Abstract
Actinobacillus actinomycetemcomitans is associated with localized aggressive periodontitis, a disease characterized by rapid loss of the alveolar bone surrounding the teeth. Receptor activator of NF-κB Ligand (RANKL) and osteoprotegerin (OPG) are two molecules that regulate osteoclast formation and bone resorption. RANKL induces osteoclast differentiation and activation, whereas OPG blocks this process by acting as a decoy receptor for RANKL. The purpose of this study was to investigate the effect of A. actinomycetemcomitans on the expression of RANKL and OPG in human gingival fibroblasts and periodontal ligament cells. RANKL mRNA expression was induced in both cell types challenged by A. actinomycetemcomitans extract, whereas OPG mRNA expression remained unaffected. Cell surface RANKL protein was also induced by A. actinomycetemcomitans, whereas there was no change in OPG protein secretion. A cytolethal distending toxin (Cdt) gene-knockout strain of A. actinomycetemcomitans did not induce RANKL expression, in contrast to its wild-type strain. Purified Cdt from Haemophilus ducreyi alone, or in combination with extract from the A. actinomycetemcomitans cdt mutant strain, induced RANKL expression. Pretreatment of A. actinomycetemcomitans wild-type extract with Cdt antiserum abolished RANKL expression. In conclusion, A. actinomycetemcomitans induces RANKL expression in periodontal connective tissue cells. Cdt is crucial for this induction and may therefore be involved in the pathological bone resorption during the process of localized aggressive periodontitis.
Periodontitis is an infectious disease that leads to the destruction of the tooth-supporting tissues, i.e., the periodontal ligament (PDL), the gingival connective tissue, and the alveolar bone. The PDL is a connective tissue consisting mainly of type I collagen fibers, which mediate the functional attachment of teeth to alveolar bone (25). The gingival connective tissue is lining the alveolar bone, protecting it from external stimuli of the oral environment and is partially mediating the attachment of tooth to alveolar bone (3). Although gingival fibroblasts (GF) and PDL cells may exhibit distinct phenotypic characteristics (13), both cell types establish a dynamic balance between tissue formation and degradation at the tooth-bone interface. Local inflammatory processes cause a shift in this balance and periodontitis may eventually occur.
Localized aggressive periodontitis is a form of periodontal disease that occurs more often in otherwise healthy adolescents and young adults and is characterized by accelerated breakdown of connective tissue and alveolar bone. The gram-negative, facultative anaerobe Actinobacillus actinomycetemcomitans is considered a major agent of localized aggressive periodontitis (12, 29, 49). Studies on the pathogenic mechanisms involved in this disease have focused to a large extent on the leukotoxic properties of A. actinomycetemcomitans toward human polymorphonuclear leukocytes and monocytes (20, 51). The capacity of A. actinomycetemcomitans to kill human leukocytes due to the expression of its leukotoxin is considered a virulence feature associated with the occurrence of localized aggressive periodontitis (12). A. actinomycetemcomitans also has the capacity to produce a cytolethal distending toxin (Cdt) (30, 47, 50), which is a member of a family of protein exotoxins, produced by several unrelated gram-negative bacteria. A. actinomycetemcomitans is the only oral bacterium known to produce this toxin and ca. 85% of its strains possess the cdt genes (63). The Cdt holotoxin, consisting of three subunits (CdtA, CdtB, and CdtC), is known to cause growth arrest or apoptosis in mammalian cells via mechanisms involving the activation of DNA damage checkpoint responses (9). Therefore, the possibility exists that the Cdt may be a virulence factor of A. actinomycetemcomitans.
A number of studies have examined the in vitro and the in vivo bone resorbing capacity of proteinaceous components and lipopolysaccharide (LPS) from A. actinomycetemcomitans (22, 32, 36-38, 41, 42, 58, 61). Local formation of osteoclasts in inflammatory diseases, such as periodontitis and rheumatoid arthritis, is suggested to occur via the enhanced expression of osteoclast-stimulating cytokines in the inflamed tissue (10, 52). Thus, T cells, B cells, GF, and PDL cells can express a wide variety of cytokines (1) with the capacity to either inhibit or induce osteoclastogenesis. Three recently described molecules, members of the tumor necrosis factor ligand and receptor superfamilies, regulate osteoclast formation via cell-to-cell interaction (5). Receptor activator of NF-κB ligand (RANKL), a surface-associated ligand on hematopoietic bone marrow stromal cells, and periosteal tissue osteoblasts (24), as well as T lymphocytes (23, 52), activate its cognate receptor RANK on osteoclast progenitor cells (15). The downstream events lead to differentiation of these mononucleated precursor cells, which then fuse and finally become multinucleated bone-resorbing osteoclasts (53). Osteoprotegerin (OPG), a decoy receptor with homology to RANK, is released from stromal cells and osteoblasts to inhibit the interaction between RANKL and RANK (48). The ratio of RANKL/OPG expression determines the amount of osteoclasts formed. Most hormones and cytokines stimulating osteoclast formation regulate the expression of RANKL and OPG in stromal cells and osteoblasts (26). PDL cells have also been shown to express both RANKL and OPG and may therefore be instrumental in mediating bone resorption via the RANKL/RANK interaction (14, 18, 44). RANKL expression in the periodontium has been implicated in physiological tooth eruption and orthodontic tooth movement (11, 28, 40), as well as in the pathological process of periodontitis (27, 33).
The aims of the present study were to explore if A. actinomycetemcomitans can regulate the expression of RANKL and OPG in GF and PDL cells in vitro and to evaluate the putative involvement of its Cdt.
MATERIALS AND METHODS
Materials.
Dulbecco modified Eagle medium, fetal bovine serum (FBS), penicillin-streptomycin, l-glutamine, and trypsin were purchased from Gibco-BRL. The DAPI (4′,6′-diamidino-2-phenylindole) fluorescence staining kit for mycoplasma detection was from Sigma (St. Louis, Mo.). The MicroBCA assay reagent kit for total protein determination was from Pierce (Cheshire, United Kingdom). The Limulus amoebocyte lysate-based assay used for LPS determination was Coamatic Chromo-LAL from Chromogenix (Mölndal, Sweden). TRIzol reagent was obtained from Life Technologies/Gibco-BRL. The First-Strand cDNA synthesis kit for reverse transcription-PCR (RT-PCR) with avian myeloblastosis virus reverse transcriptase and the PCR core kit for cDNA amplification were from Roche (Mannheim, Germany). A Hot-Start Taq DNA polymerase kit was obtained from Qiagen, Hilden, Germany. An enzyme-linked immunosorbent assay (ELISA) kit for OPG protein quantification was purchased from Immundiagnostik (Bensheim, Germany). For fluorescence-activated cell sorting analysis, recombinant human (rh) OPG-Fc chimeric protein, as well as Fc fragments of rh immunoglobulin G (IgG) used as a negative control were obtained from R&D Systems (Abington, United Kingdom). Polyclonal rabbit anti-human RANKL antibody was obtained from Santa Cruz Biotechnology, and rabbit IgG (used as negative control) was from Dakopats, A/S (Glostrup, Denmark). Fluorescein isothiocyanate (FITC)-conjugated goat antibody to human Fc, which was used as a secondary antibody to rhOPG-Fc, was purchased from Southern Biotechnology (Birmingham, Ala.), and FITC-conjugated sheep antibody to rabbit IgG, which was used as a secondary antibody to the RANKL antibody, was obtained from Dakopats.
Cell cultures.
Human GF and PDL cells lines were established as previously described (4). The PDL and gingival tissue biopsies used were obtained from four healthy young individuals (ages, 13 to 16 years), who were about to have their first premolars removed in the course of orthodontic treatment. Informed consent was given by all subjects, and approval was granted by the Human Studies Ethical Committee of Umeå University, Sweden (§68/03, dnr 03-029). The cells were cultured in Dulbecco modified Eagle medium supplemented with 10% FBS, 20 U of penicillin/ml, 20 μg of streptomycin/ml, and 200 mM l-glutamine. All cell lines were confirmed to be free of mycoplasma infections by using DAPI fluorescence staining. Cells used in all experiments were between passages 4 and 7. For the experiments, GF or PDL cells were seeded at a concentration of 25 × 103 cells/cm2 and allowed to attach for 24 h, maintaining a subconfluent status. Thereafter, the cells were cultured in the presence or absence of bacterial components.
Preparation of the bacterial extract.
Outer membrane components from A. actinomycetemcomitans were extracted as previously described (4, 16). Briefly, bacteria were harvested from cultures of A. actinomycetemcomitans HK1519, D7SS-smooth wild-type strain, and its derivative cdtABC deletion mutant, grown on bacteroides agar containing horse laked blood, for 2 days at 37°C in the presence of 5% CO2. The bacteria were suspended in phosphate-buffered (10 mM [pH 7.2]) saline (PBS) containing 30% FBS to a final concentration of 6 × 109 bacteria/ml. This suspension was gently agitated at 4°C for 1 h to extract bacterial components. After centrifugation (4°C, 10,000 × g, 15 min), the supernatant was aspirated and frozen at −80°C until use. Before the initiation of each experiment, this preparation was diluted in the cell culture medium. The extract concentration used in each experiment is expressed as the percentage (vol/vol) of the above-described stock extract dilution in the culture medium.
To investigate the role of Cdt, the D7SS-smooth and its derivative D7SS-smooth ΔcdtABC strains were used. The D7SS-smooth cdtABC deletion mutant was constructed, characterized, and used in functional assays as previously described (4, 35, 60). In the present study, the D7SS-smooth strain is designated D7SS wild type, whereas the D7SS-smooth ΔcdtABC strain is referred to as D7SS cdt mutant.
Purification of A. actinomycetemcomitans LPS and leukotoxin and analysis of their concentrations in bacterial extract.
An LPS-enriched fraction of A. actinomycetemcomitans HK1519 was obtained by liquid chromatography. The chromatographic separation was run in an ÄKTA system equipped with a Superose 12 column (Pharmacia, Uppsala, Sweden), with 300 mM NaCl as an elution buffer. This LPS fraction contained <1% (wt/vol) protein, as determined by the MicroBCA protein assay. To avoid possible effects from the remaining proteinaceous material, the LPS preparation was heated at 65°C for 30 min. The concentration of LPS in the bacterial extracts was determined by a Limulus amoebocyte lysate-based assay.
Leukotoxin was purified and quantified from A. actinomycetemcomitans HK1519, as previously described (16, 17). The concentrations of LPS and leukotoxin in the bacterial extracts are given in Table 1.
TABLE 1.
Concentrations of leukotoxin and LPS present in 1% of A. actinomycetemcomitans extracts
| Bacterial strain | Concn (μg/ml)a
|
|
|---|---|---|
| Leukotoxin | LPS | |
| HK1519 | 1.73 | 0.54 |
| D7SS wild type | 0.43 | 0.73 |
| D7SS cdt mutant | 0.47 | 0.72 |
Values represent the concentrations of leukotoxin and LPS present in a 1% (vol/vol) dilution of the stock extract in cell culture medium. This dilution is most regularly used in the experimental assays of this study. Therefore, the concentrations of LPS and leukotoxin indicated are present in the final experimental culture medium.
Haemophilus ducreyi Cdt.
Recombinant Cdt holotoxin from H. ducreyi (kind gift by T. Lagergård) was purified as previously described (62). The Cdts of H. ducreyi and A. actinomycetemcomitans exhibit 95% amino acid sequence homology (47).
Cdt antiserum: analysis of binding capacity to A. actinomycetemcomitans proteins and neutralization of Cdt.
A rabbit antiserum against H. ducreyi Cdt holotoxin was incubated with the bacterial extract in order to neutralize Cdt activity at a dilution of 1:100. The antibodies to H. ducreyi Cdt recognize A. actinomycetemcomitans Cdt as well (30, 31). This antiserum, as well as antisera to the individual Cdt subunits, were raised in rabbits by injection of the purified components, as previously described (62) (kindly provided by T. Lagergård).
To determine the binding capacity of the H. ducreyi Cdt antisera to the Cdt peptides in the D7SS wild type and to confirm their absence in the D7SS cdt mutant, immunoblot analysis was performed with bacterial lysates of the respective strains. The bacterial cells were heat treated at 100°C for 5 min and centrifuged at 10,000 × g for 5 min, and the supernatant was collected for immunoblot analysis. The methodology was similar to that for the detection of leukotoxin (16), but the proteins were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis, and the different Cdt antisera were used as primary antibodies at a 1:1,000 dilution.
RNA isolation, first-strand cDNA synthesis, semiquantitative PCRs, and quantitative real-time PCR.
At the end of the experiments, the cell cultures were rinsed three times with 1 ml of PBS each time, and total RNA was extracted from 5 × 105 cells with TRIzol reagent according to the manufacturer's protocol. The RNA was quantified spectrophotometrically, and the integrity of the RNA preparations was examined by agarose gel electrophoresis. A total of 1 μg of total RNA was reverse transcribed into single-stranded cDNA by using oligo(dT) primers (25°C for 10 min, 42°C for 60 min, and 99°C for 5 min). The cDNA was kept in a volume of 20 μl at −80°C until used for the PCR.
Aliquots of 0.5 μl of the synthesized cDNA in a total volume of 25 μl of Hot-Start Taq DNA polymerase master mix were amplified in a thermal cycler (Mastergradient Cycler; Eppendorf, Hamburg, Germany). The conditions for PCR were as follows: activation of the polymerase at 95°C for 15 min during the first cycle, followed by denaturation at 94°C for 35 s, annealing at variable temperatures (GAPDH [glyceraldehyde-3-phosphate dehydrogenase] at 57°C, RANKL at 66°C, OPG at 55°C, and type I [α1] procollagen at 59°C) for 35 s, followed by elongation at 72°C for 45 s. After completion of all cycles, an additional step of elongation at 72°C for 10 min followed. The expression of these genes was compared at the logarithmic increase phase of the PCR. The PCR products were electrophoretically size fractionated on 1.5% agarose gels and visualized by using ethidium bromide. PCR of samples without the preceding RT step did not result in any products, demonstrating that there was no genomic DNA amplification. The sequence of primers used, the GenBank accession numbers, and the sizes of the PCR products are provided in Table 2. The identity of the PCR products was confirmed by using a DYEnamic ET terminator cycle sequencing kit (Amersham Biosciences, Amersham, United Kingdom). The nucleotide sequence was determined on an ABMI 377 XL DNA sequencer (PE Applied Biosystems, Foster City, Calif.).
TABLE 2.
Primers used in semiquantitative and real-time PCR
| RT-PCR and gene | Primer or probe orientation and sequencea | Accession no. | Product size (bp) |
|---|---|---|---|
| Semiquantitative RT-PCR | |||
| RANKL | Fwd, 5′-CAGCACATCAGAGCAGAGAAAGC-3′ | AF019047 | 517 |
| Rev, 5′-CCCCAAAGTATGTTGCATCCTG-3′ | |||
| OPG | Fwd, 5′-GTACGTCAAGCAGGAGTGCAATC-3′ | U94332 | 472 |
| Rev, 5′-TTCTTGTGAGCTGTGTTGCCG-3′ | |||
| Type I (α1) procollagen | Fwd, 5′-CTGACCTCCTGCGCCTGATGTCC-3′ | NM000088 | 298 |
| Rev, 5′-GTCTGGGGCACCAACGTCCAAGGG-3′ | |||
| GAPDH | Fwd, 5′-CAACTACATGGTTTACATGTTC-3′ | NM002046 | 163 |
| Rev, 5′-GCCAGTGGACTCCACGAC-3′ | |||
| Quantitative real-time PCR | |||
| OPG | Fwd, 5′-AGTACGTCAAGCAGGAGTGCAAT-3′ | U94332 | 129 |
| Rev, 5′-CCAGCTTGCACCACTCCAA-3′ | |||
| Probe, 5′-(VIC)-AGGTAGCGCCCTTCCTTGCATTCG-(TAMRA)-3′ | |||
| RPL13a | Fwd, 5′-CCGCTCTGGACCGTCTCAA-3′ | NM12423 | 123 |
| Rev, 5′-CCTGGTACTTCCAGCCAACCT-3′ | |||
| Probe, 5′-(VIC)-TGACGGCATCCCACCGCCCT-(TAMRA)-3′ |
Fwd, forward; Rev, reverse.
Quantitative real-time PCR analyses of human OPG and RPL13a mRNA were performed by using the TaqMan Universal PCR Master Mix kit and ABI Prism 7900 HT Sequence Detection System and software (PE Applied Biosystems) as previously described (2). The concentration of the primers for OPG and RPL13a was 300 nM, and that of the probes was 100 nM. The sequences of primers and probes, the GenBank accession numbers and the predicted PCR products are given in Table 2. To control for variability in amplification due to differences in starting mRNA concentrations, RPL13a was used as an internal standard. The relative expression of target OPG mRNA was computed from the target OPG Ct values and RPL13a Ct values by using the standard curve method (User Bulletin 2; PE Applied Biosystems).
Flow cytometric analysis for RANKL protein.
After 48 h of challenge, GF and PDL cells were detached (0.1% EDTA, 0.025% trypsin) and fixed (2% paraformaldehyde). RANKL on the cell surface was detected with either 10 μg of rhOPG-Fc chimeric protein or 2 μg of rabbit anti-human RANKL antibody/ml. As negative controls, 10 μg of Fc fragments of rhIgG or 2 μg of rabbit IgG/ml was used, respectively. As secondary antibodies, 10 μg of FITC-conjugated goat antibody to human Fc or 2 μg of FITC-conjugated sheep antibodies to rabbit IgG/ml were used for the OPG-Fc and the RANKL antibody, respectively. Finally, the cells were analyzed with a fluorescence-activated cell sorter (Facstarplus) by using CellQuest software (Becton Dickinson Immunocytometry Systems). Ten thousand cells were analyzed from each sample, and the data were plotted as the distribution of cells according to their degree of fluorescence. The mean fluorescence value of the cells in each sample was then calculated.
Protein analysis for OPG.
The amount of secreted OPG protein by GF and PDL cells in response to A. actinomycetemcomitans extract was quantified by enzyme-linked immunosorbent assay (ELISA) after 48 h of challenge.
Statistical analysis.
The significance of differences between the groups was assessed by one-way analysis of variance by using the SPSS software (SPSS, Inc., Chicago, Ill.). P values of ≤0.05 were considered indicative of a statistically significant difference. The data are expressed as means ± the standard error of means (SEM).
RESULTS
Effects of A. actinomycetemcomitans on RANKL and OPG mRNA expression.
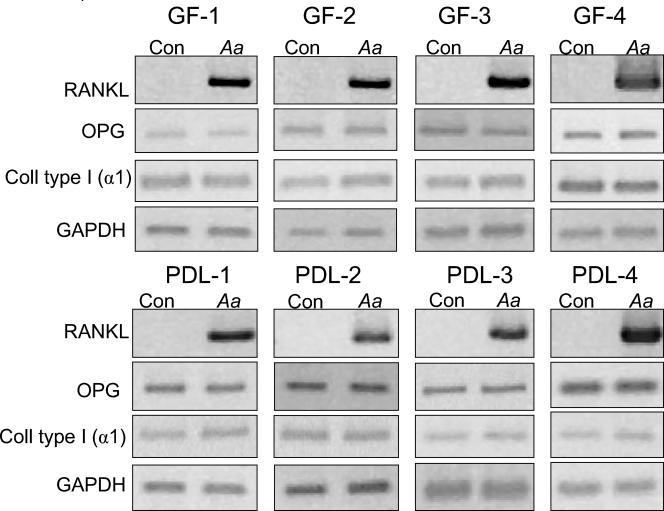
Gingival fibroblasts and PDL cells were isolated from four different healthy subjects. These cells all exhibited a spindle-shaped, elongated morphology and expressed type I (α1) procollagen mRNA, demonstrating their fibroblastic phenotype (data not shown). When GF and PDL cell cultures were challenged with A. actinomycetemcomitans HK1519 extract for 24 h, all cell lines expressed RANKL mRNA, whereas the unchallenged cells did not (Fig. 1). Upregulation of RANKL mRNA by A. actinomycetemcomitans was observed in 16 of 16 experiments with GF and PDL cells from all subjects. In contrast, OPG mRNA was expressed in all unchallenged GF and PDL cell cultures, and the expression remained unaffected after bacterial challenge, as assessed by semiquantitative RT-PCR (Fig. 1). In agreement with these data, quantification by real-time PCR also demonstrated that the bacterial extract did not affect OPG mRNA expression in GF and PDL cells (Table 3). Similarly, the mRNA expression of type I (α1) procollagen was not affected by the bacterial extract (Fig. 1).
FIG. 1.
RT-PCR analysis for RANKL, OPG, and type I (α1) procollagen mRNA expression in four GF and four PDL cell lines challenged with A. actinomycetemcomitans extract. GF or PDL cells were cultured in the absence (Con) or presence of 1% A. actinomycetemcomitans HK1519 extract (Aa) for 24 h. Number of RT-PCR cycles: RANKL, 38; OPG, 22; type I (α1) procollagen, 18; GAPDH, 20.
TABLE 3.
Effect of A. actinomycetemcomitans extract on OPG mRNA expression and protein production by GF and PDL cells
| Group | Relative OPG mRNA expression (%) ± SEM in A. actinomycetemcomitansa | Mean OPG concn (pmol/liter) ± SEM in culture mediumb
|
Relative OPG mRNA expression (%) ± SEMc
|
||
|---|---|---|---|---|---|
| Control | A. actinomycetemcomitans | D7SS wild type | D7SS cdt mutant | ||
| GF | 96.5 ± 11 | 295 ± 18 | 242 ± 16 | 104.5 ± 6 | 106 ± 8 |
| PDL cells | 104.5 ± 12 | 46 ± 7 | 53 ± 9 | 95 ± 13 | 93 ± 14 |
As determined by real-time PCR: The values are expressed as the percentage of the unchallenged controls (i.e., 100%) and represent the mean ± the SEM of OPG expression (calibrated to housekeeping gene RPL 13a) from four independent experiments in GF and PDL cells. The cells were cultured for 24 h in the presence of 1% extract from A. actinomycetemcomitans HK1519. There was no statistically significant difference between the challenged cells and their corresponding controls (P > 0.05).
As determined by ELISA: The values represent the mean ± the SEM of the OPG concentrations in the culture media from four independent experiments with GF and PDL cells. The cells were cultured in the absence (control) or presence (A. actinomycetemcomitans) of 1% extract from strain HK1519 for 48 h. The differences between the challenged cells and their corresponding controls were not statistically significant (P > 0.05).
As determined by real-time PCR. The values are expressed as a percentage of the unchallenged controls (i.e., 100%) and represent the mean ± the SEM of OPG expression (calibrated to housekeeping gene RPL13a) from four independent experiments in GF and PDL cells. The cells were cultured for 24 h in the presence of 1% extract from A. actinomycetemcomitans strains D7SS (wild type) and D7SS (cdt mutant). There were no statistically significant differences between unchallenged controls and either D7SS wild type or D7SS cdt mutant-challenged cells (P > 0.05).
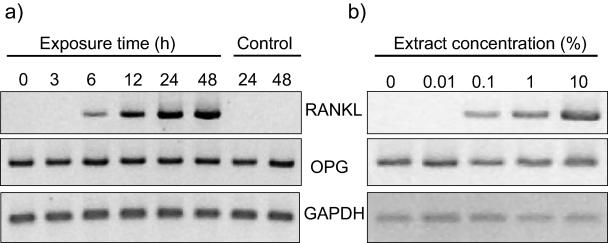
The increased mRNA expression of RANKL was dependent on time of challenge, with an initial response seen at 6 h and a maximal effect at 48 h (Fig. 2a). The induction of RANKL mRNA was also dependent on the concentration of the A. actinomycetemcomitans extract used (Fig. 2b).
FIG. 2.
Time and concentration dependency of RANKL mRNA expression in response to A. actinomycetemcomitans. (a) GF cultures were challenged with 1% A. actinomycetemcomitans HK1519 extract for up to 48 h. Two time points were selected for unchallenged (control) cultures (24 and 48 h). RT-PCR analysis was then performed for RANKL and OPG expression. (b) GF cultures were challenged with A. actinomycetemcomitans HK1519 extract with a concentration range between 0.01 and 10% for 24 h. RT-PCR analysis was then performed for RANKL and OPG expression. Number of RT-PCR cycles: RANKL, 38; OPG, 22; GAPDH, 20.
Effects of A. actinomycetemcomitans on RANKL and OPG protein expression.
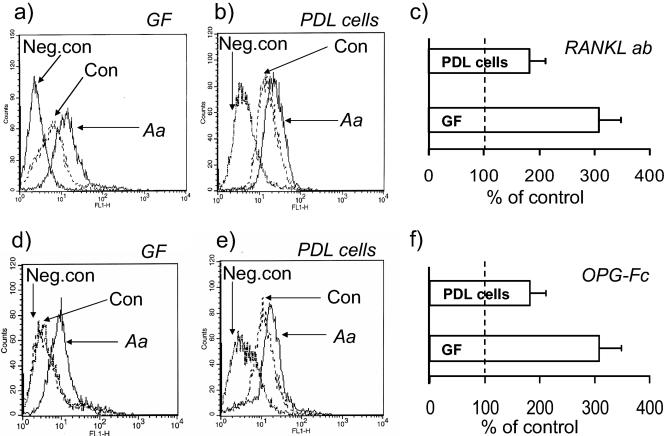
Flow cytometric analysis, using primary antibodies recognizing human RANKL, showed that substantially more PDL cells and GF expressed RANKL protein at their cell surfaces when challenged with A. actinomycetemcomitans for 48 h (Fig. 3a and b). Statistical analysis of data obtained in four independent experiments showed that A. actinomycetemcomitans caused significant (P < 0.05) 2.0- and 3.0-fold stimulation of RANKL protein in PDL cells and GF, respectively (Fig. 3).
FIG. 3.
Flow cytometric analysis for RANKL protein expression on GF and PDL cell surface. GF (a and d) or PDL cells (b and e) were cultured in the absence (Con) or presence of 1% A. actinomycetemcomitans HK1519 extract (Aa) for 48 h. The cells were collected, washed, and stained with either anti-human RANKL antibody (a and b) or OPG-Fc (d and e). The results were confirmed in all of the four GF and PDL cell lines studied. Negative controls (Neg. Con) represent unchallenged cells treated with IgG or Fc fragments instead of anti-RANKL or OPG-Fc, respectively. Representative results obtained with both methods in one GF and one PDL cell line, are provided. (c and f) The bars represent the percentage of fluorescence levels (mean ± the SEM) in A. actinomycetemcomitans-challenged GF and PDL cells relative to the fluorescence levels of unchallenged controls. The dotted line represents the fluorescence level of the unchallenged controls (i.e., 100%). These results were obtained from four experiments in each of these cell two lines, with the antibody-based method (c) or the OPG-Fc-based method (f).
We also took advantage of the fact that OPG binds to RANKL and by using chimeric OPG-Fc we could show, similarly to the data obtained with RANKL antibodies, that challenge of PDL cells and GF for 48 h with A. actinomycetemcomitans resulted in an increased number of cells binding OPG (Fig. 3d and e). Accumulated data from four independent experiments showed that A. actinomycetemcomitans caused statistically significant (P < 0.05), 1.8- and 3.1-fold stimulation of OPG binding in PDL cells and GF, respectively (Fig. 3f).
The effect of A. actinomycetemcomitans on OPG protein production by GF and PDL cells was investigated after 48 h of challenge by using an ELISA kit. The results from four independent experiments with GF and four independent experiments with PDL cells showed that A. actinomycetemcomitans did not cause any statistically significant (P > 0.05) regulation of OPG protein (Table 3).
Involvement of Cdt in RANKL induction.
When exploring the possibility that the Cdt was responsible for RANKL induction, three types of experiments were performed. First, cells were exposed to an extract from a strain of A. actinomycetemcomitans in which the cdt gene had been deleted (D7SSΔcdtABC). Second, purified recombinant Cdt holotoxin from H. ducreyi was used to challenge the cells. Third, a Cdt holotoxin antiserum was added to the extract from the D7SS wild-type strain.
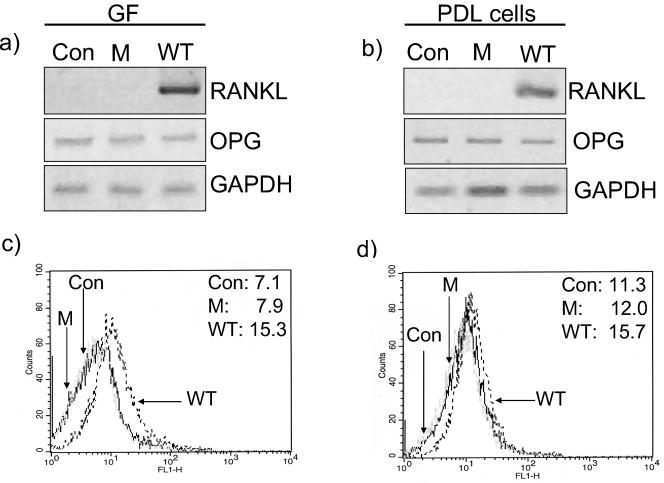
In contrast to extract from the D7SS wild-type strain, extract from the D7SS cdt mutant did not induce RANKL mRNA in GF (Fig. 4a), or in PDL cells (Fig. 4b). In agreement with the RT-PCR analysis, challenge of GF or PDL cells with extract from cdt mutant did not enhance RANKL protein expression in GF (Fig. 4c) or in PDL cells (Fig. 4d), as assessed by flow cytometric analysis. Neither the D7SS wild type nor its derivative cdt mutant affected OPG mRNA expression in relation to the controls as assessed by semiquantitative RT-PCR (Fig. 4a and b). In agreement with RT-PCR analysis, quantitative real-time PCR demonstrated that neither of these two extracts caused any significant changes in OPG expression (Table 3c).
FIG. 4.
Effect of an A. actinomycetemcomitans cdt mutant strain on RANKL expression. GF (a) and PDL cells (b) were challenged for 24 h with 1% D7SS wild-type (WT) or 1% D7SS cdt mutant (M) extracts or were cultured in the absence of bacterial challenge (Con) for 24 h. The mRNA expression of RANKL and OPG was then analyzed by RT-PCR. The results provided here are representative from one GF (a) and one PDL cell line (b) of eight experiments performed. Number of RT-PCR cycles: RANKL, 38; OPG, 22; GAPDH, 20. Flow cytometric analysis for RANKL protein analysis is also provided in one GF (c) and one PDL cell line (d). The cells were challenged for 48 h with 1% D7SS wild type (WT) or 1% D7SS cdt mutant (M) extract or were cultured in the absence of bacterial challenge (Con) (light gray line). The numbers in the upper right corner of the fluorescence-activated cell sorting charts indicate the mean fluorescence values of the cells in each sample.
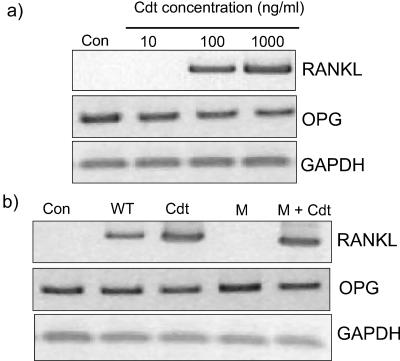
Purified recombinant Cdt from H. ducreyi induced RANKL mRNA expression by GF in a concentration-dependent manner (Fig. 5a). When H. ducreyi Cdt was added to PDL cells in combination with extract from the D7SS cdt mutant, RANKL expression was induced similarly to purified Cdt (Fig. 5b). Analysis of OPG mRNA expression by RT-PCR indicated a downregulation of its expression in Cdt-challenged GF (Fig. 5a) or PDL cells (Fig. 5b); this was also confirmed by quantitative real-time PCR. The latter analysis showed that purified Cdt (1,000 ng/ml) significantly decreased OPG mRNA expression levels in GF to 61.5 ± 10% of the unchallenged control (n = 4, mean ± the SEM; P < 0.05).
FIG. 5.
Effect of purified Cdt from H. ducreyi on RANKL and OPG expression. (a) GF cells were cultured in the absence (Con) or presence of increasing concentrations (from 10 to 1,000 ng/ml) of H. ducreyi Cdt for 24 h. RANKL and OPG mRNA expression was then analyzed by RT-PCR (b) PDL cells were challenged with 1% D7SS wild-type (WT) extract, 1% D7SS cdt mutant (M) extract, 1,000 ng of Cdt/ml (Cdt), and 1% D7SS cdt mutant extract including 1,000 ng of Cdt/ml (M+Cdt) for 24 h. RANKL and OPG mRNA levels were then analyzed by RT-PCR. Number of cycles: RANKL, 38; OPG, 22; GAPDH, 20.
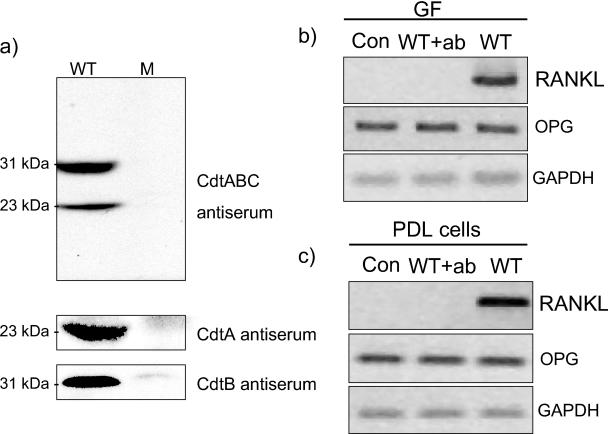
The Cdt antiserum used, raised against the Cdt holotoxin of H. ducreyi, recognized two bands in the D7SS wild-type bacterial lysate at 31 and 23 kDa that were not present in the D7SS cdt mutant lysate (Fig. 6a). Antiserum raised to the H. ducreyi CdtA and CdtB subunits recognized the 23- and the 31-kDa bands, respectively (Fig. 6a). The bacterial lysate preparations did not show reactivity to rabbit nonimmune serum (data not shown). Pretreatment of the D7SS wild-type extract with the Cdt antiserum abolished the A. actinomycetemcomitans-induced RANKL mRNA expression and did not affect OPG expression in GF (Fig. 6b) or PDL cells (Fig. 6c). In contrast, rabbit nonimmune serum did not affect the RANKL mRNA expression induced by A. actinomycetemcomitans (data not shown).
FIG. 6.
Cdt-antiserum binding specificity to A. actinomycetemcomitans' proteins and effect on RANKL and OPG expression. (a) Western blot analysis was performed on bacterial cell lysates from the D7SS wild-type (WT) and D7SS cdt mutant (M) strains with antiserum raised to H. ducreyi CdtABC, CdtA, and CdtB. GF (b) or PDL cells (c) were challenged with 1% D7SS wild-type extract alone (WT) or 1% D7SS wild-type extract that had been pretreated with CdtABC antiserum (WT+ab) for 30 min. After 24 h of bacterial challenge, the mRNA expression of RANKL and OPG were investigated by RT-PCR. Number of cycles: RANKL, 38; OPG, 22; GAPDH, 20.
Involvement of LPS and leukotoxin in RANKL induction.
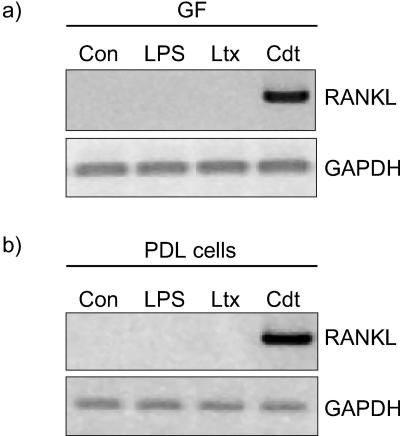
The possible role of A. actinomycetemcomitans LPS and leukotoxin in RANKL induction was also investigated. In contrast to Cdt (0.3 μg/ml), purified LPS (1 μg/ml) or leukotoxin (2 μg/ml) did not induce RANKL mRNA expression by GF (Fig. 7a) or PDL cells (Fig. 7b).
FIG. 7.
Effect of purified A. actinomycetemcomitans LPS, leukotoxin (Ltx), and H. ducreyi Cdt on RANKL expression. GF (a) or PDL cells (b) were challenged for 24 h with a final concentration of 1 μg of LPS, 2 μg of Ltx, or 0.3 μg of Cdt/ml. The expression of RANKL was then analyzed by RT-PCR. Number of cycles: RANKL, 38; GAPDH, 20.
DISCUSSION
The present study demonstrates that A. actinomycetemcomitans induces mRNA expression of RANKL in GF and PDL cells obtained from periodontally healthy individuals. The mRNA expression resulted in enhanced RANKL protein on the surface of the cells, as assessed by flow cytometric analysis. The use of either RANKL antibody or OPG-Fc indicated similar levels of RANKL upregulation, and the analysis revealed that GF were more responsive to A. actinomycetemcomitans than were PDL cells. In contrast, the mRNA and protein expression of OPG was not affected by A. actinomycetemcomitans, suggesting that GF and PDL cells exposed to this bacterium express an increased ratio of RANKL to OPG. This indicates that the bacterially challenged cells may have an increased capacity for inducing osteoclastogenesis via cell-to-cell interaction with osteoclast progenitor cells by the RANKL-RANK-dependent pathway due to enhanced RANKL expression (53). Interestingly, the biosynthesis of extracellular matrix proteins, assessed by the mRNA expression of collagen type I (α1) procollagen, was unaffected by exposure of GF and PDL cells to A. actinomycetemcomitans.
The role of the RANKL-RANK-OPG system in inflammation-induced bone loss is demonstrated by the observation that OPG treatment decreases local bone loss in experimentally induced periodontitis and arthritis (23, 43, 46, 55, 59). In inflammation-induced local bone resorption, it has been suggested that RANKL is expressed by resident osteoblasts, stimulated by proinflammatory cytokines, such as interleukin-1, tumor necrosis factor alpha, or cytokines of the interleukin-6 family (5, 26). Alternatively, the possibility exists that RANKL is expressed by infiltrating activated T lymphocytes (23, 52). This is demonstrated by the finding that A. actinomycetemcomitans antigen-specific Th1 cells obtained from experimental periodontitis rats express higher levels of RANKL compared to the healthy controls (59) and further supported by the finding that A. actinomycetemcomitans extract enhances RANKL mRNA expression in CD4+ T-lymphocytes (54, 55). We show here that A. actinomycetemcomitans stimulates RANKL mRNA and protein expression in periodontal connective tissue cells as well.
By using the cdt knockout strain of A. actinomycetemcomitans we could show the crucial role of Cdt expression for RANKL induction by this bacterium. Moreover, an antiserum to Cdt blocked RANKL stimulation, induced by the A. actinomycetemcomitans wild-type extract. The antiserum was raised against the H. ducreyi Cdt holotoxin but was found to neutralize the activity of A. actinomycetemcomitans Cdt. Immunoblot analysis showed that this antiserum bound to a 23- and a 31-kDa protein in the D7SS wild-type strain but not in the D7SS cdt mutant strain. These are likely to represent A. actinomycetemcomitans CdtA and CdtB subunits, since CdtA- and CdtB-specific antisera recognized the 23- and 31-kDa proteins, respectively. The observation that antibodies raised against H. ducreyi Cdt also identify A. actinomycetemcomitans Cdt is in agreement with a previous report (31). The induction of RANKL was also demonstrated by challenging the cells with purified Cdt holotoxin from H. ducreyi, which exhibits 95% amino acid sequence homology with A. actinomycetemcomitans Cdt (47). This toxin solely was sufficient to induce RANKL expression but also able to downregulate OPG mRNA expression. The most likely reason for OPG downregulation by purified Cdt (1,000 ng/ml), in contrast to lack of effect by the bacterial extract, is that the extract may contain less Cdt protein.
Leukotoxin or LPS, at concentrations higher than those present in the bacterial extracts, could not induce RANKL expression by the challenged cells. In addition, the extracts from the D7SS wild type and its derivative D7SS cdt mutant strains contained similar amounts of LPS and leukotoxin (Table 1), but only the D7SS wild-type could induce RANKL. Collectively, these findings indicate that Cdt alone can induce RANKL expression and that synergism with other extractable components, such as LPS, is not required. A recent study demonstrated that Porphyromonas gingivalis induces RANKL expression by mouse primary osteoblasts. By using an isogenic gingipain-deficient mutant strain, RANKL induction by P. gingivalis was attributed to the gingipains rather than to its LPS (39). However, other reports have demonstrated RANKL mRNA induction in murine osteoblasts or stromal cells by LPS from Treponema denticola, A. actinomycetemcomitans and Escherichia coli (6, 21, 45). Regarding periodontal connective tissue cells, it has been demonstrated that E. coli or Salmonella enterica serovar Abortusequi LPS were not able to induce RANKL expression by human GF (34) or PDL cells (57), respectively, although E. coli LPS upregulated OPG expression (34). One explanation for the divergent results attained with LPS in the present and other studies may be related to differences in target cell populations. Another explanation may be the molecular diversity of the LPS among various gram-negative bacteria. In the case of A. actinomycetemcomitans, it has been demonstrated that outer membrane components from this bacterium are more potent inducers of cytokine induction (42) and bone resorption (41, 61) than its LPS. Moreover, in contrast to enteropathogenic LPS that shows osteolytic activity at concentrations of nanograms/milliliter, A. actinomycetemcomitans LPS is active at concentrations of micrograms/milliliter (41).
A. actinomycetemcomitans has been found at high levels in subgingival biofilms and occasionally in periodontal connective tissues (7) of localized aggressive periodontitis patients. Its toxins can be released from its surface (4, 16) and are likely to diffuse into the deeper periodontal tissues, a possibility supported by the serum antibody immune response of periodontitis patients to leukotoxin (56) and Cdt (31). Therefore, the direct interaction of diffused toxins with cells of the periodontal tissues may induce the expression of molecules related to osteoclastogenesis at the front of alveolar bone resorption. In experimental periodontitis models, profound alveolar bone loss can be caused by transplantation of A. actinomycetemcomitans antigen-specific Th1 cells in rats (19, 59) or by transplantation of blood mononuclear leukocytes from patients with localized aggressive periodontitis to NOD/SCID mice and intraoral inoculation with A. actinomycetemcomitans (54, 55). Previous studies have demonstrated that proteinaceous surface-associated components (22, 32, 41, 42, 61) or the LPS (36-38, 58) of A. actinomycetemcomitans can activate bone resorption and osteoclastogenesis. The Cdt of A. actinomycetemcomitans is known to cause growth arrest and eventually apoptosis in mammalian cells (4, 8, 35, 47, 50), phenomena that are not related to the initiation of bone resorption. The present study demonstrates a possible virulence mechanism by which the Cdt of A. actinomycetemcomitans may be involved in osteoclastogenesis via the RANKL-RANK-dependent pathway. Moreover, the induction of RANKL by A. actinomycetemcomitans may occur in resident periodontal connective tissue cells, in addition to T cells. Since GF and PDL cells reside in close proximity to the bone resorption sites in patients with aggressive periodontitis, induction of RANKL by the Cdt in these tissues may favor the differentiation of osteoclast progenitor cells into mature osteoclasts.
Acknowledgments
This study has been supported in part by the Swedish Research Council (project 7525), the County Council of Västerbotten, the Swedish Dental Society, the Swedish Patent Revenue Fund, the Swedish Rheumatism Association, the Royal 80 Year Fund of King Gustav V, and NIDCR grant RO1 DE12212 (C. Chen).
We thank Teresa Lagergård (Department of Medical Microbiology and Immunology, Gothenburg University, Gothenburg, Sweden) for kindly providing purified H. ducreyi Cdt and Cdt antisera. We also thank Inger Lundgren for technical assistance with the real-time PCR.
Editor: J. D. Clements
REFERENCES
- 1.AAP. 1999. Informational paper: the pathogenesis of periodontal diseases. J. Periodontol. 70:457-470. [DOI] [PubMed] [Google Scholar]
- 2.Ahlen, J., S. Andersson, H. Mukohyama, C. Roth, A. Backman, H. H. Conaway, and U. H. Lerner. 2002. Characterization of the bone-resorptive effect of interleukin-11 in cultured mouse calvarial bones. Bone 31:242-251. [DOI] [PubMed] [Google Scholar]
- 3.Bartold, P. M., L. J. Walsh, and A. S. Narayanan. 2000. Molecular and cell biology of the gingiva. Periodontology 24:28-55. [DOI] [PubMed] [Google Scholar]
- 4.Belibasakis, G., A. Johansson, Y. Wang, R. Claesson, C. Chen, S. Asikainen, and S. Kalfas. 2002. Inhibited proliferation of human periodontal ligament cells and gingival fibroblasts by Actinobacillus actinomycetemcomitans: involvement of the cytolethal distending toxin. Eur. J. Oral Sci. 110:336-373. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, W. J., W. S. Simonet, and D. L. Lacey. 2003. Osteoclast differentiation and activation. Nature 423:337-342. [DOI] [PubMed] [Google Scholar]
- 6.Choi, B. K., H. J. Lee, J. H. Kang, G. J. Jeong, C. K. Min, and Y. J. Yoo. 2003. Induction of osteoclastogenesis and matrix metalloproteinase expression by the lipooligosaccharide of Treponema denticola. Infect. Immun. 71:226-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christersson, L. A., B. Albini, J. J. Zambon, U. M. Wikesjo, and R. J. Genco. 1987. Tissue localization of Actinobacillus actinomycetemcomitans in human periodontitis. I. Light, immunofluorescence, and electron microscopic studies. J. Periodontol. 58:529-539. [DOI] [PubMed] [Google Scholar]
- 8.DiRienzo, J. M., M. Song, L. S. Wan, and R. P. Ellen. 2002. Kinetics of KB and HEp-2 cell responses to an invasive, cytolethal distending toxin-producing strain of Actinobacillus actinomycetemcomitans. Oral Microbiol. Immunol. 17:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreyfus, L. 2003. Cytolethal distending toxin, p. 257-270. In D. Burns, J. Barbieri, B. Iglewski, and R. Rappuoli (ed.), Bacterial protein toxins. ASM Press, Washington, D.C.
- 10.Firestein, G. S. 2003. Evolving concepts of rheumatoid arthritis. Nature 423:356-361. [DOI] [PubMed] [Google Scholar]
- 11.Fukushima, H., H. Kajiya, K. Takada, F. Okamoto, and K. Okabe. 2003. Expression and role of RANKL in periodontal ligament cells during physiological root-resorption in human deciduous teeth. Eur. J. Oral Sci. 111:346-352. [DOI] [PubMed] [Google Scholar]
- 12.Guthmiller, J. M., E. T. Lally, and J. Korostoff. 2001. Beyond the specific plaque hypothesis: are highly leukotoxic strains of Actinobacillus actinomycetemcomitans a paradigm for periodontal pathogenesis? Crit. Rev. Oral Biol. Med. 12:116-124. [DOI] [PubMed] [Google Scholar]
- 13.Han, X., and S. Amar. 2002. Identification of genes differentially expressed in cultured human periodontal ligament fibroblasts versus human gingival fibroblasts by DNA microarray analysis. J. Dent Res. 81:399-405. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa, T., T. Kikuiri, S. Takeyama, Y. Yoshimura, M. Mitome, H. Oguchi, and T. Shirakawa. 2002. Human periodontal ligament cells derived from deciduous teeth induce osteoclastogenesis in vitro. Tissue Cell 34:44-51. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, H., D. L. Lacey, C. R. Dunstan, I. Solovyev, A. Colombero, E. Timms, H. L. Tan, G. Elliott, M. J. Kelley, I. Sarosi, L. Wang, X. Z. Xia, R. Elliott, L. Chiu, T. Black, S. Scully, C. Capparelli, S. Morony, G. Shimamoto, M. B. Bass, and W. J. Boyle. 1999. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc. Natl. Acad. Sci. USA 96:3540-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson, A., R. Claesson, L. Hanstrom, and S. Kalfas. 2003. Serum-mediated release of leukotoxin from the cell surface of the periodontal pathogen Actinobacillus actinomycetemcomitans. Eur. J. Oral Sci. 111:209-215. [DOI] [PubMed] [Google Scholar]
- 17.Johansson, A., L. Hanstrom, and S. Kalfas. 2000. Inhibition of Actinobacillus actinomycetemcomitans leukotoxicity by bacteria from the subgingival flora. Oral Microbiol. Immunol. 15:218-225. [DOI] [PubMed] [Google Scholar]
- 18.Kanzaki, H., M. Chiba, Y. Shimizu, and H. Mitani. 2001. Dual regulation of osteoclast differentiation by periodontal ligament cells through RANKL stimulation and OPG inhibition. J. Dent. Res. 80:887-891. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, T., R. Eisen-Lev, M. Seki, J. W. Eastcott, M. E. Wilson, and M. A. Taubman. 2000. Requirement of B7 costimulation for Th1-mediated inflammatory bone resorption in experimental periodontal disease. J. Immunol. 164:2102-2109. [DOI] [PubMed] [Google Scholar]
- 20.Kelk, P., A. Johansson, R. Claesson, L. Hanstrom, and S. Kalfas. 2003. Caspase 1 involvement in human monocyte lysis induced by Actinobacillus actinomycetemcomitans leukotoxin. Infect. Immun. 71:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikuchi, T., T. Matsuguchi, N. Tsuboi, A. Mitani, S. Tanaka, M. Matsuoka, G. Yamamoto, T. Hishikawa, T. Noguchi, and Y. Yoshikai. 2001. Gene expression of osteoclast differentiation factor is induced by lipopolysaccharide in mouse osteoblasts via Toll-like receptors. J. Immunol. 166:3574-3579. [DOI] [PubMed] [Google Scholar]
- 22.Kirby, A. C., S. Meghji, S. P. Nair, P. White, K. Reddi, T. Nishihara, K. Nakashima, A. C. Willis, R. Sim, M. Wilson, et al. 1995. The potent bone-resorbing mediator of Actinobacillus actinomycetemcomitans is homologous to the molecular chaperone GroEL. J. Clin. Investig. 96:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong, Y. Y., U. Feige, I. Sarosi, B. Bolon, A. Tafuri, S. Morony, C. Capparelli, J. Li, R. Elliott, S. McCabe, T. Wong, G. Campagnuolo, E. Moran, E. R. Bogoch, G. Van, L. T. Nguyen, P. S. Ohashi, D. L. Lacey, E. Fish, W. J. Boyle, and J. M. Penninger. 1999. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature 402:304-309. [DOI] [PubMed] [Google Scholar]
- 24.Lacey, D. L., E. Timms, H. L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y. X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165-176. [DOI] [PubMed] [Google Scholar]
- 25.Lekic, P., and C. A. McCulloch. 1996. Periodontal ligament cell population: the central role of fibroblasts in creating a unique tissue. Anat. Rec. 245:327-341. [DOI] [PubMed] [Google Scholar]
- 26.Lerner, U. H. 2004. New molecules in the tumor necrosis factor ligand and receptor superfamilies with importance for physiological and pathological bone resorption. Crit. Rev. Oral Biol. Med. 15:64-81. [DOI] [PubMed] [Google Scholar]
- 27.Liu, D., J. K. Xu, L. Figliomeni, L. Huang, N. J. Pavlos, M. Rogers, A. Tan, P. Price, and M. H. Zheng. 2003. Expression of RANKL and OPG mRNA in periodontal disease: possible involvement in bone destruction. Int. J. Mol. Med. 11:17-21. [DOI] [PubMed] [Google Scholar]
- 28.Lossdorfer, S., W. Gotz, and A. Jager. 2002. Immunohistochemical localization of receptor activator of nuclear factor κB (RANK) and its ligand (RANKL) in human deciduous teeth. Calcif. Tissue Int. 71:45-52. [DOI] [PubMed] [Google Scholar]
- 29.Mandell, R. L., J. L. Ebersole, and S. S. Socransky. 1987. Clinical immunologic and microbiologic features of active disease sites in juvenile periodontitis. J. Clin. Periodontol. 14:534-540. [DOI] [PubMed] [Google Scholar]
- 30.Mayer, M. P., L. C. Bueno, E. J. Hansen, and J. M. DiRienzo. 1999. Identification of a cytolethal distending toxin gene locus and features of a virulence-associated region in Actinobacillus actinomycetemcomitans. Infect. Immun. 67:1227-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mbwana, J., H. J. Ahmed, K. Ahlman, V. Sundaeus, G. Dahlen, E. Lyamuya, and T. Lagergard. 2003. Specificity of antibodies directed against the cytolethal distending toxin of Haemophilus ducreyi in patients with chancroid. Microb. Pathog. 35:133-137. [DOI] [PubMed] [Google Scholar]
- 32.Meghji, S., M. Wilson, P. Barber, and B. Henderson. 1994. Bone resorbing activity of surface-associated material from Actinobacillus actinomycetemcomitans and Eikenella corrodens. J. Med. Microbiol. 41:197-203. [DOI] [PubMed] [Google Scholar]
- 33.Mogi, M., J. Otogoto, N. Ota, and A. Togari. 2004. Differential expression of RANKL and osteoprotegerin in gingival crevicular fluid of patients with periodontitis. J. Dent. Res. 83:166-169. [DOI] [PubMed] [Google Scholar]
- 34.Nagasawa, T., H. Kobayashi, M. Kiji, M. Aramaki, R. Mahanonda, T. Kojima, Y. Murakami, M. Saito, Y. Morotome, and I. Ishikawa. 2002. LPS-stimulated human gingival fibroblasts inhibit the differentiation of monocytes into osteoclasts through the production of osteoprotegerin. Clin. Exp. Immunol. 130:338-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nalbant, A., C. Chen, Y. Wang, and H. H. Zadeh. 2003. Induction of T-cell apoptosis by Actinobacillus actinomycetemcomitans mutants with deletion of ltxA and cdtABC genes: possible activity of GroEL-like molecule. Oral Microbiol. Immunol. 18:339-349. [DOI] [PubMed] [Google Scholar]
- 36.Nishida, E., Y. Hara, T. Kaneko, Y. Ikeda, T. Ukai, and I. Kato. 2001. Bone resorption and local interleukin-1α and interleukin-1β synthesis induced by Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis lipopolysaccharide. J. Periodontal Res. 36:1-8. [DOI] [PubMed] [Google Scholar]
- 37.Nishihara, T., Y. Ishihara, T. Koseki, E. A. Boutsi, H. Senpuku, and N. Hanada. 1995. Membrane-associated interleukin-1 on macrophages stimulated with Actinobacillus actinomycetemcomitans lipopolysaccharide induces osteoclastic bone resorption in vivo. Cytobios 81:229-237. [PubMed] [Google Scholar]
- 38.Nishihara, T., N. Ueda, K. Amano, Y. Ishihara, H. Hayakawa, T. Kuroyanagi, Y. Ohsaki, K. Nagata, and T. Noguchi. 1995. Actinobacillus actinomycetemcomitans Y4 capsular-polysaccharide-like polysaccharide promotes osteoclast-like cell formation by interleukin-1 alpha production in mouse marrow cultures. Infect. Immun. 63:1893-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okahashi, N., H. Inaba, I. Nakagawa, T. Yamamura, M. Kuboniwa, K. Nakayama, S. Hamada, and A. Amano. 2004. Porphyromonas gingivalis induces receptor activator of NF-κB ligand expression in osteoblasts through the activator protein 1 pathway. Infect. Immun. 72:1706-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshiro, T., A. Shiotani, Y. Shibasaki, and T. Sasaki. 2002. Osteoclast induction in periodontal tissue during experimental movement of incisors in osteoprotegerin-deficient mice. Anat. Rec. 266:218-225. [DOI] [PubMed] [Google Scholar]
- 41.Reddi, K., S. Meghji, M. Wilson, and B. Henderson. 1995. Comparison of the osteolytic activity of surface-associated proteins of bacteria implicated in periodontal disease. Oral Dis. 1:26-31. [DOI] [PubMed] [Google Scholar]
- 42.Reddi, K., M. Wilson, S. Poole, S. Meghji, and B. Henderson. 1995. Relative cytokine-stimulating activities of surface components of the oral periodontopathogenic bacterium Actinobacillus actinomycetemcomitans. Cytokine 7:534-541. [DOI] [PubMed] [Google Scholar]
- 43.Romas, E., M. T. Gillespie, and T. J. Martin. 2002. Involvement of receptor activator of NFκB ligand and tumor necrosis factor-alpha in bone destruction in rheumatoid arthritis. Bone 30:340-346. [DOI] [PubMed] [Google Scholar]
- 44.Sakata, M., H. Shiba, H. Komatsuzawa, T. Fujita, K. Ohta, M. Sugai, H. Suginaka, and H. Kurihara. 1999. Expression of osteoprotegerin (osteoclastogenesis inhibitory factor) in cultures of human dental mesenchymal cells and epithelial cells. J. Bone Miner. Res. 14:1486-1492. [DOI] [PubMed] [Google Scholar]
- 45.Sakuma, Y., K. Tanaka, M. Suda, A. Yasoda, K. Natsui, I. Tanaka, F. Ushikubi, S. Narumiya, E. Segi, Y. Sugimoto, A. Ichikawa, and K. Nakao. 2000. Crucial involvement of the EP4 subtype of prostaglandin E receptor in osteoclast formation by proinflammatory cytokines and lipopolysaccharide. J. Bone Miner. Res. 15:218-227. [DOI] [PubMed] [Google Scholar]
- 46.Sakurai, A., N. Okahashi, I. Nakagawa, S. Kawabata, A. Amano, T. Ooshima, and S. Hamada. 2003. Streptococcus pyogenes infection induces septic arthritis with increased production of the receptor activator of the NF-κB ligand. Infect. Immun. 71:6019-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shenker, B. J., T. McKay, S. Datar, M. Miller, R. Chowhan, and D. Demuth. 1999. Actinobacillus actinomycetemcomitans immunosuppressive protein is a member of the family of cytolethal distending toxins capable of causing a G2 arrest in human T cells. J. Immunol. 162:4773-4780. [PubMed] [Google Scholar]
- 48.Simonet, W. S., D. L. Lacey, C. R. Dunstan, M. Kelley, M. S. Chang, R. Luthy, H. Q. Nguyen, S. Wooden, L. Bennett, T. Boone, G. Shimamoto, M. DeRose, R. Elliott, A. Colombero, H. L. Tan, G. Trail, J. Sullivan, E. Davy, N. Bucay, L. Renshaw-Gegg, T. M. Hughes, D. Hill, W. Pattison, P. Campbell, W. J. Boyle, et al. 1997. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89:309-319. [DOI] [PubMed] [Google Scholar]
- 49.Slots, J., and M. Ting. 1999. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol. 2000. 20:82-121. [DOI] [PubMed] [Google Scholar]
- 50.Sugai, M., T. Kawamoto, S. Y. Peres, Y. Ueno, H. Komatsuzawa, T. Fujiwara, H. Kurihara, H. Suginaka, and E. Oswald. 1998. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitans is a cytolethal distending toxin. Infect. Immun. 66:5008-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taichman, N. S., R. T. Dean, and C. J. Sanderson. 1980. Biochemical and morphological characterization of the killing of human monocytes by a leukotoxin derived from Actinobacillus actinomycetemcomitans. Infect. Immun. 28:258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taubman, M. A., and T. Kawai. 2001. Involvement of T-lymphocytes in periodontal disease and in direct and indirect induction of bone resorption. Crit. Rev. Oral Biol. Med. 12:125-135. [DOI] [PubMed] [Google Scholar]
- 53.Teitelbaum, S. L., and F. P. Ross. 2003. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4:638-649. [DOI] [PubMed] [Google Scholar]
- 54.Teng, Y. T. 2002. Mixed periodontal Th1-Th2 cytokine profile in Actinobacillus actinomycetemcomitans-specific osteoprotegerin ligand (or RANK-L)-mediated alveolar bone destruction in vivo. Infect. Immun. 70:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teng, Y. T., H. Nguyen, X. Gao, Y. Y. Kong, R. M. Gorczynski, B. Singh, R. P. Ellen, and J. M. Penninger. 2000. Functional human T-cell immunity and osteoprotegerin ligand control alveolar bone destruction in periodontal infection. J. Clin. Investig. 106:R59-R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai, C. C., W. P. McArthur, P. C. Baehni, C. Evian, R. J. Genco, and N. S. Taichman. 1981. Serum neutralizing activity against Actinobacillus actinomycetemcomitans leukotoxin in juvenile periodontitis. J. Clin. Periodontol. 8:338-348. [DOI] [PubMed] [Google Scholar]
- 57.Tsuji, K., K. Uno, G. X. Zhang, and M. Tamura. 2004. Periodontal ligament cells under intermittent tensile stress regulate mRNA expression of osteoprotegerin and tissue inhibitor of matrix metalloprotease-1 and -2. J. Bone Miner. Metab. 22:94-103. [DOI] [PubMed] [Google Scholar]
- 58.Ueda, N., T. Nishihara, Y. Ishihara, K. Amano, T. Kuroyanagi, and T. Noguchi. 1995. Role of prostaglandin in the formation of osteoclasts induced by capsular-like polysaccharide antigen of Actinobacillus actinomycetemcomitans strain Y4. Oral Microbiol. Immunol. 10:69-75. [DOI] [PubMed] [Google Scholar]
- 59.Valverde, P., T. Kawai, and M. A. Taubman. 2004. Selective blockade of voltage-gated potassium channels reduces inflammatory bone resorption in experimental periodontal disease. J. Bone Miner. Res. 19:155-164. [DOI] [PubMed] [Google Scholar]
- 60.Wang, Y., S. D. Goodman, R. J. Redfield, and C. Chen. 2002. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol. 184:3442-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson, M., S. Kamin, and W. Harvey. 1985. Bone resorbing activity of purified capsular material from Actinobacillus actinomycetemcomitans. J. Periodontal Res. 20:484-491. [DOI] [PubMed] [Google Scholar]
- 62.Wising, C., L. A. Svensson, H. J. Ahmed, V. Sundaeus, K. Ahlman, I. M. Jonsson, L. Molne, and T. Lagergard. 2002. Toxicity and immunogenicity of purified Haemophilus ducreyi cytolethal distending toxin in a rabbit model. Microb. Pathog. 33:49-62. [DOI] [PubMed] [Google Scholar]
- 63.Yamano, R., M. Ohara, S. Nishikubo, T. Fujiwara, T. Kawamoto, Y. Ueno, H. Komatsuzawa, K. Okuda, H. Kurihara, H. Suginaka, E. Oswald, K. Tanne, and M. Sugai. 2003. Prevalence of cytolethal distending toxin production in periodontopathogenic bacteria. J. Clin. Microbiol. 41:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]