectamine RNAiMAX Transfection reagent (ThermoFisher Scientific), according to the vendor's instructions. After 24 to 48 hours, cells were harvested and transfection efficiency was assessed by measuring miR-24 TaqMan miRNA expression. Menin protein expression was assessed using fluorescence-activated cell sorting. Cells were also assessed for proliferation via CellTiter assay, migration via wound healing,36 invasion via Boyden chamber, and expression of Ki-67 and angiogenic factors by real-time PCR.
In Vivo Studies
Male BALB/c 8-week-old nude (nu/nu) mice were kept in a temperature- and light-controlled environment with free access to drinking water and rodent chow.37 Mz-ChA-1 cells were suspended in extracellular matrix gel and s.c. injected into the left rear flanks of these nude mice. The experimental group received miR-24 hairpin inhibitor (GE Dharmacon; IH-300495-08-0005) injected directly into tumor at 4 ng/mm3 three times per week, whereas the control group received a hairpin inhibitor scramble. Tumor growth was measured three times a week using an electronic caliper, and volume was determined as follows: tumor volume (mm3) = π/6 × length (mm) × width (mm) × height (mm). Tumors were allowed to grow until the maximum allowable tumor burden was reached, as set forth by the Baylor Scott & White Healthcare Institutional Animal Care and Use Committee tumor burden policy (http://researchers.sw.org/imedris/imedris, last accessed April 7, 2016). After 8 weeks, mice were euthanized with sodium pentobarbital (50 mg/kg body weight i.p.).
We evaluated biliary proliferation in CCA tumor sections via immunohistochemistry for CK-19, a cholangiocyte-specific marker, Ki-67, and Sirius Red, a collagen-specific dye. In CCA tumor tissue, we measured the expression of menin, biliary proliferation (by real-time PCR for Ki-67), and the expression of select angiogenic factors (CD31) as well as p-extracellular signal regulated kinase.
Statistical Analysis
All data are expressed as means ± SEM. Differences between groups were analyzed by unpaired t-test when two groups were analyzed and analysis of variance when more than two groups were analyzed, followed by an appropriate post hoc test.
Results
The Expression of Menin Is Decreased in CCA
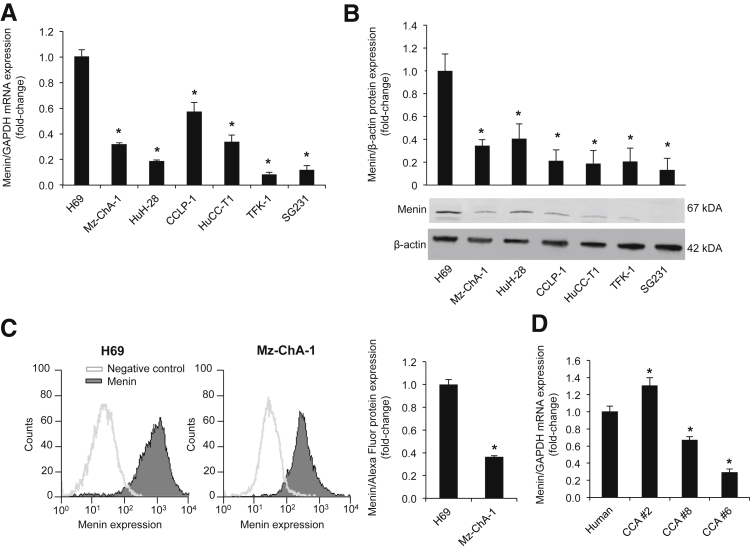
By real-time PCR and immunoblots, we demonstrated that MEN1 mRNA and protein expression was significantly decreased in all selected CCA lines compared to H69 cells (Figure 1, A and B). Similarly, by flow cytometry, we demonstrated that menin expression was decreased in Mz-ChA-1 cells compared to H69 cells (Figure 1C). By real-time PCR, there was reduced mRNA expression of menin in samples from human biopsy specimens from patients with advanced grade CCA compared to control nonmalignant samples (Figure 1D).
Figure 1.
Menin is down-regulated in CCA. A and B: By real-time PCR and immunoblots, menin expression is decreased in CCA cell lines compared to H69. Significance is shown versus H69 cells. C: Flow cytometry analysis demonstrated a decrease in menin protein expression in Mz-ChA-1 cells compared to H69 cells. D: By real-time PCR, menin expression decreased in advanced-stage human CCA tissue biopsy specimens compared with normal control. Data are expressed as means ± SEM performed in triplicate (A–D). n = 3 independent samples (A and B). ∗P < 0.05 versus H69 or human control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Menin Regulates CCA Cell Growth
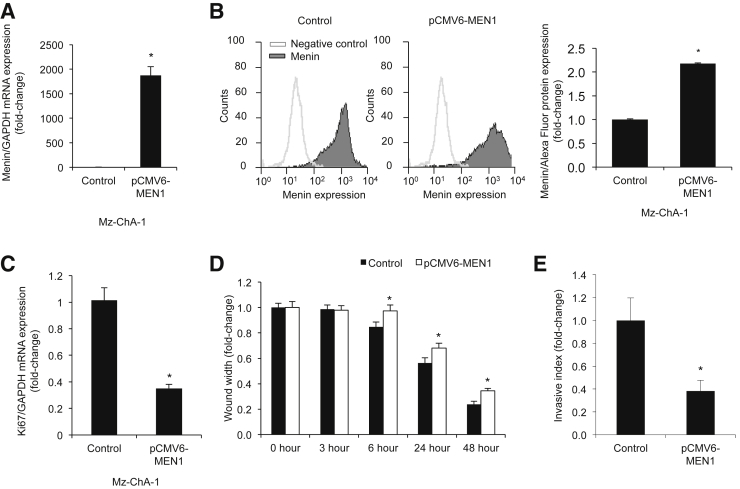
To evaluate the role of menin in the regulation of CCA proliferation, we manipulated menin expression in Mz-ChA-1 cells. Menin expression was first decreased by MEN1 siRNA in Mz-ChA-1 cells (Supplemental Figure S1, A and B). On the basis of nuclear Ki-67 protein expression, menin knockdown resulted in increased Mz-ChA-1 cell proliferation compared to controls (Supplemental Figure S1C). In contrast, overexpression of menin, by transfection with a pCMV6-Entry vector containing the MEN1 gene insert, resulted in decreased Mz-ChA-1 cell proliferation compared to controls (Figure 2, A–C). Menin overexpression resulted in decreased cell migration and invasion by Boyden chamber and wound healing assays (Figure 2, D and E, and Supplemental Figure S2).
Figure 2.
Increased menin expression decreases proliferation. Mz-ChA-1 cells overexpressing menin with pCMV6-MEN1 vector exhibit a decrease in Ki-67 proliferative marker expression. A–C: Increased menin expression in pCMV6-MEN1 Mz-ChA-1 cells by real-time PCR (A) and flow cytometry (B) decreased Ki-67 proliferative marker expression by real-time PCR (C). D: Decreased cell migration as measured by wound healing assay. E: Decreased cell invasion as measured by Boyden chamber assay in pCMV6-MEN1 Mz-ChA-1 cells. Data are expressed as means ± SEM performed in triplicate (A–E). ∗P < 0.05 versus Mz-ChA-1 control cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Effect of Changes on Menin Expression on Angiogenic and Proliferative Factors
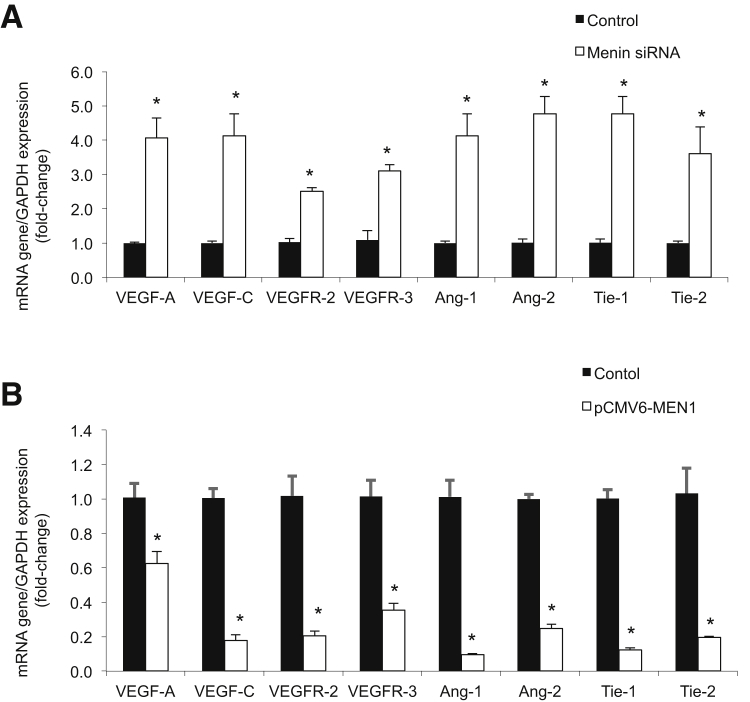
Expression of the angiogenic factors VEGF-A/C, angiopoietins-1/2 (ANG-1/2), and the corresponding receptors VEGFR-2/3 (for VEGFs) and TIE1/2 (for angiopoietins-1/2) were determined by real-time PCR after changes in MEN1 expression. A decrease in MEN1 expression in Mz-ChA-1 MEN1 siRNA cells increased expression of angiogenic factors (Figure 3A), whereas an increase in MEN1 expression in pCMV6-MEN1 Mz-ChA-1 cells decreased expression of angiogenic factors (Figure 3B).
Figure 3.
Menin expression negatively regulates angiogenesis. A: By real-time PCR, Mz-ChA-1 MEN1 knockout cells increased expression of angiogenic factors compared to Mz-ChA-1 control cells. B: By real-time PCR, pCMV6-MEN1 Mz-ChA-1 cells decreased expression of angiogenic factors compared to Mz-ChA-1 control cells. Data are expressed as means ± SEM performed in triplicate (A and B). ∗P < 0.05 versus Mz-ChA-1 control cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
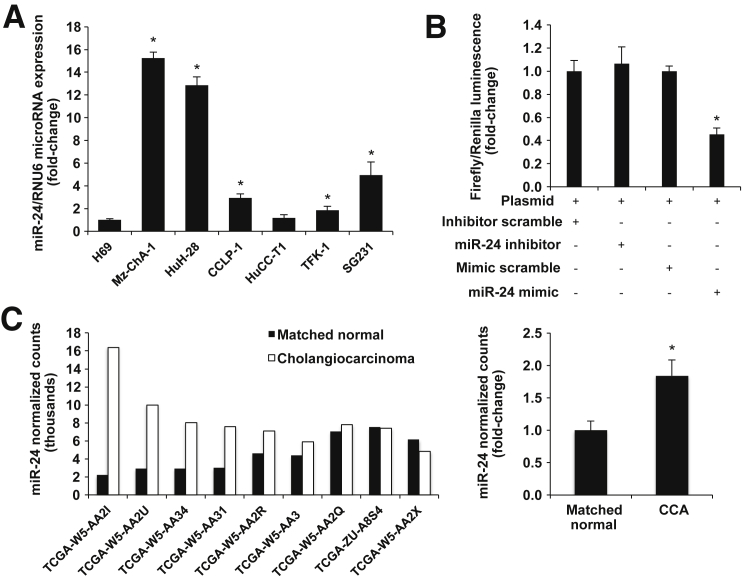
Negative Regulation of Menin Expression by miR-24
We tested the hypothesis that miR-24 acts as a negative regulator of menin in cholangiocytes. We evaluated miR-24 expression in H69 and CCA cell lines by real-time PCR and demonstrated that miR-24 is overexpressed in the selected CCA lines compared to H69 cells (Figure 4A). We transfected pmirGLO-MEN1 vector into Mz-ChA-1 cells with a miR-24 mimic, hairpin inhibitor, or negative control and measured luminescence, which demonstrated that miR-24 negatively regulates menin expression (Figure 4B). Finally, we obtained miRNA-sequencing data from nine human CCA patients from the The Cancer Genome Atlas Research Network (http://cancergenome.nih.gov, last accessed April 7, 2016). We showed that miR-24 expression is increased compared to matched normal tissue (Figure 4C).
Figure 4.
miR-24 negatively regulates menin. A: Real-time PCR evaluation of miR-24 expression in CCA and H69 cell lines demonstrates increased levels in CCA lines compared to H69 cells. B: Luciferase luminescence shows decreased menin expression with miR-24 mimic treatment. C: Left panel: miRNA-sequence data demonstrate increased expression of miR-24 in human CCA tumors compared with matched normal tissue. Right panel: Statistical significance of increased miR-24 expression is validated with an unpaired t-test. Data are expressed as means ± SEM (A–C). n = 3 (A and B); n = 9 (C). ∗P < 0.05 versus normal matched human control.
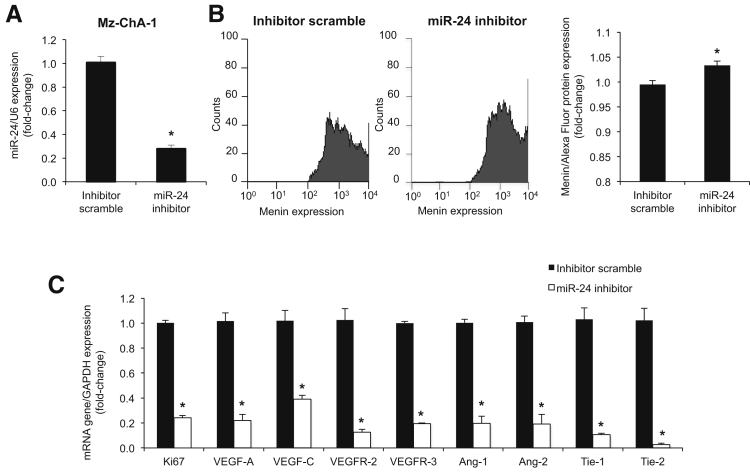
We modulated miR-24 expression in Mz-ChA-1 cells and measured the expression of menin by fluorescence-activated cell sorting, angiogenic factors by real-time PCR, cell proliferation by CellTiter assays, cell migration by wound healing assay, and cell invasion by Boyden chamber assay. We knocked down miR-24 expression in Mz-ChA-1 cells using a hairpin inhibitor (Figure 5A). miR-24 down-regulation correlated with increased menin expression (Figure 5B), decreased expression of angiogenic/proliferative factors (Figure 5C), decreased cell proliferation (Supplemental Figure S3A), and decreased migration and invasion (Supplemental Figure S3, B and C).
Figure 5.
miR-24 drives proliferation. A: Real-time PCR confirmed knockdown of miR-24 in Mz-ChA-1 cells by hairpin inhibitor. miR-24 knockdown increased menin expression via fluorescence-activated cell sorting (B) and decreased expression of angiogenic factors via real-time PCR (C). Data are expressed as means ± SEM performed in triplicate unless otherwise stated (A–C). ∗P < 0.05 versus Mz-ChA-1 control cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Effect of miR-24 Inhibitor on the Growth of Mz-ChA-1 Cells in a Xenograft in Vivo Model
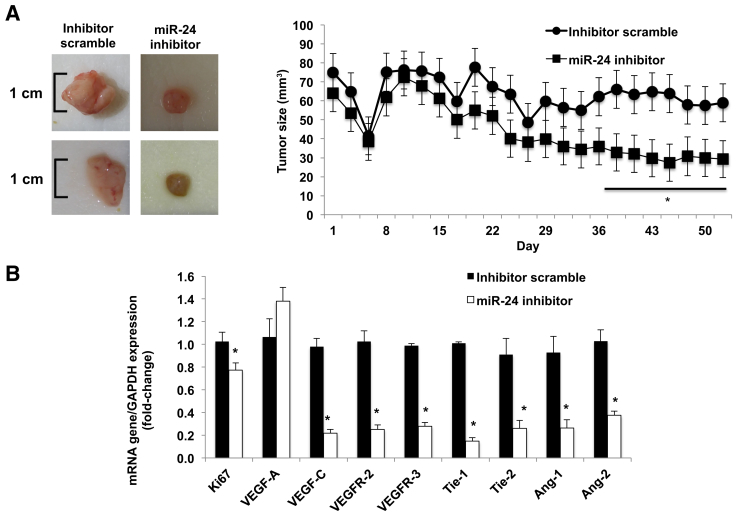
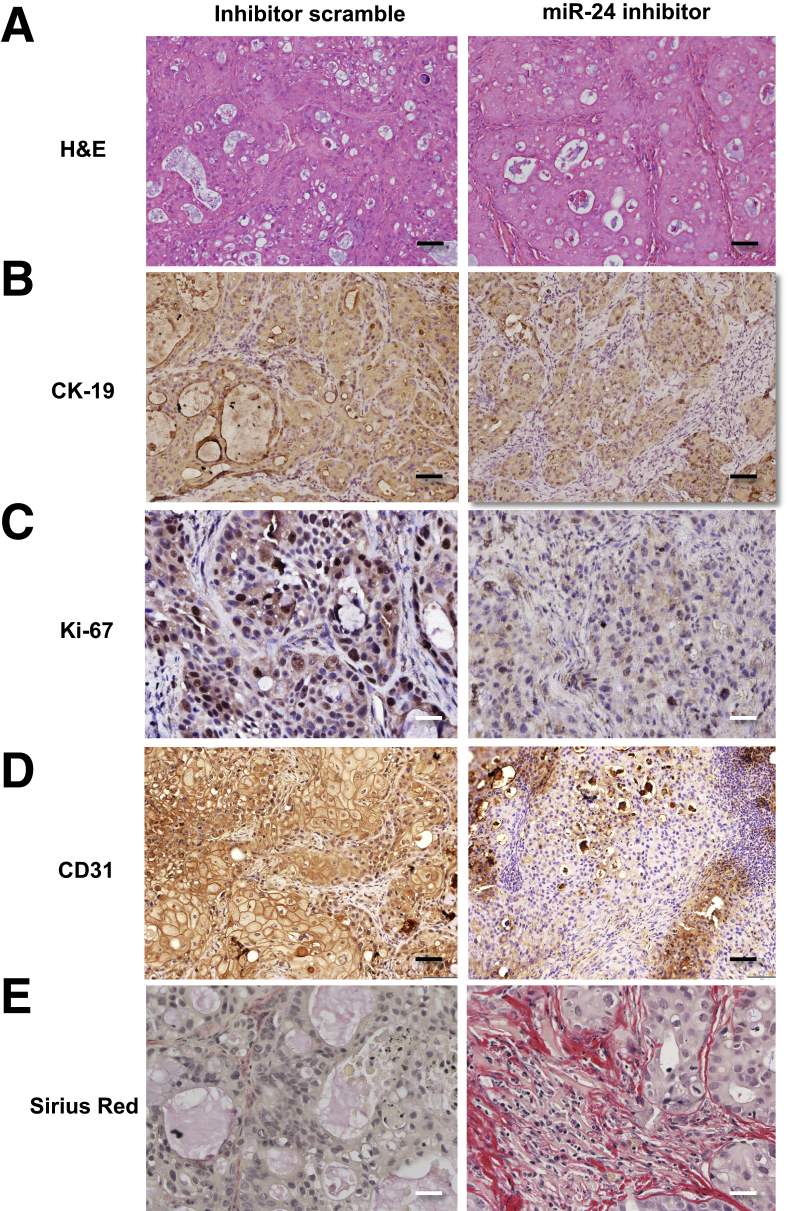
At the end of the treatment (10 weeks), as validation of our in vivo model, there was enhanced expression of menin in the tumor samples from the athymic mice treated with miR-24 hairpin inhibitor compared to scramble mismatch-treated mice (Supplemental Figure S4A). Ten weeks after Mz-ChA-1 implantation into the flanks of nude mice, there was a significant reduction in tumor size in athymic mice treated with the miR-24 hairpin inhibitor compared to mice treated with mismatch scramble (Figure 6A). No significant differences in body weight, liver weight, and liver/body weight ratio were observed between the two groups of mice (Supplemental Table S1). Real-time PCR performed on total RNA isolated from both tumor groups revealed decreased expression of proliferative and angiogenic factors (Figure 6B). Histological examination (Figure 7) demonstrated decreased proliferation via nuclear Ki-67 staining, decreased angiogenesis with CD31 staining, increased fibrosis via Sirius Red staining, and decreased p-extracellular signal regulated kinase levels (Supplemental Figure S4, B–E).
Figure 6.
In vivo miR-24 inhibition decreases growth. A: Ten weeks after Mz-ChA-1 implantation into the flanks of nude mice, there was a significant reduction in tumor size in athymic mice treated with the miR-24 hairpin inhibitor compared to scramble mismatch-treated mice. Significance is shown versus mice treated with scramble mismatch. B: Real-time PCR performed on total RNA isolated from both tumor groups revealed decreased expression of proliferative and angiogenic factors. Significance is shown versus tumor samples from mismatch-treated mice. Data are expressed as means ± SEM of tumor size evaluations. n = 4 mice per each group (A); n = 3 experiments (B). ∗P < 0.05 versus scramble mismatch control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Figure 7.
In vivo miR-24 inhibition decreases growth. A and B: Immunohistochemical images of formalin-fixed, paraffin-embedded sections confirm expected histological depictions of Mz-ChA-1 CCA tumor via hematoxylin and eosin (H&E) and CK-19 staining. C–E: miR-24 inhibition decreased nuclear Ki-67 staining from 1 ± 0.15 inhibitor scramble to 0.4 ± 0.05 miR-24 inhibition (P = 0.001; C), decreased CD31 staining from 1 ± 0.29 inhibitor scramble to 0.1 ± 0.05 miR-24 inhibition (P = 0.02; D), and increased Sirius Red staining from 1 ± 0.47 inhibitor scramble to 10.7 ± 1.68 miR-24 inhibition (P = 0.003; E). Data are expressed as means ± SEM. n = 3 independent samples (A–E). P < 0.05 versus control inhibitor scramble. Original magnification: ×20 (A, B, and D); ×40 (C and E).
Discussion
This novel study demonstrated that menin acts as a tumor suppressor in malignant cholangiocytes and is down-regulated in human CCA. Overexpressing menin in vitro decreases migration, invasiveness, proliferation, and angiogenesis, whereas knocking down menin expression in vitro increases proliferation and angiogenesis. Furthermore, we show that miR-24 acts as an oncogene partly through its negative regulation of menin expression via binding to its 3′-untranslated region. Knocking down miR-24 expression with a hairpin inhibitor increases menin expression and decreases proliferation, migration, invasiveness, and angiogenesis in both in vitro and in vivo xenograft models.
In addition to menin's canonical role in the MEN1 familial cancer syndrome, it is associated with a host of other cancers and proliferative pathologies. For example, loss of menin drives G-cell proliferation in the stomach, leading to hyperplasia and hypergastrinemia.13 Menin suppresses proliferation and migration in lung carcinoid tumors, non–small-cell lung cancer, and lung adenocarcinoma.12 Yan et al38 revealed that menin interacts with scaffold protein IQGAP1 in the β-cell cytoplasm to decrease cell motility and increase cell adhesion through enhanced E-cadherin and β-catenin intercellular junctions. Dreijerink et al15 reported mutations in the MEN1 gene disrupted menin's coactivation of estrogen receptor–mediated transcription in breast cancer cells. However, menin is not necessarily tumor suppressive in all tissues. Menin overexpression is correlated with prostate tumors.16 Functional menin expression is required for leukemogenesis in MLL to drive transcription of mutant MLL-fusion protein targets.39
Menin's role in HCC formation is controversial. One study showed that menin interacts with Sirt1, a type III histone deacetylase, to inhibit NF-κB mediated transactivation of inflammatory cytokines in HCC.14 A contradicting study showed that the menin-MLL interaction epigenetically enhances Yap1 transcription factor expression to drive HCC formation.17 However, no information has previously been published regarding the role of menin and CCA formation and progression.
Menin mainly acts in concert with scaffold proteins to drive histone methylation or deacetylation, depending on its binding partners and its tissue of origin; however, other histone marks have been associated with menin activity. In G cells, menin binds to and inhibits JunD transcriptional activation by recruiting histone deacetylases to JunD target sites and removing transcriptionally activating histone acetylation marks.13 In pancreatic β-cells, the menin-MLL complex enables transcriptionally activating H3K4 trimethylating marks at MLL target genes. In particular, this drives expression of CDKi proteins p27Kip1 and p18INK4c to suppress proliferation.40
In this study, we show that menin expression negatively regulates angiogenic and proliferation markers in cholangiocytes. Cholangiocytes secrete and respond to VEGF in an autocrine and paracrine pathway to promote proliferation.41 No evidence to date points to a role for menin in angiogenesis. However, histone acetylation dynamics are closely connected with epigenetic regulation of angiogenesis.42 In particular, JunD plays a major role in combatting hepatic ischemic/reperfusion injury through antagonizing other transcription factors to regulate hypoxia-inducible factor-1α and VEGF-A expression.43
In this study, we decreased miR-24 expression with a hairpin inhibitor to increase menin protein expression and inhibit proliferation, angiogenesis, migration, and invasion in human Mz-ChA-1 cell lines in both in vitro and in vivo xenograft models. miR-24 is up-regulated in a host of gastrointestinal cancers, and pancreatic cancer xenograft models overexpressing lentiviral miR-24 grew larger tumors compared to controls.20, 21, 22, 23 However, miR-24 has not yet been characterized in biliary disease. miR-24 is implicated in many cellular processes, including cell cycle, apoptosis, DNA repair,44 proliferation, migration, and angiogenesis.45
Melatonin negatively regulates miR-24 expression in several cancers to inhibit tumor proliferation and migration, and lower melatonin signaling in cholangiocytes leads to increased VEGF expression.44 miR-24 expression is increased in HCC, and its inhibition reduces proliferation, migration, and invasion.46
Menin expression can be modulated by other factors as well. Prolactin and glucose stimulation decreases menin expression and increases proliferation in β cells, whereas insulin signaling decreases menin expression.47 Transforming growth factor-β and somatostatin signaling increases menin expression in hepatocytes and the duodenum, respectively.47 Small-molecule inhibitors can block pathway-specific menin functions without altering its overall expression levels. For example, histone deacetylase inhibitor class 1/2 trichlorostatin A reverses menin-dependent transcriptional repression at JunD target sites.48, 49 The menin-MLL inhibitor can block menin-dependent MLL histone trimethylation.
In summary, oncogene miR-24 and tumor-suppressor menin work together in a negative feedback loop to maintain cholangiocyte homeostasis. Dysregulation of this network alters expression of cell cycle and apoptotic genes, leading to increased proliferation, migration, and angiogenesis (Supplemental Figure S5). Targeting miR-24 expression represents a novel means to increase menin expression and reduce biliary tumor growth. Other means of targeting menin expression and/or function include hormonal therapy or small-molecule inhibitors, such as menin-MLL or histone deacetylase inhibitors.
Acknowledgments
We thank Dr. Judy S. Crabtree (Louisiana State University) for graciously donating the MEN1 luciferase plasmid for our experiments; Dr. Vincent VanBuren (Texas A&M University) for guidance on how to extract meaningful data from the The Cancer Genome Atlas Research Network; and Andre Ehrlich and Dr. Elena Ehrlich for aid in file manipulation.
Footnotes
Supported by the Dr. Nicholas C. Hightower Centennial Chair of Gastroenterology from Scott & White; a VA Research Career Scientist Award; VA Merit awards 5I01BX000574 (G.A.), 5I01BX002192 (S.G.), and 1I01BX001724 (F.M.); and NIH grants DK107310 (G.A., F.M., and S.G.), DK58411 (G.A., F.M., and S.G.), DK07698 (G.A., F.M., and S.G.), and DK062975 (G.A., F.M., and S.G.). This material is the result of work supported by resources at the Central Texas Veterans Health Care System.
T.C.L. and S.G. contributed equally to this work as senior authors.
Disclosures: None declared.
The views expressed herein are those of the authors and do not necessarily represent the views of the Department of Veterans Affairs.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.10.021.
Supplemental Data
Decreased menin expression increases proliferation. Decreased menin expression in Men1 knockout Mz-ChA-1 cells by flow cytometry (A) and real-time PCR (B) increased Ki-67 proliferative marker expression by real-time PCR (C). Data are expressed as means ± SEM performed in triplicate (A–C). ∗P < 0.05 versus Mz-ChA-1 control cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Menin overexpression decreases cell migration. Images of wound healing assay show that Mz-ChA-1 cells overexpressing menin (bottom row) migrate slower than control Mz-ChA-1 cells (top row). Original magnification, ×10.
miR-24 decreases growth. miR-24 inhibited Mz-ChA-1 cells demonstrated decreased proliferation via CellTiter assay (A), decreased cell migration via wound healing assay (B), and decreased cell invasion via Boyden chamber assay (C). Data are expressed as means ± SEM (A–C). ∗P < 0.05 versus Mz-ChA-1 control cells. n = 7 (A and C); n = 6 (B). Original magnification, ×10.
In vivo miR-24 inhibition decreases growth. A: At the end of the treatment (10 weeks), there was enhanced expression of menin in the tumor samples from the athymic mice treated with miR-24 hairpin inhibitor compared to scramble mismatch-treated mice. B–E: miR-24 inhibition decreased CCA proliferation as measured by nuclear Ki-67 staining (B), decreased angiogenesis as measured by CD31 staining (C), increased fibrosis as quantified by Sirius Red staining (D), and decreased p-extracellular signal regulated kinase (ERK) levels (E). Data are expressed as means ± SEM (A–E). n = 3 experiments (A–E). ∗P < 0.05 versus tumor samples from mismatch-treated mice. Original magnification, ×20.
Network analysis. Network analysis predicts how menin can mediate fibrosis and angiogenesis through histone deacetylase (HDAC) and DNA methyltransferase mechanisms. QIAGEN's Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, Redwood City, CA; http://www.ingenuity.com) was used. ERK, extracellular signal regulated kinase; TGF, transforming growth factor.
References
- 1.Razumilava N., Gores G.J. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komuta M., Govaere O., Vandecaveye V., Akiba J., Van Steenbergen W., Verslype C., Laleman W., Pirenne J., Aerts R., Yano H., Nevens F., Topal B., Roskams T. Histological diversity in cholangiocellular carcinoma reflects the different cholangiocyte phenotypes. Hepatology. 2012;55:1876–1888. doi: 10.1002/hep.25595. [DOI] [PubMed] [Google Scholar]
- 3.Francis H., Alpini G., DeMorrow S. Recent advances in the regulation of cholangiocarcinoma growth. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1–G9. doi: 10.1152/ajpgi.00114.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvaro D., Mancino M.G., Glaser S., Gaudio E., Marzioni M., Francis H., Alpini G. Proliferating cholangiocytes: a neuroendocrine compartment in the diseased liver. Gastroenterology. 2007;132:415–431. doi: 10.1053/j.gastro.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Chandrasekharappa S.C., Guru S.C., Manickam P., Olufemi S.-E., Collins F.S., Emmert-Buck M.R., Debelenko L.V., Zhuang Z., Lubensky I.A., Liotta L.A., Crabtree J.S., Wang Y., Roe B.A., Weisemann J., Boguski M.S., Agarwal S.K., Kester M.B., Kim Y.S., Heppner C., Dong Q., Spiegel A.M., Burns A.L., Marx S.J. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 6.Huang J., Gurung B., Wan B., Matkar S., Veniaminova N.A., Wan K., Merchant J.L., Hua X., Lei M. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482:542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwal S.K., Jothi R. Genome-wide characterization of menin-dependent H3K4me3 reveals a specific role for menin in the regulation of genes implicated in MEN1-like tumors. PLoS One. 2012;7:e37952. doi: 10.1371/journal.pone.0037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H., Lee J.E., Cho E.J., Liu J.O., Youn H.D. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 2003;63:6135–6139. [PubMed] [Google Scholar]
- 9.Jin S., Mao H., Schnepp R.W., Sykes S.M., Silva A.C., D'Andrea A.D., Hua X. Menin associates with FANCD2, a protein involved in repair of DNA damage. Cancer Res. 2003;63:4204–4210. [PubMed] [Google Scholar]
- 10.Gallo A., Agnese S., Esposito I., Galgani M., Avvedimento V.E. Menin stimulates homology-directed DNA repair. FEBS Lett. 2010;584:4531–4536. doi: 10.1016/j.febslet.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 11.Walls G.V. Multiple endocrine neoplasia (MEN) syndromes. Semin Pediatr Surg. 2014;23:96–101. doi: 10.1053/j.sempedsurg.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y., Feng Z.J., Gao S.B., Matkar S., Xu B., Duan H.B., Lin X., Li S.H., Hua X., Jin G.H. Interplay between menin and K-Ras in regulating lung adenocarcinoma. J Biol Chem. 2012;287:40003–40011. doi: 10.1074/jbc.M112.382416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veniaminova N.A., Hayes M.M., Varney J.M., Merchant J.L. Conditional deletion of menin results in antral G cell hyperplasia and hypergastrinemia. Am J Physiol Gastrointest Liver Physiol. 2012;303:G752–G764. doi: 10.1152/ajpgi.00109.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gang D., Hongwei H., Hedai L., Ming Z., Qian H., Zhijun L. The tumor suppressor protein menin inhibits NF-kappaB-mediated transactivation through recruitment of Sirt1 in hepatocellular carcinoma. Mol Biol Rep. 2013;40:2461–2466. doi: 10.1007/s11033-012-2326-0. [DOI] [PubMed] [Google Scholar]
- 15.Dreijerink K.M.A., Mulder K.W., Winkler G.S., Höppener J.W.M., Lips C.J.M., Timmers H.T.M. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 16.Paris P.L., Sridharan S., Hittelman A.B., Kobayashi Y., Perner S., Huang G., Simko J., Carroll P., Rubin M.A., Collins C. An oncogenic role for the multiple endocrine neoplasia type 1 gene in prostate cancer. Prostate Cancer Prostatic Dis. 2009;12:184–191. doi: 10.1038/pcan.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu B., Li S.H., Zheng R., Gao S.B., Ding L.H., Yin Z.Y., Lin X., Feng Z.J., Zhang S., Wang X.M., Jin G.H. Menin promotes hepatocellular carcinogenesis and epigenetically up-regulates Yap1 transcription. Proc Natl Acad Sci U S A. 2013;110:17480–17485. doi: 10.1073/pnas.1312022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijayaraghavan J., Maggi E.C., Crabtree J.S. miR-24 regulates menin in the endocrine pancreas. Am J Physiol Endocrinol Metab. 2014;307:E84–E92. doi: 10.1152/ajpendo.00542.2013. [DOI] [PubMed] [Google Scholar]
- 19.Luzi E., Marini F., Giusti F., Galli G., Cavalli L., Brandi M.L. The negative feedback-loop between the oncomir Mir-24-1 and menin modulates the Men1 tumorigenesis by mimicking the “Knudson's second hit.”. PLoS One. 2012;7:e39767. doi: 10.1371/journal.pone.0039767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng F.L., Wang W., Jia W.D. Diagnostic and prognostic significance of serum miR-24-3p in HBV-related hepatocellular carcinoma. Med Oncol. 2014;31:177. doi: 10.1007/s12032-014-0177-3. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y.X., Long X.D., Xi Z.F., Ma Y., Huang X.Y., Yao J.G., Wang C., Xing T.Y., Xia Q. MicroRNA-24 modulates aflatoxin B1-related hepatocellular carcinoma prognosis and tumorigenesis. Biomed Res Int. 2014;2014:482926. doi: 10.1155/2014/482926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong W., Li B., Wang Z., Zhang Z., Wang J. Clinical significance of microRNA-24 expression in esophageal squamous cell carcinoma. Neoplasma. 2015;62:250–258. doi: 10.4149/neo_2015_030. [DOI] [PubMed] [Google Scholar]
- 23.Naito Y., Oue N., Pham T.T., Yamamoto M., Fujihara M., Ishida T., Mukai S., Sentani K., Sakamoto N., Hida E., Sasaki H., Yasui W. Characteristic miR-24 expression in gastric cancers among atomic bomb survivors. Pathobiology. 2015;82:68–75. doi: 10.1159/000398809. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu Y., Demetris A.J., Gollin S.M., Storto P.D., Bedford H.M., Altarac S., Iwatsuki S., Herberman R.B., Whiteside T.L. Two new human cholangiocarcinoma cell lines and their cytogenetics and responses to growth factors, hormones, cytokines or immunologic effector cells. Int J Cancer. 1992;52:252–260. doi: 10.1002/ijc.2910520217. [DOI] [PubMed] [Google Scholar]
- 25.Miyagiwa M., Ichida T., Tokiwa T., Sato J., Sasaki H. A new human cholangiocellular carcinoma cell line (HuCC-T1) producing carbohydrate antigen 19/9 in serum-free medium. In Vitro Cell Dev Biol. 1989;25:503–510. doi: 10.1007/BF02623562. [DOI] [PubMed] [Google Scholar]
- 26.Storto P.D., Saidman S.L., Demetris A.J., Letessier E., Whiteside T.L., Gollin S.M. Chromosomal breakpoints in cholangiocarcinoma cell lines. Genes Chromosomes Cancer. 1990;2:300–310. doi: 10.1002/gcc.2870020408. [DOI] [PubMed] [Google Scholar]
- 27.Knuth A., Gabbert H., Dippold W., Klein O., Sachsse W., Bitter-Suermann D., Prellwitz W., Meyer zum Büschenfelde K.H. Biliary adenocarcinoma: characterisation of three new human tumor cell lines. J Hepatol. 1985;1:579–596. doi: 10.1016/s0168-8278(85)80002-7. [DOI] [PubMed] [Google Scholar]
- 28.Kusaka Y., Tokiwa T., Sato J. Establishment and characterization of a cell line from a human cholangiocellular carcinoma. Res Exp Med (Berl) 1988;188:367–375. doi: 10.1007/BF01851205. [DOI] [PubMed] [Google Scholar]
- 29.Saijyo S., Kudo T., Suzuki M., Katayose Y., Shinoda M., Muto T., Fukuhara K., Suzuki T., Matsuno S. Establishment of a new extrahepatic bile duct carcinoma cell line, TFK-1. Tohoku J Exp Med. 1995;177:61–71. doi: 10.1620/tjem.177.61. [DOI] [PubMed] [Google Scholar]
- 30.Kanno N., Glaser S., Chowdhury U., Phinizy J.L., Baiocchi L., Francis H., LeSage G., Alpini G. Gastrin inhibits cholangiocarcinoma growth through increased apoptosis by activation of Ca2+-dependent protein kinase C-alpha. J Hepatol. 2001;34:284–291. doi: 10.1016/s0168-8278(00)00025-8. [DOI] [PubMed] [Google Scholar]
- 31.Francis H., DeMorrow S., Venter J., Onori P., White M., Gaudio E., Francis T., Greene J.F., Jr., Tran S., Meininger C.J., Alpini G. Inhibition of histidine decarboxylase ablates the autocrine tumorigenic effects of histamine in human cholangiocarcinoma. Gut. 2012;61:753–764. doi: 10.1136/gutjnl-2011-300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y., Demorrow S., Invernizzi P., Jing Q., Glaser S., Renzi A., Meng F., Venter J., Bernuzzi F., White M., Francis H., Lleo A., Marzioni M., Onori P., Alvaro D., Torzilli G., Gaudio E., Alpini G. Melatonin exerts by an autocrine loop antiproliferative effects in cholangiocarcinoma: its synthesis is reduced favoring cholangiocarcinoma growth. Am J Physiol Gastrointest Liver Physiol. 2011;301:G623–G633. doi: 10.1152/ajpgi.00118.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeMorrow S., Glaser S., Francis H., Venter J., Vaculin B., Vaculin S., Alpini G. Opposing actions of endocannabinoids on cholangiocarcinoma growth: recruitment of Fas and Fas ligand to lipid rafts. J Biol Chem. 2007;282:13098–13113. doi: 10.1074/jbc.M608238200. [DOI] [PubMed] [Google Scholar]
- 34.Han Y., Onori P., Meng F., DeMorrow S., Venter J., Francis H., Franchitto A., Ray D., Kennedy L., Greene J., Renzi A., Mancinelli R., Gaudio E., Glaser S., Alpini G. Prolonged exposure of cholestatic rats to complete dark inhibits biliary hyperplasia and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2014;307:G894–G904. doi: 10.1152/ajpgi.00288.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francis H., Glaser S., DeMorrow S., Gaudio E., Ueno Y., Venter J., Dostal D., Onori P., Franchitto A., Marzioni M., Vaculin S., Vaculin B., Katki K., Stutes M., Savage J., Alpini G. Small mouse cholangiocytes proliferate in response to H1 histamine receptor stimulation by activation of the IP3/CaMK I/CREB pathway. Am J Physiol Cell Physiol. 2008;295:C499–C513. doi: 10.1152/ajpcell.00369.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fronza M., Heinzmann B., Hamburger M., Laufer S., Merfort I. Determination of the wound healing effect of Calendula extracts using the scratch assay with 3T3 fibroblasts. J Ethnopharmacol. 2009;126:463–467. doi: 10.1016/j.jep.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 37.Alpini G., Invernizzi P., Gaudio E., Venter J., Kopriva S., Bernuzzi F., Onori P., Franchitto A., Coufal M., Frampton G., Alvaro D., Lee S.P., Marzioni M., Benedetti A., DeMorrow S. Serotonin metabolism is dysregulated in cholangiocarcinoma, which has implications for tumor growth. Cancer Res. 2008;68:9184–9193. doi: 10.1158/0008-5472.CAN-08-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan J., Yang Y., Zhang H., King C., Kan H.M., Cai Y., Yuan C.X., Bloom G.S., Hua X. Menin interacts with IQGAP1 to enhance intercellular adhesion of beta-cells. Oncogene. 2009;28:973–982. doi: 10.1038/onc.2008.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama A. Molecular mechanisms of MLL-associated leukemia. Int J Hematol. 2015;101:352–361. doi: 10.1007/s12185-015-1774-4. [DOI] [PubMed] [Google Scholar]
- 40.Karnik S.K., Hughes C.M., Gu X., Rozenblatt-Rosen O., McLean G.W., Xiong Y., Meyerson M., Kim S.K. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27Kip1 and p18INK4c. Proc Natl Acad Sci U S A. 2005;102:14659–14664. doi: 10.1073/pnas.0503484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaudio E., Barbaro B., Alvaro D., Glaser S., Francis H., Ueno Y., Meininger C.J., Franchitto A., Onori P., Marzioni M., Taffetani S., Fava G., Stoica G., Venter J., Reichenbach R., De Morrow S., Summers R., Alpini G. Vascular endothelial growth factor stimulates rat cholangiocyte proliferation via an autocrine mechanism. Gastroenterology. 2006;130:1270–1282. doi: 10.1053/j.gastro.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 42.Turtoi A., Peixoto P., Castronovo V., Bellahcene A. Histone deacetylases and cancer-associated angiogenesis: current understanding of the biology and clinical perspectives. Crit Rev Oncog. 2015;20:119–137. doi: 10.1615/critrevoncog.2014012423. [DOI] [PubMed] [Google Scholar]
- 43.Gerald D., Berra E., Frapart Y.M., Chan D.A., Giaccia A.J., Mansuy D., Pouyssegur J., Yaniv M., Mechta-Grigoriou F. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 44.Mori F., Ferraiuolo M., Santoro R., Sacconi A., Goeman F., Pallocca M., Pulito C., Korita E., Fanciulli M., Muti P., Blandino G., Strano S. Multitargeting activity of miR-24 inhibits long-term melatonin anticancer effects. Oncotarget. 2016;7:20532–20548. doi: 10.18632/oncotarget.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu R., Zhang H., Wang X., Zhou L., Li H., Deng T., Qu Y., Duan J., Bai M., Ge S., Ning T., Zhang L., Huang D., Ba Y. The miR-24-Bim pathway promotes tumor growth and angiogenesis in pancreatic carcinoma. Oncotarget. 2015;6:43831–43842. doi: 10.18632/oncotarget.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma Y., She X.G., Ming Y.Z., Wan Q.Q. miR-24 promotes the proliferation and invasion of HCC cells by targeting SOX7. Tumour Biol. 2014;35:10731–10736. doi: 10.1007/s13277-014-2018-6. [DOI] [PubMed] [Google Scholar]
- 47.Matkar S., Thiel A., Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci. 2013;38:394–402. doi: 10.1016/j.tibs.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y.J., Song T.Y., Park J., Lee J., Lim J., Jang H., Kim Y.N., Yang J.H., Song Y., Choi A., Lee H.Y., Jo C.H., Han J.W., Kim S.T., Youn H.D., Cho E.J. Menin mediates epigenetic regulation via histone H3 lysine 9 methylation. Cell Death Dis. 2013;4:e583. doi: 10.1038/cddis.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gobl A.E., Berg M., Lopez-Egido J.R., Oberg K., Skogseid B., Westin G. Menin represses JunD-activated transcription by a histone deacetylase-dependent mechanism. Biochim Biophys Acta. 1999;1447:51–56. doi: 10.1016/s0167-4781(99)00132-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Decreased menin expression increases proliferation. Decreased menin expression in Men1 knockout Mz-ChA-1 cells by flow cytometry (A) and real-time PCR (B) increased Ki-67 proliferative marker expression by real-time PCR (C). Data are expressed as means ± SEM performed in triplicate (A–C). ∗P < 0.05 versus Mz-ChA-1 control cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Menin overexpression decreases cell migration. Images of wound healing assay show that Mz-ChA-1 cells overexpressing menin (bottom row) migrate slower than control Mz-ChA-1 cells (top row). Original magnification, ×10.
miR-24 decreases growth. miR-24 inhibited Mz-ChA-1 cells demonstrated decreased proliferation via CellTiter assay (A), decreased cell migration via wound healing assay (B), and decreased cell invasion via Boyden chamber assay (C). Data are expressed as means ± SEM (A–C). ∗P < 0.05 versus Mz-ChA-1 control cells. n = 7 (A and C); n = 6 (B). Original magnification, ×10.
In vivo miR-24 inhibition decreases growth. A: At the end of the treatment (10 weeks), there was enhanced expression of menin in the tumor samples from the athymic mice treated with miR-24 hairpin inhibitor compared to scramble mismatch-treated mice. B–E: miR-24 inhibition decreased CCA proliferation as measured by nuclear Ki-67 staining (B), decreased angiogenesis as measured by CD31 staining (C), increased fibrosis as quantified by Sirius Red staining (D), and decreased p-extracellular signal regulated kinase (ERK) levels (E). Data are expressed as means ± SEM (A–E). n = 3 experiments (A–E). ∗P < 0.05 versus tumor samples from mismatch-treated mice. Original magnification, ×20.
Network analysis. Network analysis predicts how menin can mediate fibrosis and angiogenesis through histone deacetylase (HDAC) and DNA methyltransferase mechanisms. QIAGEN's Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, Redwood City, CA; http://www.ingenuity.com) was used. ERK, extracellular signal regulated kinase; TGF, transforming growth factor.