Abstract
Ehrlichiae are tick-transmitted, gram-negative, obligately intracellular bacteria that live and replicate in cytoplasmic vacuoles, but little is known about iron acquisition mechanisms necessary for their survival. In this study, a genus-conserved immunoreactive ferric ion-binding protein (Fbp) of Ehrlichia canis was identified and its iron-binding capability was investigated. E. canis Fbp was homologous to a family of periplasmic Fbp's involved in iron acquisition and transport in gram-negative bacteria. E. canis Fbp had a molecular mass (38 kDa) consistent with those of Fbp's in other bacteria and exhibited substantial immunoreactivity in its native conformation. The predicted three-dimensional structure of E. canis Fbp demonstrated conservation of important Fbp family structural motifs: two domains linked with a polypeptide “hinge” region. Under iron-binding conditions, the recombinant Fbp exhibited an intense red color and an absorbance spectrum indicative of iron binding, and it bound Fe(III) but not Fe(II). Fbp was observed primarily in the cytoplasm of the reticulate forms of E. canis and Ehrlichia chaffeensis but was notably found on extracellular morula fibers in morulae containing dense-cored organisms. Although expression of Fbp is regulated through an operon of three functionally linked genes in other gram-negative bacteria, the absence of an intact fbp operon in Ehrlichia spp. suggests that genes involved in ehrlichial iron acquisition have been subject to reductive evolution.
Iron is a necessity for the survival of nearly all prokaryotes and eukaryotes. For the obligately intracellular bacterium Ehrlichia chaffeensis, the importance of iron for intracellular survival has been demonstrated by inhibition of ehrlichial proliferation in the presence of the intracytoplasmic iron chelator deferoxamine (8). Early endosomes containing the bacteria (morulae) upregulate and accumulate the mammalian transferrin receptor (8), suggesting that the organism has developed specific strategies for iron acquisition. The cytoplasmic vacuole where the ehrlichiae reside is acidic, which may promote the release of free iron into the compartment (8). An E. chaffeensis protein with homology to known ferric ion-binding proteins has been reported previously (37), but the mechanisms of iron acquisition by Ehrlichia spp. are unknown, and functional iron-binding properties of ehrlichial Fbp's have not been experimentally demonstrated.
Iron is involved in many key metabolic functions of the cell, but insoluble Fe(OH)3 forms at a physiological pH and thus must be stored and transferred via iron-binding proteins (5, 25). In eukaryotic systems, extracellular iron-binding proteins include transferrin and lactoferrin, while ferritin binds and sequesters intracellular iron. Protein-bound iron also inhibits the interaction of Fe(II) with O2, preventing the formation of free radicals that are damaging to cells (5, 25). The availability of iron has been demonstrated to increase the virulence of many diverse pathogenic bacteria (29). Limiting the availability of free iron is a mechanism to naturally suppress growth of bacteria. However, under the selective pressure of the limited iron available in the host, pathogenic bacteria have evolved specific iron acquisition mechanisms including iron-binding molecules and proteins.
Bacteria commonly utilize siderophores as means of iron mobilization. These molecules are nonproteinaceous iron chelators that are expressed, secreted, and bound by surface receptors that enable the transport of free iron from the environment (5, 25). Another means of iron uptake involves iron acquisition from the host iron-binding proteins transferrin and lactoferrin (5, 25). This mechanism, used by many gram-negative pathogens, involves a conserved system by which iron is competed away from transferrin or lactoferrin at the outer membrane of the bacterium and is then shuttled across the membrane and into the periplasm (5). Mobilization of iron into the cytoplasm involves three proteins that belong to the ATP-binding cassette (ABC) transporter family: the ferric ion-binding protein (Fbp), a cytoplasmic permease, and an ATP-binding protein (1, 2, 5, 14, 25), all encoded by an operon system that is shared among many diverse bacterial species including Neisseria gonorrhoeae, Haemophilus influenzae, Mannheimia (Pasteurella) haemolytica, Serratia marcescens, Salmonella enterica serovar Typhimurium, and Yersinia pestis (1, 2, 6, 9, 10, 21, 22). The operon system of N. gonorrhoeae has been demonstrated to be under the control of the iron regulatory element Fur (ferric uptake regulator) (15).
This report represents the first functional characterization of a ferric ion-binding protein (Fbp) ortholog in any obligately intracellular bacterium. We have identified an immunoreactive 38-kDa Fbp from Ehrlichia canis that shares homology with a known family of periplasmic Fbp's involved in iron acquisition extracellular bacteria. Structural modeling of the E. canis Fbp identified an iron-binding motif conserved within the Fbp family, and we demonstrated the ability of E. canis Fbp to bind free iron. E. canis Fbp was observed primarily in the cytoplasm of bacteria with reticulate morphology, while surface and extracellular distribution on morula fibers and the membranes of morulae was observed on the dense-cored ehrlichial form. Notably, E. canis fbp was not organized with other prototypical iron acquisition genes in an operon system.
MATERIALS AND METHODS
Ehrlichiae and purification.
E. canis (Jake isolate) was propagated as previously described (24). Ehrlichiae were purified by size exclusion over a Sephacryl S-1000 column (Amersham Biosciences, Piscataway, N.J.) as previously described (30). The initial fraction containing bacteria was frozen and utilized as an antigen source.
Identification of E. canis Fbp.
Fbp was identified in an E. canis HpaII genomic library screened with an anti-E. canis antiserum as previously described (24).
DNA sequencing.
Inserts were sequenced with an ABI Prism 377XL DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.) at the University of Texas Medical Branch Protein Chemistry Core Laboratory by using the forward and reverse primers (Uni1) provided with the kit (Invitrogen, Carlsbad, Calif.).
PCR amplification of the Ehrlichia fbp gene.
Regions of the E. canis fbp gene selected for PCR amplification were chosen by Primer Select software (DNAstar, Madison, Wis.). Primers (400 nM) corresponding to nucleotides 82 to 101 (5′-CAA GAT CAG AAA CAA GAA GT, forward) within the open reading frame (ORF) and a region 198 bp downstream of the ORF (5′-TAA AAT AAA ATA GAA AGA AT, reverse) were used to amplify the fbp gene. Template E. canis DNA (from the Jake isolate) was amplified by using the PCR Master Mix (F. Hoffmann-La Roche Ltd., Basel, Switzerland) with a thermal cycling profile of 95°C for 4 min; 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; a 72°C extension for 7 min; and a 4°C hold. PCR products were analyzed in 1% agarose gels. Degenerate primers were designed (forward, 5′-ATG ATG AGA TTR MTT GCT TG; reverse, 5′-CTA TCT CCA MCC ACA CTC AT [R stands for A or G; M stands for A or C]) for conserved regions in the coding regions of fbp from E. canis and E. chaffeensis corresponding to bp 1 through 20 (forward) and bp 1025 through 1044 (reverse). These primers were used to amplify the fbp gene in Ehrlichia muris to compare sequence homology.
Signal sequence determination.
E. canis Fbp was tested for the presence of a signal sequence with the computational algorithm SignalP trained on gram-negative bacteria (www.cbs.dtu.dk/services/SignalP/) (26).
Cloning and expression of recombinant E. canis Fbp.
The amplified PCR product was cloned directly into a universal TOPO TA Echo vector (pUni; Invitrogen). This vector was recombined with the acceptor vector pRSET, designed to produce proteins with a native C terminus and an N-terminal polyhistidine region for purification. Escherichia coli TOP10 (Invitrogen) was transformed with the plasmid containing the cloned fbp gene; positive transformants were screened by PCR for the presence of the insert and the correct orientation and were sequenced to confirm the correct reading frame of the fbp gene. The recombinant Fbp was then expressed in E. coli BL21(DE3)/pLysS (Invitrogen) after induction with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Bacteria were harvested by centrifugation at 5,000 × g for 20 min and resuspended in phosphate-buffered saline (PBS). Recombinant Fbp was purified under native conditions by sonicating the pelleted bacteria resuspended in PBS (Braun-Sonic 2000) twice at 40 W for 30 s on ice and pelleting the insoluble material by centrifugation at 10,000 × g for 20 min. The clarified supernatant was loaded onto an equilibrated nickel-nitrilotriacetic acid (Ni-NTA) agarose column (QIAGEN, Valencia, Calif.). The bound recombinant protein was washed with 5 column volumes of increasing concentrations (0.8, 8, and 40 mM) of imidazole (Sigma-Aldrich, St. Louis, Mo.) and eluted with PBS plus 80 mM imidazole. Purified recombinant protein was dialyzed against PBS by using Microcon centrifugal concentrators with a 30-kDa cutoff (Millipore, Billerica, Mass.).
Gel electrophoresis and immunoblotting.
Purified E. canis antigens were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotted as previously described (23), except that primary antibodies were diluted (1:500). Dot blot analyses were conducted with either native protein, protein plus 0.1% (wt/vol) SDS, protein with 50 mM dithiothreitol (Invitrogen), or a combination of SDS plus the reducing agent. Western blot analysis was performed as described above.
Rabbit immunization.
A New Zealand White rabbit (Myrtle's Rabbitry Inc., Thompson Station, Tenn.) was immunized with the recombinant E. canis Fbp protein. Recombinant protein (100 μg) in 0.5 ml was mixed with an equal volume of Freund's complete adjuvant (Sigma) for the first injection and with Freund's incomplete adjuvant for subsequent injections. The rabbit was immunized subcutaneously three times, with a 2-week interval between immunizations.
Analysis of genomic organization.
Genome sequences of E. canis (http://www.jgi.doe.gov/) and E. chaffeensis (http://www.tigr.org/) were used to search for ehrlichial genes involved in iron acquisition. Several kilobases of genome sequence flanking fbp were analyzed for neighboring ORFs by using MapDraw software (DNAstar). The ORFs were identified with the basic local alignment search tool (BLAST) for related proteins. The fbp regions of Wolbachia pipientis (NCBI accession number NC_002978), Neorickettisa sennetsu (TIGR_222891, contig 36), and Anaplasma marginale (NCBI accession number AF305077) were located with BLAST and analyzed with MapDraw. The fbpABC sequences from N. gonorrhoeae and the yfeABCD sequences from Y. pestis were from NCBI accession numbers U33937 and U50597, respectively. Fur box identification was based on similarity to the consensus Fur recognition sequence N (ATA/TAT) (16).
RT-PCR.
Reverse transcriptase PCR (RT-PCR) was conducted on RNAs extracted from E. canis-infected DH82 cells and uninfected DH82 cells by using the RNEasy kit (QIAGEN). Contaminating DNA in the RNA preparation was removed with DNase treatment (DNA-free; Ambion, Austin, Tex.). Reactions were conducted with the Superscript III one-step RT-PCR system with Platinum DNA polymerase (Invitrogen) in a total volume of 50 μl. The degenerate primers described above (400 nM) were used to amplify fbp; membrane permease primers were 5′-AAT GGG CGT ATA GTA GGA GGA T (forward) and 5′-TAA CGC AAT AAT AGA ACC AAG AAC (reverse), corresponding to bp 25 to 46 and 514 to 537, respectively; primers used to amplify the intergenic region were 5′-ATG AGT GTG GKT GGA GAT AG (forward) (bp 1025 to 1044 of fbp) and 5′-ACA ACA CAT AAT CAT ACC ATA CTA (reverse) (bp 104 to 127 of membrane permease). Template RNA (125 ng) was added to each reaction mixture, and genomic E. canis DNA (350 ng) was added to the control reaction mixture with RT-PCR reagent. Reactions were carried out as follows: 30 min at 50°C for reverse transcription; 94°C for 2 min; a thermal cycling profile of 10 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 1 min; 25 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 1 min plus 5 s for each additional cycle; and finally a 72°C extension for 7 min and a 4°C hold. PCR products were analyzed in 1% agarose gels. Control reactions without RT were carried out with Platinum PCR Supermix (Invitrogen), which had been demonstrated to amplify products in the presence of genomic DNA (data not shown).
Structural modeling.
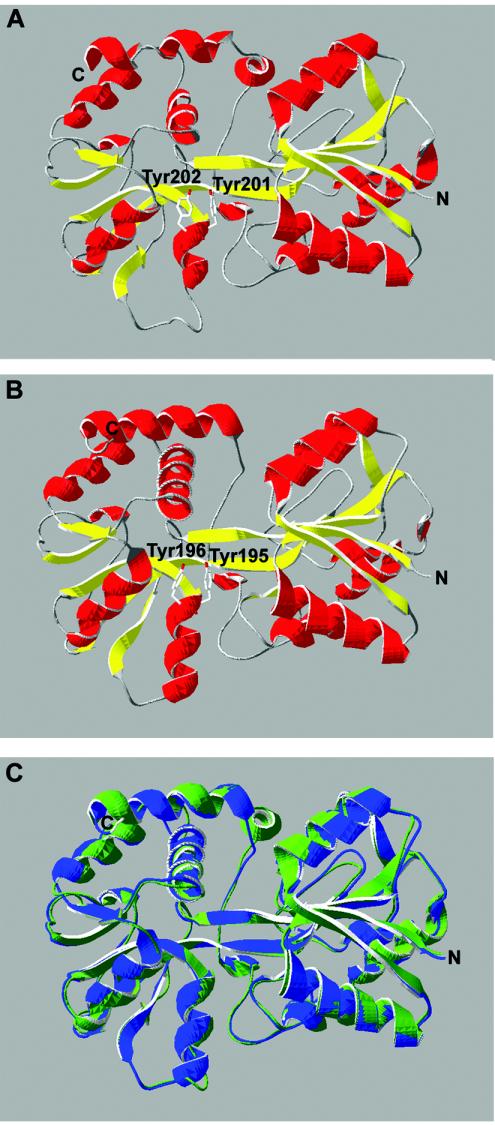
Modeling of the E. canis Fbp was conducted by using SWISS-MODEL (http://www.expasy.org/swissmod/SWISS-MODEL.html). E. canis Fbp was searched against known structures and modeled against the Fbp's from N. gonorrhoeae (PDB 1O7T and 1D9Y), H. influenzae (PDB 1MRP and 1D9V), and M. haemolytica (PDB 1Q35A). The results were visualized with Deep View (http://us.expasy.org/spdbv/). The N. gonorrhoeae Fbp structure from PDB 1O7T was used to generate the ribbon diagram in Fig. 3.
FIG. 3.
Three-dimensional structural modeling of E. canis Fbp using SWISS-MODEL. The putative structure of E. canis Fbp (A) is compared to the crystal structure of N. gonorrhoeae Fbp (B). Colors correspond to secondary structure: red, α-helix; yellow, β-pleated sheet. (C) Structural overlay of E. canis Fbp (green) with the crystal structure of N. gonorrhoeae Fbp (blue).
Iron binding determination.
Following expression of recombinant E. canis Fbp, the E. coli bacterial pellet was resuspended in a buffer containing 50 mM morpholineethanesulfonic acid and 200 mM KCl (pH 6.8), and soluble extracts were purified over a Ni-NTA column as described above. The column was washed with 1× PBS, and the matrix-bound protein turned red, indicative of iron binding. The protein was eluted from the column, and the absorbance of the purified E. canis Fbp was evaluated by a spectral scan (240 to 700 nm) with a Perkin-Elmer (Wellesley, Mass.) MBA2000 spectrophotometer to determine the absorbance wavelength indicative of iron binding. To compete off iron binding, the recombinant protein was incubated in an excess of sodium citrate or NTA to produce apo-Fbp (iron free). In addition, purified recombinant Fbp was tested for iron binding with a Ferene S stain (13). The stain consisted of 0.75 mM Ferene S, 2% glacial acetic acid, and 15 mM thioglycolic acid. The recombinant protein was combined with the Ferene S stain, dot blotted onto a nitrocellulose membrane, and observed for a blue color.
Immunoelectron microscopy.
E. canis- and E. chaffeensis-infected cells were fixed in a mixture of 2.5% formaldehyde and 0.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), postfixed in 1% OsO4 in 0.1 M cacodylate buffer, stained en bloc with 1% uranyl acetate in 0.1 M maleate buffer, and then dehydrated in ethanol and embedded in LR White (SPI Supplies, West Chester, Pa.). Ultrathin sections were cut by using a Reichert-Leica Ultracut S ultramicrotome and were placed on Formvar- and carbon-coated nickel grids. Antigen grids were treated in blocking buffer with 0.1% bovine serum albumin and 0.01 M glycerin in Tris-buffered saline for 15 min at room temperature; then they were incubated with a rabbit anti-E. canis Fbp polyclonal antibody (diluted 1:1,000 in diluting buffer, consisting of 0.05 M Tris-buffered saline with 1% bovine serum albumin) for 1 h at room temperature and then overnight at 4°C. The grids were washed in blocking buffer and incubated for 1 h at room temperature with goat anti-rabbit immunoglobulin G (heavy plus light chains) labeled with 10-nm-diameter colloidal gold particles (AutoProbe EM goat anti-rabbit IgG G15, RPN422; Amersham Life Science, Piscataway, N.J.) diluted 1:20 in diluting buffer.
Nucleotide sequence accession numbers.
The fbp sequences of E. canis and E. muris have been deposited in GenBank and assigned accession numbers AY520460 and AY520461, respectively. E. chaffeensis fbp is available in GenBank under accession number AF117273.
RESULTS
Discovery of an immunoreactive E. canis Fbp.
Expression clones from an HpaII E. canis genomic library were screened as previously described (24). A positive clone with a 3-kb insert contained a complete 1,044-bp ORF encoding a predicted protein with a molecular mass of 38.8 kDa and with homology to periplasmic ferric ion-binding proteins (Fbp) characterized in several gram-negative organisms. An N-terminal signal sequence of 27 amino acids on the E. canis Fbp was identified by the SignalP prediction server (26). The molecular mass of the mature protein is predicted to be 35.8 kDa. This 3-kb clone also contained a partial ORF upstream of fbp with homology to the E. chaffeensis surface protein p106 (37).
Identification of native E. canis Fbp protein.
Rabbit serum raised against the recombinant Fbp protein was used to identify the corresponding native protein in E. canis whole-cell lysates by Western blotting. The antiserum recognized a native E. canis protein with a molecular mass of 38 kDa, corresponding to that predicted by the amino acid sequence (Fig. 1). The protein recognized by the anti-Fbp antiserum identified an immunoreactive ehrlichial protein weakly recognized by antibodies in serum from an E. canis-infected dog (Fig. 1).
FIG. 1.
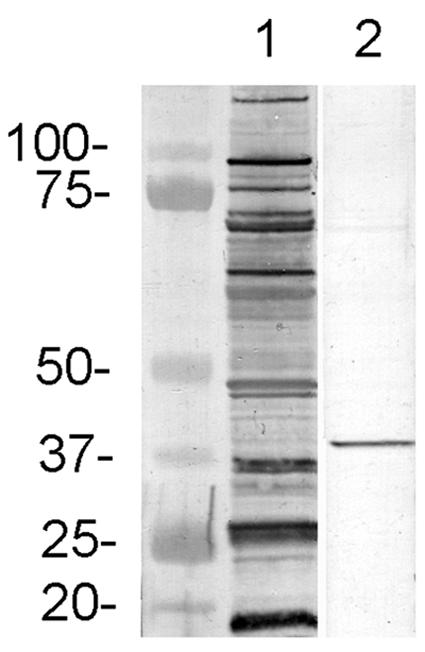
Western immunoblot of E. canis whole-cell lysates probed with serum from an E. canis-infected dog (lane 1) and a monospecific rabbit antiserum against E. canis recombinant Fbp (lane 2).
Immunoreactivity of E. canis Fbp.
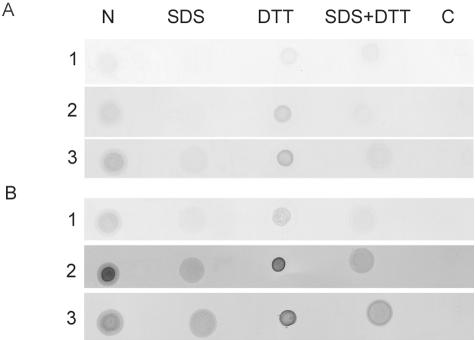
The expressed recombinant Fbp fusion protein exhibited a molecular mass of approximately 40 kDa and reacted with anti-E. canis antisera from several experimentally infected dogs (data not shown). The reactivity of sera from E. canis-infected dogs against denatured Fbp epitopes was weak. To determine if a conformational epitope contributed to the immunoreactivity that led to the identification of Fbp, recombinant Fbp was reacted with sera by dot blotting under native conditions (Fig. 2). The recombinant Fbp reacted with E. canis antisera by dot blotting both under native conditions and in the presence of reducing agents without SDS (Fig. 2). Fbp incubated in the presence of SDS exhibited reduced immunoreactivity to anti-E. canis antibodies in sera collected from dogs on day 28 (Fig. 2A) and day 42 (Fig. 2B) postinfection. Denatured (SDS-exposed) Fbp had higher reactivity with the day-42 serum than with the day-28 serum but was still substantially weaker than native Fbp, indicating that the immunoreactivity of Fbp is dependent on a conformational epitope that is sensitive to SDS and is not affected by reducing agents. Preimmune serum from each of the experimentally infected dogs tested negative for Fbp binding (data not shown). In addition, both anti-E. canis and anti-E. chaffeensis Fbp antisera exhibited cross-reactivity with the orthologous Fbp of the other species (data not shown).
FIG. 2.
Protein dot blot with recombinant Fbp from E. canis incubated under native conditions (N), under denaturing conditions with 0.1% SDS, under reducing conditions with 50 mM dithiothreitol (DTT), or in the presence of both SDS and DTT. Serum samples from E. canis-infected dogs (rows 1 to 3) were collected on day 28 (A) and day 42 (B) postinoculation. A recombinant protein expression control (C) with chloramphenicol acetyltransferase expressed by the same expression vector was used as a negative control.
Fbp homology and modeling of the Ehrlichia Fbp structure.
BLAST analysis of the protein sequence of E. canis Fbp determined that it shares homology with a family of known ferric ion-binding proteins. This family includes bacterial Fbp's from N. gonorrhoeae and H. influenzae, both of which share ∼28% homology with E. canis Fbp, and an Fbp from M. haemolytica, which shares 40% homology. The amino acid sequence of the E. canis Fbp included two adjacent tyrosines near position 200 of the mature protein which are key components of the iron-binding potential of the Fbp family (12, 34). The amino acid sequence of E. canis Fbp also maintains a basic amino acid at Lys14 that aligns with the important His9 of Fbp's from N. gonorrhoeae and H. influenzae (12). The crystal structure of M. haemolytica Fbp predicted an octahedral means of iron coordination involving eight amino acids (34), six of which (the dityrosyl motif at positions 201 and 202, another key tyrosine at position 145, and arginines at positions 13, 105, and 139) are conserved with E. canis Fbp (34). An orthologous gene from E. chaffeensis has been previously described as a putative ferric ion-binding protein but was not immunoreactive, and its iron-binding capabilities were not determined (37). The E. muris fbp gene (reported in this study) and E. chaffeensis fbp exhibited 87% homology with E. canis fbp. The crystal structures of the Fbp's from N. gonorrhoeae, H. influenzae, and M. haemolytica are known (11, 12, 33, 34), which allowed modeling of the primary sequence of E. canis Fbp against these crystal structures by using SWISS-MODEL. The model predicts that the E. canis Fbp exhibits the characteristic two-domain structure connected by a hinge region of antiparallel β-strands, as seen in the Fbp's, similar to the lobes of mammalian transferrin (12, 27) (Fig. 3). As with other bacterial Fbp's, the two domains are made up of a central β-sheet surrounded by multiple α-helices (Fig. 3).
Genomic organization of the E. canis iron acquisition proteins.
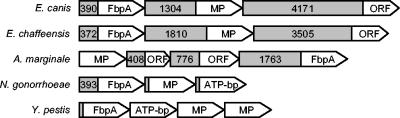
The known Fbp family members are a part of a 4-kb iron-regulated operon which includes the functionally related membrane permease and an ATP-binding protein. The members of these iron-transporting operons are separated by small intergenic sequences (56 bp between fbpA and fbpB and 21 bp between fbpB and fbpC in the N. gonorrhoeae system [1]) or by no intergenic regions (in the Y. pestis yfeABCD system [10]). Examination of the neighboring genes of fbp from the available genome of E. canis identified a putative membrane permease downstream from fbp but separated by 1.304 kb (Fig. 4). A similar study of the E. chaffeensis fbp region placed the putative membrane permease 1.81 kb downstream of fbp (Fig. 4). Interestingly, the genome of the related rickettsial organism A. marginale positions this putative membrane permease 5.5 kb upstream of the fbp gene and has two ORFs that are not homologous to other iron acquisition proteins (Fig. 4). Both genomes contain functionally unrelated ORFs on the negative-strand DNA located in the intergenic space between fbp and the putative membrane permease (data not shown). The search for fbp and neighboring genes in W. pipientis and Neorickettsia sennetsu found the fbp ortholog but no genes encoding the membrane permease or an ATP-binding protein within 5 kb of fbp. Downstream (>4 kb) from the putative ehrlichial membrane permease, there is no gene encoding an ATP-binding protein, and the closest major ORF is ∼7 kb downstream from fbp. The operon system of N. gonorrhoeae has been demonstrated to be under the control of the iron-regulatory element Fur (ferric uptake regulator) (15), and putative Fur-binding sites have been found in other Fbp systems, such as the Y. pestis YfeABCD system (10). Analysis of the sequence upstream of E. canis fbp identified a putative Fur-binding site at position −136 from the start, with a sequence of TATTATTTAATGTAATTATG that contains two copies of the E. coli Fur recognition consensus (ATA/TAT) (16). Another putative Fur box containing one copy of the consensus, with the sequence AATTATTTTGGAAAAATAAG, is located at position −85 from the beginning of the putative membrane permease gene.
FIG. 4.
Genomic organization of E. canis and E. chaffeensis iron acquisition genes compared to the operon systems of N. gonorrhoeae, Y. pestis, and the related rickettsial organism A. marginale. Abbreviations: MP, membrane permease; ATP-bp, ATP-binding protein.
Loss of fbp operon regulation in E. canis.
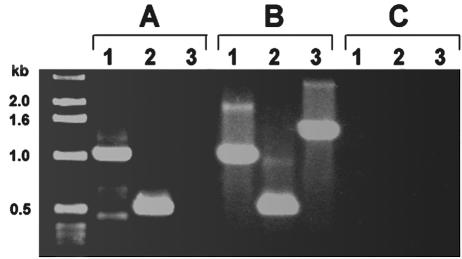
With the loss of conserved genomic organization with the associated membrane permease and ATP-binding protein genes, we hypothesized that the operon regulation had been lost in ehrlichiae. To test this hypothesis, RT-PCR was performed with primers for the 1-kb fbp gene, with primers for a 500-bp region of the downstream membrane permease, and with primers designed to span ∼1.4 kb from the 3′ end of fbp over the intergenic region and 127 bp into the membrane permease gene. If transcription was occurring in a polycistronic fashion, then all three transcripts would be detected by RT-PCR. Transcripts of fbp and membrane permease could be detected, but not a polycistronic transcript containing both genes (Fig. 5A). These transcripts were specific for E. canis-infected DH82 cells and were not seen with uninfected host DH82 cell RNA (data not shown). Controls with genomic DNA could amplify the product under the conditions used (Fig. 5B).
FIG. 5.
RT-PCR of E. canis fbp (lanes 1), E. canis membrane permease (lanes 2), and the intergenic region spanning the area from the 3′ end of fbp to the 5′ start of membrane permease (lanes 3) with either RNA from E. canis-infected DH82 cells (A), E. canis genomic DNA (B), or a control without RT (C).
Iron binding by E. canis Fbp.
Ferric ion-binding proteins exhibit a spectral peak in the 400- to 500-nm range when bound to iron (22). Following recombinant-protein purification, the Fbp exhibited a visible, intense-red color. The absorbance of the protein was determined by a spectrophotometric spectral scan, and an absorbance peak at 410 nm was observed with Fbp (Fig. 6A, top). Addition of the iron chelator sodium citrate competed away the iron, eliminating the absorbance peak (Fig. 6A, bottom). In addition, recombinant iron-bound holo-Fbp turned blue with the iron-specific Ferene S stain (13), indicative of iron binding (Fig. 6B). The iron-free apo form of Fbp did not stain positive with Ferene S (Fig. 6B). Ferene S, which is specific for iron(II), did not turn holo-Fbp blue in the absence of the reducing agent thioglycolic acid, evidence that the bound iron is the iron(III) form and not iron(II) (Fig. 6B).
FIG. 6.
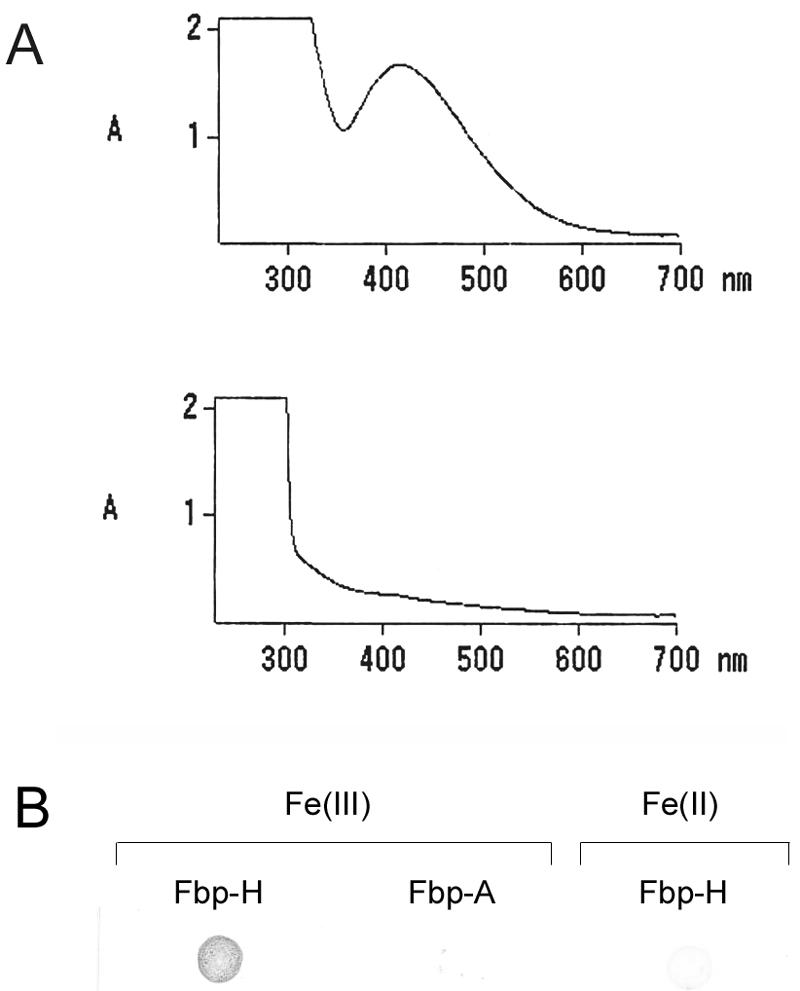
Iron binding of E. canis Fbp. (A) Absorbance of E. canis Fbp in the iron-bound (holo) (top) and iron-free (apo) (bottom) forms. (B) Positive Ferene S staining for iron of recombinant E. canis holo-Fbp (Fbp-H) but not of the apo-Fbp form (Fbp-A). In the absence of the reducing agent thioglycolic acid, holo-Fbp did not stain positive by Ferene S, demonstrating that the bound iron is Fe(III) and not Fe(II).
Cellular location of the Fbp protein.
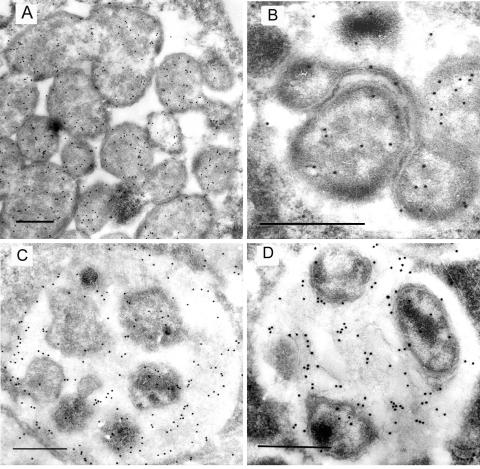
Ultrathin sections containing E. canis and E. chaffeensis in infected DH82 cells were incubated with antibodies against E. canis recombinant Fbp and visualized with colloidal gold particles by immunoelectron microscopy. Ehrlichiae exist in two distinct morphological forms: the reticulate and dense-cored forms (28). Fbp was found primarily in the cytoplasm of the reticulate form in both species, with some localization in the periplasmic space and on the bacterial surface (Fig. 7A and B). The dense-cored form of the bacteria also exhibited Fbp localization in the cytoplasm (Fig. 7C and D), but Fbp was also found extracellularly on the intramorular fibers in E. canis and E. chaffeensis morulae containing the dense-cored form (Fig. 7C and D).
FIG. 7.
Immunogold-labeled electron micrographs of E. canis and E. chaffeensis with anti-E. canis recombinant Fbp. (A and B) Localization of Fbp in the E. canis (A) and E. chaffeensis (B) reticulate morphological form. (C and D) Localization of Fbp in the E. canis (C) and E. chaffeensis (D) dense-cored morphological form. Bars, 0.5 μm.
DISCUSSION
Although iron-binding proteins have been identified in numerous extracellular bacteria, the discovery of a functional ferric ion-binding protein in E. canis represents the first such description in an obligately intracellular bacterium. Identification of ferric ion-binding proteins in ehrlichiae represents the first step in understanding the mechanisms of iron acquisition by these organisms as well as by other obligately intracellular bacteria. In the other systems, the Fbp functions as the periplasmic transporter after other proteins have acquired the iron from the extracellular iron-binding protein transferrin. Ehrlichiae are likely to obtain iron not from the extracellular milieu, but after entering the host cell. Ehrlichiae upregulate the transferrin receptor and recruit it to the intracytoplasmic vacuole (morula) (8). It is likely that iron acquisition by ehrlichiae occurs within the morula.
The presence of large intergenic sequences between fbp and the putative Ehrlichia membrane permease suggested that Ehrlichia fbp is not part of an operon iron acquisition system present in other gram-negative bacteria, a suggestion that was confirmed by RT-PCR. The absence of an ATP-binding protein in the region was also noted, although mutational analysis of the gene encoding the ATP-binding protein of the N. gonorrhoeae operon suggests that this protein is not essential for functional iron acquisition (32). Genetic analysis of fbp from the related rickettsial organisms A. marginale, W. pipientis, and Neorickettsia sennetsu also demonstrated the lack of an operon, including a change in the fbp-membrane permease gene order (Anaplasma) or the absence of the associated genes encoding the membrane permease and ATP-binding protein in the region (Wolbachia and Neorickettsia). Only one copy of the fbp gene was found in each of these genome sequences. The loss of the fbp operon in Ehrlichia spp. suggests that the ehrlichial Fbp plays a distinct and potentially more direct role in the acquisition and transport of iron.
E. canis Fbp was found to share protein sequence homology with known iron-binding proteins from other pathogens. Modeling of E. canis Fbp against the known Fbp crystal structures of N. gonorrhoeae, H. influenzae, and M. haemolytica predicted that E. canis Fbp contained the two-domain structure connected by a hinge, present in these other proteins. E. canis Fbp also contains two key tyrosines shared by all members of the Fbp family and required for iron coordination (34) and thus appears to conform well to this known iron-binding motif. Iron-binding experiments with E. canis Fbp demonstrated that it does in fact bind iron(III). This trait is shared with other Fbp's and demonstrates that ehrlichial Fbp could function by binding iron(III), dissociated from transferrin, recruited to the morulae (8). The E. canis fbp gene contained a binding site for the putative iron-regulatory element Fur, at position −136 from the start, that contained two copies of the E. coli Fur recognition consensus sequence, suggesting that fbp is likely regulated in response to the availability of iron. Another putative Fur box containing only one copy of the consensus sequence is located at position −85 from the beginning of the putative membrane permease gene, but it is not yet known whether this gene is regulated by Fur or if it plays a coordinated role with Fbp in ehrlichial iron acquisition.
By immunoelectron microscopy, we have determined that Fbp is located in the cytoplasm, in the periplasm, and on the surfaces of ehrlichiae. Yu et al. reported the localization of E. chaffeensis Fbp to the cytoplasm and the periplasmic space (37). We also report this localization in the reticulate form, but we also detected extracellular Fbp on fibers within morulae containing the dense-cored form, where it was not observed by Yu et al. With Fbp localization in all parts of the bacteria, it is possible that the protein itself can shuttle free iron into the bacteria for utilization. The E. canis Fbp protein contained a signal sequence, suggesting that it could be transported to the periplasm, transported to the outer membrane, or secreted. We have demonstrated that in morulae containing dense-cored forms of ehrlichiae, Fbp is located on the surface of the organism, in the extracellular space, and on the morula membrane. The metabolic activities of the two morphological forms of ehrlichiae have not been determined, but Fbp may be secreted extracellularly to bind to free iron and prevent toxicity while the bacteria are metabolically inactive. Several gram-negative bacteria have been discovered to produce iron-binding proteins, such as ferritin, bacterioferritin, and Dps (DNA-binding protein from starved cells), to reduce the damage from oxidative stress (3, 17, 20, 31, 35, 36, 38). The extracellular localization of the ehrlichial Fbp indicates that it functions in a manner different from that described for other gram-negative systems.
The recombinant protein was recognized by sera from experimentally infected dogs, thus confirming its immunogenicity. Sera from all dogs strongly recognized the protein primarily in a conformation-dependent manner. This conformation dependence is likely the reason that this protein is weakly reactive in denatured E. canis lysates (Fig. 1) yet was readily identified in a genomic library screened with an antibody at a high dilution factor. The presence of conformational epitopes could explain the lack of immunoreactivity of the previously reported E. chaffeensis Fbp protein (37). The dependence on native conformation for strong antibody recognition by Fbp (Fig. 2) demonstrates that proteins not identified as immunoreactive by Western blotting could potentially contain major immunoreactive epitopes. The conformational dependence of the Fbp illustrates the importance of identification of proteins with both linear and conformational epitopes and the limitations of Western blotting for comprehensive identification of all immunoreactive proteins that may be of interest for vaccine development.
Rickettsial genomes are believed to have evolved from larger genomes (4). In the course of adaptation to an obligately intracellular lifestyle, with the ability to utilize host resources, certain genes become obsolete. In the interest of minimizing energy expenditures for genome replication and transcription, these nonessential genes are mutated and subsequently deleted (4). This genome reduction process could explain the loss of the fbp operon system in ehrlichiae. The role of Fbp as a periplasmic iron transporter after extracellular competition with host proteins became unnecessary. However, the gene was conserved along with the ability of the protein to bind iron, suggesting that Fbp was adapted to serve in a novel manner.
This study determined that Ehrlichia Fbp does not remain in the periplasmic space as do other bacterial Fbp's but is found in the cytoplasm and on the surface as well. There have been reports of surface expression of Fbp in Neisseria meningiditis and of bactericidal activity of antibodies against this protein (18, 19). As E. chaffeensis was reported to be highly susceptible to an intracellular iron chelator (7), this protein may play a critical role for the survival of the organism. Learning more about iron acquisition in ehrlichiae could potentially lead to discovery of an attractive candidate for future subunit vaccine development.
Acknowledgments
We thank David Walker for review of the manuscript and Tim Mietzner and Damon Anderson for suggestions regarding iron-binding assays.
This work was supported by the Clayton Foundation for Research, the John Sealy Memorial Foundation, and the Sealy Center for Vaccine Development.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Adhikari, P., S. A. Berish, A. J. Nowalk, K. L. Veraldi, S. A. Morse, and T. A. Mietzner. 1996. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J. Bacteriol. 178:2145-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikari, P., S. D. Kirby, A. J. Nowalk, K. L. Veraldi, A. B. Schryvers, and T. A. Mietzner. 1995. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J. Biol. Chem. 270:25142-25149. [DOI] [PubMed] [Google Scholar]
- 3.Altuvia, S., M. Almiron, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, J. O., and S. G. Andersson. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664-671. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 6.Angerer, A., S. Gaisser, and V. Braun. 1990. Nucleotide sequences of the sfuA, sfuB, and sfuC genes of Serratia marcescens suggest a periplasmic-binding-protein-dependent iron transport mechanism. J. Bacteriol. 172:572-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnewall, R. E., N. Ohashi, and Y. Rikihisa. 1999. Ehrlichia chaffeensis and E. sennetsu, but not the human granulocytic erhlichiosis agent, colocalize with transferrin receptor and up-regulate transferrin receptor mRNA by activating iron-responsive protein 1. Infect. Immun. 67:2258-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnewall, R. E., Y. Rikihisa, and E. H. Lee. 1997. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 65:1455-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 10.Bearden, S. W., T. M. Staggs, and R. D. Perry. 1998. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J. Bacteriol. 180:1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruns, C. M., D. S. Anderson, K. G. Vaughan, P. A. Williams, A. J. Nowalk, D. E. McRee, and T. A. Mietzner. 2001. Crystallographic and biochemical analyses of the metal-free Haemophilus influenzae Fe3+-binding protein. Biochemistry 40:15631-15637. [DOI] [PubMed] [Google Scholar]
- 12.Bruns, C. M., A. J. Nowalk, A. S. Arvai, M. A. McTigue, K. G. Vaughan, T. A. Mietzner, and D. E. McRee. 1997. Structure of Haemophilus influenzae Fe+3-binding protein reveals convergent evolution within a superfamily. Nat. Struct. Biol. 4:919-924. [DOI] [PubMed] [Google Scholar]
- 13.Chung, M. C. 1985. A specific iron stain for iron-binding proteins in polyacrylamide gels: application to transferrin and lactoferrin. Anal. Biochem. 148:498-502. [DOI] [PubMed] [Google Scholar]
- 14.Clarke, T. E., L. W. Tari, and H. J. Vogel. 2001. Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem. 1:7-30. [DOI] [PubMed] [Google Scholar]
- 15.Desai, P. J., A. Angerer, and C. A. Genco. 1996. Analysis of Fur binding to operator sequences within the Neisseria gonorrhoeae fbpA promoter. J. Bacteriol. 178:5020-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frazier, B. A., J. D. Pfeifer, D. G. Russell, P. Falk, A. N. Olsen, M. Hammar, T. U. Westblom, and S. J. Normark. 1993. Paracrystalline inclusions of a novel ferritin containing nonheme iron, produced by the human gastric pathogen Helicobacter pylori: evidence for a third class of ferritins. J. Bacteriol. 175:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez, J. A., C. Agra, L. Ferron, N. Powell, M. Pintor, M. T. Criado, and C. M. Ferreiros. 1996. Antigenicity, cross-reactivity and surface exposure of the Neisseria meningitidis 37 kDa protein (Fbp). Vaccine 14:1340-1346. [DOI] [PubMed] [Google Scholar]
- 19.Gomez, J. A., M. T. Criado, and C. M. Ferreiros. 1998. Bactericidal activity of antibodies elicited against the Neisseria meningitidis 37-kDa ferric binding protein (FbpA) with different adjuvants. FEMS Immunol. Med. Microbiol. 20:79-86. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa, T., Y. Mizunoe, S. Kawabata, A. Takade, M. Harada, S. N. Wai, and S. Yoshida. 2003. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 185:1010-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 22.Kirby, S. D., F. A. Lainson, W. Donachie, A. Okabe, M. Tokuda, O. Hatase, and A. B. Schryvers. 1998. The Pasteurella haemolytica 35 kDa iron-regulated protein is an FbpA homologue. Microbiology 144:3425-3436. [DOI] [PubMed] [Google Scholar]
- 23.McBride, J. W., R. E. Corstvet, S. D. Gaunt, C. Boudreaux, T. Guedry, and D. H. Walker. 2003. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 71:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride, J. W., R. E. Corstvet, E. B. Breitschwerdt, and D. H. Walker. 2001. Immunodiagnosis of Ehrlichia canis infection with recombinant proteins. J. Clin. Microbiol. 39:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mietzner, T. A., S. B. Tencza, P. Adhikari, K. G. Vaughan, and A. J. Nowalk. 1998. Fe(III) periplasm-to-cytosol transporters of gram-negative pathogens. Curr. Top. Microbiol. Immunol. 225:113-135. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 27.Nowalk, A. J., S. B. Tencza, and T. A. Mietzner. 1994. Coordination of iron by the ferric iron-binding protein of pathogenic Neisseria is homologous to the transferrins. Biochemistry 33:12769-12775. [DOI] [PubMed] [Google Scholar]
- 28.Popov, V. L., S. M. Chen, H. M. Feng, and D. H. Walker. 1995. Ultrastructural variation of cultured Ehrlichia chaffeensis. J. Med. Microbiol. 43:411-421. [DOI] [PubMed] [Google Scholar]
- 29.Raymond, K. N., E. A. Dertz, and S. S. Kim. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 100:3584-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rikihisa, Y., S. A. Ewing, J. C. Fox, A. G. Siregar, F. H. Pasaribu, and M. B. Malole. 1992. Analyses of Ehrlichia canis and a canine granulocytic Ehrlichia infection. J. Clin. Microbiol. 30:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romao, C. V., M. Regalla, A. V. Xavier, M. Teixeira, M. Y. Liu, and J. Le Gall. 2000. A bacterioferritin from the strict anaerobe Desulfovibrio desulfuricans ATCC 27774. Biochemistry 39:6841-6849. [DOI] [PubMed] [Google Scholar]
- 32.Sebastian, S., and C. A. Genco. 1999. FbpC is not essential for iron acquisition in Neisseria gonorrhoeae. Infect. Immun. 67:3141-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shouldice, S. R., D. R. Dougan, R. J. Skene, L. W. Tari, D. E. McRee, R. H. Yu, and A. B. Schryvers. 2003. High resolution structure of an alternate form of the ferric ion binding protein from Haemophilus influenzae. J. Biol. Chem. 278:11513-11519. [DOI] [PubMed] [Google Scholar]
- 34.Shouldice, S. R., D. R. Dougan, P. A. Williams, R. J. Skene, G. Snell, D. Scheibe, S. Kirby, D. J. Hosfield, D. E. McRee, A. B. Schryvers, and L. W. Tari. 2003. Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins. J. Biol. Chem. 278:41093-41098. [DOI] [PubMed] [Google Scholar]
- 35.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yariv, J., A. J. Kalb, R. Sperling, E. R. Bauminger, S. G. Cohen, and S. Ofer. 1981. The composition and the structure of bacterioferritin of Escherichia coli. Biochem. J. 197:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, X. J., P. A. Crocquet-Valdes, L. C. Cullman, V. L. Popov, and D. H. Walker. 1999. Comparison of Ehrlichia chaffeensis recombinant proteins for serologic diagnosis of human monocytotropic ehrlichiosis. J. Clin. Microbiol. 37:2568-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689-27696. [DOI] [PubMed] [Google Scholar]