Abstract
The blood thinner warfarin has a narrow therapeutic range and high inter- and intra-patient variability in therapeutic doses. Several studies have shown that pharmacogenomic variants help predict stable warfarin dosing. However, retrospective and randomized controlled trials that employ dosing algorithms incorporating pharmacogenomic variants under perform in African Americans. This study sought to determine if: 1) including additional variants associated with warfarin dose in African Americans, 2) predicting within single ancestry groups rather than a combined population, or 3) using percentage African ancestry rather than observed race, would improve warfarin dosing algorithms in African Americans. Using BioVU, the Vanderbilt University Medical Center biobank linked to electronic medical records, we compared 25 modeling strategies to existing algorithms using a cohort of 2,181 warfarin users (1,928 whites, 253 blacks). We found that approaches incorporating additional variants increased model accuracy, but not in clinically significant ways. Race stratification increased model fidelity for African Americans, but the improvement was small and not likely to be clinically significant. Use of percent African ancestry improved model fit in the context of race misclassification.
1. Introduction
Warfarin is a commonly used anticoagulant with a narrow therapeutic index and high rate of significant adverse reactions from both over- and under-dosing.1 A number of pharmacogenomic variants are associated with stable warfarin dose,2 and many studies have developed dosing algorithms using these variants.1,3 Genotype-guided dosing is part of the United States Food and Drug Association (FDA) product label for warfarin.
The two largest randomized controlled trials of pharmacogenomic-guided warfarin dosing, EU-PACT4 and COAG5, yielded discordant findings on the clinical utility of incorporating pharmacogenomics into current dosing strategies. The EU-PACT study showed significantly increased percent time in therapeutic range (PTTR) over 12 weeks for the pharmacogenomic group while the COAG trial did not see a significant difference in PTTR over a 4-week time period. One of the reasons highlighted for these inconsistent findings across trials was the higher frequency of African descent individuals in COAG (27%) compared to EU-PACT (0.9%).6 In COAG, African Americans with genotype-guided dosing spent an average of 8% less time in therapeutic range than the clinical algorithm group. Studies have shown that the CYP2C9*2/*3 variants used by both COAG and EU-PACT are less frequent among those of African descent.7 There are also variants important for dosing among individuals of African descent alleles that were unaccounted for in these trials.7–11 Drozda found that failing to take into account these expanded variants resulted in significantly worse dose predictions among African Americans.12 Additionally, Limdi found that using a race stratified dosing approach resulted in significantly more dose variation explained in both whites and blacks compared to a race-combined dosing model.13
Although much work has been conducted in this area, there remain outstanding questions that need to be answered. For example, because the algorithm proposed by Drozda was developed only in African Americans, its generalizability to individuals of European descent is unknown. Additionally, clinical dosing algorithms using a stratified approach, as advocated by Limdi have not been robustly tested to determine clinical validity. Further, in other clinical predictive models, using percent African ancestry as a more nuanced and biologically accurate measure of race provided better predictive performance than categorical race.14 This study seeks to expand on previous warfarin dosing algorithm development efforts within Vanderbilt’s EMR-linked biobank15 to account for new variants associated with warfarin dose in African Americans. Additionally, we investigate whether race-stratified models or models using percent African ancestry result in clinically significant improvements (≥0.5–1mg/day) in dose prediction accuracy.
2. Methods
2.1. Study Population
Using BioVU, the Vanderbilt University biobank linked to electronic medical records (EMR), we selected all adult patients (≥18 years old) with DNA available who also had warfarin mentioned in the active prescription section of their problem list or a note from one of the hospital’s anticoagulation clinics as of July of 2015. We used two approaches to extract stable warfarin dose based on whether the patient’s warfarin was managed by an anticoagulation clinic or an individual physician.
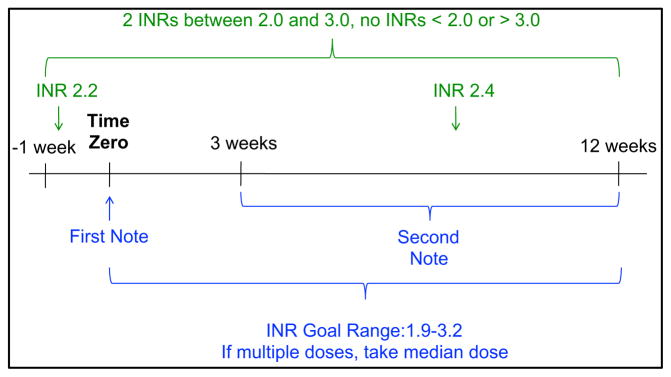
We used a previously published and validated algorithm15 to extract stable warfarin dose from patients with their dose managed by a Vanderbilt anticoagulation clinic or, for a subset of African Americans, where the dose was managed by their primary care provider. This approach identifies stable warfarin dose windows, as summarized in Figure 1. A stable dose window is defined as the presence of two or more notes from the anticoagulation clinic (or problem list entries for those managed by a primary-care provider) at least three, but not more than 12, weeks apart. During this time (from 7 days before the first note through the second note) the patient must also have two or more International Normalized Ratio (INR) measurements at least one day apart and all INR measurements in the window must be between 2 and 3. For anticoagulation clinic patients the INR goal range at the time of the stable dose window was required to be between 1.9–3.2. Primary-care managed patients were assumed to have an INR goal range of 2–3 unless otherwise specified (where ranges outside of 1.9–3.2 resulted in exclusion from the study). Warfarin dose was extracted from every anticoagulation clinic note in the window using regular expressions. The first window with identical prescribed warfarin doses throughout the window was selected as the stable warfarin dose. Patients lacking a window with identical warfarin doses throughout the window were manually reviewed to confirm accurate dose extraction. If multiple doses were prescribed during the window, the median dose was used. All primary care managed patient records were manually reviewed to extract warfarin dose and verify INR goal range because problem lists are susceptible to copy/paste redundancies and computational extraction may be invalid.
Figure 1.
Stable Warfarin Dose Window Algorithm
Clinical covariates influencing stable warfarin dose were extracted with a variety of methods. Concomitant therapies (amiodarone, carbamazepine, phenytoin, and rifampin) listed in the problem list before or during the dose window were manually reviewed to confirm the prescriptions were active during the window. Smoking status was identified combination of natural language processing (NLP) and tobacco use International Classification of Disease version 9 (ICD9) codes,16,17 followed by manual review to confirm active smoking at the time of the stable dose window. Body surface area,18 was calculated using the median height and weight across the stable dose window or the closest height and weight measurement available within 3–6 months before or after the window (extracted via manual review). Age was defined as the age at the first anticoagulation clinic note or problem list warfarin entry in the stable dose window. “EMR recorded race” is defined by the care provider, but has shown concordance with genetic ancestry.19 Indication for warfarin treatment, blood clots (i.e., deep venous thrombosis [DVT] or pulmonary embolism[PE]) or atrial fibrillation, was determined through ICD9 codes.20,21
2.2. Genotyping
This study genotyped twenty-one single nucleotide polymorphisms (SNPs) that had ever been associated with warfarin dose in European or African-descent populations and recorded in the Pharmacogenomics Knowledge Base (PharmGKB, www.pharmgkb.org).22 Three variants (rs9923231, rs1799853, rs1057910) were genotyped using a Taqman assay by the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core. A subset of white subjects had previous genotyping for these variants on the Illumina ADME assay and were not included in the Taqman assay. The remaining 17 variants were genotyped across the entire study population with a Sequenom assay performed by the VANTAGE core. Genotyping data were checked for marker efficiency and samples removed if they were missing one or more genotype calls for the tested variants. Duplicates and HapMap controls were validated.
We used existing genotyping data to calculate percent African ancestry across a subset of the population. Individuals were genotyped on one or more of the following platforms: Illumina Exome Beadchip, Human Omni Express Exome v2, Metabochip and/or OmniQuad. For each platform independently, samples with discrepant genders or sample efficiency <99% were removed. Markers with genotyping efficiencies < 99% or minor allele frequencies<5% were dropped. For the Exome chip, thresholds were set to 97% and 98% for genotyping and sample efficiency respectively as has been done previously to account for low frequency variants.23 Within each platform, percent African ancestry was calculated using the ADMIXTURE supervised learning method with HapMap Phase III CEU and YRI reference populations.24 The median estimate was used for individuals genotyped on multiple platforms.
2.3. Analysis
We fit and tested 25 different dosing models, combing 5 genetic modeling strategies (including exclusion of genetics altogether) with 5 different methods of race/ancestry adjustment. A summary of the 25 models tested are presented in Table 1. For race-stratified models, variants that were monomorphic or non-varying clinical factors were not included. To validate model summaries and prevent overfitting, we bootstrapped 1000 samples with replacement, trained a generalized linear model on each bootstrap, and tested the original dataset against each model. We calculated the mean absolute error (in mg/week) and R2 for each bootstrap model, then calculated the median and an empiric confidence interval using the 2.5th and 97.5th percentiles of the bootstrap summaries. For all combined race models, we calculated these evaluation criteria across the entire test population and then within each EMR recorded race group separately. Because there are different risks for over- and under-dosing, we also calculated these summary evaluation criteria stratified by low (<21mg/week), medium (21–49mg/week), and high (>49mg/week) stable dose across the entire test population and then within each EMR recorded race separately. To evaluate the validity of our models and compare to existing algorithms, we also calculated mean absolute error and R2 for a number of existing algorithms. The algorithms tested are summarized in Table 2.
Table 1.
Overview of Dose Prediction Models Tested
| Genetic Model
|
|||||
|---|---|---|---|---|---|
| Clinical Only | Limited Genetic | Expanded Genetic | Combined SNP | Haplotype | |
| Clinical Vars. (All) | Age (in decades); Body surface area; Smoking status; Amiodarone; Enzyme inducer | ||||
| Race Adj. (one of:) | 1) Unadjusted 2) EMR Race 3) %African Ancestry 4) White Only 5) Black Only |
||||
| Genetic Variables | - | VKORC1-1639 | VKORC1-1639 | VKORC1-1639 | VKORC1-1639 |
| CYP2C9*2 | CYP2C9*2 | CYP2C9*2 | rs2359612 | ||
| CYP2C9*3 | CYP2C9*3 | CYP2C9*3 | rs2884737 | ||
| CYP2C9*5 | rs7200749 | rs7200749 | |||
| CYP2C9*6 | rs9934438 | rs8050894 | |||
| CYP2C9*8 | rs17886199 | rs9934438 | |||
| CYP2C9*11 | rs10871454 | rs17886199 | |||
| rs2359612 | rs2108622 | rs10871454 | |||
| rs2884737 | rs11676382 | rs2108622 | |||
| rs7200749 | rs12714145 | rs11676382 | |||
| rs8050894 | rs339097 | rs12714145 | |||
| rs9934438 | rs12777823 | rs339097 | |||
| rs17886199 | VKORC1 Other1 | rs12777823 | |||
| rs10871454 | CYP2C9 Other2 | CYP2C9*1/*2 | |||
| rs2108622 | CYP2C9*1/*3 | ||||
| rs11676382 | CYP2C9*2/*2 | ||||
| rs12714145 | CYP2C9*2/*3 | ||||
| rs339097 | CYP2C9*3/*3 | ||||
| rs12777823 | CYP2C9 Other Het.3 | ||||
| CYP2C9 Other Hom.4 | |||||
If individual carries one or more minor allele at rs2359612 or rs2884737 or rs61162043 or rs8050894 then called 1, else 0;
If individual carries one or more minor allele at CYP2C9 *5/*6/*8 or *11 then call 1, else 0;
CYP2C9 *1/*11, *1/*5, *1/*6, *1/*8);
CYP2C9 *3/*8, *5/*8, *5/*11, *8/*8, *8/*11
Table 2.
Summary of Previous Algorithms Tested for Warfarin Dosing
| Algorithm | Clinical Predictors | Genetic Predictors | Notes |
|---|---|---|---|
| Fixed 35mg Weekly Dose |
- | - | - |
| FDA Dosing Table1 | - | VKORC1-1639 | Used mean of dosing range given. |
| CYP2C9*2 | |||
| CYP2C9*3 | |||
| IWPC (International Warfarin Pharmacogenetics Consortium)2 | Age (in decades) | VKORC1-1639 | - |
| Height | CYP2C9*2 | ||
| Weight | CYP2C9*3 | ||
| Asian | |||
| African American | |||
| Amiodarone | |||
| Enzyme Inducers | |||
| Ramirez et. al.3 | Age (in years) | VKORC1-1639 | - |
| Race | CYP2C9*2 | ||
| Sex | CYP2C9*3 | ||
| Body Surface Area | CYP2C9*6 | ||
| Smoking Status | CYP2C9*8 | ||
| DVT/PE | rs2108622 | ||
| Atrial Fibrillation | rs339097 | ||
| Hernandez et. al.4 | Age (in years) | VKORC1-1639 | Performed on subset of population with genotyping for rs61162043. Missing CYP2C9 rs7089580 due to probe failure. Set all patients to reference allele |
| Weight | VKORC1, | ||
| DVT/PE | rs61162043 | ||
| CYP2C9*2 | |||
| CYP2C9*3 | |||
| CYP2C9*5 | |||
| CYP2C9*8 | |||
| CYP2C9*11 | |||
| rs7089580 | |||
| rs12777823 |
Klein et. al. NEJM. 2009;
Ramirez et. al. Future Medicine. 2010;
Hernandez et. al. The Pharmacogenomics Journal. 2014.
3. Results
A total of 3,498 patients (3188 whites, 310 blacks) had a stable dose window (all features in Figure 1, except INR goal range filtering) and were genotyped on the Sequenom platform. Of these, 596 whites had VKORC1-1369 and CYP2C9*2/*3 genotypes from the ADME platform, all other individuals were genotyped via Taqman. 291 individuals were missing one or more genotypes (with exceptions of rs7089580 and rs61162043 due to poor probe performance described below) and were removed from the analysis. Of the remaining 3,207 individuals 2,419 (2,192 whites and 227 blacks) had warfarin managed by the anticoagulation clinic. Filtering this population for INR goal ranges between 1.9–3.2 removed a further 233 individuals (212 whites, 21 blacks). Manual review to confirm stable warfarin dose, height and/or weight was performed for 203 whites and 28 blacks. This review removed 52 whites and 9 blacks for missing warfarin dose, height and/or weight. A total of 56 black individuals had warfarin managed by their primary care provider and were manually reviewed to extract warfarin dose and INR goal range. Combining the anticoagulation clinic and primary care populations yielded a final population of 2,181 individuals (1,928 whites, and 253 blacks).
Population demographics are presented in Table 3. Blacks had higher warfarin doses (40.8 vs 35mg/week), were younger (60 vs 66 years), were more likely to be current smokers (16% vs 8%), were more likely to be on anticoagulants due to thromboembolic events (30.4% vs 17.5%), and less likely to be on anticoagulants due to atrial fibrillation (59% vs. 75%) than whites. All other demographics factors were similar between blacks and whites.
Table 3.
Population Demographics
| Combined (n = 2181) | Whites (n = 1928) | Blacks (n = 253) | |
|---|---|---|---|
| Weekly Warfarin Dose, mg/wk (median, sd) | 35.0 (±17.6) | 35.0 (±17.0) | 40.8 (± 19.9) |
| Age, years (mean, sd) | 66 (± 15) | 66 (± 15) | 60 (± 16) |
| Female (n, %) | 911 (41.8%) | 784 (40.7%) | 127 (50.2%) |
| African American (n, %) | 253 (11.6%) | - | - |
| % African Ancestry (median, sd)1 | 0.99 (± 31) | 0.65 (± 4.5) | 81.6 (± 10.3) |
| Height, cm (median, sd) | 173 (± 13.5) | 174 (±13.0) | 170 (±16.1) |
| Weight, kg (median, sd) | 89 (± 24.0) | 88 (± 23.9) | 91 (±24.7) |
| Body Surface Area, m2 (median, sd) | 2.0 (± 0.29) | 2.0 (± 0.29) | 2.0 (± 0.30) |
| Current Smokers (n, %) | 209 (9.6%) | 168 (8.7%) | 41 (16.2%) |
| Amiodarone (n, %) | 229 (10.5%) | 202 (10.5%) | 27 (10.7%) |
| Enzyme Inducers (n, %) | 20 (0.92%) | 15 (0.78%) | 5 (1.98%) |
| Indication | |||
| VTE (n, %) | 414 (19.0%) | 337 (17.5%) | 77 (30.4%) |
| Atrial Fibrillation (n, %) | 1592 (73%) | 1443 (75%) | 149 (59%) |
%-African ancestry available for 987 individuals (808 whites, 179 blacks)
One marker, rs7089580, failed genotyping in the Sequenom pool. Genotyping efficiency rates and minor allele frequencies are presented for the remaining 20 variants in Table 4. One variant, rs61162043 had lower genotyping efficiency (failed genotyping in 111 whites and 21 blacks) and was excluded from the expanded variants model. However, this variant was included in the VKORC1 combined variable for the Combined Variant model. A summary of the frequency of observed diplotypes for CYP2C9 is presented Table 5. The majority of both racial/ethnic populations had a *1/*1 diplotype. Homozygotes and compound heterozygotes for the *2 and *3 variants (i.e., *2/*2, *3/*3, and *2/*3) were only observed in whites. Homozygotes and compound heterozygotes of the less common *5, *6, *8, and *11 alleles were only observed in blacks.
Table 4.
Genotyping Quality Control and Minor Allele Frequencies
| Gene | SNP | Minor Allele | Call Ratea | Minor Allele Frequency (%)
|
||
|---|---|---|---|---|---|---|
| Combined (n=2181) | Whites (n=1928) | Blacks (n=253) | ||||
| VKORC1 | rs9923231 | T | 99.79b | 35.1 | 38.3 | 10.5 |
| VKORC1 | rs2359612 | A | 100 | 36.4 | 38.5 | 20.8 |
| VKORC1 | rs2884737 | C | 99.96 | 23.3 | 25.3 | 3.8 |
| VKORC1 | rs61162043 | A | 93.82 | 37.2 | 35.8 | 49.6 |
| VKORC1 | rs7200749 | A | 99.96 | 2.6 | 0.3 | 20.2 |
| VKORC1 | rs8050894 | G | 99.92 | 37.3 | 38.8 | 26.5 |
| VKORC1 | rs9934438 | A | 99.96 | 35.1 | 38.4 | 10.5 |
| VKORC1 | rs17886199 | G | 100 | 0.5 | 0 | 4.2 |
| STX4 | rs10871454 | T | 99.96 | 35.3 | 38.5 | 10.9 |
| CYP2C9*2 | rs1799853 | T | 99.79b | 13.5 | 14.9 | 2.4 |
| CYP2C9*3 | rs1057910 | C | 99.95b | 6.1 | 6.7 | 2.0 |
| CYP2C9*5 | rs28371686 | G | 100 | 0.2 | 0.05 | 1.6 |
| CYP2C9*6 | rs9332131 | del | 99.79 | 0.2 | 0 | 1.4 |
| CYP2C9*8 | rs7900194 | A | 100 | 0.9 | 0.03 | 7.3 |
| CYP2C9*11 | rs28371685 | T | 99.96 | 0.4 | 0.3 | 1.4 |
| CYP4F2 | rs2108622 | T | 100 | 28.2 | 30.4 | 11.3 |
| GGCX | rs11676382 | G | 99.96 | 8.8 | 9.7 | 2.6 |
| GGCX | rs12714145 | T | 100 | 42.1 | 41.6 | 45.8 |
| CALU | rs339097 | G | 99.83 | 1.6 | 0.2 | 12.3 |
| CYP2C-cluster | rs12777823 | A | 99.92 | 16.1 | 14.6 | 28.1 |
Call rates for completed genotyped population – not the final study population (as valid genotypes required for all but rs61162043);
Call rate for Taqman group only. ADME QC according to typical procedures.
Table 5.
CYP2C9 Diplotype Frequencies
| CYP2C9 Haplotype | Combined (n = 2181) | Whites (n = 1928) | Blacks (n = 253) |
|---|---|---|---|
| *1/*1 | 1402 (64.3%) | 1222 (63.4%) | 180 (71.2%) |
| *1/*2 | 357 (16.4%) | 345 (18%) | 12 (4.8%) |
| *1/*3 | 214 (9.8%) | 205 (10.6%) | 9 (3.6%) |
| *1/*5 | 8 (0.4%) | 2 (0.1%) | 6 (2.4%) |
| *1/*6 | 7 (0.3%) | - | 7 (2.8%) |
| *1/*8 | 28 (1.3%) | 1 (0.1%) | 27 (10.7%) |
| *1/*11 | 15 (0.7%) | 11 (0.6%) | 4 (1.6%) |
| *2/*2 | 100 (4.6%) | 100 (5.2%) | - |
| *2/*3 | 31 (1.4%) | 31 (1.6%) | - |
| *3/*3 | 11 (0.5%) | 11 (0.6%) | - |
| *3/*8 | 1 (<0.1%) | - | 1 (0.4%) |
| *5/*8 | 1 (<0.1%) | - | 1 (0.4%) |
| *5/*11 | 1 (<0.1%) | - | 1 (0.4%) |
| *8/*8 | 3 (0.1%) | - | 3 (1.2%) |
| *8/*11 | 2 (0.1%) | - | 2 (0.8%) |
Within our final study population, 978 individuals (800 whites and 178 blacks) had genome-wide data available. A total of 764 individuals were genotyped on two platforms, 98 had genotyped data from three platforms, and 5 individuals had genotyping on four platforms. Of these individuals, the majority (n=437) had a maximum difference of less than 1% between estimates across platforms. Only 7 individuals had estimates across platforms that differed by more than 5% (maximum range of 9.8%). Three individuals had an EMR-recorded race of white, but had more than 50% African ancestry. The median ancestry estimate was used for all analyses.
A summary of the mean absolute error and percent variation explained (R2) for all twenty-five fitted models, as well as the performance of existing dosing algorithms are provided in Table 6. Comparing all new and existing algorithms, the Expanded Genetic unadjusted, Expanded Genetic EMR recorded race adjusted, Haplotype unadjusted, and Haplotype EMR recorded race adjusted models had the lowest mean absolute error across the combined population, with the Haplotype models explaining slightly more dose variance (54.4% vs 54.1%). The Expanded Variant model with percent ancestry adjustment had the lowest mean absolute errors in whites, and the Expanded Genetic stratified model had the lowest mean absolute error in blacks.
Table 6.
Performance of Predictive Dosing Algorithms
| Algorithm | Mean Absolute Error (mg/week) Median (95% Confidence Interval) |
Percent Variation Explained (R2) Median (95% Confidence Interval) |
||||
|---|---|---|---|---|---|---|
| Combined | Whites | Blacks | Combined | Whites | Blacks | |
| Existing Algorithms | ||||||
|
| ||||||
| Fixed 35 mg/week | 13.5 | 13.2 | 16.1 | −2.3 | −1.1 | −23.7 |
|
| ||||||
| US FDA Table mid-range | 12.0 | 11.7 | 14.7 | 17.5 | 18.3 | 1.1 |
|
| ||||||
| IWPC | 9.5 | 9.0 | 13.4 | 42.7 | 45.2 | 20 |
|
| ||||||
| Ramirez et. al. | 9.2 | 8.8 | 12.9 | 47.5 | 49.5 | 28.7 |
|
| ||||||
| Hernandez et. al. | 10.4 | 9.9 | 14.2 | 37.3 | 39.4 | 17.2 |
|
| ||||||
| New Models | ||||||
|
| ||||||
| Clinical | ||||||
|
| ||||||
| Unadjusted | 12.0 (11.9–12.0) | 11.7 (11.7–11.8) | 14.0 (13.8–14.2) | 20.3 (19.9–20.5) | 19.6 (19.0–19.9) | 12.3 (9.9–14.9) |
|
| ||||||
| Race Adjusted | 11.9 (11.9–11.9) | 11.7 (11.7–11.7) | 13.6 (13.4–13.8) | 21.5 (21.1–21.6) | 19.8 (19.3–20.0) | 20.8 (18.9–22.1) |
|
| ||||||
| % Ancestry Adjusted | 11.5 (11.5–11.7) | 10.9 (10.8–11.0) | 14.5 (14.3–14.9) | 23.8 (22.9–24.2) | 20.6 (19.3–21.3) | 21.7 (17.9–24.2) |
|
| ||||||
| Race Stratified | - | 11.7 (11.7–11.7) | 13.4 (13.3–13.7) | - | 19.8 (19.4–20.0) | 21.7 (18.0–23.1) |
|
| ||||||
| Limited Genetic | ||||||
|
| ||||||
| Unadjusted | 9.3 (9.2–9.3) | 8.8 (8.8–8.8) | 12.9 (12.8–13.1) | 51.5 (51.2–51.6) | 53.8 (53.4–54.1) | 27.8 (25.4–29.7) |
|
| ||||||
| Race Adjusted | 9.3 (9.2–9.3) | 8.8 (8.8–8.8) | 12.8 (12.7–13.0) | 51.8 (51.5–52.0) | 54.0 (53.6–54.2) | 30.0 (27.8–31.2) |
|
| ||||||
| % Ancestry Adjusted | 9.5 (9.4–9.5) | 8.5 (8.4–8.6) | 13.7 (13.5–14.0) | 49.8 (49.0–50.2) | 52.8 (51.5–53.4) | 32.4 (28.9–34.7) |
|
| ||||||
| Race Stratified | - | 8.8 (8.8–8.8) | 12.7 (12.5–13) | - | 54.0 (53.7–54.2) | 30.9 (27.1–32.5) |
|
| ||||||
| Expanded Genetic | ||||||
|
| ||||||
| Unadjusted | 9.0 (9.0–9.1) | 8.6 (8.6–8.7) | 12.2 (11.9–12.6) | 54.1 (53.6–54.4) | 55.4 (54.9–55.7) | 37.7 (34.0–40.0) |
|
| ||||||
| Race Adjusted | 9.0 (9.0–9.1) | 8.6 (8.6–8.7) | 12.2 (11.9–12.6) | 54.1 (53.5–54.4) | 55.4 (54.9–55.8) | 37.5 (33.5–40.1) |
|
| ||||||
| % Ancestry Adjusted | 9.2 (9.1–9.3) | 8.4 (8.3–8.5) | 12.9 (12.5–13.4) | 52.5 (50.8–53.3) | 54.2 (52.5–55.1) | 39.7 (32.9–43.8) |
|
| ||||||
| Race Stratified | - | 8.6 (8.6–8.7) | 11.9 (11.6–12.4) | - | 55.7 (55.2–55.9) | 39.5 (33.8–42.5) |
|
| ||||||
| Combined SNP | ||||||
|
| ||||||
| Unadjusted | 9.9 (9.9–10.0) | 9.5 (9.5–9.6) | 12.8 (12.6–13.1) | 44.0 (43.5–44.3) | 44.7 (44.2–45.0) | 30.2 (27.2–32.5) |
|
| ||||||
| Race Adjusted | 9.9 (9.9–10.0) | 9.6 (9.5–9.6) | 12.8 (12.6–13.1) | 44.0 (43.5–44.3) | 44.7 (44.2–45.1) | 30.3 (27.0–32.6) |
|
| ||||||
| % Ancestry Adjusted | 10 (9.9–10.1) | 9.2 (9.1–9.3) | 13.6 (13.3–14.0) | 44.4 (43.3–45.0) | 44.8 (43.2–45.8) | 34.2 (29.5–37.9) |
|
| ||||||
| Race Stratified | - | 9.6 (9.5–9.6) | 12.5 (12.2–12.9) | - | 44.9 (44.3–45.1) | 33.9 (29.6–36.4) |
|
| ||||||
| Haplotype | ||||||
|
| ||||||
| Unadjusted | 9.0 (9.0–9.1) | 8.6 (8.6–8.7) | 12.1 (11.8–12.5) | 54.4 (54.0–54.7) | 55.8 (55.3–56.1) | 38.0 (34.5–40.1) |
|
| ||||||
| Race Adjusted | 9.0 (9.0–9.1) | 8.6 (8.6–8.7) | 12.1 (11.9–12.5) | 54.4 (53.9–54.7) | 55.8 (55.3–56.1) | 37.8 (34.3–40.2) |
|
| ||||||
| % Ancestry Adjusted | 9.2 (9.1–9.3) | 8.4 (8.3–8.5) | 12.7 (12.4–13.3) | 53.1 (51.6–53.9) | 54.6 (52.3–55.5) | 41.5 (36.1–44.5) |
|
| ||||||
| Race Stratified | - | 8.6 (8.5–8.6) | 12.0 (11.7–12.5) | - | 56.2 (55.7–56.4) | 39.6 (34.6–42.1) |
|
| ||||||
Bold and shaded cells indicate the best performing algorithm for each population.
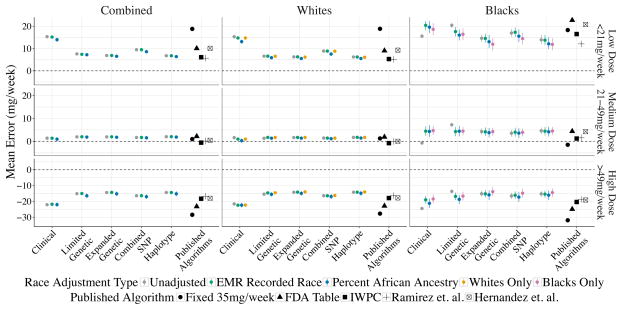
The algorithm performance with respect to mean error within low, medium, and high weekly dose groups are presented in Figure 2. When broken down by dose range 362 individuals (336 white and 26 black) had low warfarin requirements (<21mg/week), 1,313 individuals (1,173 whites and 140 blacks) had moderate warfarin requirements (21–49mg/week), and 486 individuals (402 whites and 84 blacks) had high warfarin requirements (>49mg/week). Within the medium dose requirement group (60% of the study population), dose predictions in whites were less than 5mg/week overestimated, while dose predictions in blacks were ~5mg/week overestimated. For the 17% of individuals with low warfarin dose requirements, mean dosing error was <10mg/week overestimated in whites, and 10–20mg/week overestimated in African Americans. The existing algorithm with the best performance among low-dose requiring African Americans was Ramirez et. al. (overestimating warfarin dose by 11.6mg/week). Within the high dose requirement individuals (22%), all races were consistently underestimated by 10–20mg/week.
Figure 2. Performance of Dosing Algorithms by Stable Dose Range.
This figure shows the algorithm performance (mean error in mg/week) divided by EHR recorded race and the stable dose range, e.g. patient’s stable warfarin dose is a low weekly dose (<21mg/week), medium weekly dose (21–49mg/week), or high weekly dose (>49mg/week). Mean errors greater than 0 indicate over dosing, while mean errors less than 0 indicate underdosing.
4. Discussion
The goal of this study was to: 1) account for variants associated with warfarin dose in African Americans, 2) investigate whether race-stratified dosing leads to clinically significant improved dose predictions, 3) investigate whether race adjustment using percent ancestry offers improved prediction accuracy compared to EMR recorded race. The last goal was predicated on a study of lung function predictions (a continuous trait that, like warfarin dose, differs by race) that found improved model fit when they included percent African ancestry.14 This hypothesis was bolstered by a study among Caribbean Hispanics that found adjusting for admixture improved warfarin dose prediction.25
Although this study required that individuals have DNA available in our biobank, because we took a complete cross-section of all individuals with warfarin exposure and DNA, the relative percentage of African Americans in this study (~10%) is consistent the broader Vanderbilt clinical population. As previously observed in the literature,13 our black study population had a higher incidence of DVT/PE as an indication for anticoagulation. The genetics of our population were consistent with expected allele frequencies from the HapMap populations, with African Americans having allele frequencies lying between the Yoruba in Ibadan, Nigeria (YRI) and African Americans in the Southwest USA (ASW). Ancestry estimates for the black population were as expected with African Americans having approximately 80% African ancestry,26 and allele frequencies for CYP2C9*2/*3 and VKORC1-1639 were consistent with other studies within the Vanderbilt clinical population (that are not necessarily part of the biobank population).27 Importantly, CYP2C9 *2 and *3 homozygotes and compound heterozygotes were only observed in our white population, lending support to the notion that use of only CYP2C9*2/*3 for warfarin dosing algorithms may be insufficient for African Americans.
Examining algorithm performance over the entire study population, the inclusion of additional variants associated with warfarin dose did increase dosing accuracy (mean absolute error) and percentage of dose variation explained for the combined, white and black populations. In all three populations one of the novel algorithms using SNPs independently (Expanded Genetic) or combined by CYP2C9 diplotype (Haplotype) outperformed existing algorithms, the Clinical, and the Limited Genetic models. When considering confidence intervals, the Expanded Genetic and Haplotype models performed at similar levels across all populations. This is important for future clinical implementation as algorithms such as MyDrugGenome use CYP2C9 diplotype. These diplotypes do not always have unambiguous assignments and are subject to change as the number of known genetic variants in a gene rise.28 Our results suggest that algorithms utilizing unique SNPs can perform at similar levels to those using diplotypes and may be preferable due their more stable identification.
When considering only mean absolute error, stratified dosing models outperformed combined models only in African Americans. Interesting, stratified dosing did not result in improved performance over combined models in whites. This may be due to race misclassification of the three individuals recorded as white in the EMR, but who nevertheless had greater than 50% African ancestry. We chose not to manually change these individuals’ race, as this misclassification is a real, generalizable14 problem in the clinic, and would have an effect on algorithms’ accuracy if clinically deployed. Although stratified dosing did improve algorithm performance among African Americans, it did not increase percent of warfarin dose explained by the model as has been seen in other studies.13
Correcting for race with percent ancestry yielded interesting results. Within the clinical model, percent ancestry improved model fit (lower mean absolute error, higher R2) in the combined population, but not when pharmacogenomic markers were added into the model. Interestingly, percent ancestry improved dosing among whites across all models including those with pharmacogenomic markers. It is possible that the race misclassification also affected the algorithms using percent African ancestry. While this misclassification would be an important limitation in clinical implementation, at the current time this is less important because genetic ancestry is typically unavailable in current clinical systems. However, should this information have increased clinical utility in the future, panel testing of ancestry informative markers could enable implementation of these data.
While the algorithms developed in this study outperformed existing algorithms when considering the mean absolute error of prediction, we advocate using Figure 2 to evaluate algorithm performance for desired implementation. We also caution that to determine the overall “best” algorithm, one must think within the context of clinical implementation of these algorithms. “Best” needs to be defined not just by performance, but also the generalizability and feasibility of implementation. For example, the Ramirez et. al. algorithm outperforms all algorithms for blacks with low warfarin doses and performs similarly to the best algorithms across most other race/dose requirement groups. However, the Ramirez et. al. algorithm requires the reason for anticoagulation (DVT/PE or atrial fibrillation), information typically computationally unavailable at the time of warfarin initiation. Many settings implementing prospective pharmacogenomic testing rely on automated clinical decision support and active intervention at the time of ordering to tailor the prescription. Although our overall best performing algorithm/s are not clinically significantly improved over the Ramirez et. al. algorithm, they can all be computed with information readily available in a patient’s medical record, allowing for immediate calculation of starting warfarin dose at the time of prescription.
In addition to the question of implementation one must also consider that the clinical impact of dose misclassification is not consistent across all dosing groups. Overdosing individuals with low warfarin requirements (warfarin dose <21mg/week) can lead to serious bleeding events, while under-dosing those with high warfarin requirements (doses >49mg/week) can lead to clotting events.29,30 Although the IWPC algorithm performs similarly to the highest performance algorithms, it is particularly poor at predicting doses of low dose African Americans (~4.5 mg worse than the best performing algorithms). Depending on the frequency of low dose African Americans in the health system (determined with retrospective data), the IWPC algorithm may not be the best option. However, if the health system had a significant Asian population, use of the IWPC algorithm may be preferred because it takes these variables into account even if performance among low dose African Americans is reduced.
An important limitation of this study is that one of the previously tested algorithms, Ramirez et. al. was derived on a subset of patients included in this study. Thus it is possible that the high performance of the Ramirez algorithm in our population is inflated and may not be generalizable. The novel algorithms were also likely positively biased given the lack of an external validation set. Further, the results of the Hernandez et. al. algorithm were likely negatively biased as two SNPs predicting higher dose in African Americans were not included in this study due to poor genotyping quality. This study was also limited by the small number of African Americans studied. Additionally, since these data are from a single institution the results may not generalize to other populations. Warfarin dose is highly affected by vitamin K intake and the eating habits/cultural norms in the South may not reflect other parts of the US and world. Similarly, since this study only included whites and blacks, it is not clear how well the derived algorithms will perform among other ancestry groups.
In conclusion, expanding the variants in a warfarin dosing model does increase model accuracy, but not in clinically significant ways over existing algorithms in the literature. Similarly, race stratification resulted in the best model fits for African Americans, but the difference is unlikely to be clinically significant. Finally, percent ancestry surprisingly improves model fit – especially in the context of race misspecification in EMR recorded white race. However, the improvement in model fit among the white population is not clinically significant. When determining which dosing model to use, care must be given to selecting a model that not only matches the racial distribution of the population, but is also technically and financially achievable.
Acknowledgments
Thanks to Cara Sutcliffe and Paxton Baker of the VANTAGE core and Sarah Collier of the BioVU team for their help facilitating the genotyping in this project. This project was funded by Vanderbilt University, NCI (K07CA172294), NHLBI (U01HL122904, U19HL065962), NHGRI (U01HG007253, U01HG008672), and NLM (T15LM007450). The data used for this analysis were obtained from BioVU, supported by institutional funding and funding from NCATS (UL1TR000445).
Contributor Information
LAURA K. WILEY, Div. of Biomedical Informatics and Personalized Med., University of Colorado, 13001 E. 17thPl. MS F-563 Aurora, CO 80045, USA
JACOB P. VANHOUTEN, Dept. of Biomedical Informatics, Vanderbilt University, 2525 West End Ave Ste. 1475 Nashville, TN 37203, USA
DAVID C. SAMUELS, Dept. of Mol. Physiology & Biophysics, Vanderbilt Genetics Inst., Vanderbilt University, 2215 Garland Ave. Nashville, TN 37232, USA
MELINDA C. ALDRICH, Dept. of Thoracic Surgery, Div. of Epidemiology, Vanderbilt University Medical Center, 609 Oxford House Nashville, TN 37232, USA
DAN M. RODEN, Dept. of Medicine, Vanderbilt University, 2215B Garland Ave Nashville, TN 37203, USA
JOSH F. PETERSON, Dept. of Biomedical Informatics, Dept. of Medicine, Vanderbilt University, 2525 West End Ave Ste. 1050 Nashville, TN 37203, USA
JOSHUA C. DENNY, Dept. of Biomedical Informatics, Dept. of Medicine, Vanderbilt University, 2525 West End Ave Ste. 1475 Nashville, TN 37203, USA
References
- 1.Gage BF, Eby C, Johnson JA, et al. Clin Pharmacol Ther. 2008;84:326. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson JA, Cavallari LH. Trends Cardiovasc Med. 2015;25:33. doi: 10.1016/j.tcm.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein TE, Altman RB, Eriksson N, et al. N Engl J Med. 2009;360:753. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimmel SE, French B, Kasner SE, et al. N Engl J Med. 2013;369:2283. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirmohamed M, Burnside G, Eriksson N, et al. N Engl J Med. 2013;369:2294. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 6.Scott SA, Lubitz SA. Pharmacogenomics. 2014;15:719. doi: 10.2217/pgs.14.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perera MA, Cavallari LH, Limdi NA, et al. Lancet. 2013;382:790. doi: 10.1016/S0140-6736(13)60681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera MA, Gamazon E, Cavallari LH, et al. Clin Pharmacol Ther. 2011;89:408. doi: 10.1038/clpt.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Jeong H, Takahashi H, et al. Clin Pharmacol Ther. 2012;91:660. doi: 10.1038/clpt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavallari LH, Perera M, Wadelius M, et al. Pharmacogenet Genomics. 2012;22:152. doi: 10.1097/FPC.0b013e32834f288f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daneshjou R, Gamazon ER, Burkley B, et al. Blood. 2014;124:2298. doi: 10.1182/blood-2014-04-568436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drozda K, Wong S, Patel SR, et al. Pharmacogenet Genomics. 2015;25:73. doi: 10.1097/FPC.0000000000000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limdi NA, Brown TM, Yan Q, et al. Blood. 2015;126:539. doi: 10.1182/blood-2015-02-627042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar R, Seibold MA, Aldrich MC, et al. N Engl J Med. 2010;363:321. doi: 10.1056/NEJMoa0907897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramirez AH, Shi Y, Schildcrout JS, et al. Pharmacogenomics. 2012;13:407. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Shah A, Peterson NB, et al. AMIA Annu Symp Proc. 2012 [Google Scholar]
- 17.Wiley LK, Shah A, Xu H, et al. J Am Med Inf Assoc. 2013 doi: 10.1136/amiajnl-2012-001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Bois D, Du Bois EF. Nutrition. 1989;5:303. [PubMed] [Google Scholar]
- 19.Dumitrescu L, Ritchie MD, Brown-Gentry K, et al. Genet Med. 2010;12:648. doi: 10.1097/GIM.0b013e3181efe2df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPeek Hinz ER, Bastarache L, Denny JC. AMIA Annu Symp Proc. 2013;2013:975. [PMC free article] [PubMed] [Google Scholar]
- 21.Khurshid S, Keaney J, Ellinor PT, et al. Am J Cardiol. 2016;117:221. doi: 10.1016/j.amjcard.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Clin Pharmacol Ther. 2012;92:414. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, He J, Zhao S, et al. Nat Protoc. 2014;9:2643. doi: 10.1038/nprot.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander DH, Novembre J, Lange K. Genome Res. 2009;19:1655. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duconge J, Ramos AS, Claudio-campos K, et al. PLoS One. 2016;11 doi: 10.1371/journal.pone.0145480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zakharia F, Basu A, Absher D, et al. Genome Biol. 2009;10:R141. doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Driest SL, Shi Y, Bowton EA, et al. Clin Pharmacol Ther. 2014;95:423. doi: 10.1038/clpt.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robarge JD, Li L, Desta Z, et al. Clin Pharmacol Ther. 2007;82:244. doi: 10.1038/sj.clpt.6100284. [DOI] [PubMed] [Google Scholar]
- 29.Hylek EM, Go AS, Chang Y, et al. N Engl J Med. 2003;349:1019. doi: 10.1056/NEJMoa022913. [DOI] [PubMed] [Google Scholar]
- 30.Hylek EM, Evans-Molina C, Shea C, et al. Circulation. 2007;115:2689. doi: 10.1161/CIRCULATIONAHA.106.653048. [DOI] [PubMed] [Google Scholar]