Abstract
Trigger factor is a ribosome-associated peptidyl-prolyl cis/trans isomerase that is highly conserved in most bacteria. A gene, designated ropA, encoding an apparent trigger factor homologue, was identified in Streptococcus mutans, the primary etiological agent of human dental caries. Inactivation of ropA had no major impact on growth rate in planktonic cultures under the conditions tested, although the RopA-deficient mutant formed long chains in broth. Deficiency of RopA decreased tolerance to acid killing and to oxidative stresses induced by hydrogen peroxide and paraquat, and it reduced transformation efficiency about 200-fold. Addition of synthetic competence-stimulating peptide to the culture medium enhanced transformability of both the mutant and wild-type strains, although the ropA strain did not attain levels of competence observed for the parent. Loss of RopA decreased the capacity of S. mutans to form biofilms by over 80% when cultivated in glucose, but it increased biofilm formation by over 50% when sucrose was provided as the carbohydrate source. Western blot analysis revealed that the expression of glucosyltransferases B and D was lower in the RopA-deficient mutant. These results suggest that RopA is a key regulator of acid and oxidative stress tolerance, genetic competence, and biofilm formation, all critical virulence properties of S. mutans.
Trigger factor is a ribosome-associated peptidyl-prolyl cis-trans isomerase which is a molecular chaperone that is highly conserved in most bacteria (13, 30). It is generally believed that trigger factor is central to protein biogenesis and bacterial survival of environmental insult, although the full spectrum of activities of this protein in bacterial physiology and virulence expression is not yet fully appreciated. In Escherichia coli, trigger factor, which is associated with the 50S ribosomal subunit, binds polypeptides as they emerge from the ribosome and functions in cooperation with the DnaK chaperone complex in folding of newly synthesized proteins (40). Trigger factor also interacts with GroEL, greatly promoting the affinity and binding capacity of GroEL for various denatured proteins (16-18). In E. coli, trigger factor is a protein that protects cells against low temperature (15), while DnaK is required for growth at temperatures above 37°C and below 15°C (40). The deficiency of both trigger factor and DnaK results in massive aggregation of cytosolic proteins and is lethal above 30°C, although the lethality can be overcome by overexpression of GroEL/ES (40).
In the gram-positive pathogen Streptococcus pyogenes, an apparent trigger factor homologue, RopA, has been shown to be essential for secretion and maturation of the secreted cysteine protease SpeB (29, 30). Neither the precursor nor the processed form of SpeB was readily detectable in the supernate of a RopA-deficient mutant that was generated via transposon insertion (30). In mutants that were lacking the peptidyl-prolyl isomerase activity as a result of site-specific mutations in the central domain of RopA, which is critical for substrate binding, the SpeB precursor was expressed and secreted but it took over 8 h longer for the mutants to convert the SpeB precursor to the processed form than the wild-type strain (29). The SpeB protein produced by these site-specific ropA mutants was found to have only a small fraction of the proteolytic activity of the SpeB produced by the parent, suggesting that the peptidyl-prolyl isomerase activity is essential for the establishment of an active conformation of this protease (29, 30).
Streptococcus mutans, the primary etiological agent of human dental caries, lives in biofilms on the tooth surface. S. mutans has the ability to utilize a wide variety of sugars, and the sustained production of lactic acid from glycolysis carried out by this bacterium is directly linked to demineralization of tooth enamel and caries formation (4). Therefore, the abilities to adhere to and form biofilms on tooth surfaces, to catabolize carbohydrates and generate acids, and to survive low pH and other environmental stresses are critical to the cariogenicity of this pathogen (4). Several surface-associated proteins have been shown to function as high-affinity adhesins and play a central role in initiation of biofilm formation by S. mutans (3, 41). SpaP (also called P1), a surface protein of the antigen I/II family, is critical in S. mutans for sucrose-independent adherence to the tooth (3). A gene designated brpA (for biofilm regulatory protein) has recently been identified to encode a putative surface-associated polypeptide, and loss of BrpA in S. mutans causes major defects in biofilm formation on abiotic surfaces (41). S. mutans produces three glucosyltransferases (GtfB, -C, and -D) which synthesize adhesive extracellular glucans from sucrose. Gtfs are of central importance in dental plaque formation and development of caries (33, 39). Extracellular glucans, especially the α(1,3)-linked, water-insoluble forms, facilitate adherence of S. mutans to the tooth surface and modulate cell-cell interaction by serving as binding sites for Gtf proteins and glucan-binding proteins, a group of proteins with no known enzymatic activities that contribute to sucrose-dependent adherence and biofilm cohesiveness (11, 12, 31). As a major constituent of the biofilm matrix, glucans can further influence the development and structure of oral biofilms by modulating permeability to water and nutrients and by serving as an extracellular carbon and energy source (37). In addition to specific adherence-promoting gene products, recent studies have revealed regulatory networks in S. mutans that are required for biofilm formation and stress tolerance. The cell density-dependent Com system, which is known to control genetic competence development in S. mutans and other naturally competent streptococci, is involved in biofilm formation and acid tolerance in S. mutans (25-27). Likewise, luxS and relA homologues, and components of the general stress response pathway, strongly influence biofilm development and architecture and resistance to acid and oxidative insult (22-24, 42).
Trigger factor of S. mutans was found to be up-regulated in response to deficiency of LuxS (Z. T. Wen and R. A. Burne, unpublished data), which was shown to affect acid and oxidative stress tolerance and biofilm formation (42). The expression of trigger factor was also increased in cells stimulated by the synthetic competence-stimulating peptide (CSP) (G. Svensater, personal communication) and in populations that were grown in biofilms (38). In this study, we examined an apparent trigger factor homologue in S. mutans for its role in stress tolerance, competence development, and biofilm formation.
MATERIALS AND METHODS
Bacterial cultivation.
S. mutans UA159 and its derivatives were maintained on brain heart infusion (BHI) medium, with or without addition of erythromycin (Em; 10 μg/ml). Preparation of competent cells and transformation of S. mutans were done as previously described (5, 27). For biofilm formation, S. mutans strains were grown in a semidefined biofilm medium (BM) with either 0.8% (wt/vol) glucose (BMG) or 0.5% sucrose (BMS) as the carbohydrate source (28). For growth studies, TV medium (5) or Todd-Hewitt broth (BBL; Becton Dickinson, Cockeysville, Md.) supplemented with 0.3% yeast extract (THYE) (25) was also used. All agar media were prepared similarly, with agar (Difco Laboratories, Detroit, Mich.) added at a concentration of 1.5% (wt/vol).
DNA manipulations.
Unless otherwise stated, standard recombinant DNA techniques were performed as described by Sambrook et al. (34). All restriction and modifying enzymes and reagents were purchased from Invitrogen and used as described by the supplier. For Southern hybridizations, 12 μg of genomic DNAs were digested with EcoRI and transferred to NewBond nylon membranes (NEN Life Science Products, Inc.). Probes were labeled with [α-32P]dATP using a random primers DNA labeling system from Invitrogen. Hybridizations and washes were carried out under high-stringency conditions.
Acid killing and hydrogen peroxide challenge.
The ability of cells to withstand acid challenge was determined by acid killing using the method of Belli and Marquis (1). Briefly, S. mutans strains were grown in BHI that had been adjusted to pH 7.0 with HCl. Cultures were harvested at mid-exponential phase (optical density at 600 nm [OD600], ≅0.3) by centrifugation at 3,800 × g at 4°C for 10 min, washed once with 0.1 M glycine buffer (pH 7.0), and then subjected to acid killing by incubating the cells in 0.1 M glycine buffer, pH 2.8, for 45 min. Surviving cells were appropriately diluted, plated on BHI plates in triplicate, and incubated in a 5% CO2 aerobic atmosphere at 37°C for 24 to 48 h. To evaluate oxidative stress tolerance, S. mutans strains were challenged with hydrogen peroxide (H2O2; Fisher Scientific, Fair Lawn, N.J.) or paraquat (methyl viologen; catalog no. M2254; Sigma). For paraquat challenge, aliquots from a sterile paraquat solution were added directly to bacterial cultures to give final concentrations of 25 or 50 mM, and the impact on bacterial growth was monitored using a Bioscreen C reader (Helsinki, Finland) (27). For H2O2 challenge, mid-exponential-phase cells were harvested by centrifugation and resuspended in 5 ml of 0.1 M glycine buffer, pH 7.0. Hydrogen peroxide was then added to the cell suspension to give a final concentration of 0.2% (vol/vol). A 100-μl sample was obtained right before, and 2 h after, the addition of H2O2. To inactivate hydrogen peroxide, catalase (5 mg/ml; catalog no. C-9322; Sigma) was added to samples immediately after collection. The survival rate was determined by plating in triplicate on BHI plates.
Biofilm formation assay.
Biofilm assays were done as previously described (41). Briefly, overnight cultures of S. mutans UA159 and its derivatives were transferred to prewarmed BHI and grown at 37°C in a 5% CO2 aerobic atmosphere to late exponential phase (OD600 ≅ 0.5). The cultures were then diluted 1:100 in prewarmed BM, and 200 μl of the cell suspension was inoculated into the wells of 96-well (flat-bottom) cell culture clusters (Costar 3595; Corning Inc., Corning, N.Y.). Plates were incubated at 37°C, in a 5% CO2 atmosphere for 24 h. The culture medium was then decanted, and the plates were washed twice with 200 μl of sterile distilled water to remove the planktonic and loosely bound cells. The adherent bacteria were stained with 50 μl of 0.1% crystal violet for 15 min. After rinsing twice with 200 μl of water, the bound dye was extracted from the stained cells using 200 μl of 99% ethanol, and the plates were sealed in a plastic bag and set on a shaker (Reliable Scientific) overnight to allow full release of the dye. Biofilm formation was then quantified by measuring the absorbency of the solution at 600 nm in an enzyme-linked immunosorbent assay reader (Bio-Rad).
SEM.
For scanning electron microscopy (SEM), biofilms were grown on hydroxylapatite (HA) disks that were deposited in the wells of 24-well culture clusters (Corning Inc.) (41). Briefly, late-exponential-phase cultures were grown in BHI as described above and diluted 1:100 in BMG or BMS, and 2 ml of the diluted culture was added to each well. After incubating for 24 h at 37°C in a 5% CO2 aerobic atmosphere, the HA disks were carefully rinsed by dipping twice in phosphate-buffered saline buffer (50 mM Na2PO4 [pH 7.0], 0.85% NaCl) and fixed overnight with Trump fixative solution. Following dehydration through a series of ethanol rinses, the disks were mounted and coated with gold, and the biofilms were then analyzed by SEM in the University of Florida EM core facility.
Western blot analysis.
To prepare cell extracts, mid-exponential-phase cultures were harvested by centrifugation. The cell pellets were resuspended in 10 mM potassium phosphate buffer, pH 7.0, containing phenylmethylsulfonyl fluoride and sodium azide at final concentrations of 1 mM and 0.02%, respectively. Cells were homogenized using a Bead Beater (Biospec, Bartlesville, Okla.) with glass beads (0.1 mm), as previously described (6). For extracellular enzyme analysis, cell-free supernates were recovered from late-exponential-phase cultures (OD600 ≅ 0.5) and buffered with 10 mM potassium phosphate, pH 7.0, containing phenylmethylsulfonyl fluoride and sodium azide. The supernates were then concentrated approximately 20-fold by passage through a Centricon filter with a 30-kDa molecular mass cutoff (catalog no. 4209; Millipore Co., Bedford, Mass.). Proteins were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE), blotted to polyvinylidene difluoride membrane, and probed for GtfB and -C, which share extensive homology, and GtfD using polyclonal antibodies induced against S. mutans GtfB or GtfD, respectively (43).
2-D gel analysis.
For two-dimensional (2-D) gel analysis, exponential-phase cultures (OD600 ≅ 0.5) grown in BHI were harvested by centrifugation, washed, and homogenized using a Bead Beater as previously described (42). Cell extracts were sent to Kendrick Labs, Inc. (Madison, Wis.), where 200 μg of protein was subjected to 2-D gel electrophoresis using the methods of O'Farrell (32). The first-dimensional isoelectric focusing step was carried out using 2% pH 4-to-8 ampholines, and the second dimension was carried out in an SDS-10% polyacrylamide gel. Gels were stained with Coomassie blue, and protein profiles were analyzed by densitometry using software from Alpha Innotech (San Leandro, Calif.).
RESULTS AND DISCUSSION
Construction of a RopA-deficient mutant.
The ropA gene was identified by a BLAST search of the S. mutans genome database (www.genome.ou.edu/smutans) with the S. pyogenes RopA amino acid sequence. S. mutans ropA (Smu0082 in the Oral Pathogens database [http://www.stdgen.lanl.gov/oragen/]) is a 1,281-bp open reading frame encoding a polypeptide of 427 amino acid residues with 74% identity to RopA of S. pyogenes (accession no. AAC82391), an apparent homologue of trigger factor (13, 30, 36). Analysis of the flanking region revealed that the ropA gene of S. mutans is immediately upstream of genes that are predicted to code for transposase fragments and genes for nitrite transport and hypothetical proteins upstream, but they are all transcribed in the opposite orientation to that of ropA.
To analyze the role of RopA in S. mutans, a ropA mutant was constructed by replacing nucleotide 56 to nucleotide 1205 of the coding sequence with an erythromycin resistance (erm) gene (7) using a PCR-ligation-mutation strategy (20). Specifically, a 415-bp 5′ fragment and a 512-bp 3′ fragment were amplified by PCR using gene-specific primers (Table 1) to incorporate an SstI site at the 3′ end of the 5′ fragment and a HindIII site at the 5′ end of the 3′ fragment. The fragments were then digested with restriction enzymes and ligated with the erm gene that was released with the same enzymes (41). The resulting ligation mixture was then used to transform S. mutans UA159. A mutant with an inactivated ropA gene, strain TW90, was isolated on BHI plates with erythromycin (10 μg/ml) and confirmed by PCR and Southern blot analysis (data not shown). Another deletional mutant, strain SMUTig, which has nucleotides 34 to 1177 of the coding sequence replaced with an erythromycin resistance element, was generated from S. mutans UA159 using a similar strategy. An insertional mutant, TW90A, was generated by single-crossover insertion using a suicide vector, pSU20Erm:ropA, which contains an internal fragment of ropA amplified by PCR (Table 1) to transform S. mutans UA159. Strain TW90A was used during the early stage of this study but was later replaced by the double-crossover mutant TW90 for acid and oxidative stress tolerance and biofilm formation. Similar experiments were also run with strain SMUTig and yielded similar results. In these cases, only the data generated from strain TW90 are presented. For transformation efficiency assays, only strain SMUTig was used.
TABLE 1.
Primers used for construction of mutants
| Primer | DNA sequence (5′ → 3′)a | Application | Amplicon (bp) |
|---|---|---|---|
| ropA55′ | ACTATGTCGTGACACCAGCATC | 5′ fragment for TW90 | 415 |
| ropA53′ | TAGAGCTCTTCTTTCACCAGATATGCTC | 5′ fragment for TW90 | |
| ropA35′ | ATAAGCTTATCGTAAATGTAACAACACCAC | 3′ fragment for TW90 | 512 |
| ropA33′ | AGCAAACATGGAACGCAGAGC | 3′ fragment for TW90 | |
| TF-P1 | AAAGAAGTTTTGCCAATCGC | 5′ fragment for SMUTig | 781 |
| TF-P2 | GGCGCGCCTCGTAAATGTAACAACACCACG | 5′ fragment for SMUTig | |
| TF-P3 | GGCCGGCCCATGGAAGCTGAGCAAGTGCG | 3′ fragment for SMUTig | 718 |
| TF-P4 | ATTTATCAAGGCATGACGGGG | 3′ fragment for SMUTig | |
| TF5′ | ATGGATCCGGCTGCAACTACACCGACTGC | Internal fragment for TW90A | 595 |
| TF3′ | GAGTCGACCCGATTCTACCGAAGAGCTCGGTATC | Internal fragment for TW90A |
The incorporated restriction sites are underlined.
RopA deficiency diminishes tolerance to stresses.
Strain TW90 formed mucoid, smooth, and shiny colonies on BHI agar plates, in contrast to the rough, dry colonies formed by the parent strain, UA159. When grown in BHI, BM, THYE, or TV broth, strain TW90 displayed no major differences in growth rates compared to UA159 (data not shown). However, TW90 did have a tendency to form longer chains in THYE, BHI, and BMG broth cultures, with an average of 26 ± 7 cells per chain for the mutant versus 11 ± 4 for strain UA159.
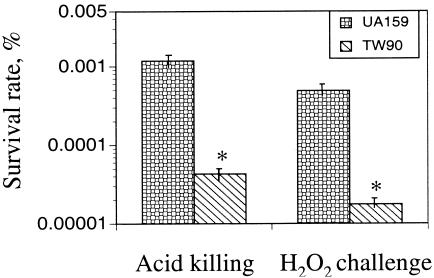
Compared to UA159, TW90 took significantly longer to form visible colonies on agar plates that were adjusted to pH 5.0 (data not shown) or to obtain similar optical densities in broth media that had been adjusted to pH 5.5. In this acidified medium, strain UA159 had a doubling time of 84.4 ± 1.2 min compared with 107.9 ± 2.5 for the mutant, indicative of defects in acid tolerance. To further analyze the impact of RopA deficiency on acid tolerance, mid-exponential-phase cultures of TW90 that were grown in BHI were subjected to acid killing (42). When incubated in buffer at pH 2.8 for 45 min, TW90 demonstrated a decrease of more than 45-fold in survival rate compared to UA159 (Fig. 1). When incubated in medium containing 25 mM paraquat, strain TW90 also displayed a lag phase that was more than 2 h longer than that of the wild type. Growth was further inhibited when the concentration of paraquat was increased to 50 mM, with a doubling time of 484.2 ± 67.3 min for TW90 compared to 199.2 ± 19.6 min for UA159 (data not shown). Similarly, when mid-exponential-phase cultures were challenged with 0.2% (vol/vol) hydrogen peroxide for 2 h, TW90 showed more than a 28-fold decrease in survival rate compared to UA159 (Fig. 1).
FIG. 1.
Acid killing and hydrogen peroxide challenge assays. For acid tolerance, all S. mutans strains were grown in BHI until mid-exponential phase (OD600 ≅ 0.3), harvested by centrifugation, washed once with 0.1 M glycine buffer, pH 7.0, and then subjected to acid killing at pH 2.8 for 45 min. For hydrogen peroxide challenge, the mid-exponential-phase cultures were incubated in buffer containing 0.2% (vol/vol) hydrogen peroxide for 2 h. The surviving cells were plated on BHI agar plates in triplicate, and results are expressed as survival rate over time. See text for more details. Data presented are means ± standard deviations (error bars) of three independent experiments. *, P < 0.01.
There are a variety of reasons that the RopA-deficient mutants may be more sensitive to low pH and oxidative stress. In S. mutans and other mutans streptococci, ATP-dependent proton extrusion mediated by the membrane-bound F-ATPase is the primary mechanism that these bacteria employ to maintain intracellular pH homeostasis (2). Ffh, a homologue of eukaryotic signal recognition particle protein, has also been recently reported as a major determinant in acid tolerance in S. mutans (8, 10). Considering the fact that trigger factor in E. coli and S. pyogenes is critical to folding of nascent proteins and establishment of active conformation of functional proteins (13, 29, 30, 40), RopA deficiency may cause defects in membrane integrity and alter expression or distribution of membrane-associated proteins, such as F-ATPase and Ffh. As detailed in the following sections, defects in the regulatory Com system could also contribute to the impaired acid tolerance of the RopA-deficient mutants.
The ropA mutant is less competent for genetic transformation.
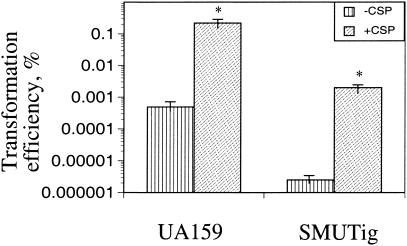
In S. mutans and other naturally competent streptococci, the ability to transport and incorporate foreign DNA, genetic competence, is primarily controlled by ComCDE, with ComC being the genetic competence factor (21, 26). The competence factor peptide, a quorum-sensing pheromone, is processed and secreted into the environment (21, 27). When the concentration of the processed peptide reaches a certain threshold, it is detected by ComD, a transmembrane histidine kinase, which then phosphorylates ComE, a response regulator, triggering transcription of a set of genes that are required for DNA uptake and the related processes. Consistent with the findings in 2-D gel analyses of S. mutans (G. Svensater, personal communication), inclusion of synthetic CSP (1 μg/μl) in the culture medium of an S. mutans comC-defective strain was also found to increase ropA transcription by 1.4-fold, suggesting RopA is involved in the ComC-mediated regulatory functions. To assess the impact of RopA deficiency on competence development, transformation efficiency was measured in the RopA-deficient strain using a procedure described elsewhere (27). It was found that the transformation efficiency was approximately 200-fold lower for the ropA mutant than for the wild-type strain (Fig. 2). However, addition of CSP at a level of 500 ng μl−1 was able to enhance the transformation efficiency of strain SMUTig by about 3 logs. Still, the transformation efficiency of the mutant could not be restored to the level of the parental strain that had been treated with CSP.
FIG. 2.
Transformation efficiency. S. mutans strains were evaluated for their ability to transform foreign DNA (plasmid) with or without inclusion of the synthetic CSP (500 ng μl−1). The bar graph represents the average of at least three independent experiments, with standard deviations denoted by the error bars. *, P < 0.01.
One interpretation of the observed defects in transformation of SMUTig is that RopA is involved in processing or secretion of the competence factor peptide and that lack of RopA causes defects in production of competence factor. An inability to fully restore competence by addition of CSP suggests that detection of the mature peptide by the cells may also be affected by the loss of RopA. Given the established role of trigger factor as a critical factor in processing, secretion, and presentation of membrane-associated and extracellular proteins (13, 29, 30, 40), deficiency of RopA in S. mutans could cause defects in extracellular and membrane-associated proteins that control competence factor sensing or DNA uptake. Similar to the S. mutans strains that are defective in either production or detection of competence factor (27), the RopA-deficient mutant also formed longer chains that were about three times the cell number of the parent, and the mutant cells appeared to be a little shorter and fatter than the parental cells under SEM (data not shown), further suggesting that there may be defects in cell division or membrane biogenesis from RopA deficiency.
Deficiency of RopA causes alterations in biofilm formation.
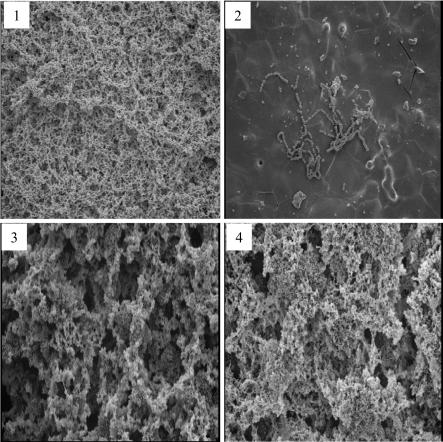
As measured by the amount of crystal violet-stained, adherent cells on a polystyrene surface, the capacity of TW90 to form biofilms was decreased by over 80% when glucose was the carbohydrate source, compared to the wild-type strain grown under the same conditions. The biofilm formation defect could be overcome by growth in sucrose-containing medium. In fact, when grown in BM with sucrose, the RopA-deficient strain displayed a markedly better adherence to the sides of culture tubes (data not shown) and actually formed over 50% more biofilms than did the parent. Similar trends were observed under SEM of biofilms formed on HA surfaces (Fig. 3). When grown on HA disks with glucose as the carbohydrate source, TW90 was able to bind to the HA surfaces, but in a diminished capacity compared to UA159. On HA disks, it also formed longer chains than did the wild type, but failed to develop into 3-D biofilms (Fig. 3). In contrast, when sucrose was present, TW90 accumulated and formed mature biofilms with multicellular clusters, and the biofilms formed by TW90 appeared to have larger, more compact cell clusters surrounded by relatively smaller gaps than those formed by strain UA159. As supported by counting of CFU (data no shown), the enhanced biofilm formation by TW90 in sucrose appears to be mainly due to increases in cell number.
FIG. 3.
SEM analysis of biofilms. The 24-h biofilms of S. mutans strains UA159 (panels 1 and 3) and TW90 (panels 2 and 4) were grown on HA disks that were deposited in 24-well cell culture clusters in semidefined BM medium with glucose (panels 1 and 2) or sucrose (panels 3 and 4) as the carbohydrate source. Magnification in all images, ×1,000.
The basis for the decrease in biofilm formation is yet to be established, but a likely factor is that the ropA strains have altered expression or presentation of surface proteins. Relatively little is known about the role of specific surface structures in the formation of S. mutans biofilms, and so further characterization of the surface-associated proteins in the RopA-deficient strains is under way to identify critical surface-anchored and secreted gene products required for biofilm maturation. Formation of longer chains by the ropA mutant may also make it difficult for the bacterial cells to remain attached by the relatively weak forces that mediate adhesion to the polystyrene when cells are grown in glucose.
Another explanation for the decrease in biofilm formation may be related to the defects in the competence regulon. Recent studies in S. mutans by Li et al. have shown that the Com system is involved in acid tolerance and biofilm formation (25, 27). S. mutans strains that were defective in the comC, comD, or comE genes, which encode the major components of the cell density-dependent regulatory system, demonstrated a diminished acid tolerance, and such defects could be overcome by addition of synthetic CSP to the culture medium (25). Compared to the parental strain, these com-deficient strains formed biofilms with less biomass and abnormal architecture. Inclusion of synthetic CSP in the culture medium or complementation with a wild-type comC gene in a shuttle vector partially restored the wild-type biofilm architecture (27). As demonstrated by transformation efficiency (Fig. 2), RopA deficiency causes defects in genetic competence. Therefore, the observed impairments in biofilm formation as well as acid tolerance by TW90 could be, at least in part, attributed to the defects in expression and/or function of the Com system.
RopA deficiency affects Gtf expression.
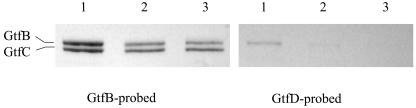
To examine whether changes in expression of the Gtfs could be responsible for the changes in sucrose-dependent biofilm formation, cell extracts and cell-free supernates of UA159 and TW90 grown in BHI were analyzed by Western blotting using polyclonal antisera induced against GtfB and GtfD, respectively (43). In S. mutans, GtfB and GtfC show extensive homology (9), so the polyclonal anti-GtfB antibody reacts strongly with both GtfB and GtfC. As shown in Fig. 4, the bands reactive with anti-GtfB antibody in samples prepared from strain TW90 were significantly less intense than those prepared from UA159. Such changes were even more apparent in the cell-free supernates. Interestingly, in contrast to GtfB, the bands corresponding to GtfC in TW90 were just slightly decreased. When probed with anti-GtfD antibody, the band reactive with anti-GtfD was virtually undetectable in cell extracts of TW90.
FIG. 4.
Western blot analysis. Whole-cell extracts (20 μg of total protein) of UA159 (lanes 1), TW90A (lanes 2), and TW90 (lane 3) were separated by SDS-10% PAGE. The separated proteins were then blotted to a polyvinylidene difluoride membrane and probed with anti-GtfB (A) and anti-GtfD (B) polyclonal antibodies.
One of the major contributors to sucrose-dependent bacterial adherence by S. mutans is the production of glucans through the cooperative functions of the glucosyltransferases (GtfB, -C, and -D). Although more complex than presented, GtfB generally assembles glucans rich in α(1,3) linkages, GtfD synthesizes α(1,6)-linked glucans, and GtfC makes both α(1,3)- and α(1,6)-linked glucans. Both α(1,3)-linked water-insoluble and α(1,6)-linked water-soluble glucans are important in cell-cell and cell-surface adhesive interactions, while the α(1,3)-rich, water-insoluble glucans have been shown to be the major contributor to bacterial adherence to hard surfaces (19, 35, 39). A recent study by Ooshima et al. (33) has shown that the presence of all three enzymes in an optimal ratio is necessary for efficient sucrose-dependent adherence. Down-regulation of Gtf enzymes, especially GtfB and GtfD in the RopA-deficient strain, would likely alter the ratio of these enzymes and, consequently, the composition of the glucans synthesized, which in turn would alter biofilm accumulation and architecture. Most likely, with very low levels of GtfD, the polymers made by the RopA-deficient mutant could be predominantly the water-insoluble, α(1,3)-linked glucans, which better support adherence, potentially enhancing biofilm formation. Other possible reasons for the enhancement of sucrose-dependent biofilm formation could include altered expression or activity of glucan-binding proteins, including the glucan-binding protein activity of the Gtf enzymes.
Further evidence of the relationship between stress tolerance and biofilm formation.
In an attempt to uncover the factors that are involved in altered stress tolerance and biofilm formation as a result of RopA deficiency, whole-cell extracts were analyzed in 2-D gels. Comparison of the protein profiles stained with Coomassie blue revealed that loss of RopA altered the expression of at least 33 proteins, 22 of which were up-regulated and 11 were down-regulated (data not shown), with the role and identity of the majority of the altered proteins being unknown. Still, among the up-regulated proteins that could be identified were DnaK, GroEL, and GroES, molecular chaperones that are inducible by heat shock and acid stress and are believed to play a central role in the ability of oral bacteria to cope with frequent stresses encountered in the oral cavity (14, 24). Given the fact that trigger factor in E. coli is associated with GroEL/ES in promoting the affinity of these chaperones and binding capacity to various stress-damaged proteins (16, 17, 40), deficiency of RopA would likely diminish the capacity of the organism to deal with stresses imposed by low pH, oxygen, and other detrimental conditions. The up-regulation of molecular chaperones DnaK, GroEL, and GroES in the ropA mutant is therefore consistent with the weakened tolerance to acid and oxidative stresses by TW90 (Fig. 1).
Summary.
S. mutans, an obligate biofilm-forming bacterium and the primary etiological agent of human dental caries, possesses a gene that is predicted to encode a homologue of bacterial trigger factor. The role for RopA in S. mutans virulence expression appears to be substantial. The results suggest that RopA in S. mutans may be involved in adherence to, and formation of biofilms on, tooth surfaces and that the protein is essential for tolerance of low pH, toxic oxygen radicals, and other adverse conditions in the oral cavity. The underlying molecular basis for the observed defects in the RopA-deficient mutant is under investigation.
Acknowledgments
This work was supported by National Institute for Dental and Craniofacial Research grants DE13239 and DE12236 to R.A.B. and DE013230 to D.G.C.
We also thank F. L. Bennett and the EM Core lab of the University of Florida for their technical assistance with the SEM analysis.
Editor: V. J. DiRita
REFERENCES
- 1.Belli, W. A., and R. E. Marquis. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57:1134-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowden, G. H., and I. R. Hamilton. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 9:54-85. [DOI] [PubMed] [Google Scholar]
- 3.Bowen, W. H., K. Schilling, E. Giertsen, S. Pearson, S. F. Lee, A. Bleiweis, and D. Beeman. 1991. Role of a cell surface-associated protein in adherence and dental caries. Infect. Immun. 59:4604-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burne, R. A. 1998. Oral streptococci products of their environment. J. Dent. Res. 77:45-452. [DOI] [PubMed] [Google Scholar]
- 5.Burne, R. A., Z. T. Wen, Y. M. Chen, and J. E. C. Penders. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y. M., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.1 urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 8.Crowley, P. J., G. Svensater, J. L. Snoep, A. S. Bleiweis, and L. J. Brady. 2004. An ffh mutant of Streptococcus mutans is viable and able to physiologically adapt to low pH in continuous culture. FEMS Microbiol. Lett. 234:315-324. [DOI] [PubMed] [Google Scholar]
- 9.Fujiwara, T., Y. Terao, T. Hoshino, S. Kawabata, T. Ooshima, S. Sobue, S. Kimura, and S. Hamada. 1998. Molecular analyses of glucosyltransferase genes among strains of Streptococcus mutans. FEMS Microbiol. Lett. 161:331-336. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazlett, K. R., J. E. Mazurkiewicz, and J. A. Banas. 1999. Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect. Immun. 67:3909-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazlett, K. R., S. M. Michalek, and J. A. Banas. 1998. Inactivation of the gbpA gene of Streptococcus mutans increases virulence and promotes in vivo accumulation of recombinations between the glucosyltransferase B and C genes. Infect. Immun. 66:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hesterkamp, T., and B. Bukau. 1996. The Escherichia coli trigger factor. FEBS Lett. 389:32-34. [DOI] [PubMed] [Google Scholar]
- 14.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 15.Kandror, O., and A. L. Goldberg. 1997. Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc. Natl. Acad. Sci. USA 94:4978-4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kandror, O., M. Sherman, and A. Goldberg. 1999. Rapid degradation of an abnormal protein in Escherichia coli proceeds through repeated cycles of association with GroEL. J. Biol. Chem. 274:37743-37749. [DOI] [PubMed] [Google Scholar]
- 17.Kandror, O., M. Sherman, R. Moerschell, and A. L. Goldberg. 1997. Trigger factor associates with GroEL in vivo and promotes its binding to certain polypeptides. J. Biol. Chem. 272:1730-1734. [DOI] [PubMed] [Google Scholar]
- 18.Kandror, O., M. Sherman, M. Rhode, and A. L. Goldberg. 1995. Trigger factor is involved in GroEL-dependent protein degradation in Escherichia coli and promotes binding of GroEL to unfolded proteins. EMBO J. 14:6021-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopec, L. K., A. M. Vacca-Smith, and W. H. Bowen. 1997. Structural aspects of glucans formed in solution and on the surface of hydroxyapatite. Glycobiology 7:929-934. [DOI] [PubMed] [Google Scholar]
- 20.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 21.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemos, J. A., T. A. Brown, Jr., and R. A. Burne. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72:1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemos, J. A., Y. Y. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y. H., M. N. Hanna, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyon, W. R., and M. G. Caparon. 2003. Trigger factor-mediated prolyl isomerization influences maturation of the Streptococcus pyogenes cysteine protease. J. Bacteriol. 185:3661-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumura, M., T. Izumi, M. Matsumoto, M. Tsuji, T. Fujiwara, and T. Ooshima. 2003. The role of glucan-binding proteins in the cariogenicity of Streptococcus mutans. Microbiol. Immunol. 47:213-215. [DOI] [PubMed] [Google Scholar]
- 32.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 33.Ooshima, T., M. Matsumura, T. Hoshino, S. Kawabata, S. Sobue, and T. Fujiwara. 2001. Contributions of three glycosyltransferases to sucrose-dependent adherence of Streptococcus mutans. J. Dent. Res. 80:1672-1677. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schilling, K. M., and W. H. Bowen. 1992. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect. Immun. 60:284-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoller, G., T. Tradler, K. P. Rucknagel, J. U. Rahfeld, and G. Fischer. 1996. An 11.8 kDa proteolytic fragment of the Escherichia coli trigger factor represents the domain carrying the peptidyl-prolyl cis/trans isomerase activity. FEBS Lett. 384:117-122. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 38.Svensater, G., J. Welin, J. C. Wilkins, D. Beighton, and I. R. Hamilton. 2001. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 205:139-146. [DOI] [PubMed] [Google Scholar]
- 39.Tsumori, H., and H. Kuramitsu. 1997. The role of the Streptococcus mutans glucosyltransferases in the sucrose-dependent attachment to smooth surfaces: essential role of the GtfC enzyme. Oral Microbiol. Immunol. 12:274-280. [DOI] [PubMed] [Google Scholar]
- 40.Vorderwulbecke, S., G. Kramer, F. Merz, T. A. Kurz, T. Rauch, B. Zachmann-Brand, B. Bukau, and E. Deuerling. 2004. Low temperature or GroEL/ES overproduction permits growth of Escherichia coli cells lacking trigger factor and DnaK. FEBS Lett. 559:181-187. [DOI] [PubMed] [Google Scholar]
- 41.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen, Z. T., and R. A. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wunder, D., and W. H. Bowen. 2000. Effects of antibodies to glucosyltransferase on soluble and insolubilized enzymes. Oral Dis. 6:289-296. [DOI] [PubMed] [Google Scholar]