Abstract
Rapamycin is a naturally occurring macrolide whose target is at the core of nutrient and stress regulation in a wide range of species. Despite well-established roles as an inhibitor of cap-dependent mRNA translation, relatively little is known about its effects on other modes of RNA processing. Here, we characterize the landscape of rapamycin-induced post-transcriptional gene regulation. Transcriptome analysis of rapamycin-treated cells reveals genome-wide changes in alternative mRNA splicing and pronounced changes in NMD-sensitive isoforms. We demonstrate that despite well-documented attenuation of cap-dependent mRNA translation, rapamycin can augment NMD of certain transcripts. Rapamycin-treatment significantly reduces the levels of both endogenous and exogenous Premature Termination Codon (PTC)-containing mRNA isoforms and its effects are dose-, UPF1- and 4EBP-dependent. The PTC-containing SRSF6 transcript exhibits a shorter half-life upon rapamycin-treatment as compared to the non-PTC isoform. Rapamycin-treatment also causes depletion of PTC-containing mRNA isoforms from polyribosomes, underscoring the functional relationship between translation and NMD. Enhanced NMD activity also correlates with an enrichment of the nuclear Cap Binding Complex (CBC) in rapamycin-treated cells. Our data demonstrate that rapamycin modulates global RNA homeostasis by NMD.
INTRODUCTION
Nonsense Mediated Decay (NMD) is a conserved RNA surveillance system which recognizes and eliminates transcripts containing Premature Termination Codons (PTCs) (1). The NMD system prevents the synthesis of potentially toxic proteins arising from transcripts containing nonsense mutations, frame shifts and those generated by alternative splicing. NMD is known to regulate the expression of many genes, including those involved in the NMD pathway itself (2,3).
The constellation of proteins associated with RNA transcripts is dynamic and reflects the diverse cellular landscapes traversed by the mRNA. Virtually all human genes undergo alternative splicing generating different mRNA isoforms (4). All newly synthesized and spliced mRNAs are bound by the nuclear Cap Binding Complex (CBC), the Exon Junction Complex (EJC) and the nuclear poly(A) binding protein (PABPN1) (5–7). By contrast, mRNAs engaged in steady-state translation are depleted of EJC proteins, and have exchanged CBC and PABPN1 at the cap and poly(A) tail for the eukaryotic translation initiation factor 4E (eIF4E) and the cytoplasmic poly(A) binding protein (PABPC), respectively (7–9). Termination codons located more than 50–55 nt upstream from an Exon Junction Complex (EJC) are strong NMD signals (10). During mRNA processing, the SURF complex (SMG1, UPF1, eRF1 and eRF3) associates with the downstream EJC and triggers SMG1-dependent phosphorylation of UPF1, which is required for inhibition of translation initiation and subsequent nucleolytic decay of the targeted mRNA (11–13). Additionally, shorter 3΄UTRs are thought to suppress NMD and presence of cis-acting elements in long 3΄UTRs can trigger NMD evasion (14,15).
NMD efficiency may be linked to the composition of messenger ribonucleoprotein particles (mRNPs), from those acquired during splicing to the cap-binding factors. The CBC is displaced by eIF4E in a translation-independent manner (9,16). This exchange mechanism is driven by interactions of CBP80 with the nuclear import receptor Importin β (IMPb) (9). It is possible that this exchange may also be regulated by mass action due to the high cytoplasmic concentrations of eIF4E (17). These rearrangements have important consequences for NMD; for example the interaction of CBC with UPF1 promotes assembly of the SURF complex and is important for activating NMD (18).
The mechanistic Target of Rapamycin Complex 1 (mTORC1) is central to the cellular nutrient/stress sensing pathway and is inhibited by the naturally occurring macrolide rapamycin, which promotes autophagy and extends lifespan in a number of species including yeast, worms, flies and mammals (19,20). One important branch of the mTORC1 signaling pathway modulates initiation of cap-dependent translation through phosphorylation of targets such as 4EBPs (eIF4E-Binding proteins) and S6K1 (Ribosomal Protein S6 Kinase B1). The 4EBP family functions as inhibitor of cap dependent translation by inducing nuclear sequestration of eIF4E (17). Active mTORC1 phosphorylates 4EBPs and counteracts their inhibitory effects on eIF4E (21) thereby stimulating translation. In addition to regulating the translation of messages bound by the cytoplasmic cap, mTORC1 also promotes the translation of CBC-bound transcripts (22). Given that NMD and translation are functionally linked in mammalian cells (9,23), inhibitors of cap-dependent translations such as rapamycin are predicted to inhibit NMD.
Here, we characterize the landscape of rapamycin-induced post-transcriptional gene regulation in HEK 293 cells. Exposure of cells to either rapamycin or the translation elongation inhibitor emetine induced a pattern of similar effects on genome-wide alternative splicing. Our results demonstrate that rapamycin modulates NMD. As expected we observed rapamycin-dependent inhibition of NMD. However, we also made the surprising observation that a proportion of established NMD isoforms were depleted after rapamycin treatment. We studied a subset of endogenous and exogenous PTC-containing mRNA isoforms, and demonstrated that rapamycin enhances the efficiency of NMD in a dose-, UPF1- and 4EBP-dependent manner. Moreover, we found that PTC-containing mRNA isoforms accumulate in polyribosomes when mTORC1 is active, but are depleted when cells are treated with rapamycin. Because rapamycin-treatment induces nuclear accumulation of eIF4E, we investigated the rapamycin-dependent dynamics of CBC- and eIF4E-cap binding. We observed the expected rapamycin-dependent nuclear accumulation of eIF4E, and consequent depletion of eIF4E from cytosolic mRNPs. Interestingly, we also found that CBC-association with cytosolic mRNPs moderately increased with rapamycin, suggesting accumulation of pioneer-like mRNPs that may facilitate NMD. Taken together, our results demonstrate that mammalian cells can dynamically regulate NMD efficiency of messenger RNA associated with polyribosomes depending on cellular need.
MATERIALS AND METHODS
Cell culture and rapamycin treatment
Human Embryonic Kidney (HEK) 293 T cells were cultured in DMEM 10% FBS (Santa Cruz Biotech) at 37°C 5% CO2. For rapamycin (Cell Signaling) experiments, cells were starved for 24 h in Glutamine deficient media DMEM 10%FBS. Following this, they were incubated in 22 nM rapamycin or vehicle (DMSO) for 30 min prior to feeding with complete medium containing DMSO or rapamycin. For polyribosome experiments, cells were harvested 8 h-post rapamycin stimulation. For dose-response assays and RNA-seq cells were harvested 72 h at the indicated concentrations diluted in vehicle (DMSO). 5 mg/ml of actinomycin D was added after 48 h rapamycin exposure and cells harvested at the indicated time points. For the RNA-seq experiments, emetine (SIGMA) was added at 50 ng/ml after 72 h during 4 h to avoid long-term effects and all cells (± rapamycin, ± emetine) were harvested at the same time.
HEK 293T cells shControl or shUPF1 were generated employing pRS vector against human UPF1 (RENT1) gene or Control (OriGene, TR308482) and sub-cloning it with its U6 promoter in pLVTHM (Tronolab) between EcoRI and MluI sites. siRNA against 4EBP1 and 4EBP2 were transfected employing Dharmafect according to manufacturer's instructions.
Immunofluorescence
HEK 293 T cells were seeded at low density on coverslips overnight. Cells were treated as described above, fixed with 3.2% paraformaldehyde for 15 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 3 min. After blocking in 1% BSA + PBS 1h at room temperature they were incubated with eIF4E-FITC primary conjugated antibody (Santa Cruz Biotechnology) overnight at 4°C and washed. After washing three times with 0.02% tween PBS, coverslips were mounted using DAPI-mounting media.
Microscopy and quantification
Immunofluorescence was performed using a Leica DM2700 white field microscope at magnification 63X (oil immersion objective). Quantification of fluorescence was performed using ImageJ.
Cell fractionation and polyribosome extraction
Cells were incubated in the presence of 100 μg/ml cycloheximide 10 min at 37°C, 5%CO2 prior harvesting. Cell lysis was performed in 0.5% NP40, 20 mM Tris–HCl pH 7.5, 100 mM KCl and 10 mM MgCl2. Lysates were passed three times through a 23 G needle and incubated on ice 7 min. Extracts were then centrifuged at 10K rpm at 4°C 7 min. Cytosolic extracts were laid on 15–45% sucrose gradients prepared in 20 mM Tris–HCl pH 7.5, 100 mM KCl and 10 mM MgCl2 using a Gradient Station Master (Biocomp). Gradients were ultracentrifuged at 40K rpm 4°C for 1 h 20 min using SW41 rotor in a Beckman Ultracentrifuge. Polyribosome monitoring and extraction of discrete complexes were conducted using a Gradient Station Master (Biocomp).
Western blotting
Cell extracts were prepared in Lysis Buffer as described above. For 4E-BP and P-4E-BP blots, cell extracts were resolved in 12% bis/acrylamide gels. For CBP80/eIF4E blots, extracts were resolved in 10% Bis/acrylamide gels. Antibodies used were: Phospho-S6 Ribosomal Protein (D57.2.2E, Cell Signaling) 4E-BP1 (53H11, Cell Signaling), CBP80 E-7 (Santa Cruz Biotech), iEF4E FL-217 (Santa Cruz Biotech), p-4E-BP1/2/3 (Thr 45)-R (Santa Cruz Biotech) and U2AF65 MC3 (Santa Cruz Biotech).
Cap binding experiments
Cells were stimulated with rapamycin for 24 h and extracts prepared in lysis buffer as described above. Oligo dT cellulose (Ambion) was resuspended in lysis buffer twice before incubating it in the presence of cell extracts on a rotating wheel at 4°C during 2 h. Cellulose-bound extracts were washed five times. RNAse treatment was done using 1/100 RNAse A/T1 cocktail (Ambion) at 37°C during 13 min shaking. Extracts were washed once more and cellulose was pelleted and resuspended in lysis buffer, boiled and loaded onto bis/acrylamide gels for CBP80/eIF4E blotting.
Luciferase assays
HEK 293 T cells were starved for 24 h in the absence of glutamine and then co-transfected with a normalizer firefly luciferase vector (pGL3, Promega) and pCI-Renilla/β-globin wt (WT) or NS39 (MUT) kindly gifted by Prof. A.E. Kulozik (24). Transfected cells were then treated with vehicle (Control), 22 nM of rapamycin (Rapa) or Emetine and chemiluminiscence was measured using the Dual-Luciferase Reporter (DLR) Assay (Promega) following manufacturer's instructions.
RT-PCR and real-time PCR (qPCR)
Total cytoplasmic RNA and polyribosome-bound RNA was extracted using TRIzol LS following manufacturer's instructions. In the case of polyribosomal-bound RNA, RNA was extracted from discrete polyribosome fractions and then pooled prior to reverse transcription with random hexamers (Thermo Fisher Scientific) following manufacturer's instructions. For actinomycin D experiments, oligodT primers were used in reverse transcription. PCR was performed using Taq Titanium (Clontech). In the case of total to polyribosome comparisons of CCAR1 the rate of inclusion was calculated using the following formula %inclusion = included/(skipped + included) in nmol as determined using an Agilent 2001 Bioanalyzer. Statistics were done using a Mann–Whitney one tailed U-test. For qPCR experiments, Sybr Green qPCR for −PTC and +PTC was done using primers described previously in (25). Our primers were designed using the Universal Probe Library System Software (Roche) and blasted in UCSC Genome Browser tool In silico PCR to hg19 assembly:
SRSF6_All_FOR: tgg aag cag atc cag gtc tc
SRSF6_All_REV: gcg act ttt tga gat act tcg ag
CCAR1_All_FOR: CAA AAA GAA GAA CAG AAG GAG TTA GAG
CCAR1_All_REV: TCG TCT TCA GAT TTC CTA TCA TCA
CCAR1_−PTC_FOR: AAC CCT CTC CCG AGG ATA CA
CCAR1_−PTC_REV: TCA TCC TTC TCT TCT TCA TCC TG
CCAR1_+PTC_FOR: TGG ACC AGA CCC AGA AAA AG
CCAR1_+PTC_REV: TTC TTC ATC CAT TGT GTG C.
Levels of each of each isoform (‘−PTC’ or ‘+PTC’) were normalized to total transcript levels (‘All’) which accounts for the total gene expression of each gene and analyzed using the DDCt method. Total levels of transcript were normalized against SDHA. Statistics were done using a two-tailed t-test and calculated as fold rapamycin-treated over control.
Primers employed in the RNA-seq validations are listed in Supplementary Table S1.
RNA-seq
RNA was extracted from cytoplasmic extracts employing TRIzol LS following manufacturer's instructions, and re-purified and DNAse treated employing miRNeasy Micro Kit (Qiagen) following manufacturer's instructions. Libraries were done employing TruSeq Stranded mRNA Library Prep Kit (Illumina) following manufacturer's instructions. RNA-seq was performed on a HiSeq 4000 (UCSF) and sequencing was performed employing 100 bp paired-end reads, generating a minimum of 37M reads per sample (Supplementary Table S2). Reads were mapped to the genome (hg19) using STAR v2.5.1b (26). An expanded transcript annotation was generated using StringTie v1.2.3 (27) with GENCODE v19 as a reference annotation, and alternative splicing events were identified and quantified using SplAdder v1.0.0 (28). Paired differential splicing tests were performed using rMATS-STAT (29) with thresholds of ΔΨ ≥ 0.1, ΔΨ ≥ 0.05 and ΔΨ > 0, and the resulting p-values were corrected for multiple hypothesis testing using the method of Benjamini and Hochberg (30). Differential gene expression analysis was conducted using DESeq2 (31). Additional data manipulation and visualization was performed using custom R and Python scripts, and BEDTools v2.25 (32). Visualization of specific alternative events was performed using Sashimi plots (33). Gene set enrichment analysis was performed using Enrichr (34–37).
RESULTS
Rapamycin modulates levels of NMD mRNA isoforms
Given that mTORC1 interacts with splicing factors and modulates their activity (22,38) we performed RNA-seq on cytoplasmic extracts from HEK 293T cells treated with rapamycin or vehicle, in order to characterize the genome-wide impact of mTORC1 in alternative splicing (AS). As expected, the phosphorylation of the ribosomal protein S6 (P-S6K) diminished when rapamycin was present in the medium (Supplementary Figure S1A). We used SplAdder (28) to identify and quantify alternative splicing events, and performed paired differential testing using rMATS-STAT (29) to identify events modulated by rapamycin (Supplementary Tables S3–S11). Our data revealed an extensive rapamycin-dependent alternative splicing program spanning much of the diversity of event types with noticeable higher fractions of skipped exons and retained introns (Figure 1A). Moreover, Gene Set Enrichment Analysis (GSEA) of differentially spliced genes (34–36) revealed ontologies associated with splicing and RNA processing among the most significantly enriched categories (Supplementary Tables S12 and S13).
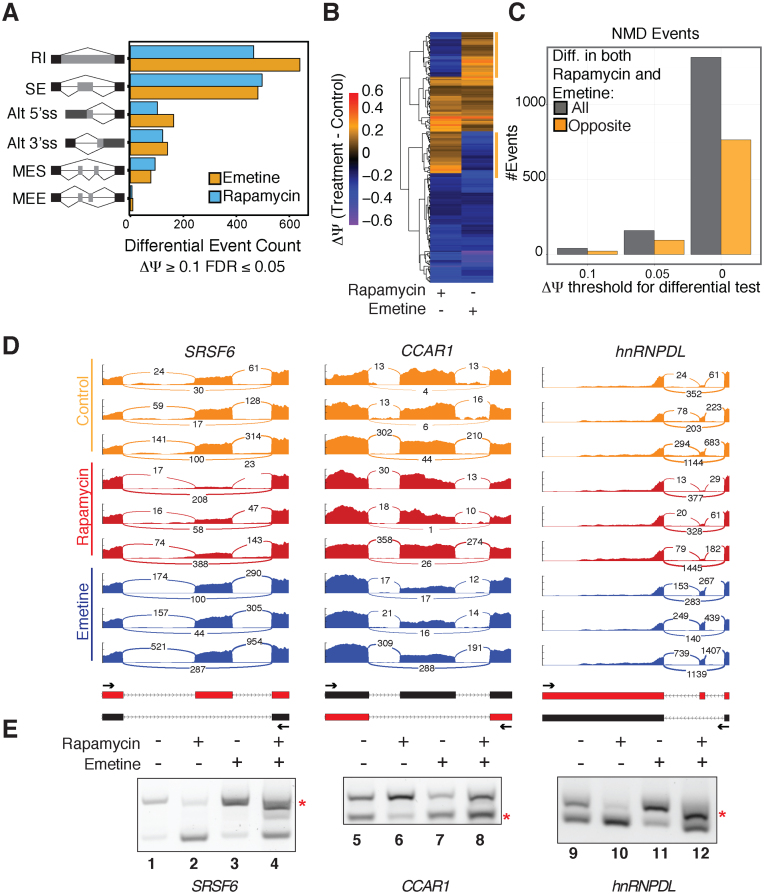
Figure 1.
Genome-wide effects of rapamycin in alternative splicing and NMD-sensitive isoforms. (A) Alternative splicing events predominantly regulated by emetine and rapamycin (ΔΨ ≥ 0.1, FDR ≤ 0.05) in HEK 293T cells. RI: retained introns; SE: skipped exons; Alt 5΄ss: Alternative 5΄ Splice Site; Alt 3΄ss: Alternative 3΄ Splice Site; MES: Multiple Exon Skipping; MEE: Mutually Exclusive Exons. (B) Heatmap depicting the directionality of those alternative splicing events showing significant changes by rapamycin or emetine treatment compared to control. (C) Bar graph showing the fraction of events with NMD annotation triggered by rapamycin (All, dark grey) and those that showed opposite behavior to emetine (orange). (D) Sashimi plots representing the mapped reads of our triplicate RNA-seq samples for SRSF6, CCAR1 and hnRNPDL genes in Control (orange), Rapamycin (red) and Emetine (blue). The Y-axis units are arbitrary, as plots were normalised by sample in the event window. Schematics of the events and the position of primers are depicted underneath. (E) Representative gel of RT-PCR analysis of SRSF6, CCAR1 and hnRNPDL PTC-containing (red asterisk) and non-PTC mRNAs for each of the genes.
The enrichment of RNA processing pathways in rapamycin-sensitive differentially spliced transcripts together with the high abundance of regulated intron retention events (Figure 1A, Supplementary Tables S12 and S13) suggested that rapamycin may modulate NMD activity (3,25). To test our hypothesis that mTORC1 inhibition alters NMD and to understand which of these AS events may be linked to RNA surveillance we employed emetine, in order to seek for overlap with rapamycin-sensitive events similarly to (39). Emetine is a translational inhibitor, and thus increases the half-life of NMD-sensitive mRNAs given the dependency of NMD on ribosomal scanning (9,23). Intriguingly, alternative events perturbed by emetine were enriched in similar categories and the overlap between differential events in the rapamycin and the emetine condition is significant (hypergeometric test, P-value < 9e−56 for ΔΨ ≥ 0.1, FDR ≤ 0.05, P-value < 2e−167 for ΔΨ ≥ 0.05, FDR ≤ 0.05, P-value < 3e−321 for ΔΨ > 0, FDR ≤ 0.05). Of these events, we find the majority to be changing in the same direction (e.g. 144 of 220 common events for ΔΨ > 0.1, FDR < 0.05) (Figure 1B). While this supports a broad notion of a common post-transcriptional response to translation inhibitors, it raises the issue of explaining the behavior of the significant number of events (e.g. 76 of 220 common events for ΔΨ > 0.1, FDR < 0.05) changing in opposite directions.
To examine the data specifically for evidence of NMD perturbation, we restricted our analysis to events overlapping transcripts already annotated as NMD targets in GENECODE v19. Moreover, we further restricted the set of events to those behaving as expected in the presence of emetine (i.e. the NMD-sensitive form must increase). In examining the behavior of these isoforms in the presence of rapamycin, we found that, of those that significantly changed, at every ΔΨ threshold tested, more than half exhibited behavior opposite to their emetine-dependent behavior (Figure 1C and Supplementary Tables S14 to S16). We also examined overall gene expression level employing DESeq2 (31) and performed GSEA, which revealed splicing-related terms to be among the most significant in both control versus rapamycin, and control versus emetine comparisons (Supplementary Tables S17 to S20). These results suggested the intriguing idea that rapamycin may conditionally enhance the efficiency of NMD.
To explore the possibility of rapamycin-dependent NMD augmentation, we began by confirming the behaviour of several emetine- and rapamycin-sensitive mRNAs exhibiting this pattern in the RNA-seq data (Figure 1D, E and Supplementary Figure S1B). We chose three candidates (CCAR1, HNRNPDL and SRSF6 genes) for RT-PCR validation, each with different modalities of AS-NMD: a) skipping of exon 13 from CCAR1 mRNA which leads to a frameshift that triggers NMD b) inclusion of exon 8 in the HNRNPDL mRNA introduces a PTC that induces NMD and c) inclusion of exon 3 in the SRSF6 mRNA which introduces a PTC and induces NMD. Importantly, these three genes encode for functionally different proteins, and thus are likely to be regulated by different processes: CCAR1 (Cell Division Cycle And Apoptosis Regulator 1) encodes for a protein that acts as a signal transductor, HNRNPDL (Heterogeneous Nuclear Ribonucleoprotein D Like) encodes for an RNA binding protein that complexes with heterogeneous nuclear RNA and SRSF6 (Serine/Arginine-Rich Splicing Factor 6) encodes one member of the SR protein family and is a splicing regulator. Figure 1D depicts the Sashimi plots (33) representing the alternative splicing pattern triggered by rapamycin and emetine compared to control, demonstrating the effects of rapamycin on the NMD-sensitive isoform according to our RNA-seq data. Figure 1E shows representative gels demonstrating that rapamycin was able to diminish the levels of the NMD-sensitive candidates (compare lanes 1 and 2, 5 and 6, and 9 and 10), an effect that was abrogated when emetine was added (compare lanes 3 and 4, 7 and 8, and 11 and 12). These data validate the RNA-seq results and bolster the hypothesis that rapamycin-treatment may augment NMD.
Depletion of PTC-containing mRNA isoforms by rapamycin is dose dependent
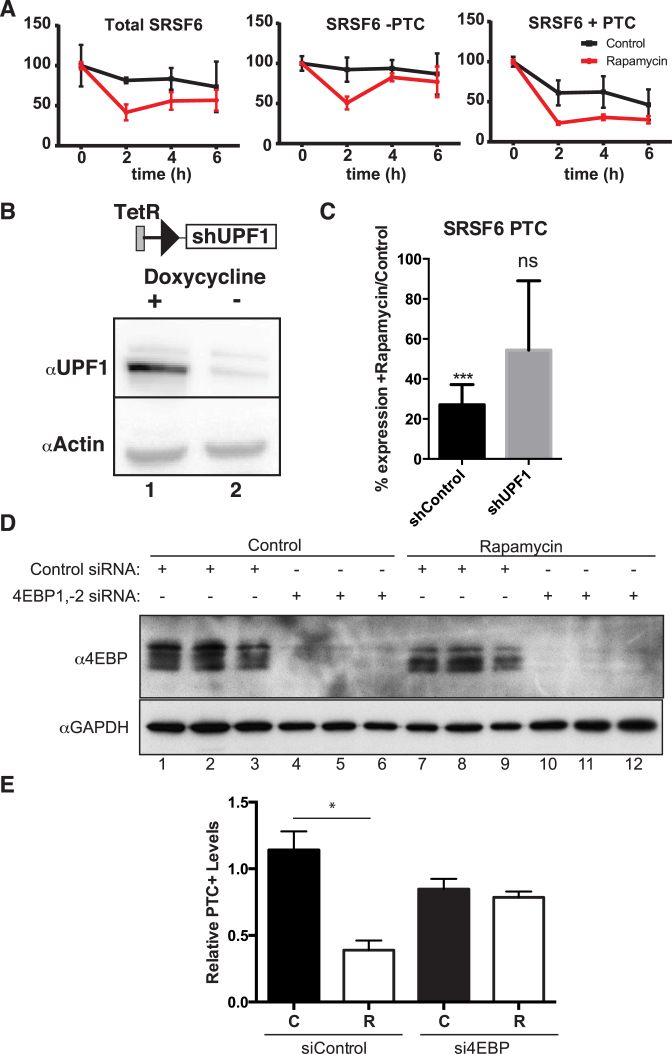
The RNA-seq analysis described above revealed a subset of isoforms that exhibited opposite expression patterns in rapamycin-treated and emetine-treated cells. These data suggest the intriguing hypothesis that rapamycin may increase nonsense mediated decay. To assess the effects of different doses of rapamycin on NMD substrates we assayed the abundance of NMD sensitive and insensitive mRNA isoforms over a broad rapamycin concentration range by RT-qPCR in HEK 293T cells. If the effects we observed in Figure 1 are dependent on rapamycin, our expectation was that decreasing the concentration of rapamycin may have an effect on the abundance of PTC-containing mRNAs accordingly. Following our starvation protocol as in Figure 1, we assayed the phosphorylation of another rapamycin downstream target, 4EBP (17). As expected, phosphorylation of 4EBP decreases with increasing rapamycin concentrations, suggesting that the activity of the mTORC1 pathway is being modulated in a dose-dependent manner (Figure 2A). We firstly set out to test the qPCR quantification of PTC-containing and non-PTC mRNA levels of a well-characterised NMD-candidate, SRSF6 gene, according to Laureau et al. (25) in our RNA-seq datasets, which showed a significant decrease in the PTC-containing isoform upon rapamycin treatment (Supplementary Figure S2). We then assayed the expression of PTC-containing (PTC+) and non-PTC-containing (PTC-) isoforms for SRSF6 and CCAR1 genes employing a range of rapamycin concentrations. Figures 2B and 2C show that PTC-containing isoforms of SRSF6 and CCAR1 genes, respectively, significantly diminish with increasing concentrations of rapamycin. This decrease is statistically significant at the highest doses, 22nM and 2.2nM of rapamycin. We also assayed the levels of total gene expression for SRSF6 and CCAR1 employing primers to amplify constitutive exons in each gene and using SDHA for normalization as described in (25). Our data for SRSF6 and CCAR1 suggest that rapamycin causes an overall increase in the total mRNA expression of both genes (Supplementary Figure S3).
Figure 2.
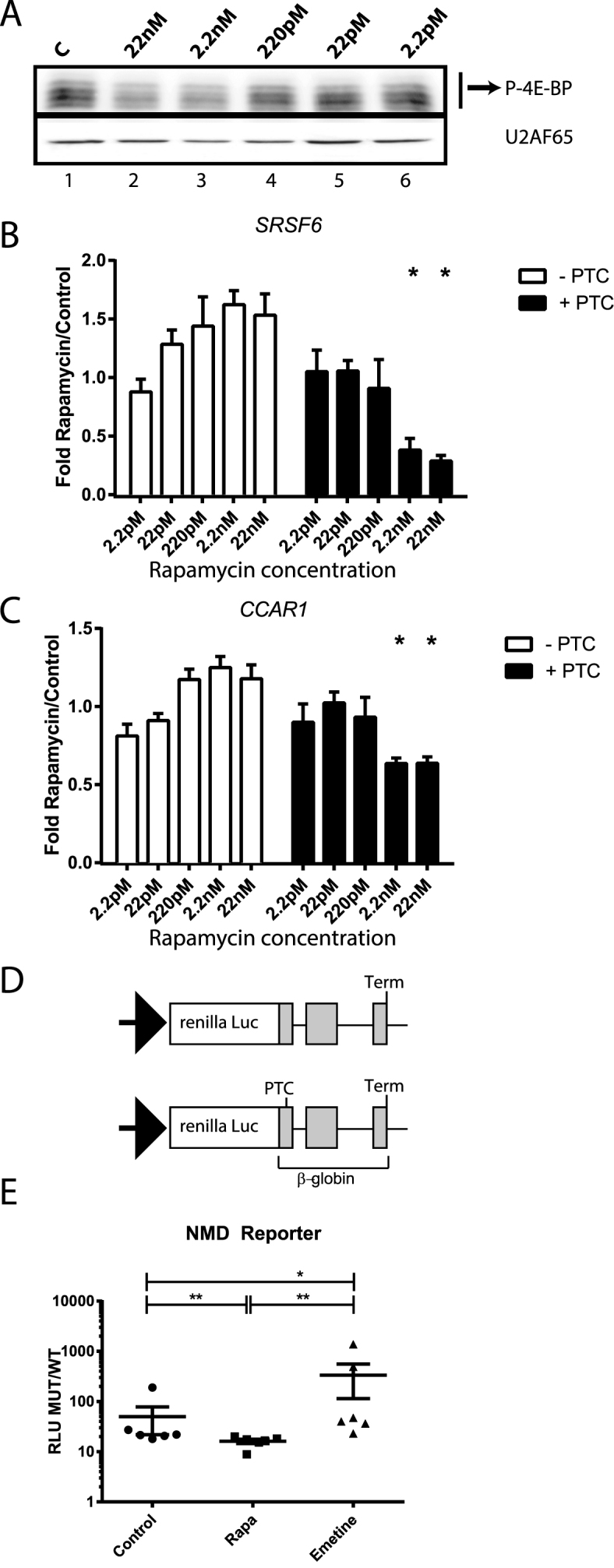
PTC-containing mRNA isoforms respond to rapamycin in a dose-dependent manner. (A) Western blot analysis of 4EBP and U2AF65 (upper and lower panel) from HEK 293T cell lysates treated with decreasing concentrations of rapamycin. (B) Relative RT-qPCR analysis of SRSF6 mRNA isoform (−/+ PTC) levels (normalized to total SRSF6 levels, paired two-tailed t-test, n = 5) from cytoplasmic extracts of HEK 293T cells treated with rapamycin as described above. Note: the data is paired and represented as mean + standard deviation of the mean. (C) RT-qPCR analysis of CCAR1 mRNA isoforms (−/+, normalized to total CCAR1 mRNA levels, paired two-tailed t-test, n = 5) as described above. (D) Schematic for the luciferase reporter (24) employed in 1E. (E) Luciferase assays for rapamycin effects on an NMD-reporter. Y axis represents the ratio of Relative Luminescence Units of the normalized renilla NMD-reporter compared to the normalized WT-reporter (RLU MUT/WT). X axis represents the different treatments: vehicle (Control), 22 nM rapamycin (Rapa) or emetine (Emetine) (n = 3, 2 biological replicates/sample, Mann–Whitney two-tailed U-test).
To determine the effects of rapamycin in exogenous PTC-containing transcripts we employed a previously developed and validated NMD reporter (24). We co-transfected HEK 293T cells with a reporter vector containing a wild type—β-globin open reading frame (WT) or an NMD-triggering mutation-globin (MUT) fused to a renilla luciferase gene (schematics in Figure 2D) together with a normalizer firefly luciferase vector. Transfected cells were then treated with vehicle (Control), 22 nM of rapamycin (Rapa) or emetine and chemiluminiscence was measured. Figure 2E shows our results as Relative Luminescence Units of the normalized renilla NMD-reporter compared to the normalized WT-reporter (RLU MUT/WT) as in (24). Our data indicate that the expression of the NMD reporter was significantly diminished (Mann–Whitney two-tailed U-test) in cells treated with rapamycin compared to vehicle (compare Rapa to Control). As expected, emetine treatment increased the stability of the NMD reporter. Taken together, our data support a mechanism for rapamycin triggering depletion of PTC-containing mRNAs.
Effects of rapamycin on SRSF6 PTC-containing mRNA isoforms require UPF1 and 4EBPs
The previous experiments suggested that rapamycin augments NMD activity and decreases levels of PTC-containing isoforms in HEK cells. In order to confirm that the observed change in the ratio of PTC-containing/non-PTC isoforms is due to RNA decay and not merely AS regulation, we assayed the half-life of PTC-containing and non-PTC isoforms of SRSF6 in a rapamycin dependent manner using RT-qPCR. Figure 3A shows that endogenous SRSF6 PTC-containing isoform displayed accelerated decay kinetics in rapamycin-treated cells as compared to the non-PTC isoform, suggestive of an increased rate of NMD rather than regulation of AS.
Figure 3.
Rapamycin alters the stability of PTC-containing isoforms and its effects are UPF1- and 4EBP-dependent. (A) Relative RT-qPCR analysis of constitutive SRSF6, SRSF6 non-PTC isoforms (SRSF6 −PTC) and PTC-containing (SRSF6 +PTC), after rapamycin treatment and translational arrest with actinomycin D to measure mRNA half-life (n = 5) in HEK 293T cells. (B) Representative western blot of the inducible Tet-on shUPF1 cell line and schematic of the Tet-on system. C. RT-qPCR analysis of SRSF6 PTC+ mRNA isoforms in control or UPF1-depleted cells treated with or without rapamycin (n = 4; ***P-value < 0.001, ns: non-significant). (D) Western blot analysis of 4EBP and GAPDH in cells transfected with siRNAs against 4EBP1-2 or control and stimulated with/without rapamycin. (E) RT-qPCR analysis of SRSF6 PTC-containing mRNA in cells from D (n = 3). C: Control (black bars), R: Rapamycin (white bars).
To test the direct role of NMD machinery in rapamycin effects, we compared the levels of PTC-containing SRSF6 mRNA isoforms in UPF1-depleted HEK cells treated with or without rapamycin. Depletion of UPF1 inhibits NMD in a variety of model systems (39,40). We generated stably transduced HEK cell lines with an inducible Tet-On shRNA against UPF1 or a scramble control (41) (Figure 3B). As previously observed, rapamycin stimulation decreased the levels of PTC-containing isoforms of endogenous SRSF6 in control cells. However, this effect was attenuated in the UPF1 depleted cells (Figure 3C). These data demonstrate that the depletion of PTC-containing isoforms by rapamycin is dependent on UPF1 and supports our findings that rapamycin enhances NMD.
Given that rapamycin effects on translation rely on 4EBP modulation, we predicted that depletion of 4EBPs should attenuate the effect of rapamycin on NMD. To test this hypothesis, we transfected HEK 293T cells with a combination of siRNAs against 4EBP1 and 4EBP2 or scrambled control and treated these cells with rapamycin. As expected, rapamycin treatment decreased the phosphorylation of 4EBP relative to control cells (Figure 3D, compare lanes 1–3 with 7–9). Additionally, siRNAs against 4EBP1 and 4EBP2 substantially reduced 4EBP protein levels relative to control siRNAs (Figure 3D, compare lanes 1–3 and 7–9 with lanes 4–6 and 10–12). RT-qPCR revealed that rapamycin significantly decreased SRSF6 PTC-containing mRNA isoforms relative to total SRSF6 mRNA in cells transfected with control siRNA, but had no effect on levels of 4EBP-depleted cells (Figure 3E). Taken together, these data demonstrate that rapamycin decreases levels of PTC-containing isoforms in a UPF1- and 4EBP-dependent manner, suggesting a role for rapamycin in augmentation of NMD through 4EBP-mediated modulation of translation.
Rapamycin decreases levels of PTC-containing isoforms bound to polyribosomes
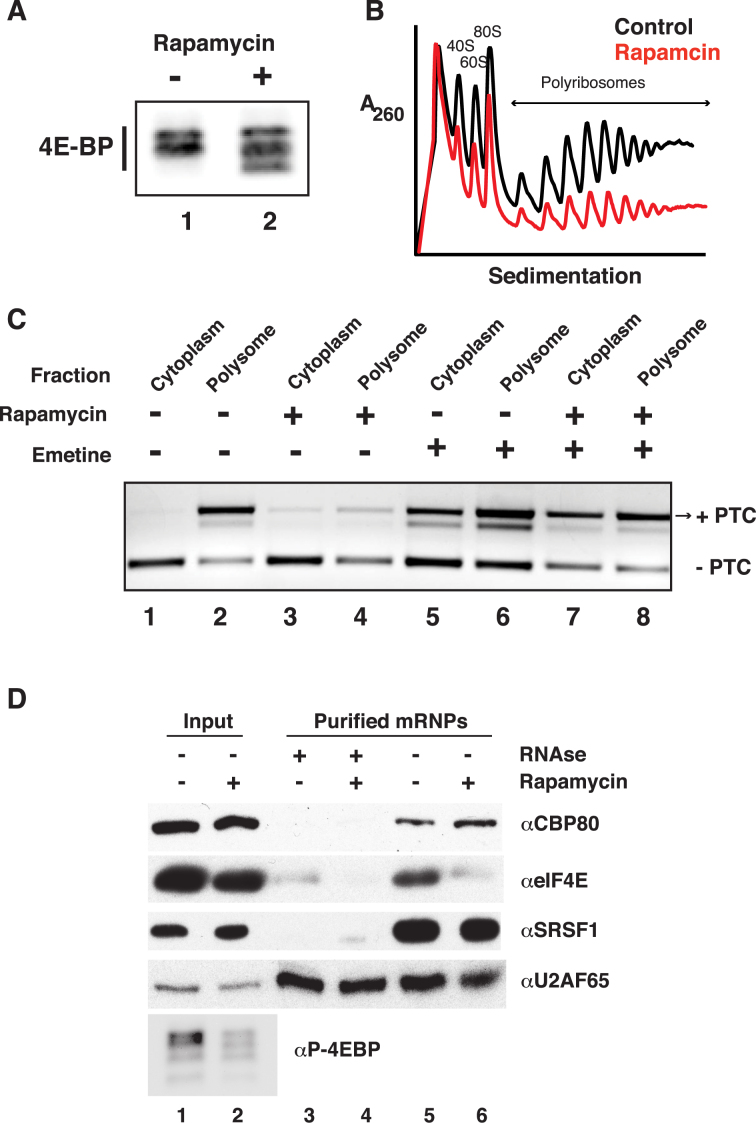
NMD can happen not only during pioneer translation but also during steady translation, given that eIF4E-bound mRNAs are substrates of NMD (42). Moreover, Durand et al. demonstrated that eIF4F-bound transcripts are susceptible to NMD (43). A prediction from these experiments is that NMD efficiency of polyribosome-associated mRNAs may be dynamically regulated. Given that NMD in mammals is translation-dependent (9,44–46), that rapamycin inhibits eIF4E-dependent translation of capped mRNAs, and that rapamycin effects are dependent on the presence of 4EBPs (Figure 3E), we hypothesized that rapamycin-treatment might enhance the decay of polyribosome-bound PTC-containing transcripts. We investigated this hypothesis in cells treated with or without 22nM rapamycin. As expected, rapamycin-treatment increased the electrophoretic mobility of 4EBP on SDS-PAGE relative to control cells (Figure 4A), indicative of 4EBP hypophosphorylation. We then isolated polyribosomes from control or rapamycin treated cells. We observed that rapamycin treated cells showed the distinctive characteristics of global translational inhibition when compared to control cells: an increase in the monosomal fraction (80S, peak) and a decrease of the polyribosome fractions (Figure 4B shows representative polyribosome profiles of rapamycin versus control cells). We then analyzed the presence of PTC-containing and non-PTC isoforms in the polyribosomal fractions, excluding the 80S monosomal peak. In control cells we observed an enrichment of the SRSF6 PTC-containing isoform in the polyribosome-associated mRNA fraction as compared to total cytoplasmic mRNAs (Figure 4C, compare lanes 1 and 2). Rapamycin-treatment dramatically reduced levels of the PTC-containing SRSF6 isoform in polyribosome-associated mRNA fractions (Figure 4C, compare lanes 2 and 4). To rule out alternative splicing as the underlying cause of PTC-containing isoform depletion from polyribosomes in rapamycin-treated cells, we stimulated cells with both rapamycin and afterwards with the translation elongation inhibitor emetine. We expected that if rapamycin effects were splicing dependent, then emetine treatment would not alter the levels of PTC-containing isoforms observed in polyribosomal-bound fractions between control and rapamycin treated cells. As expected, we observed that the effect of rapamycin was attenuated by emetine (compare lanes 6 and 8 in Figure 4C). Taken together these data suggest that rapamycin alters the stability of PTC-containing isoforms in polyribosome complexes rather than the AS of their pre-mRNA.
Figure 4.
Rapamycin alters the levels of polyribosome associated PTC-containing mRNA isoforms. (A) Representative western blot of 4EBP levels from control or rapamycin-treated HEK 293T cells. (B) Representative 260 nm UV absorbance across 15–45% sucrose gradients of cytoplasmic extracts from cells treated with or without rapamycin (n = 3). (C) Endpoint RT-PCR analysis of PTC+/− SRSF6 mRNA isoforms in either cytoplasmic (T) or polyribosome associated (P) extracts treated with or without rapamycin and emetine. (D) Western blot analysis of proteins co-purified with poly A+ RNA from cytoplasmic extracts of control or rapamycin-treated cells.
We further assayed the abundance of PTC-containing and non-PTC isoforms of SRSF6 and CCAR1 in total and polyribosome-bound mRNA fractions at 7 h post-rapamycin stimulation. Rapamycin significantly diminished the presence of the PTC-containing isoforms in polyribosomal fractions of both SRSF6 and CCAR1 (Supplementary Figure S4) genes whereas the effects were not detectable when comparing the total pool of mRNAs. Together these data suggest that rapamycin diminishes the abundance of PTC-containing transcripts through a translation-dependent mechanism.
Previous studies demonstrated that rapamycin treatment induces nuclear accumulation of eIF4E through de-phosphorylation and de-repression of 4EBP (17,21). We confirmed these data in HEK 293 T cells: upon glutamine starvation we observed rapamycin-dependent modest nuclear accumulation of eIF4E whereas it localized predominantly to the cytoplasm in control cells. In contrast, in rapamycin-treated cells we observed strong accumulation of eIF4E in the nucleus (Supplementary Figure S5). Our previous results suggest a role for rapamycin in the degradation of PTC-containing isoforms that is 4EBP- dependent and correlates with polyribosome binding. Given that rapamycin modulates translation of both eIF4E- and CBC- bound transcripts (21,22), and because rapamycin treatment caused nuclear sequestration of eIF4E in our experiments, we hypothesized that exchange of cap binding proteins on mRNAs may also be dynamically modulated by rapamycin. We thus purified cytoplasmic mRNPs from control or rapamycin-treated cells and assayed eIF4E and CBP80 association in an RNA-dependent manner. Figure 4D shows a representative western blot of eIF4E, CBP80, SRSF1 and U2AF65 in whole cell extracts (lanes 1 and 2) and RNase-treated (lanes 3 and 4) or untreated (lanes 5 and 6) eluates from oligo dT cellulose affinity chromatography. We observed that rapamycin-treatment increased the electrophoretic mobility of 4EBP, consistent with de-phosphorylation. With the exception of U2AF65, which binds directly to oligo dT cellulose, SRSF1, CBP80 and eIF4E were all RNase-sensitive (lanes 3–4), suggesting that their binding to oligo dT cellulose is RNA-dependent. Unlike SRSF1 protein, which bound to polyA-mRNAs equally in the presence or absence of rapamycin, CBP80 and eIF4E mRNPs levels were inversely correlated (lanes 5–6). Treatment with rapamycin resulted in a decrease of eIF4E association with polyadenylated mRNPs and a modest but reproducible increase in CBP80 association.
Taken together, our data suggest that rapamycin-treatment impairs the normal exchange of CBP80 with eIF4E on cytoplasmic mRNPs, rendering them more susceptible to nonsense mediated decay and subsequent increased degradation of PTC-containing mRNAs.
DISCUSSION
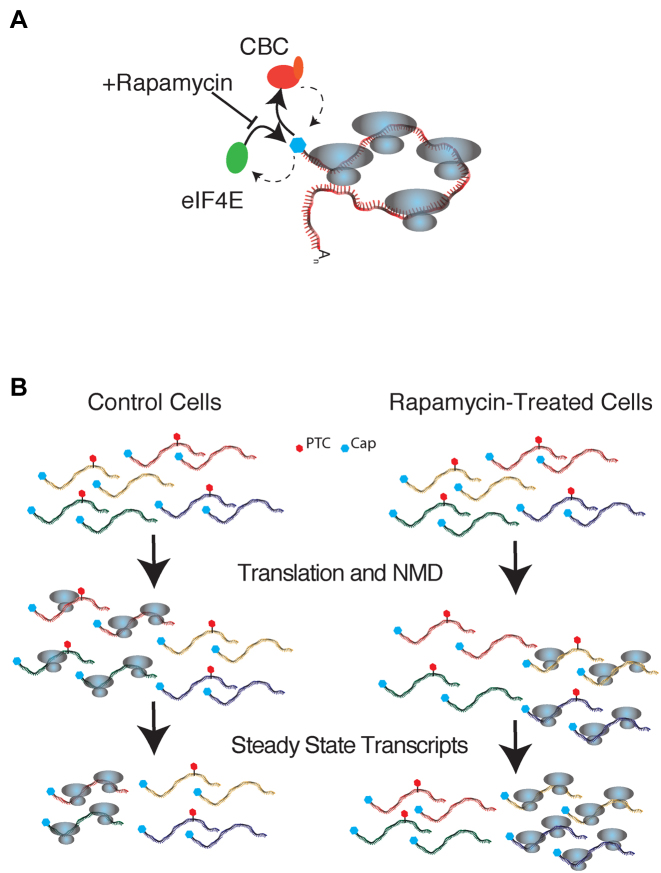
Our data demonstrate a complex relationship between rapamycin, alternative splicing and NMD. We find that rapamycin treatment can increase the abundance of PTC-containing transcripts, but also diminish expression of a significant number of putative NMD substrates. We propose at least two mechanisms that can account for these observations (Figure 5). One possibility is that rapamycin-treatment modulates the exchange of CBC and eIF4E on the caps of cytoplasmic mRNAs. The increased association of CBC with cytoplasmic transcripts observed in Figure 4D may promote decay of PTC-containing transcripts. By contrast, it is possible that rapamycin may influence the competition of transcripts for ribosomes as previously described in budding yeast (47) and favor translation of canonical mRNA isoforms while NMD-sensitive transcripts are degraded. In this case, augmentation of NMD, as observed for transcripts such as SRSF6 and CCAR1, would correlate with up-regulated translation in the presence of rapamycin.
Figure 5.
Our proposed models for rapamycin effects on NMD. Blue hexagons: cap, red hexagons: PTC. (A) Rapamycin decreases association of eIF4E rendering higher association of CBC binding and increased degradation of CBC-bound NMD-transcripts. (B) Rapamycin-dependent changes in translational efficiency for different subsets of transcripts. Rapamycin positively and negatively modulates translation of different transcripts populations. In this model augmentation of NMD is coupled to enhanced translation of NMD-insensitive transcripts in rapamycin-treated cells.
In mammalian cells, nonsense mediated decays occurs predominantly during a pioneering round of mRNA translation (48). The ribonucleoprotein context of this initial round of translation is characterized by the presence of EJC proteins, PABPN1 and the CBC (5,6). Translating ribosomes remodel the mRNP by displacing EJC factors (8,9,49). Our work demonstrates that rapamycin influences the composition of cytoplasmic mRNPs by increasing the association of CBC with cytoplasmic polyadenylated transcripts while concomitantly decreasing eIF4E association (Figure 4D). These findings are consistent with previous work showing that the mTOR inhibitors decrease the association of the eIF4F components eIF4A and eIF4G with immobilized m7G cap in mammalian cells (50). Because CBC stimulates NMD activity (18) it may be possible that elevated CBC levels on cytoplasmic mRNPs could augment NMD. Thus, rapamycin could promote a pioneer-like context for some cytoplasmic mRNPs by maintaining CBC association.
NMD is dependent upon translation and can occur in association with polyribosomes (44,51). In yeast, rapamycin has complex impacts on the association of mRNAs with polyribosomes (47). Whereas the polyribosome association of a majority of transcripts is only modestly affected by rapamycin, there is a significant fraction exhibiting increased or decreased association. In many cases these changes are positively correlated with changes in steady state mRNA levels, suggesting coupling of transcriptional and translational responses to rapamycin (47). Our data cannot exclude the possibility that augmentation of NMD by rapamycin may occur because transcripts from specific genes (such as SRSF6 or CCAR1) are preferentially associated to polyribosomes in rapamycin-treated cells relative to control. Likewise, mRNA isoforms with reduced NMD efficiency may correspond to transcripts that are translationally compromised by rapamycin-treatment. These data could be consistent with a model in which rapamycin influences competition between transcripts for the translation machinery. A similar trans-competition control mechanism has been described for rapamycin-dependent splicing regulation in yeast (52).
Rapamycin's effects on NMD may also rely on post-translational modifications (such as phosphorylation) of NMD factors in addition to augmentation of CBC-binding to mRNPs (Figure 4D). Work by Pal et al. has shown that phosphorylation of UPF1 is sensitive to rapamycin (53); however the effect of lowered UPF1 phosphorylation on PTC-containing mRNAs was not tested. Moreover, although rapamycin concentration was similar to our working dose (25nM and 22nM, respectively), cells were only exposed for a short time (30min vs 72h in our case). Similarly, Yamashita et al. (54) showed no effect of rapamycin on a β-globin reporter but exposure time (2 h) and doses (60 nM and 600 nM) were significantly different to those employed in our system. Importantly, our RNA-seq revealed that the mRNA of only one NMD factor, SMG6 gene, was up-regulated by exposure to rapamycin. We do not exclude the possibility that prolonged exposure to rapamycin may result in phosphorylation changes in some of NMD factors (or their regulators), and that these, in addition to mRNP remodeling, may contribute to the overall effect that we observe in our system.
We propose that augmentation of NMD by rapamycin could have important implications for cell biology and physiology. Given that rapamycin increases lifespan in a wide variety of model systems, including mammals (19,20), it will be important to determine how rapamycin modulates NMD efficiency and if this may affect ageing-related processes, for example, its potential role in telomere protection given the presence of NMD factors that protect chromosomes from telomere loss (55). If rapamycin does indeed augment NMD then perhaps improved transcript surveillance could facilitate degradation of potentially deleterious mRNAs, similarly to recent work suggesting that RNA decay mechanisms and ribosomes are central to degrading oxidized RNA (56), linked to neurodegenerative diseases (57). We believe our study presents novel experimental evidence for the role of rapamycin in NMD and that further exploration may give insight into RNA metabolism by extracellular signals in a wide set of biological contexts.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Javier Caceres, Luiz Penalva, Alan Zahler and Aishwarya Griselda Jacob for critical input to the manuscript. The authors also thank Prof. A.E. Kulozik for kindly providing pCI-Renilla/β-globin wt (WT) or NS39 (MUT) to perform the luciferase experiments.
Footnotes
Present address: Rocio Teresa Martinez-Nunez, Clinical and Experimental Sciences, Faculty of Medicine, University of Southampton, Southampton, Hampshire SO16 6YD, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health and Ellison Medical Research Foundation [GM109146, AG042003 and AG-NS-0623-09 to J.R.S.]; AAIR Charity (to RTMN); RTMN was supported by Post-doctoral Career track award from the Faculty of Medicine, University of Southampton; Biotechnology and Biological Sciences Research Council (BBSRC) [BB/L007576/1 to J.B. and K.D.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Popp M.W., Maquat L.E.. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 2013; 47:139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang L., Lou C.H., Chan W., Shum E.Y., Shao A., Stone E., Karam R., Song H.W., Wilkinson M.F.. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell. 2011; 43:950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O'Brien G., Shiue L., Clark T.A., Blume J.E., Ares M. Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007; 21:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J.. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008; 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- 5. Ishigaki Y., Li X., Serin G., Maquat L.E.. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001; 106:607–617. [DOI] [PubMed] [Google Scholar]
- 6. Le Hir H., Izaurralde E., Maquat L.E., Moore M.J.. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000; 19:6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lejeune F., Ishigaki Y., Li X., Maquat L.E.. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002; 21:3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dostie J., Dreyfuss G.. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 2002; 12:1060–1067. [DOI] [PubMed] [Google Scholar]
- 9. Sato H., Maquat L.E.. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 2009; 23:2537–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagy E., Maquat L.E.. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998; 23:198–199. [DOI] [PubMed] [Google Scholar]
- 11. Chiu S.Y., Lejeune F., Ranganathan A.C., Maquat L.E.. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004; 18:745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S.. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006; 20:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohnishi T., Yamashita A., Kashima I., Schell T., Anders K.R., Grimson A., Hachiya T., Hentze M.W., Anderson P., Ohno S.. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell. 2003; 12:1187–1200. [DOI] [PubMed] [Google Scholar]
- 14. Eberle A.B., Stalder L., Mathys H., Orozco R.Z., Muhlemann O.. Posttranscriptional gene regulation by spatial rearrangement of the 3΄ untranslated region. PLoS Biol. 2008; 6:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toma K.G., Rebbapragada I., Durand S., Lykke-Andersen J.. Identification of elements in human long 3΄ UTRs that inhibit nonsense-mediated decay. RNA. 2015; 21:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dias S.M., Wilson K.F., Rojas K.S., Ambrosio A.L., Cerione R.A.. The molecular basis for the regulation of the cap-binding complex by the importins. Nat. Struct. Mol. Biol. 2009; 16:930–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rong L., Livingstone M., Sukarieh R., Petroulakis E., Gingras A.C., Crosby K., Smith B., Polakiewicz R.D., Pelletier J., Ferraiuolo M.A. et al. Control of eIF4E cellular localization by eIF4E-binding proteins, 4E-BPs. RNA. 2008; 14:1318–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hosoda N., Kim Y.K., Lejeune F., Maquat L.E.. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005; 12:893–901. [DOI] [PubMed] [Google Scholar]
- 19. Kim S.G., Buel G.R., Blenis J.. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol. Cells. 2013; 35:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laplante M., Sabatini D.M.. mTOR signaling in growth control and disease. Cell. 2012; 149:274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gingras A.C., Raught B., Gygi S.P., Niedzwiecka A., Miron M., Burley S.K., Polakiewicz R.D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N.. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001; 15:2852–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma X.M., Yoon S.O., Richardson C.J., Julich K., Blenis J.. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008; 133:303–313. [DOI] [PubMed] [Google Scholar]
- 23. Hu W., Petzold C., Coller J., Baker K.E.. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2010; 17:244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boelz S., Neu-Yilik G., Gehring N.H., Hentze M.W., Kulozik A.E.. A chemiluminescence-based reporter system to monitor nonsense-mediated mRNA decay. Biochem. Biophys. Res. Commun. 2006; 349:186–191. [DOI] [PubMed] [Google Scholar]
- 25. Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E.. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007; 446:926–929. [DOI] [PubMed] [Google Scholar]
- 26. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L.. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015; 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kahles A., Ong C.S., Zhong Y., Ratsch G.. SplAdder: identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics. 2016; 32:1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995; 289–300. [Google Scholar]
- 31. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katz Y., Wang E.T., Silterra J., Schwartz S., Wong B., Thorvaldsdottir H., Robinson J.T., Mesirov J.P., Airoldi E.M., Burge C.B.. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics. 2015; 31:2400–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A.. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Croft D., Mundo A.F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M.R. et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014; 42:D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016; 44:D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015; 43:D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. White E.S., Sagana R.L., Booth A.J., Yan M., Cornett A.M., Bloomheart C.A., Tsui J.L., Wilke C.A., Moore B.B., Ritzenthaler J.D. et al. Control of fibroblast fibronectin expression and alternative splicing via the PI3K/Akt/mTOR pathway. Exp Cell Res. 2010; 316:2644–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hurt J.A., Robertson A.D., Burge C.B.. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013; 23:1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leeds P., Peltz S.W., Jacobson A., Culbertson M.R.. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991; 5:2303–2314. [DOI] [PubMed] [Google Scholar]
- 41. Bulliard Y., Wiznerowicz M., Barde I., Trono D.. KRAB can repress lentivirus proviral transcription independently of integration site. J. Biol. Chem. 2006; 281:35742–35746. [DOI] [PubMed] [Google Scholar]
- 42. Rufener S.C., Muhlemann O.. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2013; 20:710–717. [DOI] [PubMed] [Google Scholar]
- 43. Durand S., Lykke-Andersen J.. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat. Struct. Mol. Biol. 2013; 20:702–709. [DOI] [PubMed] [Google Scholar]
- 44. Belgrader P., Cheng J., Maquat L.E.. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang S., Welch E.M., Hogan K., Brown A.H., Peltz S.W., Jacobson A.. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997; 3:234–244. [PMC free article] [PubMed] [Google Scholar]
- 46. Hu W., Sweet T.J., Chamnongpol S., Baker K.E., Coller J.. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009; 461:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Preiss T., Baron-Benhamou J., Ansorge W., Hentze M.W.. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat. Struct. Biol. 2003; 10:1039–1047. [DOI] [PubMed] [Google Scholar]
- 48. Maquat L.E., Tarn W.Y., Isken O.. The pioneer round of translation: features and functions. Cell. 2010; 142:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gehring N.H., Lamprinaki S., Hentze M.W., Kulozik A.E.. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009; 7:e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tcherkezian J., Cargnello M., Romeo Y., Huttlin E.L., Lavoie G., Gygi S.P., Roux P.P.. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5΄TOP mRNA translation. Genes Dev. 2014; 28:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Atkin A.L., Altamura N., Leeds P., Culbertson M.R.. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell. 1995; 6:611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Munding E.M., Shiue L., Katzman S., Donohue J.P., Ares M. Jr. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol. Cell. 2013; 51:338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pal M., Ishigaki Y., Nagy E., Maquat L.E.. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001; 7:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamashita A., Ohnishi T., Kashima I., Taya Y., Ohno S.. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001; 15:2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 56. Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S.. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014; 9:1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J., Perry G., Smith M.A., Robertson D., Olson S.J., Graham D.G., Montine T.J.. Parkinson's disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am. J. Pathol. 1999; 154:1423–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.