Abstract
Sodium bisulphite treatment of DNA combined with next generation sequencing (NGS) is a powerful combination for the interrogation of genome-wide DNA methylation profiles. Library preparation for whole genome bisulphite sequencing (WGBS) is challenging due to side effects of the bisulphite treatment, which leads to extensive DNA damage. Recently, a new generation of methods for bisulphite sequencing library preparation have been devised. They are based on initial bisulphite treatment of the DNA, followed by adaptor tagging of single stranded DNA fragments, and enable WGBS using low quantities of input DNA. In this study, we present a novel approach for quick and cost effective WGBS library preparation that is based on splinted adaptor tagging (SPLAT) of bisulphite-converted single-stranded DNA. Moreover, we validate SPLAT against three commercially available WGBS library preparation techniques, two of which are based on bisulphite treatment prior to adaptor tagging and one is a conventional WGBS method.
INTRODUCTION
Methylation of cytosine (5-mC) residues in CpG dinucleotides is an epigenetic modification that plays a pivotal role in the establishment of cellular identity by influencing gene expression. In somatic mammalian cells, the majority of CpG sites are methylated. However, CpG sites located in regions of increased CG density, known as CpG islands, generally have low levels of CpG methylation (1). On the molecular level, it is well known that CpG methylation leads to X-chromosome inactivation, genomic imprinting, regulation of gene expression and suppression of transposable elements. Disruption of DNA methylation patterns is associated with disease, and particularly with cancer (2). These findings have spurred the development of technologies for genome wide DNA methylation profiling. The HumanMethylation450 BeadChip assay (450k Bead Arrays, Illumina) has so far been the most frequently used platform for human studies. However, with the advent of high throughput sequencing techniques there has been a rapid development of methods that interrogate a larger proportion of the CpG sites in any genome, which can interrogate from a few million CpG sites up to the whole methylome, which in humans consists of 28 million CpG sites. Sodium bisulphite treatment of DNA converts non-methylated cytosines into uracils, whilst methylated cytosines remain unchanged (3). Hence, methylation status at individual CpG sites can be read out by sequencing or genotyping (4–6). Methods for genome-wide DNA methylation analysis that rely on bisulphite conversion of DNA include BeadArrays (7,8) reduced-representation-bisulphite-sequencing (RRBS) (9–12), targeted capture methods (13–16), and whole genome bisulphite sequencing (WGBS). The 450k and 850k (EPIC) BeadArrays interrogate 2–4% of human CpG sites that are preselected based on various annotated features such as genes, promoters, CpG islands and regulatory elements such as enhancers. Targeted capture methods may be designed to interrogate any region of the genome using e.g bisulphite padlock probes (13,14) or in solution hybridisation protocols (15–17). The latter is implemented in several commercial kits, such as SureSelect Methyl-Seq (Agilent Technologies) and SeqCap Epi systems (Roche NimbleGen). RRBS uses restriction enzyme digestion to enrich for regions of high CpG content, such as CpG islands and promoters in any genome (9–12).
WGBS is the ideal choice for many DNA methylation studies since it allows for unbiased genome-wide profiling at single-base resolution. However, a drawback of WGBS is that it is still costly to perform on large sample sets. Moreover, conventional sample preparation methods for WGBS typically require microgram amounts of DNA, which may be prohibitive for many human disease applications and sample types. The strand breaking side effect of sodium bisulphite treatment renders the majority of sequencing library constructs unamplifiable during PCR and sequencing cluster generation. Using low amounts of input DNA is feasible, albeit at the expense of low complexity of the sequencing libraries and high redundancy of the obtained sequence reads caused by the large number of amplification cycles required. Tagmentation-based whole genome bisulphite sequencing (T-WGBS) (18) was developed for low DNA quantity library construction, but a major drawback to this approach is that the tagmentation is performed prior to bisulphite conversion and thus extensive amplification may still be required.
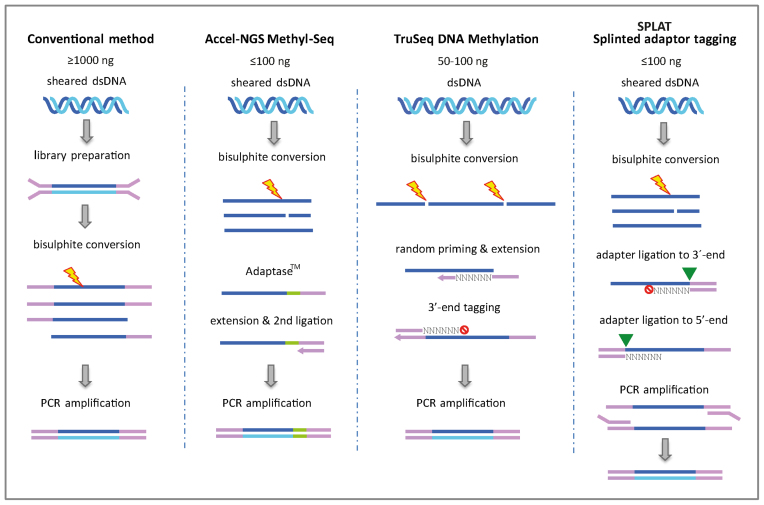
In order to circumvent the damage of library constructs, methods have been designed in which sequencing adapters are incorporated after bisulphite treatment (hereafter referred to as ‘post bisulphite’ methods) (19,20). In the pioneering post bisulphite method PBAT (post bisulphite adapter tagging), sequencing adapters are attached by two rounds of random primer extension using the bisulphite converted ssDNA as the initial template. PBAT enabled PCR-free WGBS libraries to be constructed from DNA quantities in the nanogram range (19) and has paved the way for further developments, such as Illumina's TruSeq DNA Methylation kit (Figure 1). Notably, a modified version of the PBAT protocol was recently used for single cell methylation profiling (21). An alternative approach for post bisulphite library preparation is implemented in the AccelNGS Methyl-Seq protocol (Swift Biosciences) whereby a low complexity sequence tag is added to the 3΄ end of the ssDNA in order to serve as a scaffold for the attachment of sequencing adapters (Figure 1).
Figure 1.
Principles of library preparation methods for whole genome bisulphite sequencing. In the conventional workflow (MethylC-seq) methylated adapters are ligated to double stranded sheared DNA fragments. The constructs are then bisulphite converted prior to amplification with a uracil reading PCR polymerase. The Accel-NGS Methyl-Seq uses the proprietary Adaptase™ technology to attach a low complexity sequence tail to the 3΄-termini of pre-sheared and bisulphite-converted DNA, and an adapter sequence. After an extension step a second adapter is ligated and the libraries are PCR amplified. The TruSeq DNA Methylation method (formerly EpiGnome) uses random hexamer tagged oligonucleotides to simultaneously copy the bisulphite-converted strand and add a 5΄-terminal adaptor sequence. In a subsequent step, a 3΄-terminal adapter is tagged, also by using a random sequence oligonucleotide. In the SPLAT protocol adapters with a protruding random hexamer are annealed to the 3΄-termini of the single stranded DNA. The random hexamer acts as a ‘splint’ and the adapter sequence is ligated to the 3΄-termini of single stranded DNA using standard T4 DNA ligation. A modification of the last 3΄- residue of the random hexamer is required to prevent self-ligation of the adapter. In a second step, adapters with a 5΄-terminal random hexamer overhang is annealed to ligate the 5΄-termini of the single stranded DNA, also using T4 DNA ligase. Finally the SPLAT libraries are PCR amplified using a uracil reading polymerase.
Herein, we describe splinted adapter tagging (SPLAT), which is a novel protocol for library preparation from single-stranded bisulphite-treated DNA fragments. Prompted by the insufficient knowledge of how post bisulphite WGBS methods perform with respect to the quality of sequencing data they generate, we evaluated the performance of SPLAT against two commercial post bisulphite kits (TruSeq DNA Methylation, and Accel-NGS Methyl-Seq), one conventional WGBS method and those obtained from other popular targeted approaches (RRBS, target capture by SureSelect Methyl-Seq, and 450k BeadArrays) to determine the concordance of DNA methylation data across the different methods.
MATERIALS AND METHODS
Sample source
Human genomic DNA from a lymphoblastoid B- cell line (NA10860) was obtained from the Coriell Institute for Medical Research. Genomic DNA from the pre-B acute lymphoblastoid leukemia cell line REH (22) was isolated using the AllPrep Universal kit (Qiagen). DNA was quantified using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific).
Splinted adaptor tagging (SPLAT) protocol for whole genome bisulphite sequencing library preparation
Adapter annealing
Oligonucleotides were purchased from Integrated DNA Technologies. The 3΄-adapter; ss1 (5΄GACGTGTGCTCTTCCGATCTNNNNNN-3΄-amino-modifier and 5΄P-AGATCGGAAGAGCACACGTC and 5΄-adapter; ss2 (5΄-ACACGACGCTCTTCCGATCT and 5΄-NNNNNNAGATCGGAAGAGCGTCGTGT) were prepared by mixing the two oligonucleotides in the adapter pair to a final concentration of 100 μM in 50 μl of 100 mM NaCl,10 mM Tris–HCl (pH 8.0), 0.5 mM EDTA. The oligonucleotides were annealed by heating the mixture to 95°C and slowly decreasing the temperature to 10°C.
Library construction
Genomic DNA was sheared to 300–400 bp (Covaris E220 System) and treated with sodium bisulphite (EZ DNA Methylation Gold, Zymo Research). The converted DNA was first treated with 5 units of polynucleotide kinase (Thermo Fisher Scientific), in T4 DNA ligase buffer (see buffer composition below) in a total volume 15 μl, for 15 min at 37°C. The reaction mixture was heated to 95°C for 3–5 min in a thermal cycler with a heated lid and then cooled on an ice/water bath. For the 3΄-end ligation; adapter ss1 (final conc 10 μM), T4 DNA ligase buffer (40 mM Tris–HCl pH 7.8,10 mM MgCl2, 10 mM DTT, 0.5 mM ATP), PEG4000 (5% w/v) and 30 units T4 DNA ligase (Thermo Fisher Scientific) and nuclease free water was added to the sample on ice, in a total volume of 30 μl. Ligation reaction mixtures were incubated at 20°C for 1 h, and subsequently purified using AMPure XP (BeckmanCoulter) beads in a 2:1 bead to sample ratio and eluted with 10 μl nuclease free water. The eluted DNA was heated to 95°C for 3–5 min in a thermal cycler with a heated lid and then cooled on an ice/water bath. For the 5΄-end ligation; ss2 (final conc 10 μM), T4 DNA ligation buffer, PEG4000 (5%, w/v) and 30 units T4 DNA ligase (Thermo Fisher Scientific) and nuclease free H2O was added to the sample on ice, in a total volume of 20 μl. Ligation reaction mixtures were incubated at 20°C for 1 h, purified using AMPureXP beads in a 2:1 bead to sample ratio and eluted in 10 μl nuclease free water. The libraries were amplified for 4 cycles using KAPA HiFi Uracil+ ReadyMix (KAPA Biosystems) and oligonucleotides:
5΄-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3΄, 5΄-CAAGCAGAAGACGGCATACGAGATX6GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3΄ where X6 denotes the Illumina index barcode sequence. The final libraries were purified twice with AMPure XP beads using a 1:1 bead to sample ratio. See supplementary information for more details.
Sequencing libraries
The EZ DNA Methylation Gold kit was used for sodium bisulphite conversion of DNA according to the specifications specific for each library type as listed below.
Conventional WGBS libraries were prepared using the NEBNextUltra kit. Briefly, 1 μg of gDNA was sheared to 300–400 bp (Covaris E220 system). End repair, A-tailing and ligation of methylated adapters was performed according to the manufacturer's protocol with the following minor modifications: ligated samples were cleaned using AMPure XP beads at a 2:1 bead to sample ratio prior to bisulphite conversion and PCR amplified for six cycles using the KAPA HiFi Uracil+ PCR polymerase .
The ‘post-bisulphite’ conversion WGBS libraries were prepared according to the Accel-NGS Methyl-Seq DNA library kit (Swift BioSciences) or the TruSeq DNA Methylation Kit (Illumina) according to the manufacturers’ protocols. The Accel-NGS libraries were generated from 100 ng of sheared (350 bp, Covaris E220 System) gDNA that was subsequently bisulphite converted. Four cycles of PCR were used to amplify the Accel-NGS Methyl-Seq libraries. The TruSeq DNA Methylation libraries were generated from 100 ng of sodium bisulfite treated gDNA without pre-shearing. Nine cycles of PCR were used to amplify the TruSeq DNA Methylation libraries.
RRBS libraries were prepared by a single-tube workflow using NEBNextUltra reagents (NEBNextUltra EndRep/A-tailing module, Ligation module and Methylated Adapters, New England Biolabs). Briefly, 200 ng of gDNA in a total volume of 25 μl was digested for 16 h using 50 units of MspI in NEB2 buffer (New England Biolabs). After the incubation the reaction mixture was transferred to an end repair/A-tailing reaction mixture. End repair, A-tailing and ligation of methylated adapters were performed as recommended in the NEBNextUltra kit protocol. The adapter concentrations were reduced 10-fold in the ligation mixes (50 nM final concentration) compared to the manufacturer's instruction to avoid excessive formation of adapter dimers. After ligation the samples were purified with AMPure XP beads applying a 2:1 bead to sample ratio, prior to bisulphite treatment. Bisulphite-converted libraries were PCR amplified for eleven cycles using the Pfx Turbo Cx polymerase (Agilent Technologies). Two consecutive bead purifications with a 0.9:1 bead to sample ratio were performed to clear the PCR-amplified libraries from oligonucleotides and adapter dimers.
SureSelect Methyl-Seq (Agilent Technologies) libraries were prepared from 3 μg of gDNA following the manufacturer's protocol.
Sequencing and data analysis
Paired end sequencing (2 × 125 bp) was performed on a HiSeq2500 system using the TruSeq v4 chemistry (Illumina). The specific parameters for the programs used for mapping, quality assessment, and DNA methylation calling are provided in detail in Supplementary Data. Briefly, quality control of sequencing data was performed with the FastQC tool. Sequence reads were quality filtered and adapters were trimmed using TrimGalore. Alignment to the human reference assembly GRCh37 and methylation calling was performed with the Bismark v 0.14 software (23) and the pipeline tool ClusterFlow v 0.4 http://www.clusterflow.io. GC bias metrics in sequencing data was analysed with Picard Tools (http://broadinstitute.github.io/picard/). Sequencing coverage across the whole genome and in genomic features was determined with Qualimap (24) and BEDTools (25). CpG site coverage and concordance of methylation calls was computed from the Bismark methylation extractor output files using custom R scripts. The cytosine methylation status was called per individual CpG sites as #C/(#C + #T) for CpG sites with at least 5x coverage. Thus methylation was detected if at least one methylated read was observed. CpG sites were annotated using BEDTools. Annotation files were downloaded from the UCSC Genome Browser or prepared from the GENCODE hg19 annotation by methods similar to those described in; https://www.gitbook.com/book/ycl6/methylation-sequencing-analysis. Hypomethylated regions were identified using MethylSeekR (26) and overlapping hypomethylated regions were determined using the ‘findoverlaps’ function in the R package GenomicRanges using default settings (27).
Processing of Accel-NGS Methyl-Seq reads
A low complexity tag of varying length is added to the 3΄ termini of the ssDNA during the Accel-NGS Methyl-Seq library preparation (Figure 1, Supplementary Data) and these sequence tails are present at the beginning of all second read (R2) sequences. Since paired-end sequencing read lengths of 125 base pairs are close to the insert sizes of the libraries, the additional sequence tails in the Accel-NGS Methyl-Seq libraries are present at the end of many of sequences from the first read. The kit vendor suggests that trimming off 10 residues at the beginning of each R2 sequence and end of each R1 sequence for read lengths over 100 bp should be sufficient to remove most of the artificial sequence in the data. However, based on per base sequence content plots (see Supplementary Data for per base sequence content plots for all WGBS methods) we recognised that in our Accel-NGS Methyl-Seq data, the low complexity sequence tag may be up to 15–18 nucleotides in length. Thus trimming the first 18 residues of each R2 sequence and the end of each R1 sequence was performed to avoid methylation artefacts and improve alignment efficiency.
DNA methylation analysis using 450k bead arrays
Genomic DNA (500 ng) was treated with sodium bisulphite (EZ DNA Methylation Gold) and the DNA methylation levels were measured using the Infinium HumanMethylation 450k BeadChip Array (Illumina) according to the manufacturer's instructions. Raw beta-values were extracted from the arrays using the Genome Studio Methylation Module (Illumina) and methylation data from probes that hybridize to more than one genomic location and from probes with SNPs in the target regions of their 3΄-ends were filtered out as previously described (28).
RESULTS
Splinted adaptor tagging (SPLAT) for whole genome bisulphite sequencing library preparation
In the current study we developed SPLAT, which is an alternative method that introduces a new concept for efficient ligation of sequencing adapters to bisulphite converted single stranded DNA fragments. The SPLAT method takes advantage of splint oligonucleotides (29) to create short stretches of dsDNA fragments that allow subsequent ligation of sequencing adapters using standard dsDNA ligation with T4 DNA ligase, which is more efficient than ligation of ssDNA. SPLAT, which is outlined in Figure 1 (together with the other library methods for WGBS used in this study) is a sensitive method, which uses affordable off-the-shelf enzymes. Using SPLAT with 100 ng of input DNA subjected to only four PCR amplification cycles (using KAPA HiFi Uracil+ polymerase) yields sufficient amounts of library for whole genome bisulphite sequencing (WGBS). Library size profiles and the results from library quantifications are shown in Supplementary Data. After bisulphite conversion, short double stranded adapters (20 nucleotides) comprising a random 3΄ overhang are annealed to the 3΄ ends of the ssDNA and ligated using T4 DNA ligase. Similarly in a second step, the 5΄ ends of the ssDNA are ligated using adapters comprising a random 5΄ overhang. To prevent self-ligation of adapters in the first ligation step, an amino modification (3΄ Amino Modifier, Integrated DNA Technologies) is added to the 3΄-terminal nucleotide in the random hexamer oligo. In the second ligation step the oligo modification is not required. The libraries are subsequently amplified by PCR making use of the KAPA HiFi Uracil+ polymerase, which is capable of reading the uracil base and is compatible with oligos that contain Illumina flow cell binding and indexing sequences.
We validated SPLAT by performing WGBS of DNA from two different sources, namely the lymphoblastoid B-cell line NA10860 and the B-cell leukemic cell line REH (22). These two cell lines, an immortalized B-cell line and a cancer cell line were chosen to mirror samples with differing methylation profiles. Two SPLAT libraries were prepared from each cell line and each library was sequenced on one lane, PE125 base pairs using a HiSeq2500 machine (in total four SPLAT libraries). Data yields and mapping efficiencies were in the expected range for WGBS and the PCR duplication levels were very low (1–2%) in all four SPLAT libraries. Sequencing information, mapping efficiencies and PCR duplication levels are listed in Table 1. Data from the two technical replicates per cell type were merged prior to analysis and the final average read coverage was 16x and 22× in NA10860 and REH data, respectively. At CpG dinucleotide positions specifically, the average coverage was 13× and 17× in NA10860 and REH SPLAT data, respectively (Table 2). The SPLAT libraries from both cell types displayed uniform genome coverage, albeit coverage decreases in CpG islands and in very GC rich promoter regions (Figure 3B and Supplementary Data). This is a commonly observed phenomenon in next generation sequencing (NGS) data and is thought to be a consequence of low PCR efficiency for GC-rich regions. In summary, WGBS libraries prepared with the SPLAT method displayed excellent global performance metrics such as data yield, mapping efficiency, PCR duplication rates and uniformity of coverage.
Table 1. Sequence metrics for whole genome bisulfite libraries.
| Method & DNA quantity | Cell line | PCR cycles | Mapping efficiency (%)a | Aligned read pairs (M)a | PCR duplicates (%) | ||
|---|---|---|---|---|---|---|---|
| TruSeq/ | NA10860 | 9 | 76 | 77 | 133 | 126 | 13–17 |
| 100 ng | REH | 9 | 78 | 78 | 133 | 141 | 12–15 |
| Accel-NGS/ | NA10860 | 4 | 83 | 82 | 129 | 131 | 1–2 |
| 100 ng | REH | 4 | 82 | 82 | 100 | 115 | 1–2 |
| SPLAT/ | NA10860 | 4 | 70 | 70 | 92 | 128 | 1–2 |
| 100 ng | REH | 4 | 83 | 77 | 136 | 176 | 1–2 |
| NEBNextUltra/ | NA10860 | 6 | 75 | 75 | 131 | 102 | 1–2 |
| 1000 ng | REH | 6 | 79 | 79 | 129 | 125 | 1–2 |
afor each individual sequenced library (technical replicates).
Table 2. Read coverage and correlation of methylation between SPLAT and high coverage reference data.
| WGBS data | Average read coverage | Average CpG coverage | Correlation of methylation (Pearsons R) | |
|---|---|---|---|---|
| NA10860 SPLAT | 16× | 13× | NA10860; SPLAT versus reference | 0.94 |
| NA10860 high coverage reference | 49× | 36× | ||
| REH SPLAT | 22× | 17× | REH; SPLAT versus reference | 0.97 |
| REH high coverage reference | 49× | 36× |
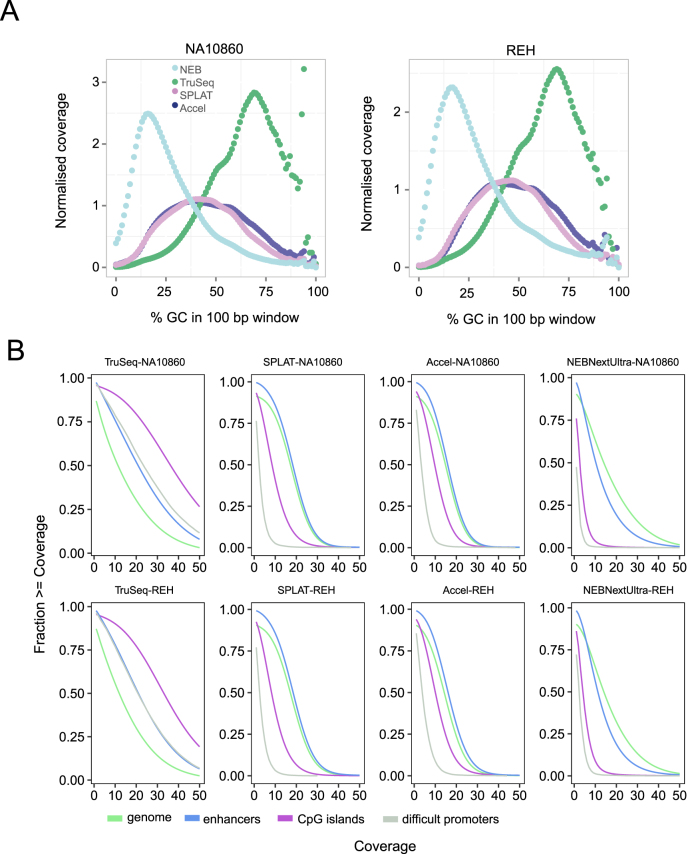
Figure 3.
Sequence bias in whole genome bisulphite sequencing libraries. (A) GC bias observed for the different methods. The plots show the normalised coverage in 100 bp windows of increasing GC content in the human reference genome. NEBNextUltra libraries had higher read coverage in AT rich regions, whilst TruSeq DNA Methylation had increased read coverage of GC rich regions. The AccelNGS Methyl-Seq and SPLAT libraries displayed a lower GC bias, however regions with extreme GC content were not well represented. (B) Cumulative coverage of different genomic regions. Coverage plots for the whole genome is shown in in green, CpG islands in purple, FANTOM5 enhancer regions in blue and a set of 1000 promoter regions that are difficult to sequence due to high GC content in grey.
Validation of methylation calls in SPLAT data against high coverage reference data sets
To obtain high coverage ‘reference’ methylation maps for both cell lines NA10860 and REH, we merged WGBS data from six sequencing libraries that were prepared using three different commercial kits (TruSeq DNA Methylation, Accel-NGS Methyl- Seq, NEBNextUltra). In this way we created two high coverage WGBS data sets with average read coverages of ∼50× for use as reference data sets for the results from SPLAT (Table 2). We investigated the concordance of methylation profiles between SPLAT and the reference data sets by comparing methylation levels computed across reads at individual CpG sites. To measure how the read coverage in SPLAT affects the accuracy of DNA methylation calls we binned CpG sites by coverage and measured the correlation with the methylation levels called in the high coverage reference data set for CpG sites with ≥20x coverage (Supplementary Data). As expected, the methylation calls were more accurate with increasing read-depth, however still at low coverage the correlation coefficient was >0.9. When comparing all CpG sites interrogated by five or more reads Pearson's R was 0.94 for NA10860 and 0.97 for REH. We performed pairwise comparisons of methylation levels between SPLAT and the reference data sets from NA10860 and REH for CpGs located in CpG islands, putative enhancer regions and known repetitive elements (Figure 2A). The mean read coverage of the CpG sites in these regions is shown in Figure 2B. To investigate regions with intermediate methylation levels we also compared methylation levels at individual CpG sites located within 36 known imprinted regions where one of the alleles is expected to be fully methylated while the other is fully unmethylated (30). In the lymphoblastoid cell line (NA10860) a large fraction of the CpG sites located in the known imprinted regions displayed the expected methylation levels close to 50%. However, in the cancer cell line (REH) aberrant DNA methylation profiles that are indicative of loss of imprinting were observed (Supplementary Data). This is a common phenotype of cancer cells (31).
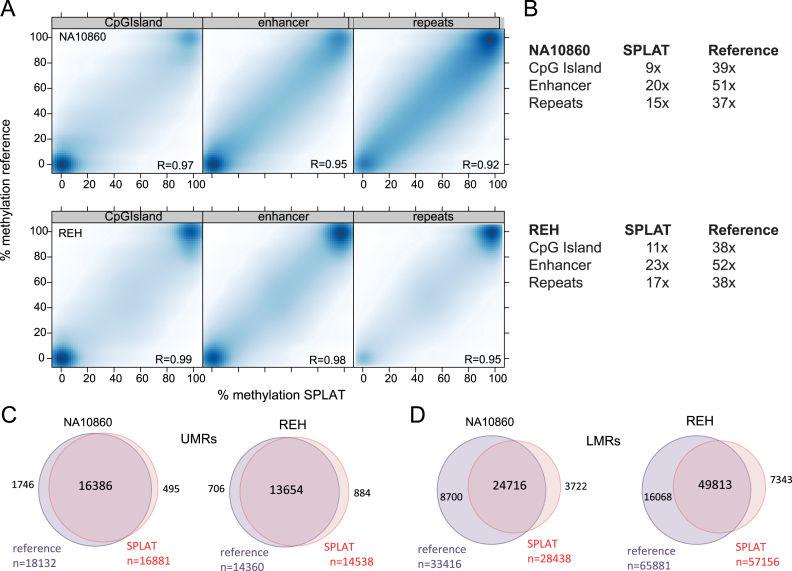
Figure 2.
Comparison of SPLAT with a high coverage reference dataset. (A) Pairwise comparison of methylation levels at individual CpG sites in CpG islands, enhancers, and repetitive regions determined by SPLAT (x-axis) and the high-coverage reference WGBS data (y-axis) obtained by combining the data from TruSeq DNA Methylation, Accel-NGS and NEBNextUltra libraries. Pearson's correlation coefficients are shown in the lower right corner of each scatter plot. (B) The mean read coverage of the CpG sites in CpG islands, enhancers and repeats for SPLAT and the high coverage reference data set. (C) Comparison of the numbers and overlaps of un-methylated regions (UMRs) between SPLAT and high coverage reference WGBS data. (D) Comparison of the numbers and overlaps of lowly methylated regions (LMRs) in SPLAT and high coverage reference WGBS data.
Hypomethylated regions in whole genome bisulfite sequencing libraries prepared with SPLAT
An important aspect for determining the quality of a WGBS experiment is the ability for unbiased, genome-wide detection of regulatory regions. Therefore, as a second approach to validate the data from SPLAT, we used the MethylSeekR software for identification of regions belonging to one of two distinct classes: CpG-rich, completely unmethylated regions that correspond to proximal regulatory sites including promoters (UMRs) and CpG-poor, low-methylated regions that correspond to distal regulatory sites (LMRs) (26). The UMRs detected in the SPLAT libraries and the high coverage reference data set were highly overlapping (Figure 2C). In total, 97% and 93% of the UMRs detected in NA10860 and REH cells, respectively, overlapped with the high coverage reference data set. Unlike UMRs, which contain regions with a high density of CpG sites that typically are completely unmethylated (mean methylation level 6.5%), LMRs are short regions with few CpG sites with more intermediate methylation levels (mean 18–22%) that are thought to be associated with TF binding. In total 74% and 76% of the LMRs identified overlapped between SPLAT and the high coverage reference data set in NA10860 and REH cells, respectively (Figure 2D). As expected, more LMRs were detected in the high coverage data than in SPLAT. Notably however, the fractions of LMRs that were uniquely identified in either data set was higher than that observed for the UMRs. For instance, in NA10860 cells 3722 LMRs were unique to the SPLAT data and 8700 LMRs were unique to the high coverage reference data. Because the uncertainty in estimation of methylation levels is dependent on coverage, especially so in the regions that display intermediate methylation levels, we looked specifically at the sequencing coverage these regions. The average coverage of the regions with overlapping LMRs was 13–17× in the SPLAT libraries and 36× in the high coverage reference datasets. Despite this, when looking at the regions specifically called in the high coverage data set, but not in the SPLAT libraries these were covered at the same average sequence depth as the overlapping regions. When inspecting the non-overlapping regions in more detail we found that the mean methylation levels were generally higher in the uniquely identified regions than in the overlapping LMRs (mean 18–22% methylation in overlapping LMRs and 26–29% methylation in unique LMRs). Thus methylation levels across unique LMRs deviated upwards towards software's upper limit cut-off for the defining a LMR (50% methylation), which may at least in part explain why these regions are called in one data set, but not in the other.
Comparison of SPLAT to commercial whole genome bisulphite sequencing methods
Next, we assessed the performance of SPLAT in individual comparisons with the three commercial methods for bisulphite sequencing library preparation. In two of the methods the DNA is bisulphite treated prior to adapter ligation using reagents from the TruSeq DNA Methylation (formerly EpiGnome) kit and the Accel-NGS Methyl-Seq kit, and a conventional method (NEBNextUltra), which is based on adapter ligation prior to bisulphite treatment (Figure 1). The TruSeq DNA Methylation protocol makes use of the PBAT concept with oligos with degenerate 3΄ ends for adapter tagging, whilst the strategy for adapter tagging in Accel NGS Methyl-Seq is based on Adaptase™ technology (Figure 1). The two types of ‘post bisulphite’ WGBS libraries were prepared using 100 ng of input DNA, whilst 1000 ng of input DNA was used for the conventional NEBNextUltra libraries.
Sequencing metrics
WGBS libraries were sequenced in one lane per technical replicate using a Hiseq2500 instrument and 125 bp paired-end reads with v4 sequencing chemistry. Sequence reads from all libraries were pre-processed prior to alignment using the same parameters, with the exception of the Accel-NGS Methyl-Seq data that required additional processing to remove the low complexity sequence tags that are introduced during library preparation. To avoid biases in the comparison, the same parameters were used for alignment of all libraries with the Bismark software (23). Mapping rates were between 70% and 83% for each of the WGBS methods, which is acceptable as the reduced complexity of the reads is expected to have a negative impact on the alignment efficiency (Table 1). The highest mapping efficiency (83%) was achieved for the SPLAT and Accel-NGS Methyl-Seq libraries. The PCR duplication rate was low for SPLAT, Accel-NGS Methyl-Seq and NEBNextUltra (1–2% per lane), while TruSeq DNA Methylation libraries displayed higher PCR duplication rates (12–17% per lane) (Table 1).
Data from two technical replicates were merged prior to downstream analysis. The amount of sequence data generated, the data yield after alignment and removal of PCR duplicates, the average coverage, and the mean library insert sizes in the WGBS methods are listed in Supplementary Data. WGBS libraries have relatively short insert sizes and consequently suffer from significant adapter contamination, particularly when read lengths are ≥100 bp. We observed the shortest mean insert sizes of ∼160 base pairs in the TruSeq DNA Methylation libraries and mean insert sizes between 170 and 195 bp for the other protocols (Supplementary Data). Lower mapping efficiencies, high duplication rates, and short insert sizes are factors that all contribute to considerable loss of data in WGBS. The data yield relative to the amount of raw sequencing data generated was lowest for the TruSeq DNA Methylation libraries, where only 55–58% of data was retained after pre-processing, alignment and de-duplication. The Accel-NGS Methyl-seq and NEBNextUltra libraries retained 65–71% of data (mean 68%). The SPLAT method retained 65–76% of the data, which on average was the method that retained the most data after filtering (Supplementary Data). Therefore, the average genome coverage ranged from 14× to 22× in the WGBS data sets. Prior to performing subsequent analyses, each of the WGBS data sets was down-sampled to approximately the same number of read pairs (218–219 M for NA10860 libraries, 211–216 M for REH libraries) to enable cross-method comparisons with the same amount of starting data.
Read coverage of WGBS libraries
Most currently used next generation sequencing library preparation methods result in an under-representation of GC-rich regions, which originates at least in part, from PCR amplification (32,33). In WGBS data, the GC bias tends to be even more pronounced than in standard sequencing libraries and thus important GC rich regions (CpG islands) that are targets of DNA methylation, e.g. in cancer can be significantly underrepresented. We determined the normalized read coverage in 100 bp windows of increasing GC content in the human reference genome, and found that the methods for sequencing library preparation generate distinct biased GC profiles (Figure 3A). In the NEBNextUltra libraries, coverage was skewed towards AT rich regions, while GC rich regions were poorly covered. In contrast, in TruSeq DNA Methylation libraries coverage was skewed towards GC rich regions. Accel-NGS Methyl-Seq and SPLAT libraries had more uniform GC bias profiles, where regions with extreme base compositions, either AT or GC rich, are underrepresented.
Next we analyzed the read coverage distribution generated by the WGBS library preparation methods between genomic regions. Cumulative coverage plots for the whole genome, enhancer regions as defined by the FANTOM5 consortium (34), CpG islands and a set of 1000 promoters that have been characterized as ‘difficult promoters’, which have very high CG content and are notoriously difficult to sequence (35), are shown in Figure 3B for both cell types (see Supplementary Data for coverage histograms). Examples of genome browser views for the NA10860 cell type are shown in Figure 4, to further illustrate coverage and methylation profiles. As was already inferred based on the GC bias profiles, the coverage of different genomic features varied in a characteristic manner between the WGBS methods. Out of the four methods, the Accel-NGS Methyl-Seq and SPLAT libraries displayed the most uniform coverage of the whole genome and of enhancer regions. However, for both these methods the read coverage dropped in CpG islands and a substantial drop in coverage was observed in the ‘difficult promoters’. The TruSeq DNA Methylation libraries displayed uneven overall coverage of the genome. However, GC-rich promoter regions and CpG islands generally displayed higher coverage than the genome on average and this was the only method that obtained adequate coverage of the ‘difficult promoters’. The NEBNextUltra libraries on the other hand were characterised both by uneven whole genome coverage and a severe decrease in coverage in CpG islands and ‘difficult promoters’. Importantly, the GC bias and coverage profiles were very reproducible between the two different cell types for each of the WGBS methods described here, which indicates these results are robust against any biological differences in total DNA methylation levels (Figure 3).
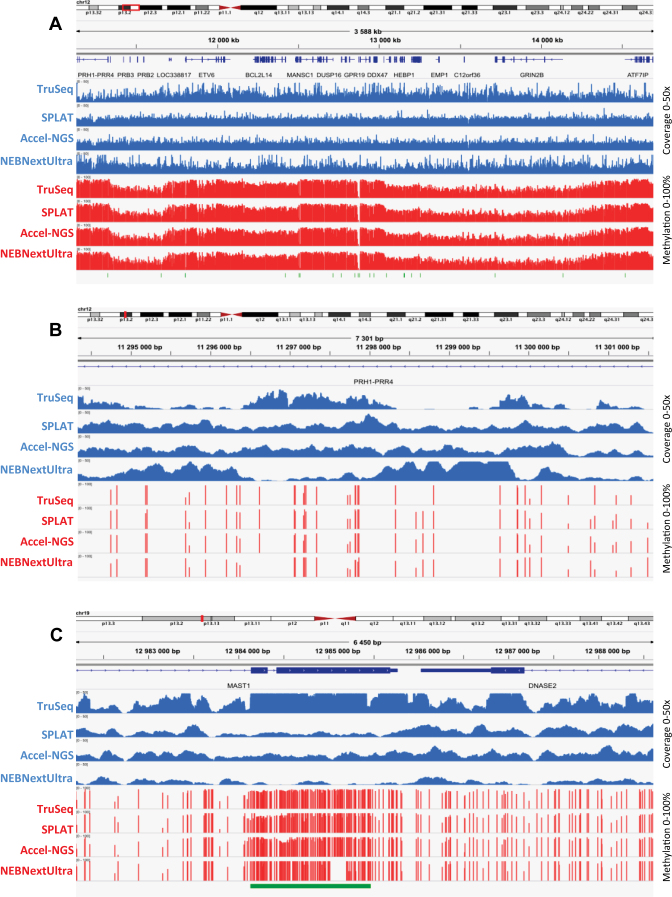
Figure 4.
Genome browser views displaying coverage and methylation tracks for the four different WGBS libraries generated from the NA10860 cell type. The blue tracks represent the read coverage, the red tracks represent the methylation levels across the regions and the green bars at the bottom of the panels represent CpG islands. (A) Read coverage and methylation levels across a 3.5 Mb region of chromosome 12. (B) Zoom-in view of a CpG poor 7.3 kb intergenic region on chromosome 12. (C) Zoom-in view of a 6.4 kb region containing a methylated CpG island on chromosome 19.
Coverage biases in public WGBS data sets
It should be noted that the findings presented herein represent observations in our own laboratory at a given point in time. To investigate if the observed biases in genomic and CpG site sequence coverage were reproducible between labs and not only characteristic to our own sequencing library preparations we downloaded human WGBS data sets from the NCBI Sequence Read Archive (SRA) from different library preparation protocols and analysed their coverage profiles (Supplementary Data). First, we assessed three TruSeq DNA Methylation libraries from the human lymphoblastoid cell line NA18507 obtained from the Blueprint consortium (GSE66285) in which we observe the same trend towards increased coverage of CpG rich regions, including difficult promoters, as in our data. Second, we evaluated five conventional human WGBS data sets originating from five various tissue types in which libraries were amplified using the two most commonly used uracil reading PCR polymerases (KAPA HiFi Uracil+ or PfuTurbo Cx) and one tagmentation-based WGBS data set (18,36–38). The coverage profiles from these libraries were very similar to those in our libraries (Figure 3B). As in our SPLAT, Accel-NGS Methyl-Seq and NEBNextUltra libraries, a common feature was a decline in coverage over CpG rich regions and difficult promoters.
Determination of DNA methylation levels
Quality control of methylation calls
Global methylation levels in the CHG and CHH (where H represents either A, T or C but not G) sequence context were low (0.3– 1%), indicating that the bisulphite conversion rates were > 99% in all libraries (Supplementary Data). An alternative method to assess bisulphite conversion rates is to determine the methylation levels in mitochondrial DNA, which is presumed to be unmethylated in human samples (39). We found that the average methylation levels across mitochondrial CpG sites were very low in the SPLAT, Accel-NGS Methyl-Seq and NEBNextUltra libraries as expected. However, the average mitochondrial CpG methylation levels originating from TruSeq DNA Methylation libraries were consistently high (Supplementary Data). The reason for this bias is unknown, but could be due to incomplete bisulphite conversion of the circular mitochondrial DNA. Furthermore, we analyzed individual reads with two or more CpG sites to determine the concordance of the methylation state of each cytosine position in a CpG context in each read (Supplementary Data). The proportion of reads with concordant methylation states was extremely similar between methods, however the proportion of reads available for such an analysis (containing ≥2 CpG sites) varied greatly between the methods, namely only 17–18% of the reads from NebNextUltra libraries, 22–26% of Accel-NGS and SPLAT libraries, and 35–36% of TruSeq DNA Methylation libraries. This result mirrors the coverage bias discussed above.
M-bias plots are frequently used for quality control of bisulphite sequencing data and are useful for detection of methylation biases at the ends of sequence reads (40). In an unbiased sequencing library the methylation levels should be independent of read position, and thus the mean methylation levels plotted against read position should form a horizontal line. Only the SPLAT libraries were unbiased in this parameter (Supplementary Data). The M-bias profiles in the TruSeq DNA Methylation libraries fluctuated, particularly at the beginning of the reads. Thus to avoid biases in methylation levels in the TruSeq DNA methylation libraries we excluded the first six residues of each read from methylation calling. The NEBNextUltra libraries exhibited a sharp decrease in methylation levels at the beginning of the second read, which is characteristic for conventional library preparation protocols where end repair of sheared dsDNA fragments is performed with unmethylated cytosines. Thus, we excluded the first two bases of each read from methylation calling. The M-bias profiles for Accel-NGS Methyl-Seq libraries (after removing the sequence tails) also displayed a small dip in the beginning of the first read and a spike at the end of the second read and accordingly these three bases were excluded from the methylation calling.
Concordance of methylation levels across methods
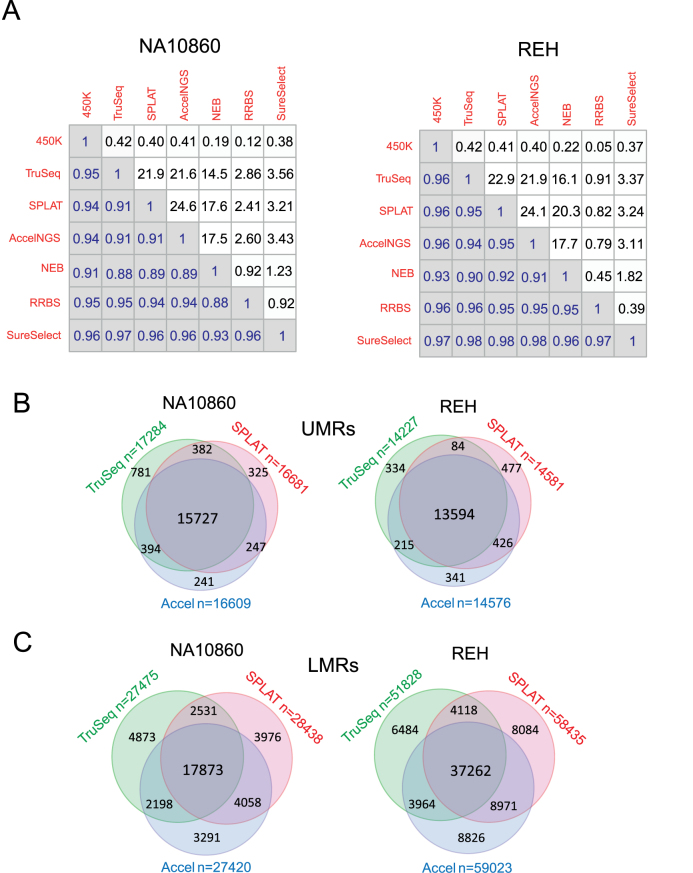
Next we performed pair-wise comparisons of the methylation levels computed across reads at individual CpG sites between methods for both cell types (Figure 5A) First, we measured the correlation between the methylation levels obtained by the WGBS methods and those measured with 450k Bead Arrays. Using a minimum of 5-fold or higher coverage in the WGBS libraries, the concordance of methylation levels between 450k Bead Arrays and all WGBS methods was excellent (Pearson's R = 0.91-0.95 for NA10860 and 0.93–0.96 for REH). Concordance of the methylation levels across the four WGBS library preparation methods was also high (Pearson's R = 0.88–0.91 for NA10860 and 0.90–0.95 for REH), with the exception of the methylation levels originating from NEBNextUltra libraries, which consistently displayed lower correlation coefficients in all pair-wise comparisons. The correlation coefficients for comparison between technical replicates for each WGBS method are given in Supplementary Data. In the same pair-wise manner, we also found a high degree of correlation between the methylation levels determined by WGBS, RRBS, and the SureSelect Methyl-Seq methods (Pearson's R = 0.93–0.97 for NA10860 and 0.95–0.98 for REH) (Supplementary Data). Generally, the correlation coefficients were higher for pair-wise comparisons in the REH sample, presumably due to the fact that a larger proportion of the of the CpG sites in REH cells, compared to the normal B-cell line, were either completely methylated or unmethylated. Differences in methylation levels at individual CpG sites may be due to differences in read coverage at the particular site, and therefore we also assessed the concordance of methylation levels across functionally different genomic regions, which is less sensitive to variance in read coverage. The concordance between regional mean methylation levels across CpG islands and FANTOM5 enhancer regions was high between WGBS methods (Pearson's R = 0.92–0.99), Supplementary Data). The mean methylation levels across CpG islands showed excellent correlations (>0.99) in all comparisons between the ‘post bisulphite’ WGBS methods. Similarly, for these methods the methylation level correlation was also high across enhancer regions (Pearson's R = 0.94–0.97). Again, lower concordance was observed in the comparisons with NEBNextUltra libraries (for enhancer regions; Pearson's R = 0.92–0.96 compared to 0.94–0.97 for comparisons solely between ‘post bisulfite’ methods).
Figure 5.
Concordance of methylation levels across methods. (A) Pairwise comparison of methylation levels at individual CpG sites, Pearson's correlation coefficients are shown to the left of the diagonal. The numbers of CpG sites (in millions) included in each comparison are shown to the right of the diagonal. (B) Venn diagrams showing the numbers and overlaps of un-methylated regions (UMRs) detected in the three ‘post-bisulphite’ library preparation methods. (C) Venn diagrams showing the numbers and overlaps of low methylated regions (LMRs) detected in the three ‘post-bisulphite’ library preparation methods.
In summary, the ‘post bisulphite’ libraries yielded overall higher concordance in methylation calls both between the WGBS libraries and each of the other non-WGBS methods than the traditional WGBS method that we used herein.
Comparison of hypomethylated regions in WGBS libraries
Next we analysed LMRs and UMRs in the different WGBS data sets. In line with the results described above, we detected as many as 25% fewer UMRs and 50% fewer LMRs in NEBNextUltra libraries compared to the other methods (Supplementary Data). Therefore we only assessed the overlap of LMRs and UMRs in the ‘post bisulphite’ WGBS libraries. Between 90% and 96% of the UMRs detected in any given library overlapped with those detected in all of the libraries (Figure 5B), therefore each of the ‘post bisulphite’ methods were accurate in detecting UMRs (methylation level mean 6%, median 4%). Comparable numbers of LMRs were identified by all of the ‘post bisulphite’ methods (27 475–28 438 for NA10860 and 51 828–59 023 for REH, Figure 5C). Similar to what we described in the previous section when comparing SPLAT to the high coverage reference data, the degree of overlap was lower (63–72%) than observed for the UMRs. The greatest number of LMRs overlapped between the SPLAT and Accel-NGS methods, presumptively due to the more even genome-coverage achieved with these methods. However, when inspecting the non-overlapping regions in more detail, despite that the coverage was similar in the regions containing unique and overlapping LMRs (12–16×) across the WGBS methods, we found that methylation levels of unique LMRs (mean methylation 26–29%) was on average 10% higher than in the overlapping LMRs (mean methylation 16–20%).
Genome-wide CpG site coverage across methods
For each sequencing-based method and cell type we measured the total number of CpG sites that were covered by five reads or more (Table 3). The SPLAT and Accel-NGS Methyl-Seq data sets gave the best overall and most even coverage of the CpG sites in the human genome with 88–90% of the total CpG sites covered by at least five reads. The corresponding proportions were 82–83% for TruSeq DNA Methylation and 62–67% and NEBNextUltra. We also compared the results from the WGBS methods to RRBS and SureSelect Methyl-seq libraries where 8–10% and 14% of the total CpG sites were covered by at least five reads, respectively (Table 3, Supplementary Data).
Table 3. CpG site coverage for genome wide bisulphite sequencing methods.
| Library | N analyzed read pairs | N CpG sites covered by zero reads | N CpG sites covered by ≥5 reads | N CpG sites covered by ≥10 reads |
|---|---|---|---|---|
| NA10860 TruSeq | 219 M | 1.04 M | 23.1 M (82%) | 17.5 M (62%) |
| REH TruSeq | 211 M | 0.92 M | 23.2 M (84%) | 17.0 M (62%) |
| NA10860 SPLAT | 218 M | 0.49 M | 25.5 M (90%) | 19.5 M (69%) |
| REH SPLAT | 213 M | 0.55 M | 24.9 M (89%) | 18.2 M (65%) |
| NA10860 Accel-NGS | 218 M | 0.53 M | 24.9 M (89%) | 16.5 M (58%) |
| REH Accel-NGS | 213 M | 0.52 M | 24.5 M (88%) | 16.1 M (58%) |
| NA10860 NEBNextUltra | 219 M | 1.59 M | 17.5 M (62%) | 9.2 M (33%) |
| REH NEBNextUltra | 216 M | 1.01 M | 18.9 M (67%) | 9.3 M (34%) |
| NA10860 RRBS | 20 M | n.d | 2.9 M | 2.2 M |
| REH RRBS | 10 M | n.d | 1.0 M | 0.2 M |
| NA10860 SureSelect | 44 M | n.d | 3.9 M | 3.2 M |
| REH SureSelect | 35 M | n.d | 3.6 M | 2.9 M |
M = million.
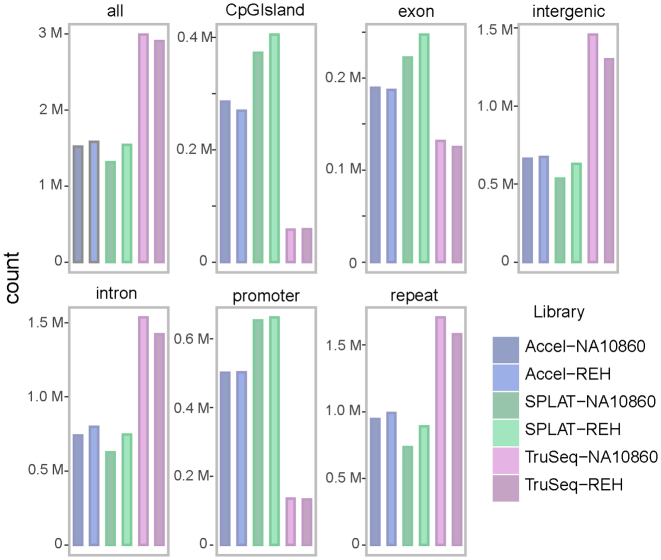
Moreover, we identified CpG sites that were under-represented in each WGBS library by applying a coverage threshold of ≤2× and annotating the regions with poor coverage (Figure 6). Approximately 0.5 million CpG sites that were below this threshold were shared between all the ‘post bisulphite’ conversion libraries and were mainly annotated to intergenic regions, introns and repeats. Overall, the 3 M CpG sites with low coverage in TruSeq DNA Methylation libraries were over-represented in introns, intergenic regions and repetitive elements. The SPLAT and Accel-NGS Methyl-Seq libraries had 1.3–1.5 and 1.5–1.6 M low coverage CpGs, respectively. As expected, in these libraries there were more low coverage sites in CpG islands and promoter regions (TSS ± 200 bp) compared to TruSeq DNA Methylation libraries. The NEBNextUltra libraries had the largest number of low covered CpG sites (4.7–6.0M) and a large proportion of them were annotated to CpG islands and promoter regions, however many sites in intergenic regions and introns were also insufficiently covered (Supplementary Data).
Figure 6.
Annotation of CpG sites with low coverage in the ‘post bisulphite’ WGBS methods. A coverage threshold of ≤2 reads was applied to identify CpG sites that were insufficiently represented by the different WGBS library methods (based on down-sampled data). In TruSeq DNA Methylation libraries, low coverage sites were mostly found in intergenic regions and introns. The SPLAT and Accel-NGS Methyl-Seq data displayed overall lower number of CpG sites with low coverage, although the number of poorly represented sites in CpG islands and promoter regions were higher compared to the TruSeq DNA methylation libraries.
The greatest total number of CpG sites covered was obtained by the SPLAT dataset followed by the Accel-NGS dataset (Table 3). Moreover, these two protocols provided a better overall genome-wide coverage of CpG sites in repetitive elements, intergenic regions and introns, which are the regions that provide the most important advantage of the WGBS approach over the other targeted methods.
DISCUSSION
Whole genome bisulphite sequencing (WGBS) is becoming a popular method to interrogate genome-wide DNA methylation levels in many cell types and species (41,42). However, to date, no systematic review of the different methods has been presented and the choice of which library preparation method to choose remains a difficult task. Herein, we present an assessment of library preparation methods for WGBS. Our aim was to identify any possible method-specific biases, to determine the concordance of methylation levels across WGBS methods and to other commonly used genome-wide DNA methylation methods, and to determine which methods are most cost efficient it terms of amount of data retained after filtering and CpG site coverage. Furthermore, we introduce SPLAT, which is an inexpensive, fast and simple method that shows excellent DNA methylation concordance with the other existing methods analysed herein.
WGBS of human samples is still costly to perform and thus the choice of sample preparation method that will generate the largest amount usable data is important. Points that may be taken into consideration are (i) DNA input amount requirements, (ii) the scientific question of the study; i.e. are some genomic regions/features of particular interest? (iii) cost efficiency, (iv) will the WGBS data also be used to assess genetic variation and/or copy number alterations? An overall summary of the method evaluation can be found in Table 4. The ‘post-bisulphite’ WGBS library preparation methods bring about a significant reduction in the required DNA input and performed better than the traditional method in all of the metrics presented herein, thus providing a more economical alternative for WGBS especially for scarce samples. In this study, the DNA input amount was set to 100 ng in order to obtain high quality methylation profiles for comparison. However, for the SPLAT and Accel-NGS Methyl-Seq methods the amount of DNA input can be substantially decreased considering that for 100 ng of input DNA, as few as four amplification cycles gave sufficient yield and that the PCR duplication rates were very low. The TruSeq DNA Methylation protocol on the other hand required more than twice as many amplification cycles for the same input amount to obtain the same yield, and thus rather high PCR duplication rates limit the flexibility with respect to input quantity.
Table 4. Summary of methods for whole genome bisulphite sequencing library preparation.
| TruSeq DNA methylation | SPLAT | Accel-NGS Methyl-Seq | NEBNextUltra | |
|---|---|---|---|---|
| Input DNA quantity | 50–100 ng | ≤100 ng | ≤100 ng | ∼1000 ng |
| Number of PCR cycles | 9 | 4 (100 ng input) | 4 (100 ng input) | 6 |
| Amount of PCR duplicates | High | Low | Low | Low |
| Genome coverage | GC biased | Relatively uniform | Relatively uniform | AT biased |
| Advantages | Excellent coverage of CpG dense regions | High total CpG site coverage | High total CpG site coverage | _ |
| Methylation highly concordant with other methods | Methylation highly concordant with other methods | Methylation highly concordant with other methods | ||
| Very low library prep cost | ||||
| Disadvantages | High proportion of data removed in pre-processing steps | Coverage in CpG dense regions is lower than the average coverage | Coverage in CpG dense regions is lower than the average coverage | Overall low coverage of CpG sites, particularly in CpG dense regions |
| Sequence tag removal is required |
One important aspect in choosing a WGBS method is bias in the regions of the genome covered. The SPLAT and Accel-NGS Methyl-Seq methods would be the best if data from the largest number of CpG sites with most even coverage over the genome is desired, especially if genetic variation (for example SNPs) will also be assessed from the WGBS data. In both SPLAT and Accel-NGS Methyl-Seq data, we found some degree of coverage decline in CpG islands and in very GC rich promoter regions, which is a common characteristic of many types WGBS data (Supplementary Data). Notably, the library preparation cost for SPLAT (<10 $/sample) is less than one tenth of the cost for any of the commercial WGBS methods, making SPLAT a particularly attractive method also for bisulphite sequencing of smaller genomes such as Arabidopsis and for applications such as ChIP-bisulphite sequencing (ChIP-BS-seq) (43,44) where the cost of library preparation per sample approaches the cost of sequencing. The post bisulphite adaptor tagging (PBAT) method, which is the only other alternative protocol for post bisulphite WGBS library preparation that has been described in a peer-reviewed journal, has the advantage of producing amplification free libraries from low amounts of DNA. However, chimeric reads formed by joining of two distinct genomic regions in PBAT libraries often result in a low mapping efficiency (see online article; https://sequencing.qcfail.com/articles/pbat-libraries-may-generate-chimaeric-read-pairs). In contrast, our results suggest that chimeric reads is an issue that occurs using the original PBAT protocol and not in the methods described here. The mapping procedure used herein does not consider chimeric read pairs as valid alignments. Hence, the high mapping efficiencies for SPLAT as well as the other two post bisulphite methods (>70%) demonstrate that the contribution of chimeric fragments to the mapping efficiency estimation in these libraries is minor.
The TruSeq DNA Methylation method exhibited the best overall coverage of CpG dense regions, such as promoters and CpG islands, however this method suffered from lower coverage of total CpG sites and more data discarded than the other methods. Thus if CpG dense regions are of particular interest, this would be the WGBS method of choice. Lastly, the conventional NEBNextUltra libraries required more input DNA, displayed poor coverage of CpG islands and promoter regions, and generally interrogated fewer CpG sites compared to the other methods and thus this method did not outperform any of the ‘post-bisulphite’ methods.
The exact cause and origin of the method specific biases are still unclear and multiple mechanisms may be involved. Differences in the efficiency of PCR amplification due to the sequence context and composition of the PCR buffer are recognized sources of coverage bias: DNA polymerases can differ in their ability to efficiently amplify regions of extreme base composition. Hence the AT skewed coverage of the NEBNextUltra libraries in this study might be attributed to the use of the KAPA HiFi Uracil+ polymerase. On the contrary, the same polymerase was used to amplify SPLAT libraries that did not display a similar type of AT bias. Apart from PCR amplification, other causes of coverage bias might be related to specific steps in the different protocols. For instance, non-random bisulphite-induced fragmentation in CpG dense regions might account for increased coverage of GC rich regions in the TruSeq DNA methylation libraries, by increasing the fraction of such regions available for efficient adaptor tagging. By contrast, biased fragmentation of GC rich regions in the pre-sheared DNA used for SPLAT and Accel-NGS protocols might lead to very short fragments that are lost in the library preparation. Moreover, secondary structure of single stranded GC rich regions might negatively affect ligation or 3΄-end tagging and contribute to the lower representation of such regions in SPLAT and Accel-NGS Methyl-Seq protocols.
The performance of WGBS methods may vary between labs and until a cross-laboratory comparison can be performed any recommendations based on data produced by one lab only should be treated with caution. However, the characteristics of the different library preparation methods were highly reproducible across the two different cell types analysed. We choose to benchmark the methods using the large human genome, an approach that is highly relevant for many researchers, although extensive benchmarking of a large number of samples and methods may be prohibited by the sequencing costs. However, by complementing our benchmarking with publicly available human WGBS data sets from several external laboratories, we observed the same type of coverage biases in our data and in the public data (Supplementary Data).
In summary, all the ‘post bisulphite’ library preparation methods performed better than the conventional library preparation method. Concordance of methylation levels across the different methods were high, both at the level of individual CpG sites and across regions. Since no method is completely free from sequence bias, the method of choice depends on the aim of the study and the scientific questions asked. Our method, SPLAT, provides a straightforward and highly cost efficient approach for WGBS that compares favourably to commercially available methods.
ACCESSION NUMBER
The sequencing data from this study are publicly available and have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) with accession number: GSE89213.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Helena Fällmar for preparing the SureSelect Methyl-Seq libraries. DNA methylation array analysis and sequencing was performed at the SNP&SEQ Technology Platform in Uppsala, which is part of the National Genomics Infrastructure (NGI) funded by the Swedish Council for Research Infrastructures and Science for Life Laboratory. Computational analysis was performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) through the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Foundation for Strategic Research [RBc08-008]; Swedish Research Council for Science and Technology [90559401]; Joint Swedish Research Councils [259-2012-23]. Funding for open access charge: Swedish Research Council for Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 2. Portela A., Esteller M.. Epigenetic modifications and human disease. Nat. Biotechnol. 2010; 28:1057–1068. [DOI] [PubMed] [Google Scholar]
- 3. Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L.. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lister R., O'Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R.. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008; 133:523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E.. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008; 452:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L. et al. . High density DNA methylation array with single CpG site resolution. Genomics. 2011; 98:288–295. [DOI] [PubMed] [Google Scholar]
- 7. Moran S., Arribas C., Esteller M.. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2015; 8:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M.A., Bibikova M., Esteller M.. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011; 6:692–702. [DOI] [PubMed] [Google Scholar]
- 9. Gu H., Smith Z.D., Bock C., Boyle P., Gnirke A., Meissner A.. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature Protocols. 2011; 6:468–481. [DOI] [PubMed] [Google Scholar]
- 10. Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B. et al. . Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008; 454:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith Z.D., Gu H., Bock C., Gnirke A., Meissner A.. High-throughput bisulfite sequencing in mammalian genomes. Methods (San Diego, Calif.). 2009; 48:226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boyle P., Clement K., Gu H., Smith Z.D., Ziller M., Fostel J.L., Holmes L., Meldrim J., Kelley F., Gnirke A. et al. . Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012; 13:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng J., Shoemaker R., Xie B., Gore A., LeProust E.M., Antosiewicz-Bourget J., Egli D., Maherali N., Park I.H., Yu J. et al. . Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009; 27:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diep D., Plongthongkum N., Gore A., Fung H.L., Shoemaker R., Zhang K.. Library-free methylation sequencing with bisulfite padlock probes. Nat. Methods. 2012; 9:270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ivanov M., Kals M., Kacevska M., Metspalu A., Ingelman-Sundberg M., Milani L.. In-solution hybrid capture of bisulfite-converted DNA for targeted bisulfite sequencing of 174 ADME genes. Nucleic Acids Res. 2013; 41:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee E.J., Pei L., Srivastava G., Joshi T., Kushwaha G., Choi J.H., Robertson K.D., Wang X., Colbourne J.K., Zhang L. et al. . Targeted bisulfite sequencing by solution hybrid selection and massively parallel sequencing. Nucleic Acids Res. 2011; 39:e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Allum F., Shao X., Guenard F., Simon M.M., Busche S., Caron M., Lambourne J., Lessard J., Tandre K., Hedman A.K. et al. . Characterization of functional methylomes by next-generation capture sequencing identifies novel disease-associated variants. Nat. Commun. 2015; 6:7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adey A., Shendure J.. Ultra-low-input, tagmentation-based whole-genome bisulfite sequencing. Genome Res. 2012; 22:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miura F., Enomoto Y., Dairiki R., Ito T.. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012; 40:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miura F., Ito T.. Highly sensitive targeted methylome sequencing by post-bisulfite adaptor tagging. DNA Res. 2015; 22:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smallwood S.A., Lee H.J., Angermueller C., Krueger F., Saadeh H., Peat J., Andrews S.R., Stegle O., Reik W., Kelsey G.. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods. 2014; 11:817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosenfeld C., Goutner A., Venuat A.M., Choquet C., Pico J.L., Dore J.F., Liabeuf A., Durandy A., Desgrange C., De The G.. An effect human leukaemic cell line: Reh. Eur. J. Cancer. 1977; 13:377–379. [DOI] [PubMed] [Google Scholar]
- 23. Krueger F., Andrews S.R.. Bioinformatics (Oxford, England). 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia-Alcalde F., Okonechnikov K., Carbonell J., Cruz L.M., Gotz S., Tarazona S., Dopazo J., Meyer T.F., Conesa A.. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics (Oxford, England). 2012; 28:2678–2679. [DOI] [PubMed] [Google Scholar]
- 25. Quinlan A.R. BEDTools: the Swiss-Army Tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014; 47, doi:10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burger L., Gaidatzis D., Schubeler D., Stadler M.B.. Identification of active regulatory regions from DNA methylation data. Nucleic Acids Res. 2013; 41:e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nordlund J., Backlin C.L., Wahlberg P., Busche S., Berglund E.C., Eloranta M.L., Flaegstad T., Forestier E., Frost B.M., Harila-Saari A. et al. . Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013; 14:r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore M.J., Query C.C.. Joining of RNAs by splinted ligation. Methods Enzymol. 2000; 317:109–123. [DOI] [PubMed] [Google Scholar]
- 30. Court F., Tayama C., Romanelli V., Martin-Trujillo A., Iglesias-Platas I., Okamura K., Sugahara N., Simon C., Moore H., Harness J.V. et al. . Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014; 24:554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uribe-Lewis S., Woodfine K., Stojic L., Murrell A.. Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert Rev. Mol. Med. 2011; 13:e2. [DOI] [PubMed] [Google Scholar]
- 32. Benjamini Y., Speed T.P.. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012; 40:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aird D., Ross M.G., Chen W.S., Danielsson M., Fennell T., Russ C., Jaffe D.B., Nusbaum C., Gnirke A.. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011; 12:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. . An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ross M.G., Russ C., Costello M., Hollinger A., Lennon N.J., Hegarty R., Nusbaum C., Jaffe D.B.. Characterizing and measuring bias in sequence data. Genome Biol. 2013; 14:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hansen K.D., Sabunciyan S., Langmead B., Nagy N., Curley R., Klein G., Klein E., Salamon D., Feinberg A.P.. Large-scale hypomethylated blocks associated with Epstein-Barr virus-induced B-cell immortalization. Genome Res. 2014; 24:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O'Malley R., Castanon R., Klugman S. et al. . Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011; 471:68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blattler A., Yao L., Witt H., Guo Y., Nicolet C.M., Berman B.P., Farnham P.J.. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014; 15:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong E.E., Okitsu C.Y., Smith A.D., Hsieh C.L.. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol. Cell. Biol. 2013; 33:2683–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hansen K.D., Langmead B., Irizarry R.A.. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012; 13:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Z., Cunningham J., Slager S., Kocher J.P.. Base resolution methylome profiling: considerations in platform selection, data preprocessing and analysis. Epigenomics. 2015; 7:813–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Plongthongkum N., Diep D.H., Zhang K.. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat. Rev. Genet. 2014; 15:647–661. [DOI] [PubMed] [Google Scholar]
- 43. Gao F., Ji G., Gao Z., Han X., Ye M., Yuan Z., Luo H., Huang X., Natarajan K., Wang J. et al. . Direct ChIP-bisulfite sequencing reveals a role of H3K27me3 mediating aberrant hypermethylation of promoter CpG islands in cancer cells. Genomics. 2014; 103:204–210. [DOI] [PubMed] [Google Scholar]
- 44. Brinkman A.B., Gu H., Bartels S.J., Zhang Y., Matarese F., Simmer F., Marks H., Bock C., Gnirke A., Meissner A. et al. . Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012; 22:1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.