Abstract
Homologous recombination (HR) is a major mechanism to repair DNA double-strand breaks (DSBs). Although tumor suppressor CtIP is critical for DSB end resection, a key initial event of HR repair, the mechanism regulating the recruitment of CtIP to DSB sites remains largely unknown. Here, we show that acidic nucleoplasmic DNA‐binding protein 1 (And‐1) forms complexes with CtIP as well as other repair proteins, and is essential for HR repair by regulating DSB end resection. Furthermore, And-1 is recruited to DNA DSB sites in a manner dependent on MDC1, BRCA1 and ATM, down-regulation of And-1 impairs end resection by reducing the recruitment of CtIP to damage sites, and considerably reduces Chk1 activation and other damage response during HR repair. These findings collectively demonstrate a hitherto unknown role of MDC1→And-1→CtIP axis that regulates CtIP-mediated DNA end resection and cellular response to DSBs.
INTRODUCTION
Maintenance of genome stability depends on efficient and accurate repair of DNA damage (1,2). DNA double-strand breaks (DSBs) are among the most lethal types of DNA damage, with the potential to cause deleterious gene mutations and chromosomal rearrangement that could lead to cancer (3). DSBs can be repaired by two main mechanisms: non-homologous end-joining (NHEJ) and homologous recombination (HR). Whereas NHEJ repair predominates in cells at the G0/G1 phase in the cell cycle and is error-prone, HR repair occurs in cells at the S and G2 phases and it ensures accurate, error free repair of DNA damage (3–6).
A key event during the initial phase of HR repair is DSB end resection, a process in which DSBs undergo nucleolytic degradation of their 5΄-ending strands to generate a 3΄-end single strand (3). This end resection is essential for HR repair because the resulting 3΄-ended single-strand DNA (ssDNA) invades a homologous template to initiate HR repair (3). It is known that following DSB generation, the highly conserved MRN protein complex consisting of Mre11, Rad50 and NBS1 is the early group of proteins recruited to DSB ends in a process dependent on Rad17 and MDC1 (7–12). Then, the MRN complex initiates checkpoint signaling through recruitment and activation of ATM (13–18). It should be noted that Mre11 has DNA binding, exonuclease and endonuclease activities which are important for DSB end resection (19–21).
The tumor suppressor protein CtIP, a CtBP (carboxy-terminal binding protein)-interacting protein, is recruited onto the DSB sites in a MRN-dependent manner (22). CtIP physically interacts with MRN at DSB ends (22–24). The recruitment of CtIP depends on its phosphorylation by ATM and Cdk2 as well as ubiquitination by BRCA1 (25–28). It has been shown that CtIP promotes DNA end resection by stimulating the nuclease activity of Mre11-Rad50 complex toward ssDNA (29). Following MRN and CtIP, the Exo1 exonuclease and BLM/DNA2 helicase are loaded onto the DSB sites to carry out further resection to generate a long 3΄-ssDNA tail for HR repair (30,31). The recruitment of CtIP onto DSB sites appears to be a key step that facilitates the transition from DSB sensing by MRN to DSB end resection (32). However, the mechanism by which CtIP is recruited onto the DSB sites remains largely unknown (32). Given that the recruitment of CtIP to DSB sites occurs 5–15 min after the recruitment of NBS1 (22), it is most likely that CtIP is not passively brought to the DSB sites through its interaction with MRN and unidentified factor(s) may be required for the recruitment CtIP to sites of DSBs (22,32). Alternatively, a platform must be first established in a time-dependent fashion on the DSB sites in the presence of MRN and possibly other proteins before CtIP could be recruited to it.
MDC1 serves as an adaptor that is recruited to DSB sites by interacting with γ-H2AX, where it promotes the recruitment of MRN complex to damage sites, as well as recruitment and activation of ATM (9,11,33–35). Once recruited to the DSB sites, MDC1 is phosphorylated by ATM and acts as an a scaffold for the recruitment of ubiquitin E3 ligases RNF8 and RNF168, which function to ubiquitinate histone H2A to create a docking site for 53BP1 and BRCA1 (36–40). BRCA1 is essential for HR repair at multiple stages of HR due to its ability to interact with several repair proteins including CtIP and MRN (23,25).
And-1 is an acidic nucleoplasmic DNA-binding protein that contains an N-terminal WD40 domain and a C-terminal HMG (High Mobility Group) domain (41). Previous studies by others and us indicate that And-1 is a replisome component for efficient DNA replication (41–47). Additionally, our studies indicate that And-1 also associates with histone acetyltransferase Gcn5 to maintain its stability (48,49). Ctf4, a yeast ortholog of And-1, was found to regulate multiple pathways including chromosome transmission fidelity, sister chromatin cohesion, DNA damage repair, DNA replication and telomere replication (46,50–55). Most recently, we also discovered that And-1 is required for efficient activation of Chk1 by regulating replisome component Claspin at stalled replication forks (56). Interestingly, using reporter assays two groups have independently demonstrated that Ctf4/And-1 is involved in HR repair in both yeast and human cells (42,57). However, it remains unknown how And-1 regulates HR repair.
Here, we demonstrate, for the first time, that And-1 forms complexes with multiple HR repair proteins including CtIP, MRN, BRCA1 and MDC1, and is essential for DSB end resection in human cells. We found that And-1 is recruited to DSB sites in the early stage of DSBs. The recruitment of And-1 to DSB sites is dependent on BRCA1, MDC1 and ATM. Significantly, down-regulation of And-1 impairs the recruitment of CtIP to DSB sites and end resection, and considerably reduces Chk1 phosphorylation. These results collectively demonstrate an important role of And-1 in HR repair by regulating CtIP-mediated DSB end resection.
MATERIALS AND METHODS
Cells and cell culture
U2OS, 293T, HCC1937 and HCC1937/BRCA1 were cultured in DMEM medium supplemented with 10% FBS at 37°C in 5% CO2. HCC1937 and HCC1937/BRCA1 were kindly provided by Dr Xiaochun Yu (City of Hope).
Plasmids
Plasmid containing full-length And-1 and And-1 mutants were described previously (56). FLAG-And-1 truncated mutants (798–1129) and (898–1129) were amplified by PCR from a plasmid containing the full-length And-1 cDNA and sub-cloned into the pEFF-FLAG vector at the BamHI and NotI sites. Plasmids expressing FLAG-tagged wild-type or mutant CtIP were gifts from Dr. Junjie Chen (MD Anderson). Plasmid transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction.
siRNA interference
The following are the sequences of the siRNA duplexes: And-1-1, 5΄-AGGAAAACAUGCCUGCCACdTdT-3΄; And-1-2, 5΄-GAAGAUGGUCAAGAAGGCAGCAdTdT-3΄; NBS1, 5΄-CCAACUAAAUUGCCAAGUAUU dTdT-3΄; MDC1, 5΄-GUCUCCCAGAAGACAGUGAdTdT-3΄; BRCA1, 5΄-GGAACCUGUCUCCACAAAGdTdT-3΄; and CtIP, 5΄-GCUAAAACAGGAACGAAUCdTdT-3΄. Control Gl2 (luciferase) siRNA was as described previously (58). Cells were transfected once with siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Cells were harvested for analyses 48 h after the siRNA transfection.
Antibodies and chemicals
The antibodies against And-1, γ-H2AX, Chk1, p-Chk1-S317, p-Chk1-S345, RPA32, FLAG, BrdU and ATR were described previously (56); mouse monoclonal antibody CtIP was a gift of Dr Richard Baer (Columbia University, New York); antibodies against p-H2AX-S139, p-Chk1-S317, RAD50, MRE11, NBS1, BRCA1, Chk2, p-Chk2-T68 were from Cell Signaling; antibody against pRPA32-S4/S8 was from Bethyl Laboratories; antibodies against actin and cyclin A were from Santa Cruz Biotechnology.
Camptothecin was from Sigma and KU-55933 was from Tocris Bioscience.
Immunofluorescence
Cells cultured on coverslips coated with 0.01% of poly-l-lysine (Sigma) were washed once with PBS, then pre-extracted with 0.5% Triton X-100 extraction buffer (10 mM PIPES pH 7.0, 100 mM NaCl, 3 mM MgCl2 and 300 mM sucrose) on ice for 5 min. After washing with PBS, cells were fixed with 4% paraformaldehyde in PBS for 15 min. Cells were then washed and incubated for 10 min in blocking buffer (PBS containing 3% BSA and 0.02% Tween-20) and subsequently incubated for 1 h with a primary antibody at room temperature. Cells were washed three times with PBS-T (PBS containing 0.02% Tween-20) and then incubated with a secondary antibody (rabbit Alexa Fluor-594 and mouse Alexa Fluor-488 were from Life Technology). After washing with PBS-T, cells were mounted with Fluoromount G (SouthernBiotech) containing DAPI. Slides were imaged using a Nikon Eclipse 80i microscope.
Irradiation
γ-Irradiation was performed with a 137Cs source at 10 Gy. Cells were harvested for analyses 3 h after the irradiation.
Protein purification
Recombinant And-1 proteins were expressed and purified as previously (41). Recombinant CtIP proteins (H00005932-P01) were purchased from Novus Biologicals.
Co-immunoprecipitation and Western blotting
Co-IP was performed as we described previously (41,48).
For in vitro immunoprecipitation, recombinant CtIP proteins were mixed with either BSA or And-1 proteins at 4°C for 2 h. The mixtures were then immunoprecipitated with protein A/G agarose beads coupled with And-1 antibody at 4°C overnight. The beads were washed and then the proteins associated with beads were eluted for immunoblotting as described previously (56).
For western blot, cells were lysed in RIPA buffer (50 mM Tris–HCl at pH 7.6, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% IGEPAL® CA-630, 1 mM EDTA, 20 mM β-glycerophosphate, 1× protease inhibitor) on ice for 15 min followed by sonication. Samples were separated by SDS-PAGE.
Mass spectrometry
293T cells were transfected with SFB-FLAG-CtIP plasmid were harvested and immunoprecipitated as described in the co-IP procedure. Mass spectrometry analysis of proteins was performed as described previously (59).
Homologous recombination assay
U2OS cells expressing HR reporter DR-GFP were gift from Dr. Maria Jason. HR repair assay was performed as described by others previously (24,60). Briefly, 48 hr after siRNA transfection, U2OS/DR-GFP cells were transfected with I-SceI expression vector (pCBASce) using Lipofectamine 2000 (Invitrogen). Cells were harvested two days after I-SceI transfection and subjected for flow cytometry analysis to determine the percentage of GFP positive cells. FACS data were analyzed using FlowJo software.
Laser micro-irradiation
DNA damage induction by laser was performed as previously described (61,62). Briefly, cells plated in a microwell dish with a glass bottom (P35GC-1.5-14-C, MatTek) were pre-sensitized with 10 μg/ml viable Hoechst dye 33258 (Sigma-Aldrich) for 10 min at 37°C. Laser micro-irradiation was performed with an inverted confocal microscope (LSM 510 Meta; Carl Zeiss) equipped with a 37°C heating chamber (PeCon, 160-800 006) and a 405 nm laser diode (25 Mw) focused through a 63× Plan-Apochromat/1.4 NA oil objective. DNA damage was generated in a restricted region with a width of 2 μm strip without major cytotoxic effects. After bleaching, the cells were fixed at the indicated time points for immunofluorescence analysis.
Clonogenic survival assay
Cells treated with siRNAs were plated in 60-mm dishes at a density of 300 cells per dish. Forty eight hours after siRNA transfection, cells were treated with camptothecin at indicated concentrations for 1 h. After washing with PBS, cells were incubated in camptothecin-free medium and left to grow for 14–15 days to allow colony formation. Cells were then fixed and stained with 0.5% crystal violet. Colonies with >50 cells were counted.
DSB induction by UVC micro-irradiation
The details of the protocol was described previously (63). Briefly, cells were labeled with 10 μM BrdU for 48–72 h. Cells were washed with PBS and covered by a micropore membrane (Isopore 5 μM, Millipore), followed by exposure to UVC lamp at a dose of 30 J/m2 (Hitachi, Germicidal lamp, GL-15). After the exposure, cells were incubated in culture medium for 30 min and then subjected to immunofluorescence.
RESULTS
And-1 associates with CtIP and is required for HR repair
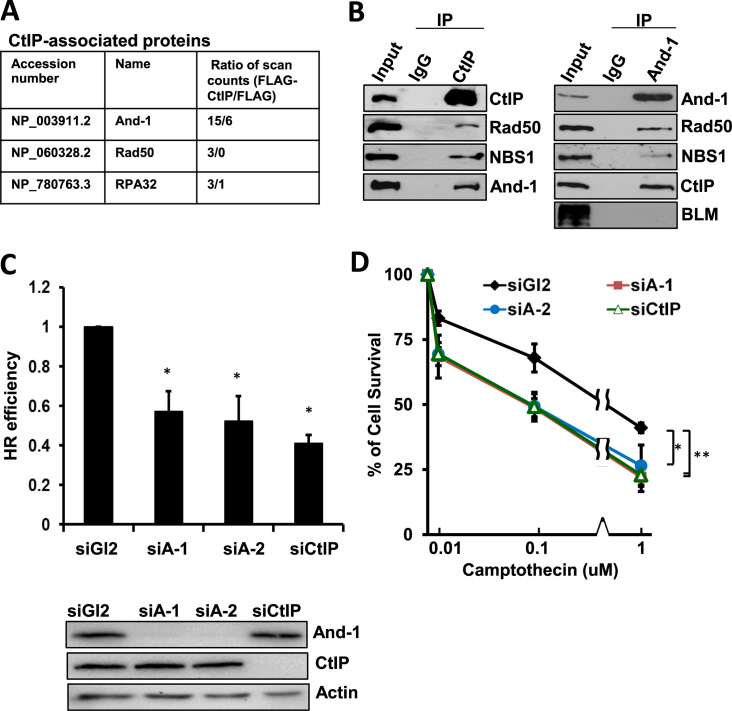
To investigate the role of CtIP in HR repair, we conducted mass spectrometry analyses to identify specific CtIP-associated proteins using an approach we previously described (59). We incubated anti-FLAG beads with cell extracts prepared from 293T cells ectopically expressing either FLAG-CtIP or FLAG to immunoprecipitate proteins. To rule out the false positive protein–protein interactions that are often due to protein–chromatin association, we included ethidium bromide and DNase I in the cell lysis buffer to disrupt protein–DNA interactions. After gel electrophoresis followed by silver staining, FLAG-CtIP-associated unique proteins were subjected to mass spectrometry analyses and the resulting data were used for the search of proteins identities in a database as we previous described (59). We identified several HR repair proteins, including Rad50 and RPA32 in the FLAG-CtIP immunoprecipitates (IPs) (Figure 1A). Interestingly, we also identified And-1 in the FLAG-CtIP IPs (Figure 1A). The presence of And-1 in the FLAG-CtIP IPs suggested a possible role of And-1 in the regulation of CtIP-mediated HR repair. To investigate whether And-1 regulates CtIP-mediated HR repair, we next conducted co-immunoprecipitation (co-IP) assays to confirm the interaction of endogenous CtIP with And-1 in HCT116 cells. From the endogenous CtIP IPs, we could identify not only Rad50 and NBS1, but also And-1 (Figure 1B). Reciprocally, we also detected CtIP, Rad50, NBS1 in endogenous And-1 IPs obtained from HCT116 cells. These results collectively demonstrated that And-1 forms a complex with HR repair proteins including CtIP, Rad50 and NBS1 in a manner independent of chromatin structure.
Figure 1.
And-1 forms complexes with HR repair proteins and is required for HR repair. (A) Identification of CtIP-associated proteins that are involved in HR repair by mass spectrometry. (B) CtIP interacts with And-1. Co-immunoprecipitation (co-IP) assays were performed using U2OS cell lines. Cell lysates were immunoprecipitated with control IgG, anti-CtIP or anti-And-1 antibodies and the IPs were then resolved by SDS-PAGE and immunoblotted for the indicated proteins. Left panel, immunoprecipitation with anti-CtIP antibody. Right panel, immunoprecipitation with anti-And-1 antibody. (C) And-1 depletion impairs HR repair. U2OS-DR-GFP cells treated with the indicated siRNAs were transfected with pCBASce plasmids 48 h post siRNA transfection. The percentage of GFP-positive cells was determined by flow cytometry 48 h after plasmid transfection. The data were normalized to those obtained from cells transfected with control siGl2 (set as 1.0). Data represent means ± SD from three independent experiments. *P ≤ 0.05. (D) U2OS cell survival after exposing cells transfected with indicated siRNAs to the indicated doses of camptothecin. Data represent means ± SD from three independent experiments. *P ≤ 0.05; **P ≤ 0.01.
Given that And-1 forms a complex with HR repair proteins, we postulated that And-1 plays a role in HR repair pathway. To test this hypothesis, we assessed HR repair efficiency using a sensitive HR reporter assay (60). Consistent with previous publications (24,29), CtIP depletion by siRNA reduced HR efficiency by 60% (Figure 1C). Strikingly, And-1 depletion by two independent siRNAs reduced HR repair efficiency by 50% (Figure 1C and Supplementary Figure S1), indicating that And-1 is as critical as CtIP for efficient HR repair.
We next investigated whether And-1 depletion affects clonogenic survival of HCT116 cells following treatment with various concentrations of the topoisomerase I inhibitor camptothecin, which is known to induce replication-dependent DSBs (64). In agreement with previous data (29), depletion of CtIP caused hypersensitivity towards camptothecin (Figure 1D). Importantly, depletion of And-1 by two independent siRNAs also induced hypersensitivity towards camptothecin that is indistinguishable from that caused by CtIP depletion (Figure 1D). Since camptothecin-induced DSBs during S phase are mainly repaired by HR repair, our data suggest that And-1 is critical for HR repair of DSBs. Consistently, we also found that And-1 depleted cells were hypersensitive to PARP1 inhibition by PARP1 inhibitor Olaparib (Supplementary Figure S1B) and chromosomal abnormalities were increased upon And-1 depletion (Supplementary Figure S1C).
And-1 is recruited to DNA DSB sites
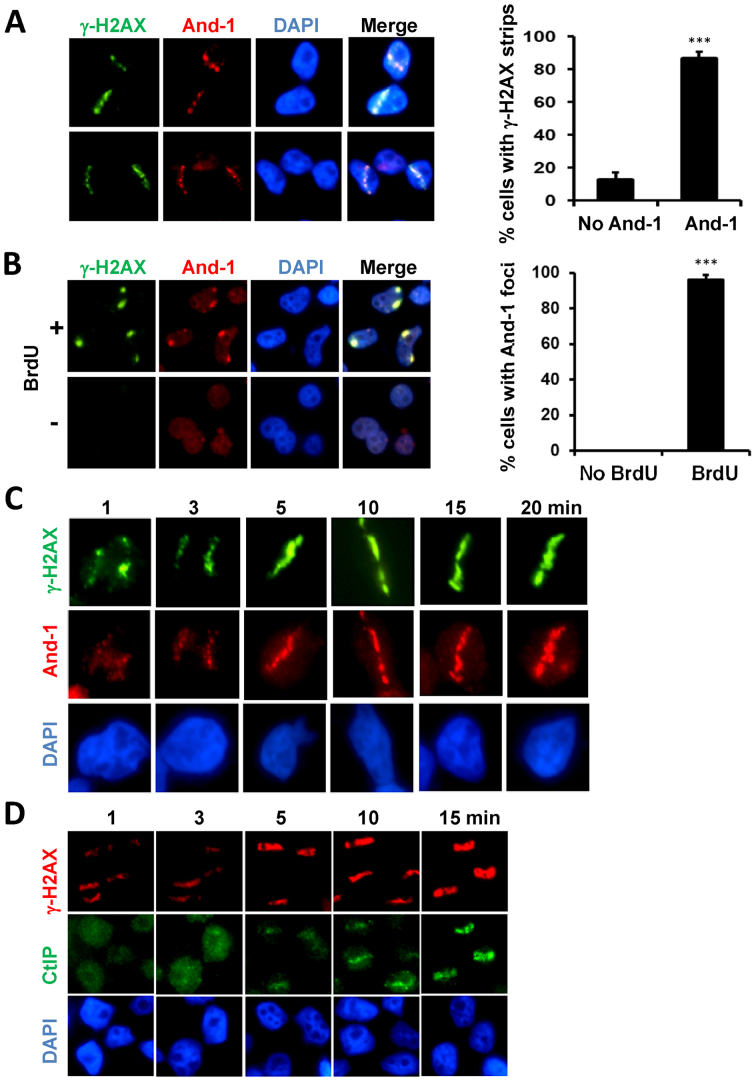
If And-1 is involved in the HR repair of DSBs, it is likely that And-1 is recruited to DSB sites. To test this possibility, we generated DSB-containing tracks in the nuclei of human U2OS cells using laser micro-irradiation as described previously (62). Ten minutes after laser micro-irradiation, we detected γ-H2AX strips on the nuclei, indicating the sites of DSBs (Figure 2A). Intriguingly, we also detected co-localization of And-1 and γ-H2AX on the same strips, suggesting that And-1 was recruited to DSB sites. To further confirm the localization of And-1 at DSB sites, we next created localized DSBs in U2OS cells using a micro-irradiation approach developed by Yamashita's group (63). Using this protocol, we induced DSBs in cells by ultraviolet C (UVC) irradiation through a porous membrane shielding the cells that had been labeled with 5-bromo-2΄-deoxyuridine (BrdU) (63). Notably, And-1 foci were detected at DSB sites as indicated by γ-H2AX foci (Figure 2B). Moreover, we also found that And-1 and γ-H2AX were present in the same foci in U2OS cells treated with 10 Gy ionizing radiation (Supplementary Figure S2). Taken together, these data suggest that And-1 is directly recruited to DSB sites.
Figure 2.
And-1 is recruited to DSB sites. (A) And-1 co-localizes with γ-H2AX at DSB sites induced by laser micro-irradiation. U2OS cells were micro-irradiated with laser and fixed for immunostaining using the indicated antibodies 10 min after irradiation. For each experiment, >50 cells were counted and the percentage of cells exhibiting γ-H2AX strips or both γ-H2AX and And-1 strips was determined. Data represent means ± SD from three independent experiments. ***P ≤ 0.001. (B) And-1 co-localizes with γ-H2AX at DSB sites induced by UVC light. U2OS cells labeled with or without 10 μM BrdU for 72 h were covered with a 5 μm polycarbonate isopore membrane filter and subjected to UVC irradiation (30J/m2). 30 min after irradiation, cells were harvested and immunostained for the indicated proteins. For each experiment, >100 cells were counted and the percentage of cells exhibiting And-1 foci was determined. Data represent means ± SD from three independent experiments. ***P ≤ 0.001. (C) And-1 recruitment to laser-induced DSB sites at the indicated time points. U2OS cells were micro-irradiated with laser and then processed at indicated time points for immunostaining for indicated proteins. (D) CtIP recruitment to laser-induced DSB sites at indicated time points. U2OS cells were micro-irradiated with laser and then processed at the indicated time points for immunostaining for indicated proteins.
Having found that And-1 is recruited to DSB sites, we next examined and compared the kinetics of the recruitment of both And-1 and CtIP to damage sites. Consistent with previous report (22,29), CtIP started to accumulate at damage sites approximately five minutes after laser micro-irradiation and its accumulation onto damage sites was slower than γ-H2AX, which was observed at one minute after micro-irradiation (Figure 2D). Interestingly, the accumulation of And-1 at DSB sites was detected one minute after the micro-irradiation (Figure 2C), indicating that the accumulation of And-1 at the damage sites is rapid and precedes the accumulation of CtIP to the damage sites. We also compared the timing of the recruitment of And-1 and NBS1 to damage sites in cell using live image analysis and our data indicated that the accumulation of And-1 at the laser-induced damage lines was detected around one minute after irradiation and this timing is similar to the recruitment of NBS1 to damage sites (Supplementary Figure S2B). Taken together, And-1 is recruited to DSB sites at the early stage of HR repair of DSB before CtIP recruitment occurs.
And-1 facilitates ssDNA generation in cells with DSBs
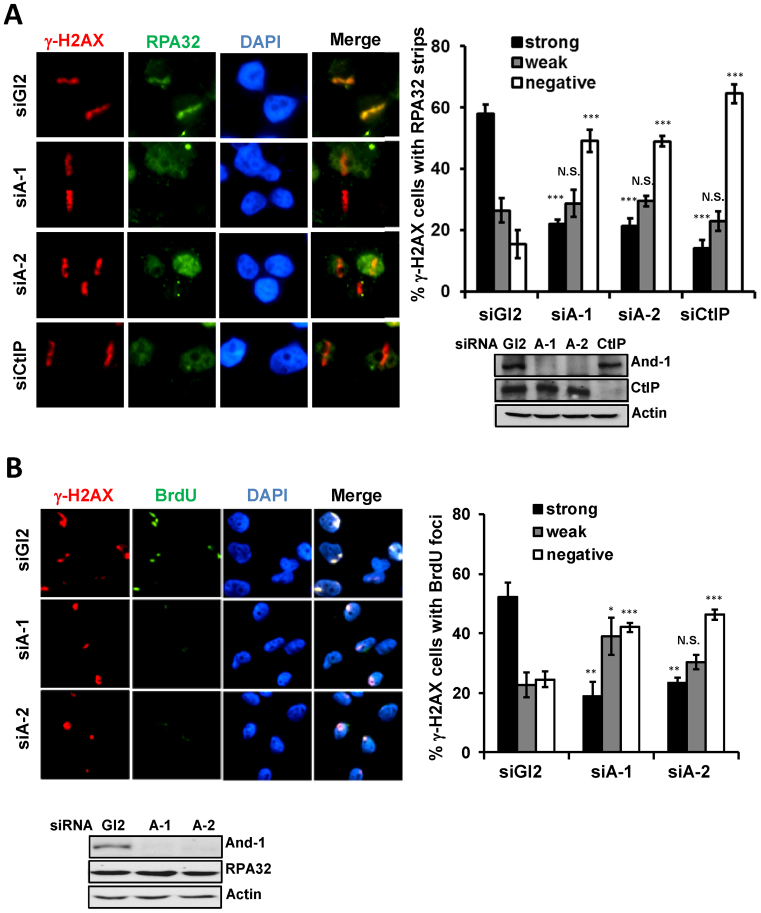
Given that And-1 is recruited to DSB sites earlier than that of CtIP and that And-1 forms complex with CtIP, we hypothesized that And-1 may regulate DSB end resection by establishing a condition or acting as a platform for CtIP recruitment to the DSB sites. To test this hypothesis, we first examined whether And-1 affects ssDNA generation by examining DSB-induced RPA focus formation. In agreement with previous report (29), depletion of CtIP significantly reduced the recruitment of RPA32 to laser-induced DSB sites (Figure 3A). Although it had no effects on γ-H2AX formation, And-1 depletion by two independent siRNAs dramatically impaired the recruitment of RPA to laser-induced DSB sites (Figure 3A), suggesting that DSB end resection is impaired. To further test that And-1 indeed affects ssDNA generation, we examined ssDNA generation by using an anti-BrdU antibody staining technique that only detects ssDNA as we described previously (56,58). To precisely examine ssDNA at DSB sites, we induced DSB by UVC microirradiation at localized area of nuclei in cells pre-labelled with BrdU and examined the generation of ssDNA at DSB sites by BrdU staining without denaturing DNA. Intriguingly, And-1 depletion by two independent siRNAs significantly reduced ssDNA generation at DSB sites as indicated by BrdU staining (Figure 3B). Taken together, these data suggest that And-1 is required for end resection by regulating ssDNA generation in cells with DSBs.
Figure 3.
And-1 depletion impairs DSB end resection. (A) And-1 is required for RPA recruitment to laser-induced DSB sites. U2OS cells treated with the control siGl2, two independent siAnd-1s (siA-1 or siA-2) or siCtIP were micro-irradiated and co-immunostained for γ-H2AX and RPA32 25 min post irradiation. For each experiment, >50 cells were counted and the percentage of γ-H2AX cells exhibiting RPA32 strips was determined. Data represent means ± SD from three independent experiments. N.S., not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (B) And-1 depletion impairs ssDNA generation. BrdU-labeled U2OS cells transfected with indicated siRNAs were treated as in Figure 2B. Cells were processed 30 min after UVC treatment and examined for ssDNA formation by a non-denaturing staining (See methods and text for detail). For each experiment, >50 cells were counted and the percentage of γ-H2AX cells exhibiting BrdU foci was determined. Data represent means ± SD from three independent experiments. N.S., not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
And-1 facilitates the recruitment of CtIP to DSB sites
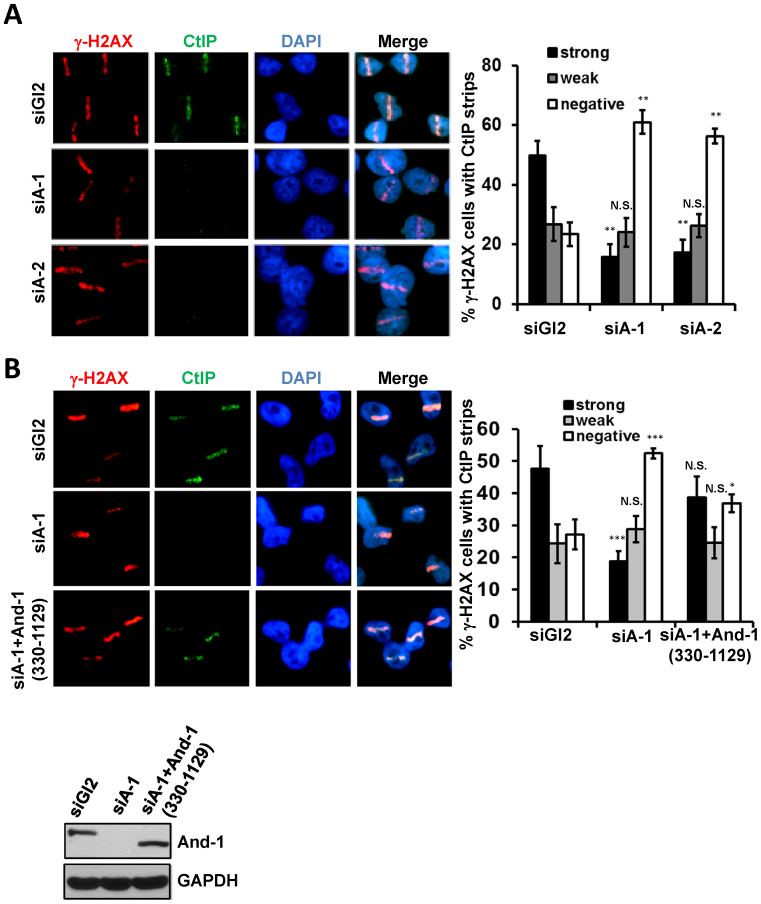
Since And-1 interacts with CtIP and is required for end resection, we next investigated whether And-1 regulates the recruitment of CtIP to DSB sites. To this end, we depleted And-1 using two independent siRNAs. In the cells treated with the control siGl2, CtIP was found to co-localize with γ-H2AX tracks following laser micro-irradiation (Figure 4A). However, depletion of And-1 by siRNAs significantly reduced the recruitment of CtIP to DSB sites as indicated by the greatly diminished co-localization of CtIP with the γ-H2AX tracks (Figure 4A). Consistently, And-1 depletion also compromised formation of Rad51 foci at damage sites (Supplementary Figure S3C). Interestingly, when we examined whether CtIP depletion could affect the recruitment of And-1 to DSB sites, we found that deletion of CtIP had no effects on the co-localization of And-1 with γ-H2AX at DSB sites (Supplementary Figure S3).
Figure 4.
And-1 is required for efficient recruitment of CtIP to DSB sites. (A) And-1 depletion impairs CtIP recruitment to DSB sites. U2OS cells treated with siGl2 or two independent siAnd-1s (siA-1 or siA-2) were micro-irradiated by laser and co-immunostained for γ-H2AX and CtIP 15 min post irradiation. For each experiment, >50 cells were counted and the percentage of γ-H2AX cells exhibiting CtIP strips was determined. Data represent means ± SD from three independent experiments. N.S., not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (B) Restoration of CtIP recruitment to DSB sites in cells expressing siRNA resistant And-1. U2OS cells expressing the mutant And-1 (330–1129) were transfected with the indicated siRNAs and subjected to treatment as described in Figure 2A. For each experiment, >50 cells were counted and the percentage of γ-H2AX cells exhibiting CtIP strips was determined. Data represent means ± SD from three independent experiments. N.S., not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
To rule out the possibility of the siRNA off-target effects, we used a siRNA to deplete endogenous And-1 in U2OS cells expressing And-1 siRNA resistant mutant And-1 (330–1129), which lacks And-1 siRNA target sequence at the N-terminus and hence is resistant to the siRNA. Although And-1 depletion reduced the recruitment of CtIP to laser-induced damage sites (Figure 4B), expression of the siRNA resistant And-1 (330–1129) was able to restore the recruitment of CtIP to DSB sites. The above findings collectively demonstrated that And-1 is indeed required for the recruitment of CtIP to DSB sites, the N-terminus of And-1 is dispensable for the recruitment of CtIP to damage sites, and the middle region and C-terminus of And-1 contain a domain(s) that is essential for And-1 to facilitate the recruitment of CtIP to DSB sites for end resection.
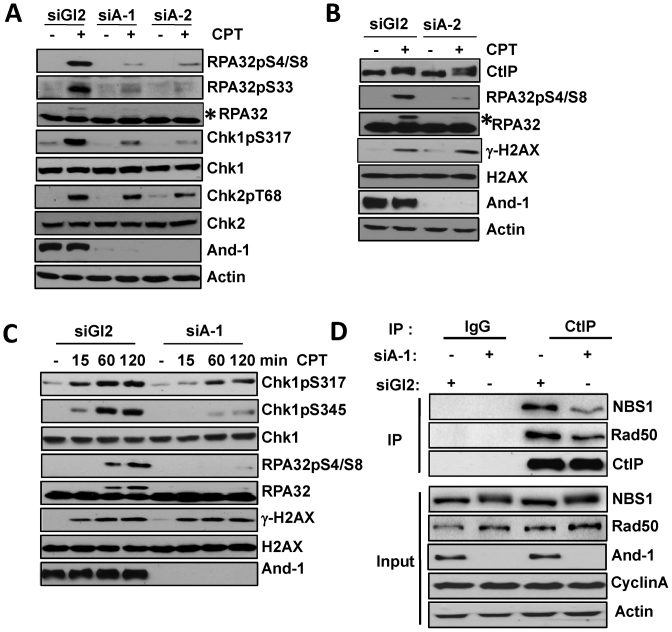
Since CtIP interacts with MRN (29), we next tested whether And-1 is required for the interaction between MRN and CtIP. To this end, we enriched siGl2 or siAnd-1 treated cells in the S/G2 phase using an approach as described previously (65). Briefly, U2OS cells transfected with siRNAs were enriched at S/G2 phase six hours after a double-thymidine block was terminated. From the endogenous CtIP IPs, we found that the interactions of CtIP with NBS1 and Rad50 were decrease upon And-1 depletion (Figure 5D), suggesting that And-1 facilitates the interaction between CtIP and MRN complex.
Figure 5.
And-1 depletion impairs DNA damage response induced by end resection. (A and B) Depletion of And-1 impairs Chk1 and RPA phophorylation but not Chk2 phosphorylation after camptothecin exposure. U2OS cells transfected with the indicated siRNAs were treated with camptothecin (1 μM) for 1 h. Cells were then harvested and immunoblotted for indicated proteins. Asterisks in A and B: hyper-phosphorylated RPA32. (C) Depletion of And-1 impairs Chk1 phosphorylation at both early and late time-points after continuous camptothecin exposure. U2OS cells transfected with the indicated siRNAs were treated with camptothecin (1 μM) and harvested at the indicated time-points and immunoblotted for the indicated proteins. (D) U2OS cells transfected with indicated siRNAs were synchronized by using a double-thymidine (2 mM) block and then released for 6h. Cells enriched in S/G2 phase were harvested for IP. CtIP or IgG IPs were immunoblotted for the indicated proteins.
It was known that CtIP phosphorylation by ATR is required for the recruitment of CtIP to damage sites and subsequent processive resection of DSB (66). We therefore examined whether inhibition of CtIP phosphorylation could interfere the interaction between And-1 and CtIP in cells with DSBs. Interestingly, in cells treated with the ATR inhibitor VE-821 there was reduced interaction between And-1 and CtIP (Supplementary Figure S6), suggesting that ATR-mediated association of CtIP with DSB sites may be dependent on its interaction with And-1.
And-1 facilitates the ATR-mediated DNA damage response in cells with DSBs
To further investigate the role of And-1 in DSB end resection, we examined DNA damage response in HCT116 cells with DSBs induced by camptothecin treatment. And-1 depletion did not change CtIP protein levels (Figure 5B), suggesting that And-1 does not regulate end resection via affecting CtIP stability or expression. Notably, And-1 depletion also did not affect the levels of γ-H2AX (Figure 5B), indicating that And-1 has no effects on the generation of DSBs. Consistently, And-1 depletion had little effects on Chk2 phosphorylation. Thus, ATM-mediated pathway is still intact in the absence of And-1 (Figure 5A). Since And-1 is involved in the generation of ssDNA during end resection, we expected that And-1 depletion may reduce ATR activation and hence Chk1 phosphorylation. Indeed, And-1 depletion markedly impaired Chk1 phosphorylation on Serine 317 (Figure 5A). In agreement with this observation, And-1 depletion significantly reduced RPA32 phosphorylation as indicated by the reduction in the amount of up-shifted phosphorylated RPA32 form, as well as phosphorylation of RPA32 on Serine 33 in cells following CPT treatment (Figure 5A). Since ATR is the major kinase that phosphorylates RPA32 at Serine 33, these data suggest that ATR activation elicited by end resection is impaired in the absence of And-1. Together, these data suggest that And-1 is not involved in the DSB detection and ATM- mediated signaling, instead, And-1 is important for the activation or propagation of ATR-mediated signaling pathway during end resection of DSBs.
To further test how And-1 regulates Chk1 phosphorylation during DSB repair, we examined the time course of Chk1 activation in And-1 depleted cells during continuous camptothecin exposure. We found that And-1 depletion reduced Chk1 phosphorylation on Ser345 not only at early time point (∼15 min) but also through 120 min after camptothecin exposure begun (Figure 5C), indicating that And-1 impairs Chk1 phosphorylation at both early and late time points during camptothecin exposure. It should be noted that it was reported that CtIP depletion has no effect on Chk1 phosphorylation soon after DSB induction but only impairs the maintenance of Chk1 phosphorylation at late time points (29). Thus, And-1 and CtIP seem to play different roles in the kinetics and maintenance of Chk1 phosphorylation. A detailed discussion on this point is included in the discussion section.
The C-terminus of And-1 is important for its interaction with CtIP
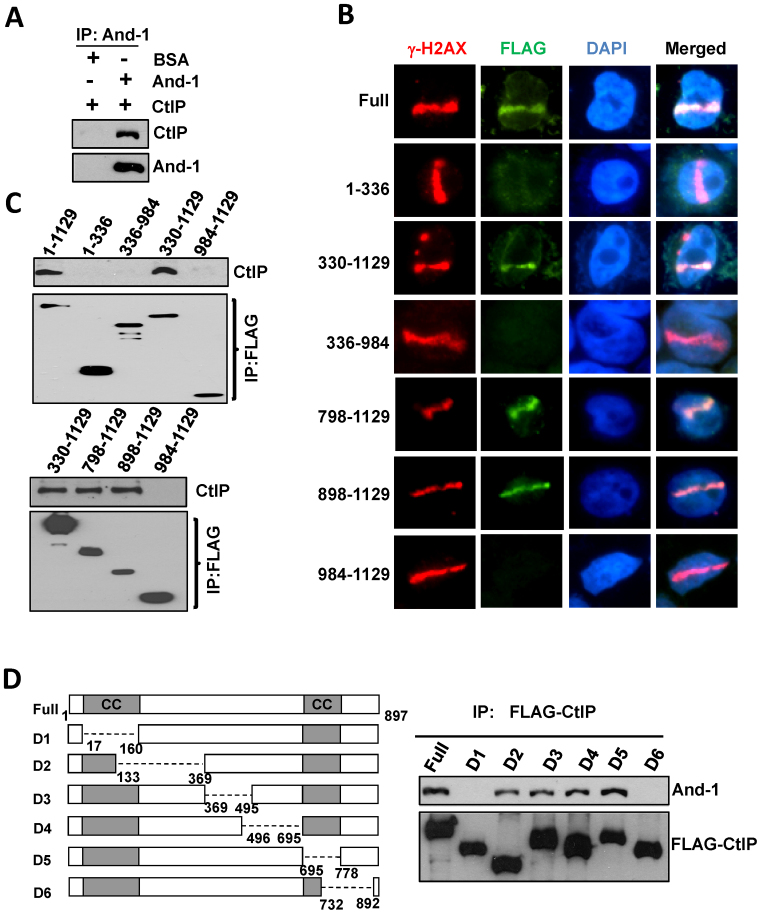
To further study the role of And-1 in the regulation of CtIP-mediated HR repair, we investigated whether And-1 directly interacts with CtIP by examining And-1-CtIP interaction in an in vitro assay. To this end, we mixed recombinant And-1 and CtIP proteins and then immunoprecipitated And-1 proteins. Intriguingly, we detected robust CtIP from the IPs of recombinant And-1 (Figure 6A), suggesting that And-1 indeed interacts with CtIP directly in vitro. And-1 contains WD40 repeats at its N-terminus, a SepB domain in the middle, and a HMG domain at its C-terminus (56). We next mapped the domains of And-1 that mediate the interaction between And-1 and CtIP. We performed co-IPs using cell extracts from 293T cells expressing full-length And-1 or its truncated mutants from transfected plasmids. The results showed that the interactions with CtIP were detected in IPs from full-length And-1 and And-1 (330–1129) (Figure 6C), indicating that SepB and HMG domains mediate the association of And-1 with CtIP. We next examined which parts of SepB and HMG domains mediate And-1–CtIP interactions and found that And-1 (898–1129) but not And-1 (984–1129) interacted with CtIP (Figure 6C), indicating that the C-terminus of And-1 is critical for its interaction with CtIP.
Figure 6.
The C-terminus of And-1 is required for its interaction with CtIP and localization to DSB sites. (A) And-1 directly interacts with CtIP. Purified recombinant CtIP proteins (400 ng) were mixed with recombinant And-1protiens (200 ng) or BSA (200 ng) as described in Method. CtIP was immunoprecipitated with anti-CtIP antibody and IPs were then resolved by SDS-PAGE and immunoblotted for the indicated proteins. (B) The associations of And-1 or its mutants with CtIP. FLAG-And-1 and its mutants were expressed in 293T cells. FLAG-IPs were resolved by SDS-PAGE and immunoblotted for the indicated proteins. (C) The localization of And-1 and its mutants at DSB sites. U2OS cells expressing the indicated And-1 or it mutants were subjected to the treatment as in Figure 2A. (D) The associations of CtIP or its mutants with And-1. FLAG-CtIP and its mutants were expressed in 293T cells. Left panel, schematic of the CtIP protein domains and deletion mutants used for protein–protein interactions; Right panel, FLAG-IPs were resolved on SDS-PAGE and immunoblotted for the indicated proteins.
Having mapped the CtIP-interacting domains of And-1, we next examined the recruitment of each of these And-1 mutants to DSB sites induced by laser micro-irradiation. Interestingly, full-length And-1 as well as mutants And-1 (330–1129), And-1 (798–1129) and And-1 (898–1129) that interact with CtIP were found to accumulate at DSBs sites, whereas And-1 mutants including And-1 (1–336), And-1 (336–984) and And-1 (984–1129) that do not interact with CtIP failed to localize at DSB sites (Figure 6B). These data suggest that the interaction between And-1 and CtIP may be required for continuous association of And-1 with DSB sites.
We also examined which regions of CtIP are responsible for its interaction with And-1. To this end, we examined the interactions of And-1 with CtIP mutants created by Dr Junjie Chen's group (24). And-1 protein was detected from IPs of CtIP mutants D2, D3, D4 and D5 but not D1 and D6 (Figure 6D), indicating that both the N-terminus and the C-terminus of CtIP are important to mediate the association of CtIP with And-1.
Recruitment of And-1 to DSB sites depends on BRCA1, MDC1 and ATM
Given that And-1 is rapidly recruited to DSB sites at the early stage of HR repair (Figure 2C), we therefore hypothesized that And-1 may interact with MDC1 and BRCA1. Indeed, from endogenous And-1 IPs, we could detect MDC1 and BRCA1 (Supplementary Figure S4). These data indicate that And-1 interacts with both MDC1 and BRCA1 and further support that And-1 participates in the early stage of HR repair.
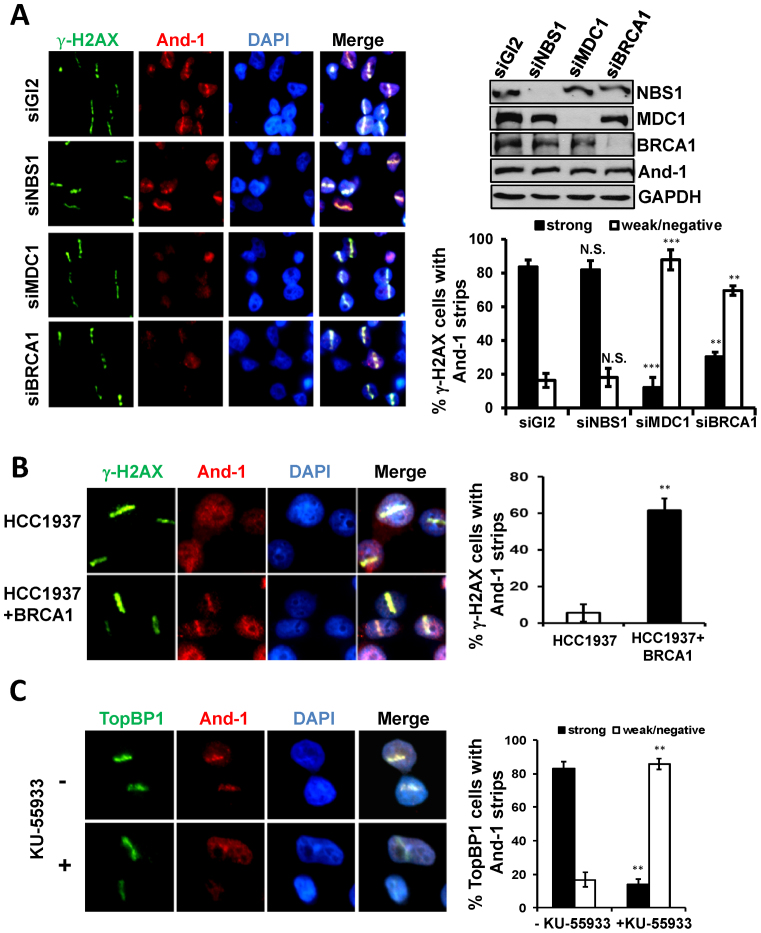
To investigate the mechanism of how And-1 is recruited to damage sites, we examined the localization of And-1 at DSB sites in cells with down-regulated early HR repair factors including MRN and MDC1 by siRNA. Although MRN complex plays a critical role in the initiation of end resection, depletion of NBS1 had no effects on the recruitment of And-1 to damage sites (Figure 7A). Intriguingly, depletion of MDC1 significantly reduced the recruitment of And-1 to DSB sites (Figure 7A). Since MDC1 regulates the localization of BRCA1 to DSB sites, we went forward to investigate whether BRCA1 regulates the recruitment of And-1 to DSB sites. Markedly, we found that the localization of And-1 to DSB sites was also impaired in cells with BRCA1 depletion (Figure 7A). To further confirm the role of BRCA1 in the regulation of And-1 recruitment to DSB sites, we examined the And-1 localization to DSBs in HCC1937 cells with deficient BRCA1. Strikingly, the recruitment of And-1 to DSB sites was significantly impaired in HCC1937 cells with deficient BRCA1. However, the localization of And-1 to DSB sites was restored in HCC1937 cells with expression of wild-type BRCA1 from a transgene (Figure 7B). Taken together, our data strongly suggest that MDC1-mediated pathway is involved in the recruitment of And-1 to DSB sites.
Figure 7.
Recruitment of And-1 to DSB sites is dependent on BRCA1, MDC1 and ATM but not on MRN complex. (A) Depletion of BRCA1, or MDC1 but not NBS1 impairs the localization of And-1 to DSB sties. U2OS cells transfected with the indicated siRNAs were treated as in Figure 2A. The percentage of γ-H2AX cells exhibiting And-1 strips was determined. Data represent means ± SD from three independent experiments. N.S., not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (B) Localization of And-1 to DSB sites is restored in HCC1937 cells expressing wild type BRCA1. HCC1937 or HCC1937 cells expressing wild-type BRCA1 were treated as in Figure 2A. The percentage of γ-H2AX cells exhibiting And-1 strips was determined. Data represent means ± SD from three independent experiments. **P ≤ 0.01. (C) Inhibition of ATM impairs the localization of And-1 at DSB sites. U2OS cells treated with KU-55933 (10 μM) for 4 h were treated as in Figure 2A. The percentage of TopBP1 cells exhibiting And-1 strips was determined. Data represent means ± SD from three independent experiments. **P ≤ 0.01.
Given that ATM kinase activity is required for the recruitment of CtIP to DSB sites (22), we next examined whether And-1 recruitment to DSB site is ATM-dependent. To inhibit ATM, we used an ATM specific inhibitor, KU-55933 (67). The results showed that the recruitment of And-1 to DSB sites was significantly compromised in cells treated with KU-55933 (Figure 7C). Thus, ATM appears to be a critical factor to regulate the recruitment of And-1 to DSB sites.
DISCUSSION
Requirement of And-1 for DSB end resection
Accumulating evidence has shown that And-1/Ctf4 is important for HR repair in yeast and human cells (42,54,57,68). Although it was predicted that And-1 is directly involved in HR repair rather than acting indirectly via other pathways (42,57), it was unknown how And-1 directly regulates HR repair. HR is a complex process involving multiple steps including DSB-induced chromatin relaxation, recruitment of HR repair proteins to DSB sites and end resection. Although CtIP has been shown to facilitate HR repair by promoting end resection (29), the mechanisms by which CtIP is recruited to DSB sites remain speculative. Here we identified a novel functional MDC1-And-1 link that is critical for end resection by regulating the recruitment of CtIP. This study not only elucidates a hitherto unknown role of And-1 in HR repair but also identifies a novel pathway to regulate end resection. During the manuscript preparation, Dr. Huadong Pei's group (National Center for Protein Sciences in China) has independently identified the same pathway (personal communication).
For the first time, we found that And-1 is localized to the DSB sites in human cells. This discovery reinforces the idea that And-1 is involved in DNA repair and also provides the evidence that And-1 acts directly at DSBs to promote HR repair. Our data demonstrate that And-1 directly interacts with CtIP in vivo and in vitro, promotes end resection by regulating CtIP recruitment to DSB sites, and activates ATR-Chk1 pathway during the end resection. We further show that And-1 is recruited to DSB sites at early stage in the HR process, similar to MRN complex but before the recruitment of CtIP. Consistent with this functional order, the recruitment of And-1 to damage sites may establish a platform for the recruitment of CtIP to damage sites to initiate end resection. To support this notion, And-1 contains a C-terminal DNA binding HMG domain (59), which usually facilitates the formation of nucleoprotein complexes on the chromatin via modulating DNA structure (69,70). It is possible that at DSB sites, And-1 binds to DNA and facilitates the assembly of HR proteins by regulating DNA structure or DSB-associated protein complexes. Interestingly, the concept that And-1 may act as a structural scaffold to facilitate loading of repair proteins at the damage sites has been proposed by others despite a lack of strong evidence (42).
Functional relationships among MDC1, BRCA1, MRN, And-1 and CtIP
MDC1 is required for the recruitment of MRN complex to damage sites by direct interaction with NBS1 (9,11). Our data show that And-1 is recruited to DSB sites at early stage of HR repair process and interacts with MRN complex (Figures 1A and 2C), but MRN is not required for its localization at DSB sites (Figure 7A). Given that And-1 depletion does not affect the recruitment of NBS1 to DSB sites (Supplementary Figure S5), it appears that the recruitment of MRN and the recruitment And-1 to DSB sites are not interdependent. However, these results do not rule out the possibility that And-1 and MRN may coordinate to regulate the recruitment of CtIP to DSB sites. Indeed, our data indicate that And-1 is required for the interaction between CtIP and MRN (Figure 5D). Thus, And-1 may function as a platform to facilitate damage site recruitment of CtIP by promoting the interaction between CtIP and MRN at damage site. Significantly, we found that BRCA1 and MDC1 are both required for the recruitment of And-1 to DSB sites. Given that MDC1 and BRCA1 recruitment to DSB sites is a very early event for the initiation of HR repair, and the timing of And-1 recruitment to DSB sites is similar to the timing of the recruitment of MRN to DSB sites, it appears that the condition established by MDC1 and BRCA1 at damage sites is required for the accumulation of both MRN and And-1 at damage sites. Although we have detected the interactions of And-1 with MDC1 and BRCA1, how MDC1 and BRCA1 recruit And-1 to DSB sites remain to be investigated in the future.
Dual roles of And-1 in ATR-Chk1 signaling during end resection
Following DSB induction, MRN/ATM-dependent DSB end resection results in the formation of ssDNA regions that promote ATR activation and subsequent Chk1 phosphorylation by ATR (71). Consistent with this notion, our data showed that And-1 depletion impairs end resection that significantly reduces the activation of ATR-Chk1 pathway. Similar results have been observed in cells with downregulated CtIP (29). These results strongly suggest that both And-1 and CtIP function in the same pathway leading to DSB end resection. Interestingly, CtIP depletion impairs Chk1 phosphorylation at late time-points but not at early time-points after continuous camptothecin exposure (29), suggesting that CtIP is involved in damage response through its regulation of end resection. However, unlike CtIP depletion, And-1 depletion not only decreases the initial Chk1 activation triggered by replication stress induced by trapped DNA topoisomerase I inhibitor, but also the late Chk1 activation induced by camptothecin-induced replication-dependent DSBs, which require resection to activate ATR-Chk1 pathway. This difference is within our expectation because we recently reported that And-1 is required for efficient ATR-Chk1 activation in cells with replication stress (56). We believe that And-1 plays dual roles in DSB end resection, one is to regulate the recruitment of CtIP to damage sites for resection, the other is to regulate initial Chk1 activation following DNA replication stress.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs J. Chen, J. Maria, X. Yu and R. Baer for reagents, Dr Chi-wei Chen for help on statistical analysis. The authors would like to thank Dr. Kristy J. Brown for her kind assistance in collection and processing of the mass spectrometry samples.
Author contributions: Experiments were performed by Y.L., R.W. and Z.L. Experimental design, interpretation of data and manuscript writing were performed by Y.L., Z.H. and W.Z.
Footnotes
Present address: Yongming Li, College of Basic Medical Sciences, Dalian Medical University, Dalian 116044, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [CA177898 and CA184717 to W.Z.]; Research Scholar Grant [RSG-13-214-01-DMC to W.Z.] from the American Cancer Society; GWU McCormick Research Award; National Natural Science Foundation of China [81402330 to Y.L.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kastan M.B., Bartek J.. Cell-cycle checkpoints and cancer. Nature. 2004; 432:316–323. [DOI] [PubMed] [Google Scholar]
- 2.Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heyer W.D., Ehmsen K.T., Liu J.. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010; 44:113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieber M.R., Ma Y., Pannicke U., Schwarz K.. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst.). 2004; 3:817–826. [DOI] [PubMed] [Google Scholar]
- 5.West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell. Biol. 2003; 4:435–445. [DOI] [PubMed] [Google Scholar]
- 6.Sung P., Klein H.. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell. Biol. 2006; 7:739–750. [DOI] [PubMed] [Google Scholar]
- 7.Lisby M., Barlow J.H., Burgess R.C., Rothstein R.. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004; 118:699–713. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q., Goldstein M., Alexander P., Wakeman T.P., Sun T., Feng J., Lou Z., Kastan M.B., Wang X.F.. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 2014; 33:862–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman J.R., Jackson S.P.. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008; 9:795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J.. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 2008; 181:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spycher C., Miller E.S., Townsend K., Pavic L., Morrice N.A., Janscak P., Stewart G.S., Stucki M.. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 2008; 181:227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L.M., Luo K.T., Lou Z.K., Chen J.J.. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:11200–11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You Z., Bailis J.M., Johnson S.A., Dilworth S.M., Hunter T.. Rapid activation of ATM on DNA flanking double-strand breaks. Nat. Cell Biol. 2007; 9:1311–1318. [DOI] [PubMed] [Google Scholar]
- 14.Difilippantonio S., Nussenzweig A.. The NBS1-ATM connection revisited. Cell Cycle. 2007; 6:2366–2370. [DOI] [PubMed] [Google Scholar]
- 15.Adams K.E., Medhurst A.L., Dart D.A., Lakin N.D.. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006; 25:3894–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuadrado M., Martinez-Pastor B., Murga M., Toledo L.I., Gutierrez-Martinez P., Lopez E., Fernandez-Capetillo O.. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 2006; 203:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C., Lukas J., Jackson S.P.. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006; 8:37–45. [DOI] [PubMed] [Google Scholar]
- 18.Myers J.S., Cortez D.. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 2006; 281:9346–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Amours D., Jackson S.P.. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 2002; 3:317–327. [DOI] [PubMed] [Google Scholar]
- 20.Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J.M., Chang S., Ferguson D.O.. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008; 135:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams R.S., Moncalian G., Williams J.S., Yamada Y., Limbo O., Shin D.S., Groocock L.M., Cahill D., Hitomi C., Guenther G. et al. . Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008; 135:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.W., Basilio A., Tonnu N., Verma I.M., Berns M.W., Hunter T.. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 2009; 36:954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L., Nievera C.J., Lee A.Y., Wu X.. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008; 283:7713–7720. [DOI] [PubMed] [Google Scholar]
- 24.Yuan J., Chen J.. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J. Biol. Chem. 2009; 284:31746–31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu X., Chen J.. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004; 24:9478–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aylon Y., Liefshitz B., Kupiec M.. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004; 23:4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huertas P., Jackson S.P.. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009; 284:9558–9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X., Fu S., Lai M., Baer R., Chen J.. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006; 20:1721–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravel S., Chapman J.R., Magill C., Jackson S.P.. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008; 22:2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G.. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008; 134:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You Z., Bailis J.M.. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010; 20:402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P.. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005; 123:1213–1226. [DOI] [PubMed] [Google Scholar]
- 34.Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M.A., Celeste A., Manis J.P., van Deursen J., Nussenzweig A., Paull T.T. et al. . MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006; 21:187–200. [DOI] [PubMed] [Google Scholar]
- 35.Stewart G.S., Wang B., Bignell C.R., Taylor A.M., Elledge S.J.. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003; 421:961–966. [DOI] [PubMed] [Google Scholar]
- 36.Huen M.S.Y., Grant R., Manke I., Minn K., Yu X.C., Yaffe M.B., Chen J.J.. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007; 131:901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J.. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007; 131:887–900. [DOI] [PubMed] [Google Scholar]
- 38.Mattiroli F., Vissers J.H.A., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., Sixma T.K.. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012; 150:1182–1195. [DOI] [PubMed] [Google Scholar]
- 39.Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J. et al. . RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009; 136:435–446. [DOI] [PubMed] [Google Scholar]
- 40.Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M. et al. . The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009; 136:420–434. [DOI] [PubMed] [Google Scholar]
- 41.Zhu W., Ukomadu C., Jha S., Senga T., Dhar S.K., Wohlschlegel J.A., Nutt L.K., Kornbluth S., Dutta A.. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007; 21:2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshizawa-Sugata N., Masai H.. Roles of human AND-1 in chromosome transactions in S phase. J. Biol. Chem. 2009; 284:20718–20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Im J.S., Ki S.H., Farina A., Jung D.S., Hurwitz J., Lee J.K.. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:15628–15632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bermudez V.P., Farina A., Tappin I., Hurwitz J.. Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J. Biol. Chem. 2010; 285:9493–9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Errico A., Cosentino C., Rivera T., Losada A., Schwob E., Hunt T., Costanzo V.. Tipin/Tim1/And1 protein complex promotes Pol alpha chromatin binding and sister chromatid cohesion. EMBO J. 2009; 28:3681–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R.C., Edmondson R.D., Calzada A., Labib K.. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009; 28:2992–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon A.C., Zhou J.C., Perera R.L., van Deursen F., Evrin C., Ivanova M.E., Kilkenny M.L., Renault L., Kjaer S., Matak-Vinkovic D. et al. . A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature. 2014; 510:293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y., Jaramillo-Lambert A.N., Yang Y., Williams R., Lee N.H., Zhu W.. And-1 is required for the stability of histone acetyltransferase Gcn5. Oncogene. 2012; 31:643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y.M., Jaramillo-Lambert A., Hao J., Yang Y., Zhu W.G.. The Stability of Histone Acetyltransferase General Control Non-derepressible (Gcn) 5 Is Regulated by Cullin4-RING E3 Ubiquitin Ligase. J. Biol. Chem. 2011; 286:41344–41352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kouprina N., Kroll E., Bannikov V., Bliskovsky V., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V, Hieter P., Spencer F.. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1992; 12:5736–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanna J.S., Kroll E.S., Lundblad V., Spencer F.A.. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 2001; 21:3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer M.L., Pot I., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W. et al. . Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell. 2004; 15:1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams D.R., McIntosh J.R.. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryotic Cell. 2002; 1:758–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsutsui Y., Morishita T., Natsume T., Yamashita K., Iwasaki H., Yamao F., Shinagawa H.. Genetic and physical interactions between Schizosaccharomyces pombe Mcl1 and Rad2, Dna2 and DNA polymerase alpha: evidence for a multifunctional role of Mcl1 in DNA replication and repair. Curr. Genet. 2005; 48:34–43. [DOI] [PubMed] [Google Scholar]
- 55.Gambus A., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K.. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006; 8:358–366. [DOI] [PubMed] [Google Scholar]
- 56.Hao J., de Renty C., Li Y., Xiao H., Kemp M.G., Han Z., DePamphilis M.L., Zhu W.. And-1 coordinates with Claspin for efficient Chk1 activation in response to replication stress. EMBO J. 2015; 34:2096–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogiwara H., Ui A., Lai M.S., Enomoto T., Seki M.. Chl1 and Ctf4 are required for damage-induced recombinations. Biochem. Biophys. Res. Commun. 2007; 354:222–226. [DOI] [PubMed] [Google Scholar]
- 58.Zhu W., Chen Y., Dutta A.. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 2004; 24:7140–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y.M., Xiao H.J., de Renty C., Jaramillo-Lambert A., Han Z.Y., DePamphilis M.L., Brown K.J., Zhu W.G.. The involvement of acidic nucleoplasmic DNA-binding protein (And-1) in the regulation of prereplicative complex (pre-RC) assembly in human cells. J. Biol. Chem. 2012; 287:42469–42479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinstock D.M., Nakanishi K., Helgadottir H.R., Jasin M.. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006; 409:524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dinant C., de Jager M., Essers J., van Cappellen W.A., Kanaar R., Houtsmuller A.B., Vermeulen W.. Activation of multiple DNA repair pathways by subnuclear damage induction methods. J. Cell Sci. 2007; 120:2731–2740. [DOI] [PubMed] [Google Scholar]
- 62.Baldeyron C., Soria G., Roche D., Cook A.J.L., Almouzni G.. HP1 alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol. 2011; 193:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki K., Yamauchi M., Oka Y., Suzuki M., Yamashita S.. Creating localized DNA double-strand breaks with microirradiation. Nat. Protoc. 2011; 6:134–139. [DOI] [PubMed] [Google Scholar]
- 64.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006; 6:789–802. [DOI] [PubMed] [Google Scholar]
- 65.Feng L., Li N., Li Y., Wang J., Gao M., Wang W., Chen J.. Cell cycle-dependent inhibition of 53BP1 signaling by BRCA1. Cell Discov. 2015; 1:15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peterson S.E., Li Y., Wu-Baer F., Chait B.T., Baer R., Yan H., Gottesman M.E., Gautier J.. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell. 2013; 49:657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hickson I., Yan Z., Richardson C.J., Green S.J., Martin N.M.B., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C.M.. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004; 64:9152–9159. [DOI] [PubMed] [Google Scholar]
- 68.Bennett C.B., Lewis L.K., Karthikeyan G., Lobachev K.S., Jin Y.H., Sterling J.F., Snipe J.R., Resnick M.A.. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 2001; 29:426–434. [DOI] [PubMed] [Google Scholar]
- 69.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 1999; 19:5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grosschedl R., Giese K., Pagel J.. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994; 10:94–100. [DOI] [PubMed] [Google Scholar]
- 71.Shiotani B., Zou L.. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009; 33:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.