Abstract
DNA polymerase β (pol β) requires nuclear localization to fulfil its DNA repair function. Although its small size has been interpreted to imply the absence of a need for active nuclear import, sequence and structural analysis suggests that a monopartite nuclear localization signal (NLS) may reside in the N-terminal lyase domain. Binding of this domain to Importin α1 (Impα1) was confirmed by gel filtration and NMR studies. Affinity was quantified by fluorescence polarization analysis of a fluorescein-tagged peptide corresponding to pol β residues 2–13. These studies indicate high affinity binding, characterized by a low micromolar Kd, that is selective for the murine Importin α1 (mImpα1) minor site, with the Kd strengthening to ∼140 nM for the full lyase domain (residues 2–87). A further reduction in Kd obtains in binding studies with human Importin α5 (hImpα5), which in some cases has been demonstrated to bind small domains connected to the NLS. The role of this NLS was confirmed by fluorescent imaging of wild-type and NLS-mutated pol β(R4S,K5S) in mouse embryonic fibroblasts lacking endogenous pol β. Together these data demonstrate that pol β contains a specific NLS sequence in the N-terminal lyase domain that promotes transport of the protein independent of its interaction partners. Active nuclear uptake allows development of a nuclear/cytosolic concentration gradient against a background of passive diffusion.
INTRODUCTION
Efficient DNA repair is dependent on the recruitment of damage-dependent polymerases to the cell nucleus. DNA polymerase β (pol β) plays a key role in base excision repair (1), as well as participating in other repair pathways (2–4) and in lesion bypass (5–12). The strong relationship between functional mutations in pol β and the development and progression of cancer is increasingly substantiated in many, although not all studies (13–21). Variations in the expression levels of pol β and other components of the base excision repair complexes have been reported to be associated with various pathologies, and particularly with cancer (22–28). In addition to dysregulated expression levels, altered subcellular distribution provides another increasingly appreciated mechanism for perturbing nuclear protein concentrations, resulting in cellular dysfunction and disease (29–32). Consistent with this mechanism, a variant form of Xeroderma Pigmentosum was recently determined to result from a mutation in the nuclear localization signal (NLS) of the translesion repair enzyme DNA polymerase η (33). Altered nuclear levels of the DNA repair proteins aprataxin and DNA ligase I that have been connected to Achalasia-Addisonianism-Alacrimia (Triple A) syndrome and other functional impairments also have been demonstrated to result from mutated or altered expression levels of the nuclear pore protein ALADIN (34–36).
In order to fulfil their roles in DNA repair, family X DNA polymerases (pol X) require nuclear localization. Among the four mammalian pol X enzymes, three: pol μ; pol λ; and terminal deoxynucleotidyl transferase contain a putative NLS, while pol β is generally thought to lack an NLS motif (37–42). Consequently, pol β nuclear localization has been thought to depend on co-transport with other repair proteins to which it binds, or to depend on its small size allowing it to diffuse through the nuclear pore without reliance on active nuclear uptake (43,44). Pol β has been reported to interact with other DNA repair proteins that contain NLS sequences (45–51). However, detailed structural information and binding affinity data are available only for the interaction with XRCC1 (52–54). XRCC1 is reported to mediate the co-transport of DNA Ligase 3α (55,56) and JWA (57) into the nucleus, and so it might also facilitate nuclear transport of other XRCC1-associated proteins, including pol β. Nevertheless, not all XRCC1 binding partners are efficiently co-transported into the nucleus, as is apparent from studies of aprataxin localization (34,36). Furthermore, there is increasing evidence that some of the repair functions of pol β do not require XRCC1 (58–60). Hence, it would make functional sense for the nuclear localization of pol β not to be completely dependent on XRCC1 binding and co-transport.
Despite the prevailing consensus that pol β lacks an NLS, the enzyme does contain a string of highly conserved, basic residues at its N-terminus, and nearly all available crystal structures indicate that ∼10 N-terminal residues are disordered (Supplementary Figure S1). These characteristics led us to conclude that pol β might possess a classical, monopartite NLS at its extreme N-terminus. To evaluate this possibility, we have undertaken studies of the interaction of the N-terminal lyase domain of pol β (residues 2–87) with murine Importin α1 (mImpα1) as well as with human Importin α5 (hImpα5). Fluorescence anisotropy studies using a fluorescein derivative of the N-terminal pol β peptide (residues 2–13) in combination with wild-type or mutated forms of mouse Impα1ΔIBB provide a quantitative description of this interaction and demonstrate specificity for the minor binding pocket of mImpα1. These in vitro results are further supported by immunofluorescent staining of cells containing pol β with the wild-type or mutated NLS (R4S,K5S), where a strong reduction in nuclear localization is seen for cells expressing the mutated NLS sequence.
MATERIALS AND METHODS
Materials
The fluorescein-labeled pol β NLS peptide: S3KRKAPQETLNGG14-Lys(FITC), used for fluorescence polarization assays, was obtained from Genscript at a purity level of > 90%. Methyl methanesulfonate (MMS) was from Sigma-Aldrich. Following a strategy used previously (61), we studied a pol β complex with a double-hairpin that forms a one-nucleotide gapped DNA substrate with the following sequence: 5΄-PGGCGAAGCCTGGTGCGAAGCACC-3΄ (underlined nucleotide is in the gap). The oligonucleotide was from Oligos Etc., Wilsonville, OR, USA. The non-hydrolyzable deoxynucleoside triphosphate 2΄-deoxyadenosine-5΄-[(α,β)-methyleno]triphosphate (dAPCPP) was obtained from Jena Bioscience.
Protein expression
The [methyl-13C]methionine-labeled wild-type pol β and the NLS variant pol β(R4S,K5S) were prepared as described previously (62) by growth of the plasmid-containing Escherichia coli on a medium containing [methyl-13C]methionine (CIL, Cambridge, MA). U-[15N] pol β lyase domain was expressed in E. coli BL21(DE3) grown in M9 minimal medium containing 15NH4Cl as the sole nitrogen source. U-[2H, 15N] pol β was expressed in E. coli BL21(DE3) grown in M9 deuterated (99% D2O) medium containing U-[2H] glycerol and 15NH4Cl as the sole carbon and nitrogen sources. The expressed proteins were prepared as described previously (17). The R4S,K5S mutations, chosen in order to preserve the hydrophilicity of the NLS residues while reducing the interaction with Impα, were generated using the QuikChange kit (Agilent Technologies). The protein concentrations were determined using 280 nm extinction coefficients of 20,088 M−1cm−1 for full-length polymerases and 3591 M−1cm−1 for the isolated lyase domains.
His-tagged mImpα1 with the Importin β binding domain (IBB) deleted (mImpα1ΔIBB) and its major pocket variant (mImpα1ΔIBB[W184R/W231R]) were prepared as described previously (56). The minor binding pocket double mutant (mImpα1ΔIBB[W357R/W399R]) was created using the QuikChange kit (Agilent). C-terminal His-tagged human Importin α5, corresponding to residues 66–512, (hImpα5ΔIBB) was purchased from Genscript and cloned into the pET30 expression vector. All clones were sequence verified. mImpα1ΔIBB[W357R/W399R] and hImpα5ΔIBB were expressed and purified by the IMAC and gel filtration protocol previously described (56). Protein concentrations were determined by the Edelhoch procedure (63).
NMR spectroscopy
For the mImpα1ΔIBB binding experiments, the samples contained 0.13 mM U-[2H,15N]pol β and 0.13 mM mImpα1ΔIBB in a buffer consisting of 50 mM Tris-d11 (pH 7.6), 150 mM KCl, 1 mM CDTA, 1 mM dithiothreitol (DTT), 0.1 mM AEBSF, 0.04% NaN3 and 50 μM DSS as an internal chemical shift standard, with 10% D2O for 1H-15N HSQC experiments and 100% D2O for 1H-13C HSQC experiments. For studies of the effects of the R4S,K5S mutation on pol β structure and function, the [methyl-13C]methionine-labeled pol β samples contained 0.1 mM pol β or the NLS-variant in the above 100% D2O buffer along with 0.11 mM of a 1-nucleotide gapped double-hairpin DNA substrate and 0.14 mM of the non-hydrolyzable nucleoside triphosphate substrate dAPCPP. Nuclear magnetic resonance (NMR) experiments were performed at 25°C on a Varian UNITY INOVA 600 or 800 MHz NMR spectrometer, using a 5 mm Varian 1H{13C,15N} triple-resonance room-temperature or Cold Probe, equipped with actively shielded Z-gradients. The 1H-13C HSQC spectra were acquired using Varian's gChsqc sequence. The spectra were processed using NMRPipe version 2.1 (64) and analyzed using NMRView version 5.0.4 (65). All spectra were processed using squared cosine bell apodization functions in all dimensions and forward-backward linear prediction in the indirect dimension (66).
Fluorescence polarization measurements
Apparent peptide dissociation constants were determined based on fluorescence polarization measurements using the fluorescein-labeled pol β NLS peptide as previously described (56). The binding constants for the interaction of pol β lyase domain with the Importin α constructs were determined by displacement of the FITC-labeled NLS peptide using a competition assay as previously described (56).
Chromatography
For an initial assessment of binding, a sample of mImpα1ΔIBB was mixed with a 2.8-fold excess of pol β lyase domain and the sample was eluted on a HiLoad 26/60 Superdex 200 column (GE Healthcare) with a buffer containing 20 mM Tris-HCl, pH 7.8, 125 mM NaCl, 2 mM DTT, 1 mM ethylenediaminetetraacetic acid. For the analytical gel filtration experiments, hImpα5ΔIBB was mixed with 2-fold excess of wild-type or the NLS variant pol β(R4S,K5S) lyase domain and the samples were eluted on a Superdex 200 10/300 GL column (GE Healthcare) with a buffer containing 20 mM HEPES, 125 mM NaCl, 5mM DTT, 1mM ethylenediaminetetraacetic acid, pH 7.4.
Cell lines and plasmids
Pol β null SV40-transformed mouse embryonic fibroblasts (MB38Δ4) have been described previously (59). These cells were maintained in Dulbecco's Modified Eagle's medium (Life Technologies, Carlsbad, CA, USA) supplemented with GlutaMAX-1 (Life Technologies), 10% fetal bovine serum (HyClone, Logan, UT, USA) and hygromycin (80 μg/ml; Roche Molecular Biochemicals, Indianapolis, IN, USA) in a 10% CO2 incubator at 34°C. Xrcc1−/− p53-deficient mouse embryonic fibroblasts were obtained from Dr Robert Tebbs (67). These cells were maintained in low glucose Dulbecco's Modified Eagle's medium (Life Technologies) supplemented with 10% fetal bovine serum in a 10% CO2 incubator at 37°C. Mycoplasma testing was performed routinely using a MycoAlert® Mycoplasma detection kit (Lonza, Rockland, ME, USA), and results were negative.
The human pol β coding sequence was amplified from pRC23 pol β (68). For transient transfection, primers containing NheI and NotI restriction sites were used to introduce the pol β coding sequence into the pCDH531vector (System Biosciences, Mountain View, CA, USA). Protein expression is controlled by the EF1 promoter and an IRES sequence mediates the coexpression of a reporter gene, RFP. Since there is no antibiotic selection in the pCDH531vector, pol β cDNA was also subcloned into the pENTR/D-Topo vector, then the pEF-DEST51 vector utilizing Gateway technology (Life Technologies). Site-directed mutagenesis with the QuikChange site-directed mutagenesis kit (Agilent) was used to create a pol β variant without the predicted NLS by replacing residues 4 and 5 with serines (R4S,K5S). This resulted in the four sequence-verified mammalian cell expression vectors used in this study: pCDH531 pol β and pol β(R4S,K5S), used for transient expression; and pEF-DEST51 pol β and pEF-DEST51 pol β(R4S,K5S), used for the generation of stable cell lines.
Stable cell lines were generated by seeding 2 × 105 pol β null cells in a six-well dish in 2 ml of growth medium. Cells were transfected with pEF-DEST51 pol β or pEF-DEST51 pol β(R4S,K5S) using Lipofectamine™ 2000 (Life Technologies). Forty-eight hours after transfection, cells were split into fresh growth medium containing blasticidin (10 μg/ml; Life Technologies), and single cell clones for wild-type pol β and pol β(R4S,K5S) were isolated and screened for pol β expression by Western blotting. The two stable cell lines selected for further study, pol β WT clone 96 and pol β(R4S,K5S) clone 18, were chosen because they have similar pol β expression levels (Supplementary Table S1).
Fluorescence Microscopy
Cells were seeded in 35 mm glass bottomed petri dishes (MatTek, Ashland, MA, USA) at 2 × 105 cells per dish and incubated in growth medium for 24 h. Initial cellular localization of wild-type pol β and the pol β(R4S,K5S) variant was conducted by transient transfection with the indicated pCDH531 construct using Lipofectamine™ 2000 24 h after cell plating. Twenty-four hours after transfection, cells were fixed with a 3.7% neutral buffered formaldehyde solution (Thermo Scientific) for 10 min at room temperature. Cells were then washed three times with phosphate-buffered saline (PBS, HyClone). After fixation, cells were permeabilized with 1% sodium dodecyl sulphate, as previously described (69). Cells were blocked in PBS + 1% bovine serum albumin for 30 min, and then incubated with anti-pol β antibody (1:200; ab 26343, Abcam) for 1 h. Cells were washed three times with PBS, then incubated in Alexa 488 conjugated anti-rabbit secondary antibody (1:2000; Life Technologies) and stained with DAPI NucBlue® Fixed Cell Stain ReadyProbes™ (Life Technologies) for 5 min. For stable wild-type pol β and pol β(R4S,K5S) expressing cell lines, cells were fixed 24 h after plating using the same procedure described for the transiently transfected cells. Immunofluorescent staining was also performed using a method similar to that given above except for use of an Alexa 546 conjugated anti-rabbit secondary antibody (1:2000; Life Technologies).
For transiently transfected cells, fluorescence images were acquired with a 40X C-Apochromat (numerical aperture 1.2) water immersion objective coupled to a Zeiss LSM510 META confocal microscope (Carl Zeiss MicroImaging). Multi-track configuration was used to ensure the absence of excitation cross-talk or emission bleed- through between channels. The 364 nm laser line was used at 3.5% maximum intensity, the 488 nm laser line was used at 10% of maximum intensity and the 543 nm laser line was used at 100% of maximum intensity. For pol β imaging, the 488 nm laser line with a 505–550 bandpass filter was used with a gain setting of 650 or less for all quantitative imaging acquisition. Gain setting was determined by examining pol β null cells stained with the pol β specific antibody (ab26343, Abcam) and the fluorescent secondary antibody. Non-specific staining was observed at gain settings higher than 700. Images of cells containing the non-specific stain at a gain setting of ∼850 showed a non-specific nuclear to cytoplasmic (βN/βC) ratio of 0.9 ± 0.01 (mean intensity ± standard error of mean). To control for this non-specific staining effect, a gain setting of 650 was chosen for imaging of the transiently transfected pol β null cells, where non-specific staining throughout the cell was no longer observed. RFP imaging was done using the 543 nm laser line with a 560–615 bandpass to confirm the presence of transfected pol β in the pol β null cells, since transfection efficiency in the cells was typically 50–60%. DAPI imaging was with the 364 nm laser line and 385–470 bandpass filter. Two-dimensional images were acquired and DAPI staining was used to select the best focal plane for nuclear imaging. Images were acquired with a pinhole of 1 airy unit, a zoom of 1.0. Zen 2009 software was used for all image acquisition.
For the stably transfected cell lines, fluorescence images were acquired with a 40X C-Apochromat (numerical aperture 1.2) water immersion objective coupled to a Zeiss LSM 780 confocal microscope. Multi-track configuration was used to ensure the absence of excitation cross-talk or emission bleed-through between channels. The 405 nm laser line with a 417–480 bandpass filter was used at 1% maximum intensity for DAPI imaging, and the 561 nm laser line with a 562–615 bandpass filter was used at 1.5% of maximum intensity for pol β imaging. A gain setting of 500 was used for all quantitative imaging acquisition. The gain setting was determined using the same non-specific staining method described for the transiently transfected cells, though non-specific staining could not be completely eliminated without significant loss of brightness in the stable cells. An optimal nuclear slice was determined, and images were acquired with a pin hole of 1 AU and a zoom of 1.0. Zen 2012 software was used for all image acquisition.
Fluorescence intensity analysis
Images of the transiently and stably transfected cells were analyzed using MetaMorph (Molecular Devices, Sunnyvale, CA, USA). Nuclear and cytoplasmic boundaries were determined using the images taken from the DAPI and Alexa 488 or 546 channels, respectively. The intensity of the pol β for each of these regions was determined from the Alexa channel. The intensity indicates the relative amount of pol β that is localized to either cellular compartment. The ratio of the nuclear intensity of Alexa 488 or 546 to the cytoplasmic intensity of Alexa 488 or 546 was taken to represent the extent of nuclear localization. A total of 40–60 cells were analyzed in this manner for the transiently expressed wild-type pol β and pol β(R4S,K5S) in the pol β null cell lines, and for the stably transfected pol β WT 96 and pol β(R4S,K5S) 18 cell lines. Nuclear:cytoplasmic ratio (βN/βC) of all cells for each construct were averaged to determine a mean value. Values that were two standard deviations above or below the mean value were determined to be outliers and were eliminated. The data were analyzed by means of one-way analysis of variance and Student's t-test.
Cytotoxicity studies
Hypersensitivity to MMS is the hallmark phenotype of pol β deficiency. Cytotoxicity was determined by growth inhibition assays as described previously (70). Stably transfected cells (pol β WT 96, pol β(R4S,K5S) 18, and pol β null) were seeded in six-well dishes at a density of 40,000 cells/well. The following day they were treated for 1 h with a range of concentrations of MMS. Cells were then washed in Hanks' balanced salt solution (Life Technologies) and fresh medium was added. Dishes were incubated for 6 to 7 days at 37°C in a 10% CO2 incubator until untreated control cells were approximately 80% confluent. Cells (triplicate wells for each MMS concentration) were counted by a cell lysis procedure (71). Results were expressed as the number of cells in MMS-treated wells relative to untreated cells (% control growth).
RESULTS
The N-terminal lyase domain of pol β binds to Impα1ΔIBB
An initial in silico analysis of pol β using the program NLS Mapper (72) identified an extended N-terminal 28 amino-acid sequence beginning at residue 3: KRKAPQETLNGGITDMLTELANFEKNVS, as a possible, though low-scoring, bipartite NLS. A bipartite type of interaction of this peptide with Impα would require complete unfolding of the N-terminal helix A (Supplementary Figure S1). Since even an L22P point mutation is sufficient to result in unfolding of the entire lyase domain (17), this NLS identification was rejected. However, further evaluation of the extensive structural database for pol β indicates that electron density is generally absent or very limited for residues preceding positions 10 or 11, indicating that the 10 N-terminal residues are disordered and readily available for additional interactions. Furthermore, helix A contains a pair of consecutive glycine residues at positions 13 and 14 that serve no obvious structural or functional role. Glycine residues are generally not particularly favorable for α-helix formation, e.g. (73,74), but consecutive glycine residues have been identified in several proteins that utilize a conformational switch (75,76). We thus considered it feasible that the entire N-terminal sequence from S2-G14 could participate in interactions with an importin carrier protein, assuming M1 will be removed by methionine aminopeptidase (77,78).
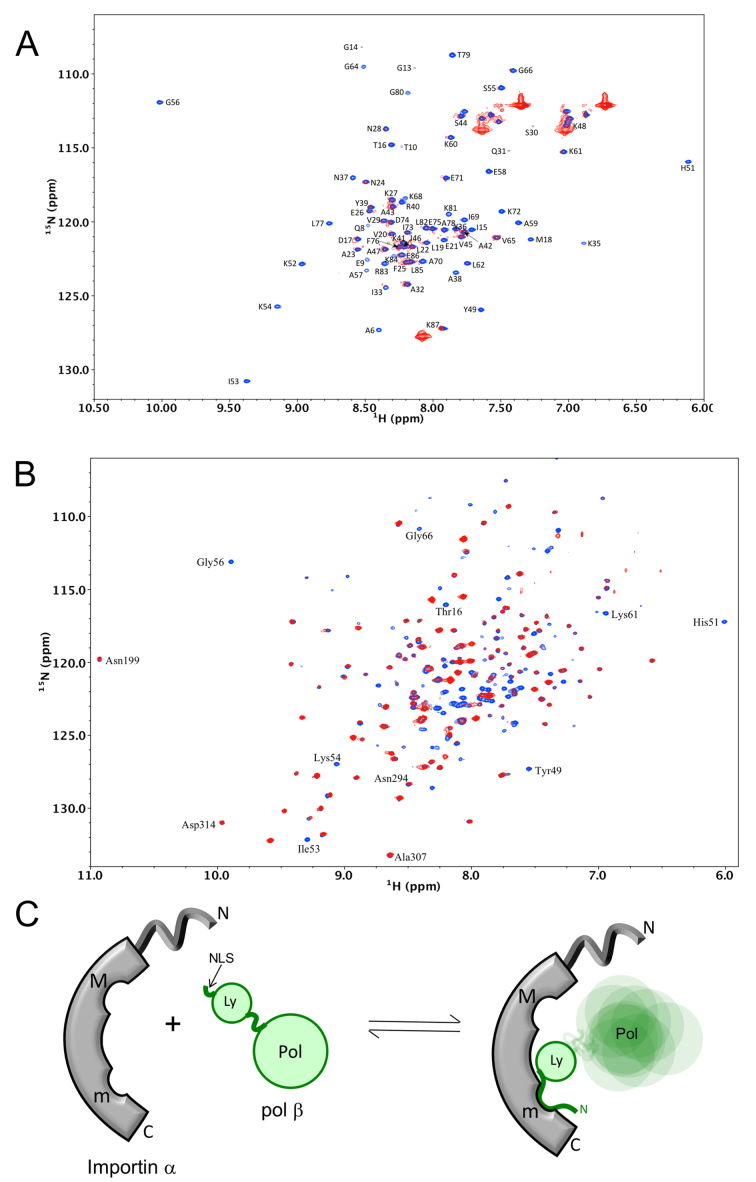
Analysis of samples containing a mixture of the pol β lyase domain (residues 2–87) and mImpα1ΔIBB by gel filtration indicated an interaction between the two proteins (Supplementary Figure S2), where the ΔIBB construct lacking the autoinhibitory Importin β binding domain was used in order to expose the NLS binding pockets, as is customary in most NLS studies (43,56,79). Note that the mouse and human wild-type pol β sequences are identical for the first 19 amino acids so their NLS interactions should also be identical. The binding result prompted us to compare the 1H-15N HSQC spectrum of U-[15N] pol β lyase domain in the absence or presence of mImpα1ΔIBB (Figure 1A). As is immediately apparent from the spectra, the binding affinity is sufficient to yield a lyase domain-mImpα1ΔIBB complex that displays broadened lyase domain amide resonances. Several less severely broadened resonances correspond to residues in mobile loop regions of the domain. The loss of intensity in the HSQC experiment is consistent with a reduction in the transverse relaxation times resulting from formation of a complex of the 8 kDa lyase domain with the 50 kDa mImpα1ΔIBB.
Figure 1.
Nuclear magnetic resonance (NMR) characterization of the pol β-Impα interaction. (A) Overlay of 1H-15N HSQC spectra of 130 μM U-[15N] pol β(1-87) in the absence (blue) or the presence (red) of an equal concentration of mImpα1ΔIBB. (B) Overlay of 1H-15N HSQC spectra of 130 μM U-[2H,15N] pol β in the absence (blue) or the presence (red) of 130 μM mImpα1ΔIBB (an expanded view illustrating the selectivity of mImpα1-induced immobilization is shown in Supplementary Figure S3). (C) Schematic illustration of the domain-selective immobilization of pol β resulting from Impα binding. The NMR buffer contained 50 mM Tris-d11 (pH 7.6), 150 mM KCl, 1 mM CDTA, 1 mM dithiothreitol (DTT), 0.1 mM AEBSF, 0.04% NaN3 and 50 μM DSS as an internal chemical shift standard, with 10% D2O for the lock.
In order to further characterize the interaction, similar studies were performed using the full-length U-[2H,15N] pol β. In this case, addition of mImpα1ΔIBB produces a domain-selective effect, reducing the intensity of most of the lyase domain amide resonances, while having a minimal effect on resonances corresponding to the polymerase domain (Figure 1B). An expanded view of the spectrum reveals a broadening pattern similar to that obtained for the isolated lyase domain (Supplementary Figure S3). These results indicate a lyase domain-specific interaction with mImpα1ΔIBB, allowing the flexibly linked polymerase domain considerable motional freedom (schematic Figure 1C).
Affinity and specificity of the pol β NLS – Impα interaction
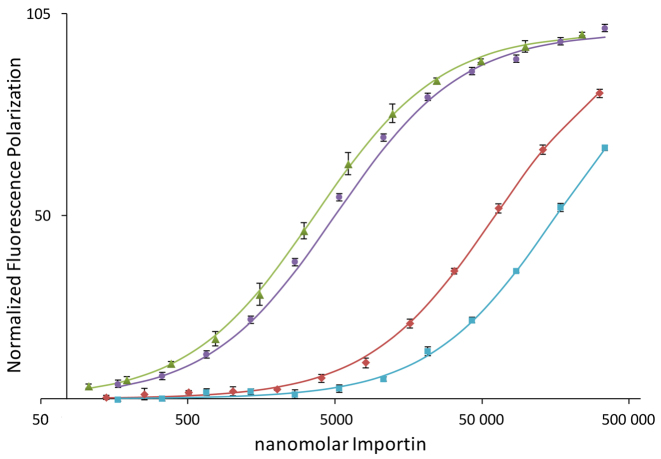
Additional insight into the pol β NLS-Impα interaction was derived from fluorescence polarization measurements utilizing a fluorescein-NLS peptide adduct. Fluorescence polarization studies were performed using full-length mImpα1, the ΔIBB form lacking the Importin β binding domain, as well as two ΔIBB constructs in which either the major or minor binding pocket was blocked: major-site blocked analog, mImpα1ΔIBB(W184R,W241R); minor-site blocked analog, mImpα1ΔIBB(W357R,W399R). For both variant constructs, a pair of Trp residues that contribute to NLS binding is replaced by a pair of Arg residues that are expected to interact unfavorably with the positively charged NLS peptides (56). Titration curves showing the normalized fluorescence polarization as a function of the concentration of the mImpα1 analogs are shown in Figure 2, and apparent Kd values are summarized in Table 1. Binding to full length Impα1 is weak, yielding a measured Kd value of 54.3 μM, while in the absence of the N-terminal IBB domain, the apparent Kd drops to 2.0 μM; this 27-fold increase in affinity demonstrates the specificity of the pol β peptide for the cargo-binding region of Impα1. The titration studies using the major and minor site-blocked mImpα1ΔIBB analogs further demonstrate that the interaction is highly selective for the minor binding pocket of mImpα1, with KdMaj = 225 μM and KdMin = 4.5 μM (Figure 2 and Table 1).
Figure 2.
Binding of the pol β nuclear localization signal (NLS) to mImpα constructs. Fluorescence polarization studies of the fluorescein-labeled pol β NLS peptide as a function of Impα concentration corresponding to: mImpα1ΔIBB (green triangles); full-length mImpα1 (red diamonds); major pocket mutant—mImpα1ΔIBB(W184R,W231R) (purple circles); or minor pocket mutant—mImpα1ΔIBB(W357R,W399R) (cyan squares). Error bars show the standard deviation.
Table 1. pol β NLS dissociation constants.
| Ligand | Target | Kd (μM) |
|---|---|---|
| SKRKAPQETLNGG[Lys(FITC)]a | mImpα1ΔIBB | 3.6 ± 0.4 |
| SKRKAPQETLNGG[Lys(FITC)]a | mImpα1 | 61.8 ± 1.7 |
| SKRKAPQETLNGG[Lys(FITC)]a | mImpα1ΔIBB (W357R,W399R) Minor site blocked | 225 ± 11 |
| SKRKAPQETLNGG[Lys(FITC)]a | mImpα1ΔIBB (W184R,W231R) Major site blocked | 4.5 ± 0.5 |
| SKRKAPQETLNGG[Lys(FITC)]a | Impα5ΔIBB | 0.720 ± 0.030 |
| Pol β lyase domainb | mImpα1ΔIBB | 0.140 ± 0.017 |
| Pol β(R4S,K5S) lyase domainb | Impα5ΔIBB | 530 ± 2 |
| Pol β lyase domainb | Impα5ΔIBB | 0.017 ± 0.006 |
aKd values determined by fluorescence polarization.
bKd determined by ligand displacement.
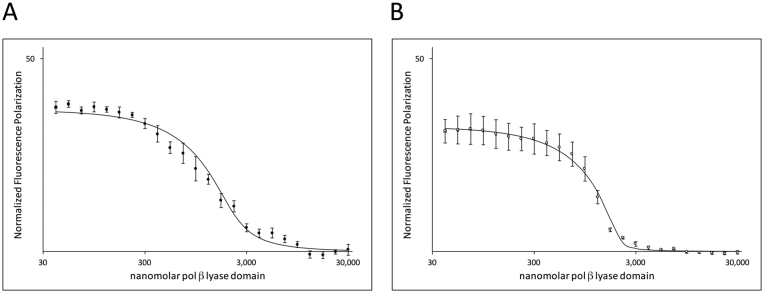
The NMR data shown in Figure 1 indicate that the entire lyase domain experiences substantial immobilization upon complex formation with mImpα1ΔIBB, suggesting the possibility that the interaction may also involve other lyase domain residues in addition to those of the N-terminal segment. This possibility is also suggested by crystallographic studies of the complexes formed between Impα and the C-terminal domain of influenza virus polymerase PB2 subunit. These complexes demonstrate an interaction that involves both the NLS peptide sequence, as well as a small globular domain adjacent to the NLS sequence (80,81). In order to evaluate whether the entire lyase domain of pol β contributes to Impα binding, we performed a fluorescence polarization ligand displacement assay, in which the lyase domain was titrated into a sample initially containing the fluorescein-labeled NLS pol β peptide plus mImpα1ΔIBB. As shown in Figure 3A, displacement of the bound fluorescein-NLS peptide by the (unlabeled) lyase domain eliminates the polarization effect that results from binding of the fluorescein peptide to mImpα1ΔIBB. Analysis of the data based on a ligand competition experiment yielded an apparent Kd = 140 nM for the full lyase domain, consistent with the conclusion that the interaction with the domain is considerably stronger than the interaction with the 13-residue N-terminal NLS peptide. A similar ligand displacement study using an unlabeled pol β lyase domain double mutant (R4S,K5S) generated an apparent Kd = 450 μM (Table 1). The (R4S,K5S) mutations preserve the hydrophilicity of the NLS but eliminate interactions that typically are required for binding affinity with Impα (43,82–84), thus confirming that the intact NLS is required for binding of the lyase domain to Impα.
Figure 3.
Binding of the pol β lyase domain to mImpα1ΔIBB and hImpα5ΔIBB. Ligand displacement studies in which the pol β lyase domain, pol β(1-87), was titrated into a sample containing 100 nM of the fluorescein labeled NLS peptide plus: (A) 2 μM murine Impα1ΔIBB or (B) 2 μM human Imp5αΔIBB. The initial fluorescence polarization value corresponds to the fraction of NLS peptide complexed with Impα, so that 100 would correspond to a fully complexed peptide. Displacement of the fluorescein-NLS peptide by the pol β lyase domain reduces the fluorescence polarization, allowing determination of the apparent dissociation constant for the interaction of the Impα construct with the lyase domain. Error bars show the standard deviation.
Involvement of another Impα family member
The NLS-binding regions of the Impα isoforms are well conserved throughout the Impα family, leading to Kd values for NLS peptides that are generally similar among family members even though there is some specificity for cargo proteins (85–88). Based on the structural results obtained by Tarendau et al. (80), we tested human Impα5ΔIBB in order to evaluate whether this family member exhibited enhanced binding to the full lyase domain. A titration yielded a Kd = 718 nM for binding of the NLS peptide to hImpα5ΔIBB, about 3-fold lower than the Kd obtained using murine Impα1ΔIBB (Table 1). Similarly, a ligand displacement study akin to that discussed above gave a Kd = 17 nM for the interaction of the pol β lyase domain with hImpα5ΔIBB (Figure 3B), corresponding to 8-fold increase in affinity relative to the value obtained using the mImpα1ΔIBB.
NLS modification abolishes complex formation between hImpα5ΔIBB and pol β lyase domain
In order to further characterize the interaction between Impα5 and the pol β lyase domain, we again made use of the R4S,K5S NLS variant of the domain. Analytical gel filtration experiments performed on mixtures of hImpα5ΔIBB and either wild-type or R4S,K5S lyase domain (Supplementary Figure S4) indicate that no stable complex of R4S,K5S lyase domain elutes from the column, while wild-type lyase domain does elute as a complex.
The conformational activation of pol β is not altered by NLS modification
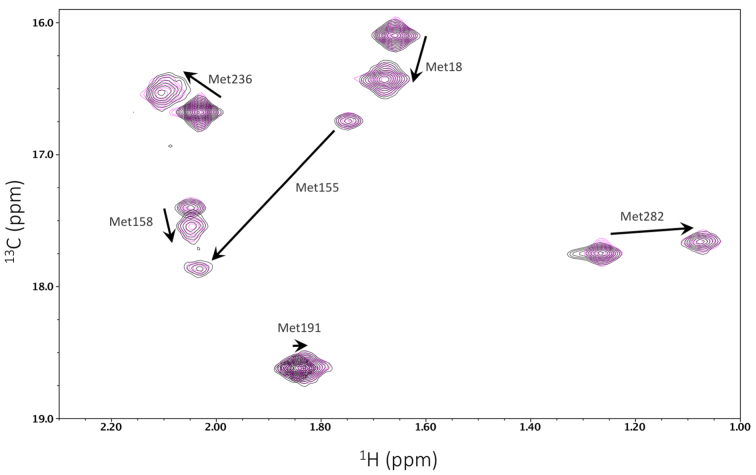
As outlined previously, pol β undergoes a ligand dependent conformational activation when an incoming dNTP that is complementary to the templating base binds to the enzyme (61,62). This activation is conveniently monitored by NMR analysis of the methionine methyl resonances of [13CH3-Met] pol β. It was anticipated that mutational variation of the disordered N-terminal segment would not significantly influence the conformational response of the enzyme. This assumption was evaluated by comparing the substrate response of [methyl-13C]methionine-labeled pol β(R4S,K5S) with the response of the wild-type enzyme. Methionine resonances have been shown to be sensitive to proper folding, substrate binding, and catalytic activation when the correct incoming nucleotide is present (61,62). The results of these studies indicate no differences between wild-type pol β and pol β(R4S,K5S) in either the uncomplexed state or in the ternary complex with one-nucleotide gapped DNA and the non-hydrolyzable nucleotide analog dAPCPP (Figure 4). Hence, as expected, the introduction of these NLS mutations has no impact on the folding or substrate-dependent conformational responses of the enzyme, and the in vivo effects of the mutations may be attributable to localization differences.
Figure 4.
Substrate-dependent conformational activation of pol β is unchanged by mutations in the NLS sequence. The 1H-13C HSQC spectra of [13CH3-Met] pol β and a ternary complex of gapped DNA•Mg-dAPCPP•pol β (black) are overlaid with spectra of [13CH3-Met] pol β(R4S,K5S) and a complex of gapped DNA•Mg-dAPCPP•pol β(R4S,K5S) (magenta). The arrows show the repositioning of the resonances in response to the addition of substrates. The N-terminal mutations apparently exert no effect on the conformational response of the enzyme to substrates.
Nuclear localization of wild-type and NLS-mutated pol β
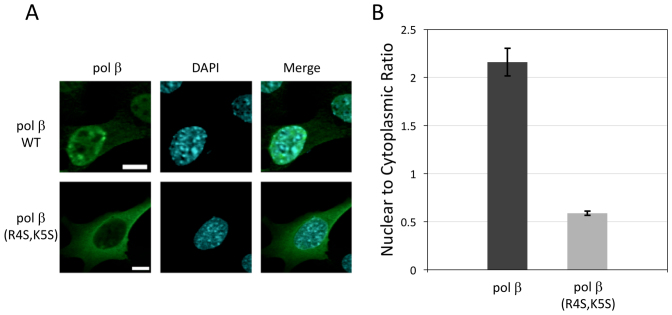
In order to evaluate the nuclear localization role of the pol β Impα-binding motif, we evaluated the cellular distribution of both wild-type and NLS variant pol β transiently expressed in pol β null mouse embryonic fibroblasts. We studied wild-type pol β and an N-terminal analog: pol β(R4S,K5S) where residues 4 and 5 are replaced by serines. Fluorescent images were obtained for cells transiently expressing wild-type or pol β(R4S,K5S) (Figure 5). Consistent with expectations based on Impα binding affinity, the pol β(R4S,K5S) variant exhibited no significant localization preference for the nucleus (Figure 5). The nuclear:cytoplasmic ratio for wild-type pol β (βN/βC) was 2.22 ± 0.15, while the localization of the R4S,K5S variant was significantly lower, with a βN/βC ratio of 0.58 ± 0.02 (P < 0.001, Figure 5B). These results indicate that the binding data summarized in Table 1 correspond to a functional NLS for pol β. Further, the results indicate that all other contributions to the nuclear localization of pol β fail to significantly increase the βN/βC ratio.
Figure 5.
Subcellular distribution of pol β. (A) The pol β (Alexa 488) panel shows the cellular distribution of wild-type pol β and the pol β(R4S,K5S) NLS variant transiently transfected into pol β null cells. The DAPI column (DNA staining) defines the cell nucleus. The Merge column superimposes the Alexa 488 stain and the nuclear stain to reveal the extent of nuclear localization for each pol β construct. Representative cells are shown. Scale bar is 10 μm. (B) Bar graph showing the effect on nuclear localization of mutations made to the pol β NLS. The data represent imaging of between 40 and 60 cells and error bars show the standard error of the mean.
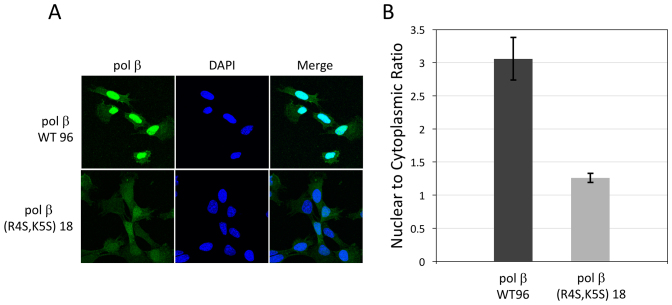
To confirm that localization was due to the NLS mutation and was not a result of the transient expression of pol β(R4S,K5S), we created stable cell lines expressing either wild-type pol β (pol β WT clone 96) or the pol β(R4S,K5S) variant (pol β(R4S,K5S) clone 18) in the pol β null background, and examined the nuclear localization by fluorescence microscopy (Figure 6). Inspection of the images reveals a localization pattern that is qualitatively similar to that observed for the transiently transfected cells. The images shown in columns 1 and 3 of Figure 6A demonstrate a decrease in the βN/βC ratio for cells expressing the pol β(R4S,K5S) variant—βN/βC ratio = 1.26 ± 0.07—relative to cells expressing pol β wild-type enzyme—βN/βC ratio = 3.06 ± 0.32 (Figure 6B).
Figure 6.
Distribution of wild-type pol β and pol β(R4S,K5S) NLS variant in stable cell lines. (A) Fluorescence images of cells stained with a pol β primary and fluorescent secondary antibody, with DAPI to indicate nuclear location and merged displays as described in Figure 5. Representative cells are shown. (B) Bar graph showing the effect on nuclear localization of mutations made to the pol β NLS. The data represent imaging of 56 and 49 cells, for pol β WT 96 and pol β(R4S,K5S) 18, respectively. Error bars show the standard error of the mean.
We also evaluated the possible contribution of XRCC1-mediated co-transport of pol β using fluorescence imaging of Xrcc1-/- cells. As shown in Supplementary Figure S5, the βN/βC ratio was within the experimental error of the value for the cells containing XRCC1. Thus, despite the ability to form a high affinity XRCC1-pol β complex (52,54), XRCC1-mediated uptake does not significantly alter the nuclear/cytoplasmic ratio of pol β.
Dependence of MMS sensitivity on a functional NLS
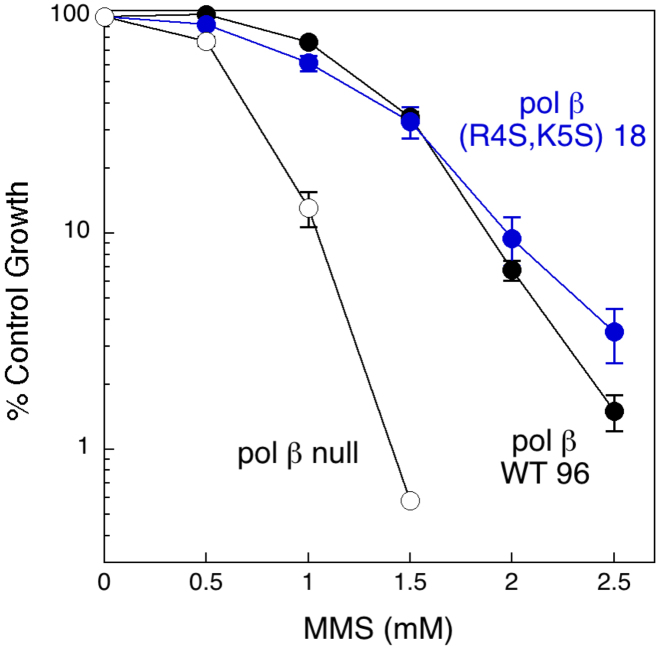
Pol β has been demonstrated to play an important role in the protection of cells against MMS-induced cytotoxic DNA damage (1,58). In order to assess the possible importance of the putative pol β NLS for the repair activity of pol β, we challenged the stable cell lines expressing the wild-type or polβ(R4S,K5S) variant for 1 h with the alkylating agent MMS and then cultured them to assess viability. No significant difference in MMS sensitivity was observed between the two cell lines (Figure 7). Given that the mutant NLS does not alter the conformational activity of pol β (Figure 4), we conclude that in this assay, the decreased nuclear level of the pol β(R4S,K5S) variant remains above the threshold required for pol β-dependent base excision repair of MMS-induced damage. Repair of the alkylation damage is thus not limited by the availability of pol β at the lower concentration achieved in cells containing the NLS-mutated enzyme. The observed growth inhibition of the cells treated with higher MMS concentrations indicates a failure of all available repair pathways to deal with the damage.
Figure 7.
Effect of NLS inactivation on sensitivity to methyl methanesulfonate (MMS). Pol β null cells (open circles), or cells expressing wild-type pol β WT 96 (black circles), or the NLS double mutant pol β(R4S,K5S) 18 (blue circles) were treated for 1 h with MMS at the concentrations indicated. Survival was analyzed by a growth inhibition assay.
DISCUSSION
The studies presented here demonstrate that: (i) pol β contains an unstructured N-terminal motif that is available for binding to Impα; (ii) the pol β-Impα interaction involves formation of a specific complex with the Impα minor site; (iii) stronger interactions are observed with hImpα5 than with mImpα1 and for the full lyase domain compared with the N-terminal NLS peptide; (iv) mutating the putative pol β NLS to a non-binding sequence eliminates the interaction with Impα and significantly reduces the nuclear accumulation of the enzyme; (v) although reduced nuclear pol β levels may significantly impair the DNA repair functions of pol β, no effect was observed in the MMS cytotoxicity assay.
Although not initially anticipated, the specific preference of the pol β NLS for the Impα minor binding pocket is consistent with results in the literature. Ligands that effectively target the Impα major site generally exhibit two characteristics: (i) a consensus binding motif defined as: K(K/R)X(K/R) (83), and (ii) a minimum of three residues N-terminal to the binding motif that make non-specific contacts with the Impα, often involving H-bond interactions with the backbone carbonyl groups (56). The pol β NLS lacks both of these characteristics; after removal of the N-terminal methionine by methionine aminopeptidase (77,78,89), only a single residue precedes the KRK motif. The pol β SKRKA sequence is, however, quite similar to other sequences shown to have high affinity for the Impα minor pocket (82,84), including the recently determined GKRKL minor site motif of the XRCC1 bipartite NLS (56). Comparative sequence analysis indicates that the KRKxP motif in pol β is highly conserved among higher eukaryotes (Supplementary Figure S6). In addition to the minor site binding motif, Pro7 ensures that the N-terminus does not form an extension of helix A1.
Mutations in the unstructured N-terminus of pol β have not been reported in studies of tumor-associated variants (15,19,90). Examination of the dbSNP indicates rare polymorphisms at positions 1, 8, and 11, but none have been reported for the critical K3RK5 residues or for the immediately flanking residues that may bind directly to the minor site of Impα. Thus, the identified polymorphisms might at most be expected to exert a very weak effect on the Impα interaction. Conversely, the fact that an unstructured N-terminal sequence is so well conserved supports the conclusion that the pol β NLS makes a significant contribution to the cellular functions of the enzyme.
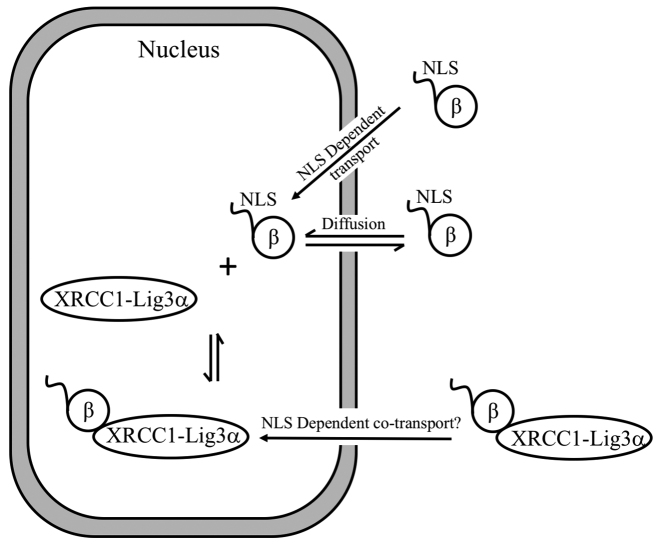
Based on our results, the cellular distribution of pol β is, in principle, dependent on at least three transport pathways: (i) passive diffusion through the nuclear pore; (ii) possible co-transport involving XRCC1 or other DNA repair proteins; (iii) active nuclear uptake by NLS-dependent classical nuclear transport (Figure 8). Since pol β is well below the size threshold for passive diffusion (44), equilibration of the nuclear and cytoplasmic pools due to diffusion is likely to be important for this enzyme. For both transiently transfected and stable cell lines containing NLS-mutated pol β(R4S,K5S), our results were close to the ratio of 1.0 expected if passive transport plays a dominant role. The similar nuclear/cytosolic pol β ratios exhibited by the Xrcc1+/+ and Xrcc1−/− cells (Supplementary Figure S5), indicate that any additional nuclear uptake resulting from XRCC1-dependent co-transport is either absent or insufficient to significantly alter this ratio. Thus, pol β appears to be sufficiently small to preclude development of a significant nuclear concentration gradient in the absence of active transport. Alternatively, a nuclear/cytosolic pol β concentration gradient is developed in the presence of an NLS-dependent active transport process.
Figure 8.
Schematic illustration of pathways contributing to the nucelar/cytoplasmic distribution of pol β. A possible co-transport pathway with XRCC1 is illustrated, although there is no evidence for a contribution by this pathway. A substantial flux due to passive diffusion of pol β will tend to equilibrate the nuclear and cytosoplasmic levels, leading to a requirement for a more active, NLS-dependent uptake mechanism to produce a concentration difference.
Given the apparent importance of pol β expression levels for optimizing genome stability (25,26,28), it is not surprising that this distribution is under the control of a functional NLS, rather than depending primarily on passive diffusion or co-transport mechanisms. Nuclear transport by the Impα/β system is bidirectional (91), so that nuclear import is determined by a combination of import and export rate constants:
 |
leading to a steady state ratio of the nuclear and cytosolic pol β pools given by:
 |
If the only mechanism for nuclear transport were passive diffusion, which presumably contributes equally to kin and kout, then the net nuclear excess of pol β would depend primarily on contributions from various binding interactions, such as the interaction with XRCC1, as well as on degradation processes, such as CHIP-mediated proteasomal degradation (60,92). Based on the schematic pathways shown in Figure 8, the lack of an effect of XRCC1 on pol β distribution (Supplementary Figure S5) is not surprising. If XRCC1 does function as a pol β co-transport protein, the nuclear XRCC1-pol β complex remains subject to dissociation, after which the small pol β molecule can exit the nucleus via passive diffusion. As in the example of pol β, histones are small enough to enter/exit the nucleus via passive diffusion, but also utilize NLS-dependent nuclear import, although involving different importins (93) as well as co-transport mechanisms (94). Thus, NLS-dependent uptake mechanisms provide a consistent basis for maintaining a net nuclear excess against a background of passive diffusion.
The position of the NLS at the N-terminus and the enhanced affinity found for the lyase domain further substantiate the important role that this domain plays in DNA repair (59). These results are also consistent with data indicating that at least a portion of the pol β repair function is independent of XRCC1 (58,59). Identification of a pol β NLS provides a new tool that can be used for altering the nuclear/cytosolic distribution of this critical repair enzyme, and for understanding the role that dysregulation of pol β subcellular distribution may play in the etiology of various diseases.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to C. Jeff Tucker and to Dr Agnes K. Janoshazi at the NIEHS Fluorescence Microscopy and Imaging Center for their expert assistance with the implementation and analysis of the localization studies, and to Scott A. Gabel and Eugene F. DeRose for assistance with the experimental studies.
Footnotes
Present addresses:
Natalie R. Gassman, University of South Alabama Mitchell Cancer Institute, Molecular & Metabolic Oncology, 1660 Springhill Ave, Mobile, AL 36604-1405, USA.
Cassandra Smith, University of Colorado School of Medicine, Anschutz Medical Campus, Campus Box: C296 AMC, Aurora, CO 80045, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the NIH, National Institute of Environmental Health Sciences [project number ZIA ES050111 to R.E.L. and project numbers Z01 ES050158 and ES050159 to S.H.W., R00ES023813 to N.R.G., in part]. Funding for open access charge: Intramural Research Program of the NIH, National Institute of Environmental Health Sciences [project number ZIA ES050111].
Conflict of interest statement. None declared.
REFERENCES
- 1.Sobol R.W., Horton J.K., Kuhn R., Gu H., Singhal R.K., Prasad R., Rajewsky K., Wilson S.H.. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996; 379:183–186. [DOI] [PubMed] [Google Scholar]
- 2.Crespan E., Czabany T., Maga G., Hubscher U.. Microhomology-mediated DNA strand annealing and elongation by human DNA polymerases λ and β on normal and repetitive DNA sequences. Nucleic Acids Res. 2012; 40:5577–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.London R.E. The structural basis of XRCC1-mediated DNA repair. DNA Repair (Amst). 2015; 30:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith C.A., Okumoto D.S.. Nature of DNA repair synthesis resistant to inhibitors of polymerase α in human cells. Biochemistry. 1984; 23:1383–1391. [DOI] [PubMed] [Google Scholar]
- 5.Belousova E.A., Maga G., Fan Y., Kubareva E.A., Romanova E.A., Lebedeva N.A., Oretskaya T.S., Lavrik O.I.. DNA Polymerases β and λ Bypass Thymine Glycol in Gapped DNA Structures. Biochemistry. 2010; 49:4695–4704. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann J.S., Pillaire M.J., Maga G., Podust V., Hübscher U., Villani G.. DNA polymerase β bypasses in vitro a single d(GpG)-cisplatin adduct placed on codon 13 of the HRAS gene. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5356–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaiswal A.S., Banerjee S., Aneja R., Sarkar F.H., Ostrov D.A., Narayan S.. DNA polymerase β as a novel target for chemotherapeutic intervention of colorectal cancer. PLoS One. 2011; 6:e16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolay N.H., Helleday T., Sharma R.A.. Biological relevance of DNA polymerase β and translesion synthesis polymerases to cancer and its treatment. Curr. Mol. Pharmacol. 2012; 5:54–67. [PubMed] [Google Scholar]
- 9.Smith L.A., Makarova A.V., Samson L., Thiesen K.E., Dhar A., Bessho T.. Bypass of a psoralen DNA interstrand cross-link by DNA polymerases β, ι, and κ in vitro. Biochemistry. 2012; 51:8931–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stachelek G.C., Dalal S., Donigan K.A., Campisi Hegan D., Sweasy J.B., Glazer P.M.. Potentiation of temozolomide cytotoxicity by inhibition of DNA polymerase β is accentuated by BRCA2 mutation. Cancer Res. 2010; 70:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang J.B., Svilar D., Trivedi R.N., Wang X.H., Goellner E.M., Moore B., Hamilton R.L., Banze L.A., Brown A.R., Sobol R.W.. N-methylpurine DNA glycosylase and DNA polymerase β modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro Oncol. 2011; 13:471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trivedi R.N., Wang X.H., Jelezcova E., Goellner E.M., Tang J.B., Sobol R.W.. Human methyl purine DNA glycosylase and DNA polymerase β expression collectively predict sensitivity to temozolomide. Mol. Pharmacol. 2008; 74:505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An C.L., Chen D., Makridakis N.M.. Systematic biochemical analysis of somatic missense mutations in DNA polymerase β found in prostate cancer reveal alteration of enzymatic function. Hum. Mutat. 2011; 32:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobashi Y., Shuin T., Tsuruga H., Uemura H., Torigoe S., Kubota Y.. DNA polymerase β gene mutation in human prostate cancer. Cancer Res. 1994; 54:2827–2829. [PubMed] [Google Scholar]
- 15.Donigan K.A., Sun K.-w., Nemec A.A., Murphy D.L., Cong X., Northrup V., Zelterman D., Sweasy J.B.. Human POLB gene is mutated in high percentage of colorectal tumors. J. Biol. Chem. 2012; 287:23830–23839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwanaga A., Ouchida M., Miyazaki K., Hori K., Mukai T.. Functional mutation of DNA polymerase β found in human gastric cancer - inability of the base excision repair in vitro. Mutat. Res. 1999; 435:121–128. [DOI] [PubMed] [Google Scholar]
- 17.Kirby T.W., DeRose E.F., Beard W.A., Shock D.D., Wilson S.H., London R.E.. Substrate rescue of DNA polymerase β containing a catastrophic L22P mutation. Biochemistry. 2014; 53:2413–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sliwinski T., Ziemba P., Morawiec Z., Kowalski M., Zadrozny M., Blasiak J.. Polymorphisms of the DNA polymerase β gene in breast cancer. Breast Cancer Res. Treat. 2007; 103:161–166. [DOI] [PubMed] [Google Scholar]
- 19.Starcevic D., Dalal S., Sweasy J.B.. Is there a link between DNA polymerase β and cancer. Cell Cycle. 2004; 3:996–999. [PubMed] [Google Scholar]
- 20.Wallace S.S., Murphy D.L., Sweasy J.B.. Base excision repair and cancer. Cancer Lett. 2012; 327:73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonelli V., D'Errico M., Palli D., Prasad R., Wilson S.H., Dogliotti E.. Characterization of DNA polymerase β splicing variants in gastric cancer: the most frequent exon 2-deleted isoform is a non-coding RNA. Mutat. Res. 2009; 670:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabelof D.C., Ikeno Y., Nyska A., Busuttil R.A., Anyangwe N., Vijg J., Matherly L.H., Tucker J.D., Wilson S.H., Richardson A. et al. . Haploinsufficiency in DNA polymerase β increases cancer risk with age and alters mortality rate. Cancer Res. 2006; 66:7460–7465. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava D.K., Husain I., Arteaga C.L., Wilson S.H.. DNA polymerase β expression differences in selected human tumors and cell lines. Carcinogenesis. 1999; 20:1049–1054. [DOI] [PubMed] [Google Scholar]
- 24.Canitrot Y., Cazaux C., Frechet M., Bouayadi K., Lesca C., Salles B., Hoffmann J.S.. Overexpression of DNA polymerase β in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12586–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel-Fatah T.M.A., Russell R., Agarwal D., Moseley P., Abayomi M.A., Perry C., Albarakati N., Ball G., Chan S., Caldas C. et al. . DNA polymerase β deficiency is linked to aggressive breast cancer: A comprehensive analysis of gene copy number, mRNA and protein expression in multiple cohorts. Mol. Oncol. 2014; 8:520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergoglio V., Pillaire M.J., Lacroix-Triki M., Raynaud-Messina B., Canitrot Y., Bieth A., Gares M., Wright M., Delsol G., Loeb L.A. et al. . Deregulated DNA polymerase β induces chromosome instability and tumorigenesis. Cancer Res. 2002; 62:3511–3514. [PubMed] [Google Scholar]
- 27.Scanlon K.J., Kashanisabet M., Miyachi H.. Differential gene-expression in human cancer-cells resistant to cisplatin. Cancer Invest. 1989; 7:581–587. [DOI] [PubMed] [Google Scholar]
- 28.Sobol R.W., Foley J.F., Nyska A., Davidson M.G., Wilson S.H.. Regulated over-expression of DNA polymerase β mediates early onset cataract in mice. DNA Repair (Amst). 2003; 2:609–622. [DOI] [PubMed] [Google Scholar]
- 29.McLane L.M., Corbett A.H.. Nuclear localization signals and human disease. IUBMB Life. 2009; 61:697–706. [DOI] [PubMed] [Google Scholar]
- 30.Hung M.C., Link W.. Protein localization in disease and therapy. J. Cell Sci. 2011; 124:3381–3392. [DOI] [PubMed] [Google Scholar]
- 31.Knudsen N.O., Andersen S.D., Lutzen A., Nielsen F.C., Rasmussen L.J.. Nuclear translocation contributes to regulation of DNA excision repair activities. DNA Repair (Amst). 2009; 8:682–689. [DOI] [PubMed] [Google Scholar]
- 32.Poletto M., Lirussi L., Wilson D.M., Tell G.. Nucleophosmin modulates stability, activity, and nucleolar accumulation of base excision repair proteins. Mol. Biol. Cell. 2014; 25:1641–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opletalova K., Bourillon A., Yang W., Pouvelle C., Armier J., Despras E., Martin L., Mateus C., Robert C., Kannouche P. et al. . Correlation of phenotype/genotype in a cohort of 23 xeroderma pigmentosum-variant patients reveals 12 new disease-causing POLH mutations. Hum. Mutat. 2014; 35:117–128. [DOI] [PubMed] [Google Scholar]
- 34.Hirano M., Furiya Y., Asai H., Yasui A., Ueno S.. ALADINI482S causes selective failure of nuclear protein import and hypersensitivity to oxidative stress in triple A syndrome. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:2298–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juhlen R., Idkowiak J., Taylor A.E., Kind B., Arlt W., Huebner A., Koehler K.. Role of ALADIN in human adrenocortical cells for oxidative stress response and steroidogenesis. PLoS One. 2015; 10:e0124582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiriyama T., Hirano M., Asai H., Ikeda M., Furiya Y., Ueno S.. Restoration of nuclear-import failure caused by triple A syndrome and oxidative stress. Biochem. Biophys. Res. Commun. 2008; 374:631–634. [DOI] [PubMed] [Google Scholar]
- 37.Shimazaki N., Yoshida K., Kobayashi T., Toji S., Tamai K., Koiwai O.. Over-expression of human DNA polymerase λ in E-coli and characterization of the recombinant enzyme. Genes Cells. 2002; 7:639–651. [DOI] [PubMed] [Google Scholar]
- 38.Bentolila L.A., Dandon M.F., Nguyen Q.T., Martinez O., Rougeon F., Doyen N.. The 2 isoforms of mouse terminal deoxynucleotidyl transferase differ in both the ability to add N-regions and subcellular-localization. EMBO J. 1995; 14:4221–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominguez O., Ruiz J.F., de Lera T.L., Garcia-Diaz M., Gonzalez M.A., Kirchhoff T., Martinez-A C., Bernad A., Blanco L.. DNA polymerase μ (Pol μ), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000; 19:1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramadan K., Shevelev I., Hubscher U.. The DNA-polymerase-X family: controllers of DNA quality. Nat. Rev. Mol. Cell Biol. 2004; 5:1038–1043. [DOI] [PubMed] [Google Scholar]
- 41.Martin M.J., Blanco L.. Chen C. Evolving DNA repair polymerases: from double—strand break repair to base excision repair and VDJ recombination. New Research Directions in DNA Repair. 2013; Rijeka: InTech. [Google Scholar]
- 42.Martin M.J., Garcia-Ortiz M.V., Gomez-Bedoya A., Esteban V., Guerra S., Blanco L.. A specific N-terminal extension of the 8 kDa domain is required for DNA end-bridging by human Pol μ and Pol λ. Nucleic Acids Res. 2013; 41:9105–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H.. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 2007; 282:5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R., Brattain M.G.. The maximal size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett. 2007; 581:3164–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett R.A., Wilson D.M. 3rd, Wong D., Demple B.. Interaction of human apurinic endonuclease and DNA polymerase β in the base excision repair pathway. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Z., Zheng L., Dai H., Zhou M., Xu H., Shen B.. Human DNA polymerase β polymorphism, Arg137Gln, impairs its polymerase activity and interaction with PCNA and the cellular base excision repair capacity. Nucleic Acids Res. 2009; 37:3431–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prasad R., Singhal R.K., Srivastava D.K., Molina J.T., Tomkinson A.E., Wilson S.H.. Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 1996; 271:16000–16007. [DOI] [PubMed] [Google Scholar]
- 48.Dimitriadis E.K., Prasad R., Vaske M.K., Chen L., Tomkinson A.E., Lewis M.S., Wilson S.H.. Thermodynamics of human DNA ligase I trimerization and association with DNA polymerase β. J. Biol. Chem. 1998; 273:20540–20550. [DOI] [PubMed] [Google Scholar]
- 49.Kedar P.S., Kim S.J., Robertson A., Hou E., Prasad R., Horton J.K., Wilson S.H.. Direct interaction between mammalian DNA polymerase β and proliferating cell nuclear antigen. J. Biol. Chem. 2002; 277:31115–31123. [DOI] [PubMed] [Google Scholar]
- 50.Prasad R., Williams J.G., Hou E.W., Wilson S.H.. Pol β associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res. 2012; 40:11571–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moor N.A., Vasil'eva I.A., Anarbaev R.O., Antson A.A., Lavrik O.I.. Quantitative characterization of protein-protein complexes involved in base excision DNA repair. Nucleic Acids Res. 2015; 43:6009–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marintchev A., Gryk M.R., Mullen G.P.. Site-directed mutagenesis analysis of the structural interaction of the single-strand-break repair protein, X-ray cross-complementing group 1, with DNA polymerase β. Nucleic Acids Res. 2003; 31:580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cuneo M.J., London R.E.. Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase β binding affinity. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:6805–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gabel S.A., Smith C.E., Cuneo M.J., Mueller G.A., Kirby T.W., DeRose E.F., Krahn J.M., London R.E.. Characterization of the redox transition of the XRCC1 N-terminal domain. Structure. 2014; 22:1754–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellenberger T., Tomkinson A.E.. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008; 77:313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirby T.W., Gassman N.R., Smith C.E., Pedersen L.C., Gabel S.A., Sobhany M., Wilson S.H., London R.E.. Nuclear localization of the DNA repair scaffold XRCC1: uncovering the functional role of a bipartite NLS. Sci. Rep. 2015; 5:13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S.Y., Gong Z.H., Chen R., Liu Y.R., Li A.P., Li G., Zhou J.W.. JWA regulates XRCC1 and functions as a novel base excision repair protein in oxidative-stress-induced DNA single-strand breaks. Nucleic Acids Res. 2009; 37:1936–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horton J.K., Gassman N.R., Dunigan B.D., Stefanick D.F., Wilson S.H.. DNA polymerase β-dependent cell survival independent of XRCC1 expression. DNA Repair (Amst). 2015; 26:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sobol R.W., Prasad R., Evenski A., Baker A., Yang X.P., Horton J.K., Wilson S.H.. The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000; 405:807–810. [DOI] [PubMed] [Google Scholar]
- 60.Fang Q.M., Inanc B., Schamus S., Wang X.H., Wei L.Z., Brown A.R., Svilar D., Sugrue K.F., Goellner E.M., Zeng X.M. et al. . HSP90 regulates DNA repair via the interaction between XRCC1 and DNA polymerase β. Nat. Commun. 2014; 5:5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirby T.W., DeRose E.F., Beard W.A., Wilson S.H., London R.E.. A thymine isostere in the templating position disrupts assembly of the closed DNA polymerase β ternary complex. Biochemistry. 2005; 44:15230–15237. [DOI] [PubMed] [Google Scholar]
- 62.Bose-Basu B., DeRose E.F., Kirby T.W., Mueller G.A., Beard W.A., Wilson S.H., London R.E.. Dynamic characterization of a DNA repair enzyme: NMR studies of [methyl-13C]methionine-labeled DNA polymerase β. Biochemistry. 2004; 43:8911–8922. [DOI] [PubMed] [Google Scholar]
- 63.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967; 6:1948–1954. [DOI] [PubMed] [Google Scholar]
- 64.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 65.Johnson B.A., Blevins R.A.. NMRView: A computer-program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994; 4:603–614. [DOI] [PubMed] [Google Scholar]
- 66.Zhu G., Bax A.. Improved linear prediction of damped NMR signals using modified forward backward linear prediction. J. Magn. Reson. 1992; 100:202–207. [Google Scholar]
- 67.Tebbs R.S., Flannery M.L., Meneses J.J., Hartmann A., Tucker J.D., Thompson L.H., Cleaver J.E., Pedersen R.A.. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 1999; 208:513–529. [DOI] [PubMed] [Google Scholar]
- 68.Beard W.A., Wilson S.H.. Methods Enzymol. 1995; 262, Cambridge: Academic Press; 98–107. [DOI] [PubMed] [Google Scholar]
- 69.Gassman N.R., Stefanick D.F., Kedar P.S., Horton J.K., Wilson S.H.. Hyperactivation of PARP triggers nonhomologous end-joining in repair-deficient mouse fibroblasts. PLoS One. 2012; 7:e49301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horton J.K., Joyce-Gray D.F., Pachkowski B.F., Swenberg J.A., Wilson S.H.. Hypersensitivity of DNA polymerase β null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair (Amst). 2003; 2:27–48. [DOI] [PubMed] [Google Scholar]
- 71.Butler W.B. Preparing nuclei from cells in monolayer-cultures suitable for counting and for following synchronized cells through the cell-cycle. Anal. Biochem. 1984; 141:70–73. [DOI] [PubMed] [Google Scholar]
- 72.Kosugi S., Hasebe M., Tomita M., Yanagawa H.. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chou P.Y., Fasman G.D.. Empirical predictions of protein conformation. Annu. Rev. Biochem. 1978; 47:251–276. [DOI] [PubMed] [Google Scholar]
- 74.Malkov S.N., Zivkovic M.V., Beljanski M.V., Hall M.B., Zaric S.D.. A reexamination of the propensities of amino acids towards a particular secondary structure: classification of amino acids based on their chemical structure. J. Mol. Model. 2008; 14:769–775. [DOI] [PubMed] [Google Scholar]
- 75.Pester O., Barrett P.J., Hornburg D., Hornburg P., Probstle R., Widmaier S., Kutzner C., Durrbaum M., Kapurniotu A., Sanders C.R. et al. . The backbone dynamics of the amyloid precursor protein transmembrane helix provides a rationale for the sequential cleavage mechanism of γ-secretase. J. Am. Chem. Soc. 2013; 135:1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou Y., Xia X.M., Lingle C.J.. Cysteine scanning and modification reveal major differences between BK channels and Kv channels in the inner pore region. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12161–12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar A., Widen S.G., Williams K.R., Kedar P., Karpel R.L., Wilson S.H.. Studies of the domain-structure of mammalian DNA polymerase-β - identification of a discrete template binding domain. J. Biol. Chem. 1990; 265:2124–2131. [PubMed] [Google Scholar]
- 78.Lowther W.T., Matthews B.W.. Structure and function of the methionine aminopeptidases. Biochim. Biophys. Acta. 2000; 1477:157–167. [DOI] [PubMed] [Google Scholar]
- 79.Fontes M.R., Teh T., Kobe B.. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 2000; 297:1183–1194. [DOI] [PubMed] [Google Scholar]
- 80.Tarendeau F., Boudet J., Guilligay D., Mas P.J., Bougault C.M., Boulo S., Baudin F., Ruigrok R.W.H., Daigle N., Ellenberg J. et al. . Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 2007; 14:229–233. [DOI] [PubMed] [Google Scholar]
- 81.Pumroy R.A., Ke S., Hart D.J., Zachariae U., Cingolani G.. Molecular determinants for nuclear import of influenza A PB2 by importin α isoforms 3 and 7. Structure. 2015; 23:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang C.-W., Counago R.M., Williams S.J., Boden M., Kobe B.. Distinctive conformation of minor site-specific nuclear localization signals bound to importin-α. Traffic. 2013; 14:1144–1154. [DOI] [PubMed] [Google Scholar]
- 83.Chelsky D., Ralph R., Jonak G.. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol. Cell. Biol. 1989; 9:2487–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pang X., Zhou H.-X.. Design rules for selective binding of nuclear localization signals to minor site of importin α. PLoS One. 2014; 9:e91025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pumroy R.A., Cingolani G.. Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem. J. 2015; 466:13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boivin S., Hart D.J.. Interaction of the influenza A virus polymerase PB2 C-terminal region with importin α isoforms provides insights into host adaptation and polymerase assembly. J. Biol. Chem. 2011; 286:10439–10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zienkiewicz J., Armitage A., Hawiger J.. Targeting nuclear import shuttles, importins/karyopherins α by a peptide mimicking the NFκB1/p50 nuclear localization sequence. J. Am. Heart Assoc. 2013; 2:e000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paterson C.P., Ayalew L.E., Tikoo S.K.. Mapping of nuclear import signal and importin α3 binding regions of 52K protein of bovine adenovirus-3. Virology. 2012; 432:63–72. [DOI] [PubMed] [Google Scholar]
- 89.Bradshaw R.A., Brickey W.W., Walker K.W.. N-terminal processing: the methionine aminopeptidase and N-α-acetyl transferase families. Trends Biochem. Sci. 1998; 23:263–267. [DOI] [PubMed] [Google Scholar]
- 90.Li M., Zang W., Wang Y., Ma Y., Xuan X., Zhao J., Liu L., Dong Z., Zhao G.. DNA polymerase β mutations and survival of patients with esophageal squamous cell carcinoma in Linzhou City, China. Tumor Biol. 2014; 35:553–559. [DOI] [PubMed] [Google Scholar]
- 91.Lolodi O., Yamazaki H., Otsuka S., Kumeta M., Yoshimura S.H.. Dissecting in vivo steady-state dynamics of karyopherin-dependent nuclear transport. Mol. Biol. Cell. 2016; 27:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parsons J.L., Tait P.S., Finch D., Dianova II, Allinson S.L., Dianov G.L.. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008; 29:477–487. [DOI] [PubMed] [Google Scholar]
- 93.Baake M., Bauerle M., Doenecke D., Albig W.. Core histones and linker histones are imported into the nucleus by different pathways. Eur. J. Cell Biol. 2001; 80:669–677. [DOI] [PubMed] [Google Scholar]
- 94.Arregi I., Falces J., Bañuelos S., Urbaneja M.a.A., Taneva S.G.. The nuclear transport machinery recognizes nucleoplasmin–histone complexes. Biochemistry. 2011; 50:7104–7110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.