Abstract
An interplay between the nucleosome binding proteins H1 and HMGN is known to affect chromatin dynamics, but the biological significance of this interplay is still not clear. We find that during embryonic stem cell differentiation loss of HMGNs leads to down regulation of genes involved in neural differentiation, and that the transcription factor OLIG2 is a central node in the affected pathway. Loss of HMGNs affects the expression of OLIG2 as well as that of OLIG1, two transcription factors that are crucial for oligodendrocyte lineage specification and nerve myelination. Loss of HMGNs increases the chromatin binding of histone H1, thereby recruiting the histone methyltransferase EZH2 and elevating H3K27me3 levels, thus conferring a repressive epigenetic signature at Olig1&2 sites. Embryonic stem cells lacking HMGNs show reduced ability to differentiate towards the oligodendrocyte lineage, and mice lacking HMGNs show reduced oligodendrocyte count and decreased spinal cord myelination, and display related neurological phenotypes. Thus, the presence of HMGN proteins is required for proper expression of neural differentiation genes during embryonic stem cell differentiation. Specifically, we demonstrate that the dynamic interplay between HMGNs and H1 in chromatin epigenetically regulates the expression of OLIG1&2, thereby affecting oligodendrocyte development and myelination, and mouse behavior.
INTRODUCTION
Chromatin dynamics play an essential role in regulating cell differentiation and tissue development. Dynamic interplay among chromosome architecture proteins, histone modifiers, nucleosome remodelers and transcription factors in chromatin establishes and maintains proper epigenetic landscape and ensures the integrity and accuracy of tissue specific transcription profiles. Histone H1 is the most abundant family of chromatin binding proteins; in almost all vertebrate nuclei most of the nucleosomes contain one molecule of H1. The binding of H1 to nucleosomes, which is dynamic and independent DNA sequence specificity, affects chromatin organization and gene expression (1–4). Similarly, HMGN is a family of chromatin binding proteins that is ubiquitously expressed in all vertebrate cells and binds dynamically to nucleosome core particles, the building block of the chromatin fiber, without any specificity for the underlying DNA sequence (5,6). Genome wide studies revealed that HMGNs play a role in the establishment and maintenance of DNase I hypersensitive sites, the hallmark of regulatory sites in chromatin, and also can affect the level of posttranslational modifications in histone tails (7–10). Imaging analyses of living cells and in vitro studies, showed that HMGN variants compete with linker H1 for chromatin binding sites and function within a dynamic network of interdependent chromatin interactions in which the binding of one variant affects the binding of other members of the protein network (11–13).
Appropriate myelination of the central nervous system (CNS) is essential for correct neural development and function (14,15). Myelination is critically dependent on the differentiation of the oligodendrocyte cell lineage that originates from neural precursor cells which are derived from embryonic ectoderm (16). Oligodendrocyte lineage differentiation is regulated by several transcription factors including OLIG1 and OLIG2, which play an essential role in both embryonic and adult oligodendrocyte development and CNS myelination (17,18). While the central role of OLIG1&2 in oligodendrocyte differentiation is well established (19–21), the processes regulating the expression of these two transcription factors are not fully known. Here we demonstrate that interplay between H1 and HMGN proteins affects OLIG1&2 expression and oligodendrocyte differentiation.
Pluripotent ESCs can be induced to form embryoid bodies (EBs), which are three-dimensional aggregates of differentiating ESCs undergoing cell specification along all the three germ layers: ectoderm, endoderm, and mesoderm (22). EBs provide a starting point to generate lineage-specific progenitor cells, including ectoderm derived neural progenitors (NPs), which can be induced to further differentiate into OLIG1&2 positive oligodendrocyte precursor cells (OPCs) (23,24). Such developmental changes, from ESCs to lineage specific cells, are known to involve significant changes in chromatin organization and transcription (25,26). Thus, during differentiation, the highly de-compacted and dynamic chromatin structure of pluripotent ESCs (27,28) is remodeled into more compact states that express lineage specific transcription profiles (29). In view of these global changes in genome organization, it is possible that nucleosome-binding proteins known to affect chromatin organization and function, such as histone H1 variants and members of the high mobility group N (HMGN), (1,3,5,30) could affect differentiation processes.
In this study, we prepared ESCs from Hmgn1/Hmgn2 double knock-out (DKO) mice, lacking the two major HMGN variants, HMGN1 and HMGN2, which bind redundantly to chromatin (9). We differentiated ESCs obtained from DKO and wild type (WT) mice into EBs and tested for differences in the transcription profile between the two genotypes, throughout eight days of EB differentiation. We found that loss of HMGN1 and HMGN2 altered the expression of genes involved in neural lineage differentiation and significantly reduced the expression level of OLIG1 and OLIG2, at every time point during EB differentiation. We focused on these two genes and discovered that their decreased expression in ESCs lacking HMGNs is due to increased chromatin binding of histone H1, which leads to epigenetic changes around the Olig1 and Olig2 locus that leads to a decreased expression of these two genes. As a consequence, the oligodendrocyte development is affected both in vitro and in vivo. The differentiation of NPs into OPCs is impaired, and HMGNs DKO mice show decreased oligodendrocytes and a reduction in myelin sheath components in their spinal cords, and display neurological phenotypes indicative of defects in myelin sheath.
MATERIALS AND METHODS
Mice, ES cell culture and in vitro differentiation
Mice were generated and maintained according to approved NIH protocols as previously described (9). Neurological phenotype screening of 15 male and 15 female DKO mice and their age and genetically matched WT controls, a total of 30 mutant and 30 control mice for each mouse line, were performed at the German Mouse Clinic using a standard battery of tests (31–34). The detailed phenotypic analysis of the mice is described in Supplemental Materials and Methods and at the German Mouse Clinic website (www.mouseclinic.de).
Hmgn1+/+N2+/+(WT) and Hmgn1−/− n2−/− (DKO) ES cell lines were derived from 3.5 days blastocysts from WT and DKO mice we previously generated (9). Three independent ESC clones (biological replicates) for each genotype were used for analysis. The ES cells were co-cultured with mitomycin-c treated mouse primary embryonic fibroblast (MEFs) feeders (Millipore) or cultured under feeder-free conditions. They were maintained in Knock-out Dulbecco's modified Eagle's medium (KO-DMEM, Invitrogen), with 20% serum replacement (SR, Invitrogen), 0.055 mM β-mercaptoethanol (Sigma), 2 mM l-glutamine (Invitrogen), 0.1 mM MEM non-essential amino acid (Invitrogen), 5000 U/ml penicillin/streptomycin (Invitrogen), 1000 U/ml LIF (Millipore) and MEK/GSK3 inhibitors (2i) (Millipore). In vitro differentiation of ES cells along oligodendrocyte lineage was performed using the Mouse Oligodendrocyte Differentiation Kit (R&D System) according to the manufacturer's instruction. Briefly, ESCs were differentiated into EBs in hanging drops, 4-day EBs were attached to Fibronectin coated dishes, and differentiated into NPs using insulin/transferrin/selenium/fibronectin (ITSF) supplemented medium (35). The selected NPs were differentiated into OPCs by growing in N-2 medium supplemented with a combination of growth factors including hFGF, hEGF and hPDGF-AA (24).
ESCs transfection
For siRNA knock-down, 25 nM ON-TARGET SMARTpool siRNA specifically targeting mouse Hmgn1 or Hmgn2 (GE Dharmacon) were transfected into ESCs using Lipofectamine RNAiMAX reagent (Invitrogen). Cells were split and transfected again after first 48 h, and harvested after another 48 h. For HMGN1/2 rescue experiments, 1 μg pCI plasmid vectors (Promega) carrying the expression cassette of human HMGN1 or HMGN2 or HMGN1SE mutant were transiently transfected into DKO ESCs using Mouse ES Cell Nucleofector Kit (Lonza) according to the manufacturer's instruction. Cells were harvested after 48 h for western blot and ChIP assay.
Antibodies
Rabbit affinity pure polyclonal anti-mouse HMGN1, anti-human HMGN1, anti-human HMGN2 and anti-calf histone H1 antibodies were generated in our laboratory (36). Anti-OLIG1, anti-OLIG2 and anti-Nestin antibodies were purchased from Millipore. Anti-H3K27me3, anti-EZH2, anti-H3, anti-PGDFR-α, anti-CNPase, anti-PLP, anti-MBP and anti-β-actin antibodies were purchased from Abcam. HRP-conjugated secondary antibodies for western blots were purchased from Pierce. Fluorescence secondary antibodies for immunostaining were purchased from Invitrogen.
Western blot
Whole cell or tissue lysates were prepared in 1× SDS PAGE sample buffer (Bio-Rad) supplemented with protease inhibitors (Roche Applied Science). The samples were fractionated on 4–20% pre-cast Criterion gels, transferred by semi-dry method to PVDF membrane, blocked with non-fat milk in TBST and probed with indicated antibodies. Chemiluminiscent detection using ECL Plus has been performed according to Amersham's recommendations. The quantitation has been performed using ChemiDoc MP System (BioRad Laboratories).
Immunostaining
For immunofluorescence staining, cells were cultured on four-well Millicell EZ slides (Millipore), and 10 μm frozen spinal cord cross sections at upper limb level were prepared from 8-weeks old WT and DKO mice. Immunofluorescence staining was performed using indicated antibodies as previously described (8). The images were taken using Zeiss LSM 710 confocal system and were processed using Zeiss software.
RNA-seq
Total RNA from ESCs or embryoid bodies was isolated by RNeasy Mini Kit (Qiagen) followed by ‘on-column’ DNase I treatment. mRNA-seq libraries were prepared from 1 μg of total RNA using the Illumina TruSeq RNA Sample Preparation Kit, following the manufacturer's instructions. Libraries were sequenced on the Illumina HiSeq 2000 with 2 × 100 bp paired-end reads. The data analysis was performed using the RNA-seq pipeline in Partek Genome Suite. A gene was considered expressed if reads per kilobase of transcript model per million mapped reads (RPKM) was ≥1.0. |Fold Change| ≥ 1.5 with P-value <0.05 was considered significantly differentially expressed between WT and DKO. Gene ontology analysis was performed using DAVID online software (https://david.ncifcrf.gov/). Genomatix Pathway System (GePS, Genomatix GmbH) was used to generate a simple network using the following criteria: organism—Mus musculus; annotation type—biological processes; subcategory—nervous system development.
ChIP-seq and ChIP-qPCR
ESCs were crosslinked directly in culture medium with 1% formaldehyde solution for 10 min at room temperature. Chromatin was fragmented to 200–300 bp by 10 cycles of sonication (30 s on/30 s off) (Bioruptor). Chromatin from 2 × 107 cells and 10 μg antibodies against HMGN1, HMGN2, H3K27me3 or EZH2 were used for each ChIP experiment. For ChIP-seq, libraries were prepared using Illumina TruSeq ChIP Sample Prep Kit with compatible indexed adaptors according to manufacturer's instruction. Equal amounts of 4–6 indexed library samples were pooled for cluster generation and sequenced on Illumina HiSeq2000 system with 2 × 100 bp paired-end reads. Data analysis was performed as previously described (9). For ChIP-qPCR, DNA purified from ChIP assays were applied for quantitative PCR reactions with Power SYBR Green Mix (Applied Biosystems) using the AB 7900HT Fast Real Time PCR system, and data were normalized to input DNA.
In vitro HMT assay
Recombinant HMGN were obtained as in (37), recombinant calf H3 was purchased from Boeringer Mannheim (Cat #84936921), and polynucleosome fraction devoid of H1/H5 was purified from chicken erythrocytes as described (38). The free histone or polynucleosome preparations were used as substrates for methylation by EZH2 complex. EZH2 (EZH2/EED/SUZ12/RbAp48/AEBP2) complex, FLAG-tag, His-tag was purchased from BPS Bioscience (Cat# 51004). Methylation assay has been carried out as in (39) with modifications. Briefly, methylation reaction was performed in a 20-μl volume containing either 1 μg H3 or 10 μg polynucleosomes, various amounts of H1 and HMGN, 20 mM phosphate buffer pH 7.4, 0.05% Tween-20, 20 μM S-adenosylmethionine as a methyl donor, and 200 ng EZH2 complex. Reaction mixture was incubated for 1 h at 30°C and terminated by the addition of SDS-PAGE sample buffer. H3K27me3 modification levels were evaluated by western blot.
Statistical analysis
Statistical analyses were performed using two-tailed equal variance Student's t-test in GraphPad Prism software and P < 0.05 was considered as statistically significant. Data are presented as mean ± SEM.
Data access
The next-generation sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRP068453.
RESULTS
Loss of HMGNs reduces the expression of neural lineage differentiation genes
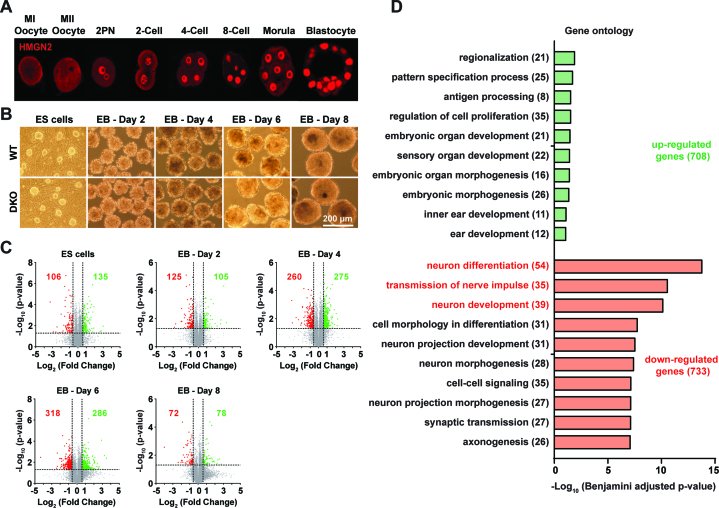
HMGN1 and HMGN2, the major variants of the HMGN protein family (40), compete for nucleosome binding sites and have similar effects on chromatin structure (9,12). HMGN1 was shown to be highly expressed throughout all mouse preimplantation stages (8). We now find that likewise, the HMGN2 variant is also highly expressed during early mouse development and can be detected at all preimplantation stages examined, from oocytes to blastocysts (Figure 1A). In view of this finding and given the functional redundancy between HMGN1 and HMGN2 (9), we established ESCs from wild type (WT) and Hmgn1−/−/n2−/−, DKO mice, and tested whether the combined loss of both HMGN1 and HMGN2 affects the differentiation potential of the ESCs.
Figure 1.
Loss of HMGNs down regulates the expression of genes involved in neural lineage differentiation. (A) HMGN2 protein expression during mouse embryonic preimplantation. Shown are immunofluorescence images at the stages indicated above each panel. (B) Morphology of WT and DKO ESCs colonies and differentiating embryoid bodies (EBs). (C) The number of significantly (fold change ≥ 1.5, P < 0.05) up (green) or down (red) regulated genes between WT and DKO ESCs and EBs at indicated stages. RNA-seq data were obtained from three independent ESC clones (biological replicates). (D) Gene ontology (GO) categories enriched for up (green) or down (red) regulated genes during ESCs differentiation. Number of changed genes in each category is shown in brackets.
The WT and DKO ESCs colonies showed clear borders, were indistinguishable in their morphological appearance (Figure 1B), and expressed similar levels of the pluripotent marker genes such as Nanog, Pou5f1 (Oct3/4), Klf4, Sox2 and Esrrb (Supplementary Figure S1, and deposited data), suggesting that loss of HMGNs did not significantly affect their pluripotency. Indeed, upon removal of LIF/2i and cultured in suspension, the WT and DKO ESCs formed morphologically indistinguishable EBs (Figure 1B). RNA-seq analysis of the differentiating ESCs, at two day intervals up to 8 days, showed that induction of differentiation leads to a rapid decrease in the levels of pluripotency marker genes and up regulation of the genes that mark the three germ layers (41), such as Nestin, Pax6 and Sox3 for ectoderm, T, Wt1 and Mixl1 for mesoderm, and Sox17, Foxa2 and Lama1 for endoderm (Supplementary Figure S1, and deposited data), indicating that loss of HMGNs does not affect the ability of the ESCs to differentiate into the major germ layers.
However, complete analyses of the transcriptomes, using three biological replicates for each differentiation point (Supplementary Figure S2) revealed differences in gene expression between WT and DKO cells. Depending on the differentiation stage, 150–600 genes showed significantly altered expression levels between WT and DKO lines, with a total of 708 genes up regulated and 733 genes down regulated (Figure 1C). Notably, GO analysis of the 708 up-regulated genes did not show significant enrichment in any specific GO category, but the 733 down-regulated genes showed highly significant enrichment in several categories, with the top three (Benjamini adjusted P-value < 1 ×10−10) classified as neural lineage related categories, including ‘neuron differentiation’, ‘transmission of nerve impulse’ and ‘neuron development’ (Figure 1D). Thus, loss of HMGN1 and HMGN2 may alter the potential of ESCs to differentiate towards the neural lineage.
HMGNs are positive regulators of OLIG1 and OLIG2 expression
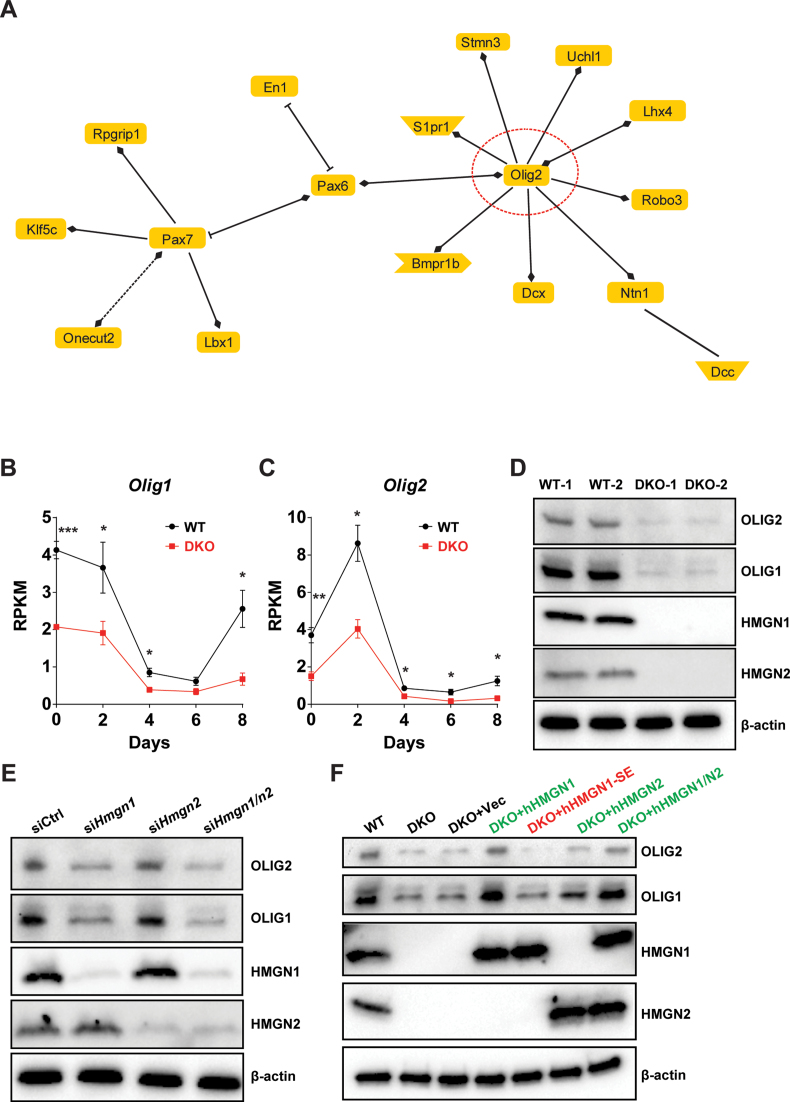
To gain insights into the functional relationship among the 54 down-regulated genes in the ‘neuron differentiation pathway’, which is the most significantly pathways down regulated due to loss of HMGNs (Figure 1D), we generated a simple network using Genomatix Pathway System (GePS). The gene regulatory network generated by this analysis suggests that the transcription factor OLIG2 is a central node in this pathway (Figure 2A). Furthermore, our RNA-seq data shows that the transcription of both Olig1 and Olig2, two transcription factors essential for oligodendrocyte lineage development and commitment (14,19), are significantly down-regulated in DKO, throughout the entire differentiation process, from ESCs to EBs (Figure 2B and C). Indeed, western blots verify that the protein levels of these two transcription factors are also significantly down-regulated in DKO ESCs (Figure 2D). Furthermore, siRNA mediated down-regulation of either HMGN1, or both HMGN1 and HMGN2, in WT ESCs verifies that the down regulation of Olig1&2 expression is indeed due to loss of HMGN expression (Figure 2E). In addition, rescue experiments in DKO cells show that re-expression of only HMGN1, or only HMGN2, or both these HMGNs in DKO cells restores OLIG1 and OLIG2 expression to levels comparable to those seen in WT cells (Figure 2F). Significantly, re-expression of the mutant protein HMGN1-S20,24E (HMGN1-SE) that cannot bind to nucleosomes (42,43), fails to rescue OLIG1&2 expression, evidence that HMGNs regulates OLIG1&2 expression by binding to chromatin (Figure 2F). Thus, the RNA-seq and western analyses, together with the siRNA mediated down-regulation and with the rescue experiments, indicate that the binding of HMGNs to chromatin facilitates the expression of OLIG1 and OLIG2, two transcription factors known to regulate oligodendrocyte differentiation.
Figure 2.
HMGNs regulate the expression of OLIG1 and OLIG2 in ESCs. (A) Regulatory network of genes down-regulated during DKO ESC differentiation and attributed to ‘Neuron Differentiation’ GO category. (B and C) RNA-seq analysis of Olig1&2 expression during ESCs differentiation. Data were obtained from three independent ESC clones (biological replicates) and presented as mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001). (D) Westerns showing OLIG1&2 protein expression in WT and DKO ESCs. (E) Westerns showing siRNA mediated depletion of HMGNs down regulated OLIG&2 expression. (F) Rescue of OLIG1&2 expression by re-introducing either wild type (green) or mutant (red) HMGN in DKO ESCs. Shown are representative blots from at least two independent experiments.
HMGNs modulate EZH2 binding and H3K27me3 levels around Olig1&2 genes
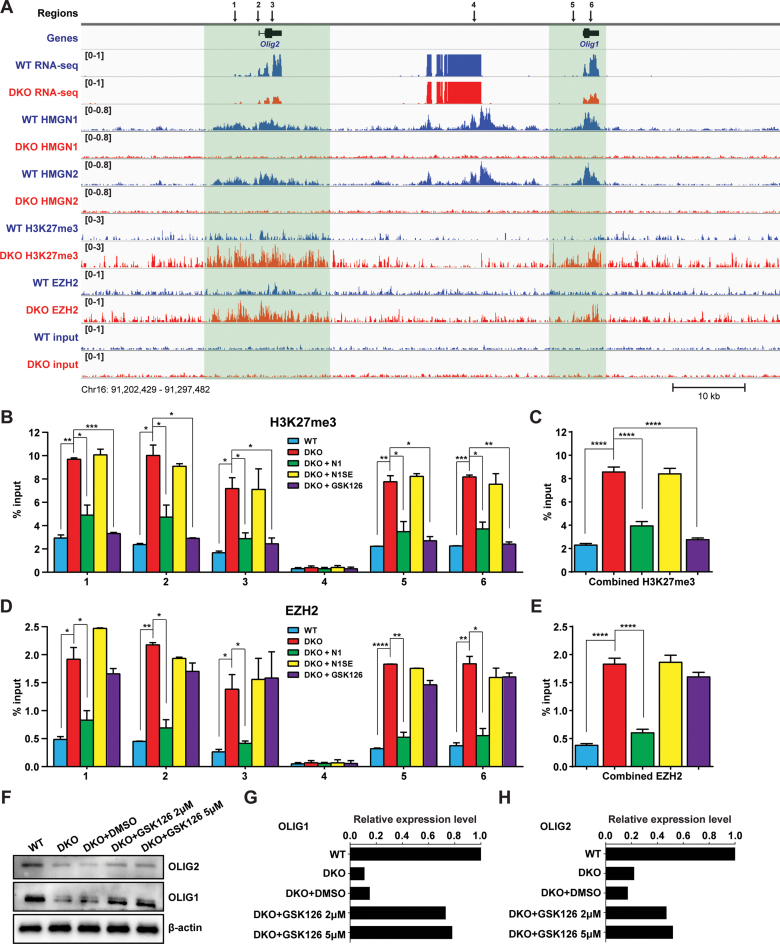
Toward understanding the molecular mechanisms whereby loss of HMGNs down-regulates OLIG1&2 expression, we mapped the genome wide distribution of HMGN1 and HMGN2, as well as several epigenetic marks known to affect gene expression. We found that the occupancy of both HMGN1 and HMGN2 is enriched in the genomic area containing Olig1&2 (Figure 3A). Importantly, in DKO cells, the levels of H3K27me3, an epigenetic mark known to be associated with gene silencing (44) are significantly increased at Olig1&2 (Figure 3A, Supplementary Figure S3). Consistent with this observation, we find that the occupancy of the histone methyltransferase EZH2, the catalytic subunit of Polycomb Repressive Complex 2 (PRC2) which is responsible for H3K27 methylation, is also significantly increased around Olig1&2 genes in DKO ESCs (Figure 3A). Thus, at Olig1&2, loss of HMGN leads to increase EZH2 occupancy and elevation in H3K27me3.
Figure 3.
HMGNs modulate EZH2 binding and H3K27me3 levels around Olig1&2 genes. (A) Genome browser snapshot visualizes decreased mRNA and increased H3K27me3 and EZH2 levels at genic regions encompassing Olig1&2 in DKO ESCs. Numbers in parenthesis indicate the scales of the y-axis, in RPM. Input lanes show number of reads in the absence of antibody. (B–E) ChIP-qPCR analysis of H3K27me3 and EZH2 at the specific loci indicated by arrows in (A). Legend shows cell type. Combined values are shown in C and E. Data were obtained from two independent ESC clones (biological replicates) and presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. IgG controls shown in supplemental Figure 3B. Primers used are listed in Supplementary Table S1. (F) Westerns showing OLIG1&2 expression in DKO ESCs treated with two concentrations of EZH2 specific inhibitor GSK126. (G–H) Quantifications of panel F, normalized to β-actin.
To verify these findings, we performed ChIP-qPCR on six selected regions at and around Olig1&2 genes (marked 1–6 in Figure 3A) in both WT and DKO cells and also in DKO cells transfected with either wild-type or mutant HMGNs. These analyses verified that indeed, in DKO cells the levels of both H3K27me3 levels and EZH2 occupancy are significantly increased in the genomic regions containing Olig1&2 (Figure 3B–E, compare the blue and red bars). Furthermore, fully consistent with the results of the rescue experiments shown in Figure 2E, we find that re-expression of wild-type HMGN1, but not of the mutant HMGN1S20, 24E in DKO ESCs, significantly decreased the binding of EZH2 (Figure 3D and E, compare the red, green and yellow bars) and lowered the level of H3K27me3 (Figure 3B, C compare the red, green and yellow bars). Thus, HMGNs regulate OLIG1/2 expression by modulating the binding of EZH2 to chromatin and the levels of H3K27me3 in nucleosomes. These epigenetic effects are contingent on the ability of HMGN to bind to nucleosomes, since a mutant HMGN that cannot bind to nucleosomes does not affect the binding of EZH2 to chromatin. In addition, treatment of DKO ESCs with GSK126, a known inhibitor of the methyltransferase activity of EZH2 (45) significantly decreases the H3K27me3 levels (Figure 3B, C, compare the red and purple bars) and increased the expression of OLIG1&2 (Figure 3F–H), without affecting the binding of EZH2 (Figure 3D and E, compare the red and purple bars). Thus, the direct cause of the down regulation in Olig1&2 expression levels is the increased levels of H3K27me3, rather than just the binding of EZH2 to chromatin.
HMGNs modulate EZH2-mediated H3K27me3 levels via interplay with linker histone H1
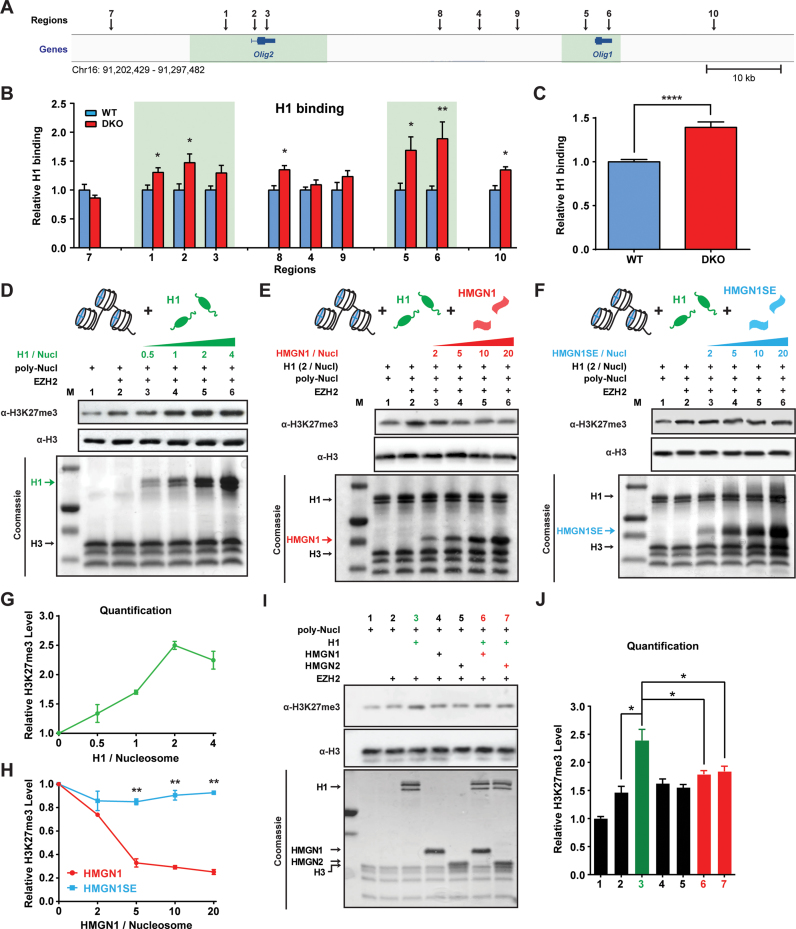
Linker histone H1 was shown to enhance the EZH2 mediated methylation of histone H3 in chromatin (46,47). Given that HMGN proteins compete with H1 for nucleosome binding and counteract the chromatin condensing activity of H1 (1,48), we tested whether an interplay between H1 and HMGNs affects the EZH2 mediated methylation of H3K27. To this end, we first used quantitative ChIP-qPCR to compare the chromatin binding of H1 in WT and DKO ESCs at 10 different regions around the Olig1&2 genes (see Figure 4A). We find that loss of HMGNs increased significantly the chromatin binding of H1, especially in the genomic regions encompassing the Olig1&2 genes (Figure 4B and C). Taken together with the data shown in Figure 3B–E, the results raise the possibility that the increased H3K27me3 at Olig1&2 are due to increased chromatin binding of H1 in DKO cells.
Figure 4.
HMGN histone H1 interplay modulates EZH2-mediated H3K27me3. (A) Genome browser snapshot shows 10 regions (indicated by arrows) near Olig1&2 genes analyzed for histone H1 binding. Sites 1–6 correspond to the same sites in Figure 3. (B) Relative enrichment in H1 occupancy at the indicated sites in DKO cells, normalized to occupancy in WT cells. Data were obtained from two ESC clones (biological replicates) in two independent experiments and presented as mean ± SEM. Primers used are listed in Supplementary Table S1. (C) Average increase in H1 occupancy at the selected sites in DKO chromatin. (D) Histone methyl transfer (HMT) assays, using H1-depleted poly-nucleosomes (poly-Nucl) as substrates show H1 dose dependent increase in EZH2 mediated H3K27me3 levels (E and F) HMT assays show that HMGN1 but not HMGN1S20,24E inhibit the H1 mediated enhancement of H3K27me3 levels (G and H) Quantification of panels D–F, the levels of H3K27me3 normalized to H3. (I) HMT assays showing effect of H1, HMGN1 and HMGN2 on H3K27me3 levels. (J) Quantification of panel I. All HMT quantification data were obtained from two independent experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001.
As a direct test whether the HMGN-H1 interplay, by itself, affects the EZH2-mediated H3K27me3 levels, we prepared H1-depleted poly-nucleosomes and quantified the effect of purified H1 (Supplementary Figure S4A) on the EZH2 directed H3K27 methylation, in either the presence or absence of purified HMGN1 (Supplementary Figure S4A), or of the purified HMGN1-S20,24E mutant that does not bind to chromatin. We find that pre-incubation of the H1-depleted poly-nucleosomes with increasing amounts of H1 stimulates the EZH2-mediated H3K27 trimethylation in a dose dependent fashion (Figure 4D and G). Importantly, addition of HMGN1 (Figure 4E), but not of HMGN1-S20, 24E mutant that does not bind to chromatin inhibits the stimulating activity of H1 (Figure 4E, F and H), an indication that HMGN1 inhibits H3K27 methylation by binding to chromatin. As expected, the effect of HMGN2 is very similar to that of HMGN1 (Figure 4I–J, compare lane 3 to lanes 6 & 7), and in the absence of H1, neither HMGN1 nor HMGN2 affect the EZH2 mediated H3K27me3 levels (Figure 4I–J, compare lanes 4 & 5 to lane 2). Finally, we note that the stimulating effects of H1 and the inhibitory effects of HMGNs occur only in the context of chromatin, neither of these nucleosome-binding proteins affects the EZH2 mediated methylation of purified, non-nucleosomal H3 (Supplementary Figure S4B).
In summary, ChIP analyses of ESCs indicate that loss of HMGNs increases the binding of H1 and elevates the levels of both EZH2 and H3K27me3 at Olig1&2 genomic sites, and in vitro assays reveal that HMGNs counteract the ability of H1 to stimulate the EZH2 mediated H3K27me3 levels in poly-nucleosomes. We conclude that depletion of HMGNs increases the chromatin binding of H1 thereby enhancing the binding of EZH2 complex and the level of H3K27me3, a modification known to be associated with gene silencing (44). Thus, the interplay between HMGNs and H1 in chromatin leads to epigenetic changes that regulate the expression of Olig1&2.
Loss of HMGNs affects oligodendrocyte development both in vitro and in vivo
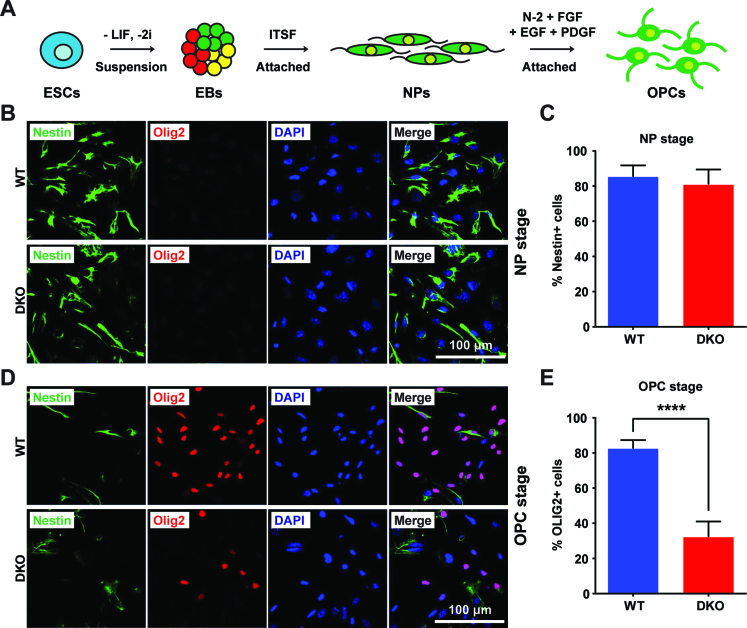
OLIG1 and OLIG2 are transcription factors known to play critical roles in oligodendrocyte specification and in regulating the oligodendrocyte lineage differentiation (14,15,21). We therefore examined whether the down regulation of Olig1/2 expression caused by HMGNs depletion is sufficient to affect the ability of ESCs to differentiate toward the oligodendrocyte lineage. We attached the 4-day EBs to Fibronectin coated dishes, selected the outgrowing Nestin+ neural progenitor cells (NPs) using ITSF supplemented medium (35), and then used N-2 supplemented medium with a combination of growth factors including hFGF, hEGF and hPDGF-AA (24) to induce the NPs to differentiate into OPCs (Figure 5A).
Figure 5.
Loss of HMGNs affects oligodendrocyte differentiation in vitro. (A) Schematic diagram of the protocol for in vitro differentiation of ESCs toward embryoid body (EB), neural progenitor (NP), and oligodendrocyte precursor cells (OPCs). (B) Representative immunofluorescence images showing equal Nestin expression in WT and DKO NPs. (C) Quantification of Nestin positive cells, over 250 cells from 10 random regions were counted. (D) Immunofluorescence showing lower frequency of OLIG2 positive cells in DKO OPCs. (E) Quantification of OLIG2 positive cells, over 250 cells from 10 random regions were counted. All the data are presented as mean ± SEM (****P < 0.0001).
The WT and DKO ESCs lines differentiated into NPs with similar efficiency with ∼80% of the cells showing strong Nestin signals (Figure 5B and C). Thus, HMGNs do not affect the ability of ESC to differentiate into NPs. However, upon differentiating the NPs into OPCs, we noted significant differences between the WT and DKO cells since 80% of WT NPs, but only 30% of DKO NPs differentiated into OLIG2-positive OPCs (Figure 5D and E). Thus, loss of HMGNs reduces the expression of Olig1/2 to levels that cause a significant decrease in the ability of cultured NPs to differentiate into OPCs.
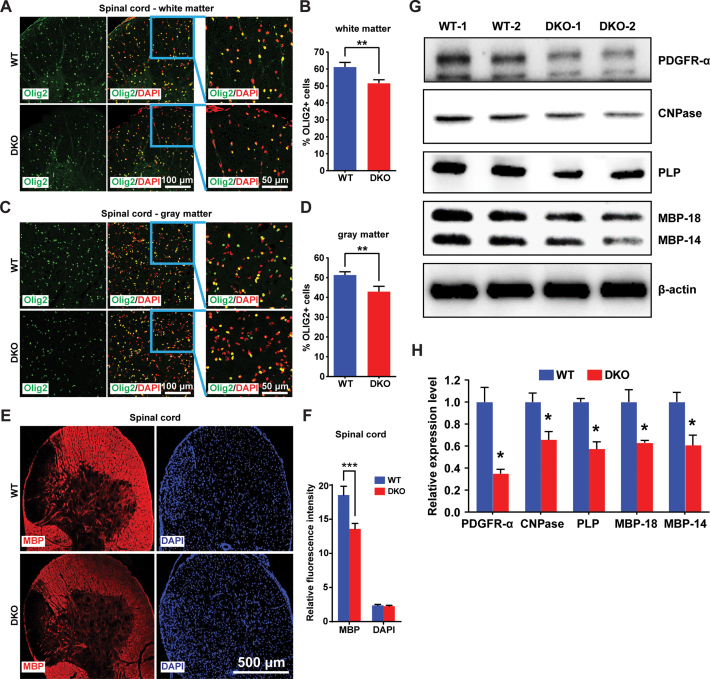
In adult mice, oligodendrocytes are enriched in both the white and gray matter of the spinal cord, and produce the myelin sheath covering neuron axons (15,49). By immunofluorescence analysis and quantification of OLIG2 positive cells of the spinal cords derived from 4 WT and 4 DKO, we find significant reduction in OLIG2-positive cells in both the white and gray matter of the spinal cords of adult DKO mice (Figure 6A–D). In agreement with these results, immunofluorescence analyses with antibody to Myelin Basic Protein (MBP), the major component of the myelin sheath, reveals significant loss of MBP signal in the spinal cord of DKO mice (Figure 6E and F). As a further test, we used western blot analyses to quantify the relative expression levels of markers for different developmental stages of oligodendrocytes in spinal cords of WT and DKO mice (50). We find that in the spinal cord of DKO mice the levels of platelet-derived growth factor α receptor (PDGFR-α), a marker for OPCs (51) was reduced by 65%, indicating significant reduction in OPC frequency. Likewise, the levels of 2΄,3΄-cyclic-nucleotide 3΄-phosphodiesterase (CNPase), a marker for mature oligodendrocytes (52), as well as the levels of MBP and proteolipid protein (PLP), known markers for myelin sheath (53), were reduced by almost 50% in DKOs (Figure 6G and H). Thus, loss of HMGN and the consequent down regulation of Olig1/2 expression impairs oligodendrocyte lineage differentiation and reduces the frequency of oligodendrocytes and of the myelin sheath components in the spinal cord of mice. These changes may be a contributing factor to the neurological phenotypes seen in DKO as measured by several tests including, the open field test (54), transfer arousal test (55), rotarod test (56,57) acoustic startle test (58) and the hot plate test (59) (Supplementary Figure S5).
Figure 6.
Loss of HMGNs affects oligodendrocyte development in mouse spinal cord. (A–D) Immunofluorescence showing decreased OLIG2 positive cells in both white matter (A) and gray matter (C) of DKO mouse spinal cords. (B, D) Quantifications of four WT and four DKO mice. For each mouse, >1500 cells from six (white matter) or four (gray matter) different regions of spinal cord sections were counted. (E) Immunofluorescence showing decreased MBP level in DKO spinal cords. (F) Quantification of panel J, eight WT and eight DKO images were quantified by ImageJ. (G) Western blots showing the expression of oligodendrocyte markers in WT and DKO mouse spinal cords. (H) Quantifications of panel L, expression levels normalized to β-actin. All the data are presented as mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001).
DISCUSSION
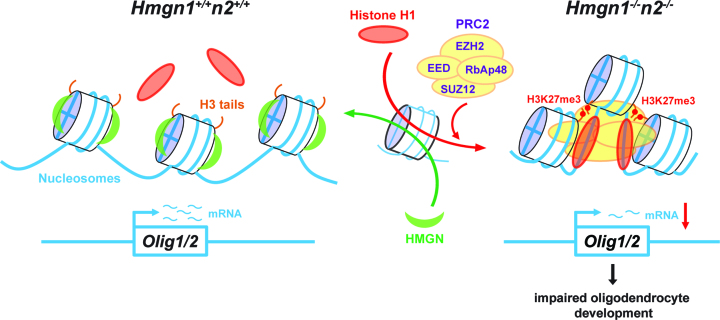
Our study reveals that at early developmental stages, in pluripotent embryonic stem cells, an interplay among two major families of chromatin architectural proteins establishes an epigenetic state that regulates the expression of Olig1 and Olig2 thereby affecting oligodendrocyte lineage differentiation and myelination (Figure 7). We delineate the molecular mechanism whereby the ubiquitous nucleosome binding protein HMGN1 and HMGN2 affect Olig1 and Olig2 expression and demonstrate that both in cell grown in tissue culture, and in genetically altered mice, loss of HMGNs leads to reduced oligodendrocyte lineage differentiation. Thus, we identify HMGNs as factors that affect oligodendrocyte differentiation and axon myelination.
Figure 7.
Model of the effect of HMGN-H1 interplay on Olig1&2 expressions. HMGN and H1 continuously compete for nucleosome binding sites (red and green arrows on the single nucleosome in the center). In WT cells (left) the presence of HMGNs reduced the binding of H1 thereby facilitating Olig1&2 expressions. In DKO cells (right) loss of HMGN enhances the binding of H1 thereby recruiting the PRC2 complex including its EZH2 histone methyltransferase, thus leading to increased levels of H3K27me3 and gene silencing at the Olig1&2 genomic loci. Reduced Olig1&2 expressions lead to impaired oligodendrocyte development, reduced myelination in the spinal cords, and altered mouse neurological behaviors.
Chromatin dynamics play a central role in the ability of regulatory factors to access their target sites and ultimately affect gene expression. The linker histone H1 and the HMGN protein family are major groups of chromatin architectural proteins known to modulate chromatin states in all vertebrate cells, yet their biological function and the role of these proteins in regulating gene expression remains a major unresolved question in chromatin biology (4,60). Quantitative changes in the levels of H1 or HMGN variants lead to both up and down regulation in the expression of numerous genes; however, the changes were not major, most likely because of compensatory effects. Thus, neither HMGNs nor H1s proteins are specific regulators of gene expression, they seem to ‘fine tune’ gene expression programs (11). Likely, changes in the proteins alter the ability of specific regulators to interact with their cognate chromatin binding sites. In agreement with studies in other cells, we find that loss of HMGNs leads to both up and down regulation of numerous gene with distinct genes being affected at distinct ESCs differentiation stages (Figure 1).
Importantly, previous in vivo and in vitro studies revealed that members H1 and HMGN proteins compete for chromatin binding sites and that the interplay between these proteins can affect chromatin compaction (48,61). However, the biological significance of this interplay and its effect on DNA related activities such as transcription is still poorly understood. Our study demonstrating that this interplay affects the levels of H3K27me3, delineates a mechanism whereby the HMGN-H1 interplay impacts an epigenetic process that affects gene expression, thereby providing a prime example of the biological significance of the H1-HMGN interplay in chromatin. We speculate that this interplay may be of particular importance at early developmental stages, when the chromatin is hyperdynamic (27), and perhaps more amenable to epigenetic remodeling. It may be relevant that differentiation processes, including oligodendrocyte differentiation, is associated with down regulation of HMGN levels (8,60,62,63) and changes in H1 variants (30). In fact, the expression of the H10 variant, which binds to chromatin with relatively high affinity (64), is a hallmark of terminal differentiation (65).
Specifically, we find that HMGN1 and HMGN2, regulate the expression of OLIG1 and OLIG2, two basic-helix-loop-helix transcription factors crucial for oligodendrocyte development. The number of OLIG2 positive cells detected in the spinal cord of the DKO mice and in the in vitro differentiating DKO NPs was significantly lower than that detected in wild type mice and cells. However, the immunofluorescence signal of OLIG2 in the differentiated WT and DKO cells was similar suggesting that the effect of HMGNs on OLIG expression in ESCs is more significant than the differentiated cells.
HMGNs affect Olig1 and Olig2 gene expression directly, by binding to chromatin since HMGN mutants that do not bind to chromatin do not affect Olig1&2 expression. In considering the molecular mechanism whereby HMGNs regulate Olig1 &2 expressions we note that in HMGN DKO cells Olig1 and Olig2 gene display an epigenetic signature characteristic of repressed genes: increased occupancy of the histone methyltransferase EZH2, and enhanced H3K27me3 levels, an epigenetic mark known to be associated with gene silencing (66). Indeed, the elevated H3K27me3 levels are directly responsible for reduced Olig1&2 expressions, since treatment of DKO cells with the EZH2 inhibitor GSK126 significantly restored the expression of OLIG1 and OLIG2. Thus, our results suggest that loss of HMGNs upregulates H3K27me3 levels, a finding that is in agreement with a previous study of mice and cells which indicated that up regulation of HMGN1 reduces H3K27me3 levels (67). Taken together, the available data raise the possibility that HMGNs are negative regulators of H3K27me3 levels.
Considering the mechanism whereby HMGNs can affect H3K27me3 levels, we note that the linker H1 has been shown to interact with components of the PRC2 complex, including EZH2, and to enhance the levels of H3K27me3 in nucleosomes by promoting chromatin compaction (46,47). In agreement, we find increased H1 occupancy at the Olig1&2 loci of DKO ESCs. Furthermore, our in vitro studies indicate that H1 enhances the EZH2 mediated levels of H3K27me3 in poly-nucleosomes in a dose dependent fashion, that wild type, but not a mutant HMGN1 that cannot bind to nucleosomes, counteracts the ability of H1 to enhance H3K27me3 levels, and that all these effects are not seen with purified H3. Thus, HMGNs modulate the levels of H3K27me3 at Olig1&2 by reducing the chromatin condensing activity of H1, a finding that is in full agreement with several in vitro and in vivo studies documenting that HMGNs reduce the chromatin binding of H1 (1,12,48,61). The 30–50% increase in chromatin binding could have a significant effect chromatin compaction and on the binding of EZH2 and complexes containing this enzyme. These effects could be larger than those observed in vitro with purified components and short oligonucleosomes whose ability to form higher order structures differs from that of the ‘native’, intact chromatin fiber inside the nucleus. Interestingly, a region between Olig1 and Olig2 loci (Region 4 in Figure 3A), that shows strong HMGN1/2 binding in WT cells, seems unusual since it does not show enhanced H3K27me3 levels and elevated EZH2 binding in the absence of HMGN1/2. We note that this region contains a non-annotated transcript which is strongly expressed in both WT and DKO cells. This region contains very low levels of H3K27me3 and EZH2, and strong active histone marks including H3K27ac, H3K4me3 and H3K4me1 in both WT and DKO. Thus, in contrast to the other regions examined, none of these marks change in response to loss of HMGNs (Supplementary Figure S3A). Likely, the transcripts in this region are tightly regulated, perhaps by a unique set of transcription factors or a specialized chromatin organization which ensure robust expression. Loss of HMGNs is not a sufficient change to alter the function of these factors, or alter chromatin conformation. Consistent with this possibility, the binding of H1 in this region is not increased in the absence of HMGNs (Figure 4A). Thus, the H1-HMGN equilibrium functions within a gene context, and as previously described, the interaction of H1 with chromatin is modulated by a diverse set of factors (1).
Interestingly, H1 depletion has been shown to impair ESCs specification into the neuroectoderm lineage without affecting the expression of core pluripotency genes (68). Together, the available data support the possibility that at the most fundamental level, Olig1&2 expressions is modulated by a dynamic interplay between HMGNs and H1 in chromatin. We are cognizant that this interplay is not the only factor that affects Olig1&2 expression. Indeed, we find that loss of HMGNs does not abolish OLIG1&2 expression but it downregulates it sufficiently to reduce oligodendrocytes differentiation and nerve myelination, processes that are important for the development of the CNS.
The factors affecting OPC differentiation are not fully known (69). The mouse Olig1 and Olig2 genes are located in chromosome 16, ∼36 kb apart. Their expressions are coordinately regulated both in space and time, but their biological functions in regulating oligodendrocyte development are only partially redundant, with OLIG2 playing a dominant role (18). We find significant Olig1 and Olig2 expression in WT ESCs, and that the levels of these transcription factors are markedly down regulated in the DKO ESCs which do not express HMGN1 and HMGN2. Consistent with these results, we find that loss of HMGNs does not affect the in vitro differentiation of ESCs into NPs, but does impair the differentiation of NPs into OPCs. In full agreement with the in vitro differentiation studies, HMGN1/N2 DKO mice show decreased OLIG 2 positive cells and decreased levels of myelin proteins in their spinal cord, an indication of impaired myelination.
Myelinating oligodendrocytes provide axon insulation and enable efficient conduction of action potential and therefore are essential for proper motor, sensory, and higher-order cognitive function (15,18). Indeed, the HMGN1/N2 DKO mice show neurological defects that could be related to the impaired oligodendrogenesis. However, it is likely that the phenotypic effects seen in mice are not solely due to the specific effect of HMGNs on Olig1 and Olig2 expression since our GO analysis shows other neural related genes are affected. Indeed, analysis by the Alan Institute for Brain Science (www.alleninstitute.org), and our previous studies also indicate prominent HMGN expressed in the CNS, especially in regions contain neural stem cells such as subventricular zone (8). In addition, HMGN proteins have been shown to modulate neural-related MeCP2 gene expression, affect Astrocyte differentiation (70) and play a role in several neurological disorders including Down syndrome and autism (71,72).
Taken together the accumulating data suggest that HMGNs play a role in regulating neural system development and neural functions. The present study reveals that by regulating OLIG1 and OLIG2 expression, HMGNs optimize neurological processes that require proper oligodendrocyte lineage differentiation and myelination. It is intriguing that both myelinating oligodendrocytes and HMGN proteins have been detected only in vertebrates (73,74).
Our study demonstrates a role for architectural chromatin binding proteins in regulating the expression of neural differentiation genes during embryonic stem cell differentiation. By focusing on the regulation of specific transcription factors, OLIG1 and OLIG2, we find that the interplay between two families of nucleosome-binding architectural proteins, H1 and HMGNs, modulates epigenetic processes that regulate oligodendrocyte lineage differentiation and affect mouse behavior. Our study provides insights into molecular processes affecting the expression of genes involved in CNS development and into the biological function of chromatin architectural proteins.
Supplementary Material
ACKNOWLEDGEMENTS
Authors' contributions: T.D. conceived the project, performed experiments, analyzed data and wrote the manuscript, Y.P., S.Z., L.G., S.M.H., L.B., I.R. performed experiments and analyzed data. V.G.D., H.F., W.W. and M.H.A. conceived and designed the phenotyping experiments. MB conceived the project, analyzed data, wrote the manuscript and coordinated the project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Programs of the Center for Cancer Research (CCR); National Cancer Institute (NCI); National Institutes of Health [ZIA BC 011154]; German Federal Ministry of Education and Research [Intrafrontier grant 01KX1012]. Funding for open access charge: NCI, CCR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Catez F., Ueda T., Bustin M.. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 2006; 13:305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woodcock C.L., Skoultchi A.I., Fan Y.. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006; 14:17–25. [DOI] [PubMed] [Google Scholar]
- 3. Hergeth S.P., Schneider R.. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015; 16:1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jordan A. Histone H1 in gene expression and development. Biochim. Biophys. Acta. 2016; 1859:429–430. [DOI] [PubMed] [Google Scholar]
- 5. Postnikov Y., Bustin M.. Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta. 2010; 1799:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kugler J.E., Deng T., Bustin M.. The HMGN family of chromatin-binding proteins: dynamic modulators of epigenetic processes. Biochim. Biophys. Acta. 2012; 1819:652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lim J.H., Catez F., Birger Y., West K.L., Prymakowska-Bosak M., Postnikov Y.V., Bustin M.. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell. 2004; 15:573–584. [DOI] [PubMed] [Google Scholar]
- 8. Deng T., Zhu Z.I., Zhang S., Leng F., Cherukuri S., Hansen L., Marino-Ramirez L., Meshorer E., Landsman D., Bustin M.. HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells. Mol. Cell.Biol. 2013; 33:3377–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deng T., Zhu Z.I., Zhang S., Postnikov Y., Huang D., Horsch M., Furusawa T., Beckers J., Rozman J., Klingenspor M. et al. . Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers. Genome Res. 2015; 25:1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martinez de Paz A, Ausio J.. HMGNs: the enhancer charmers. Bioessays. 2016; 38:226–231. [DOI] [PubMed] [Google Scholar]
- 11. Bustin M., Catez F., Lim J.H.. The dynamics of histone H1 function in chromatin. Mol. Cell. 2005; 17:617–620. [DOI] [PubMed] [Google Scholar]
- 12. Catez F., Brown D.T., Misteli T., Bustin M.. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002; 3:760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding H.F., Bustin M., Hansen U.. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol. Cell. Biol. 1997; 17:5843–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. El Waly B., Macchi M., Cayre M., Durbec P.. Oligodendrogenesis in the normal and pathological central nervous system. Front. Neurosci. 2014; 8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bercury K.K., Macklin W.B.. Dynamics and mechanisms of CNS myelination. Dev. Cell. 2015; 32:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfeiffer S.E., Warrington A.E., Bansal R.. The oligodendrocyte and its many cellular processes. Trends Cell Biol. 1993; 3:191–197. [DOI] [PubMed] [Google Scholar]
- 17. Li H., Richardson W.D.. The evolution of Olig genes and their roles in myelination. Neuron Glia Biol. 2008; 4:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meijer D.H., Kane M.F., Mehta S., Liu H., Harrington E., Taylor C.M., Stiles C.D., Rowitch D.H.. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat. Rev. Neurosci. 2012; 13:819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dai J., Bercury K.K., Ahrendsen J.T., Macklin W.B.. Olig1 function is required for oligodendrocyte differentiation in the mouse brain. J. Neurosci. 2015; 35:4386–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ligon K.L., Fancy S.P., Franklin R.J., Rowitch D.H.. Olig gene function in CNS development and disease. Glia. 2006; 54:1–10. [DOI] [PubMed] [Google Scholar]
- 21. Zhou Q., Anderson D.J.. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002; 109:61–73. [DOI] [PubMed] [Google Scholar]
- 22. Murry C.E., Keller G.. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008; 132:661–680. [DOI] [PubMed] [Google Scholar]
- 23. Liu S., Qu Y., Stewart T.J., Howard M.J., Chakrabortty S., Holekamp T.F., McDonald J.W.. Embryonic stem cells differentiate into oligodendrocytes and myelinate in culture and after spinal cord transplantation. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6126–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brustle O., Jones K.N., Learish R.D., Karram K., Choudhary K., Wiestler O.D., Duncan I.D., McKay R.D.. Embryonic stem cell-derived glial precursors: a source of myelinating transplants. Science. 1999; 285:754–756. [DOI] [PubMed] [Google Scholar]
- 25. Chen T., Dent S.Y.. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 2014; 15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keenen B., de la Serna I.L.. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J. Cell Physiol. 2009; 219:1–7. [DOI] [PubMed] [Google Scholar]
- 27. Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T.. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006; 10:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M. et al. . Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006; 8:532–538. [DOI] [PubMed] [Google Scholar]
- 29. Ho L., Crabtree G.R.. Chromatin remodelling during development. Nature. 2010; 463:474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pan C., Fan Y.. Role of H1 linker histones in mammalian development and stem cell differentiation. Biochim. Biophys. Acta. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fuchs H., Gailus-Durner V., Adler T., Aguilar-Pimentel J.A., Becker L., Calzada-Wack J., Da Silva-Buttkus P., Neff F., Gotz A., Hans W. et al. . Mouse phenotyping. Methods. 2011; 53:120–135. [DOI] [PubMed] [Google Scholar]
- 32. Fuchs H., Gailus-Durner V., Adler T., Pimentel J.A., Becker L., Bolle I., Brielmeier M., Calzada-Wack J., Dalke C., Ehrhardt N. et al. . The German Mouse Clinic: a platform for systemic phenotype analysis of mouse models. Curr. Pharm. Biotechnol. 2009; 10:236–243. [DOI] [PubMed] [Google Scholar]
- 33. Gailus-Durner V., Fuchs H., Adler T., Aguilar Pimentel A., Becker L., Bolle I., Calzada-Wack J., Dalke C., Ehrhardt N., Ferwagner B. et al. . Systemic first-line phenotyping. Methods Mol. Biol. 2009; 530:463–509. [DOI] [PubMed] [Google Scholar]
- 34. Gailus-Durner V., Fuchs H., Becker L., Bolle I., Brielmeier M., Calzada-Wack J., Elvert R., Ehrhardt N., Dalke C., Franz T.J. et al. . Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat. Methods. 2005; 2:403–404. [DOI] [PubMed] [Google Scholar]
- 35. Pachernik J., Esner M., Bryja V., Dvorak P., Hampl A.. Neural differentiation of mouse embryonic stem cells grown in monolayer. Reprod. Nutr. Dev. 2002; 42:317–326. [PubMed] [Google Scholar]
- 36. Birger Y., West K.L., Postnikov Y.V., Lim J.H., Furusawa T., Wagner J.P., Laufer C.S., Kraemer K.H., Bustin M.. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 2003; 22:1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim J.H., Catez F., Birger Y., Postnikov Y.V., Bustin M.. Preparation and functional analysis of HMGN proteins. Methods Enzymol. 2004; 375:323–342. [DOI] [PubMed] [Google Scholar]
- 38. Postnikov Y.V., Bustin M.. Reconstitution of high mobility group 14/17 proteins into nucleosomes and chromatin. Methods Enzymol. 1999; 304:133–155. [DOI] [PubMed] [Google Scholar]
- 39. Rea S., Eisenhaber F., O'Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D. et al. . Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000; 406:593–599. [DOI] [PubMed] [Google Scholar]
- 40. Bustin M. Chromatin unfolding and activation by HMGN (*) chromosomal proteins. Trends Biochem. Sci. 2001; 26:431–437. [DOI] [PubMed] [Google Scholar]
- 41. Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005; 19:1129–1155. [DOI] [PubMed] [Google Scholar]
- 42. Catez F., Lim J.H., Hock R., Postnikov Y.V., Bustin M.. HMGN dynamics and chromatin function. Biochem. Cell Biol. 2003; 81:113–122. [DOI] [PubMed] [Google Scholar]
- 43. Prymakowska-Bosak M., Misteli T., Herrera J.E., Shirakawa H., Birger Y., Garfield S., Bustin M.. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 2001; 21:5169–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Conway E., Healy E., Bracken A.P.. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr. Opin. Cell Biol. 2015; 37:42–48. [DOI] [PubMed] [Google Scholar]
- 45. McCabe M.T., Ott H.M., Ganji G., Korenchuk S., Thompson C., Van Aller G.S., Liu Y., Graves A.P., Della Pietra A. 3rd, Diaz E. et al. . EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012; 492:108–112. [DOI] [PubMed] [Google Scholar]
- 46. Martin C., Cao R., Zhang Y.. Substrate preferences of the EZH2 histone methyltransferase complex. J. Biol. Chem. 2006; 281:8365–8370. [DOI] [PubMed] [Google Scholar]
- 47. Yuan W., Wu T., Fu H., Dai C., Wu H., Liu N., Li X., Xu M., Zhang Z., Niu T. et al. . Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012; 337:971–975. [DOI] [PubMed] [Google Scholar]
- 48. Postnikov Y.V., Bustin M.. Functional interplay between histone H1 and HMG proteins in chromatin. Biochim. Biophys. Acta. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haroutunian V., Katsel P., Roussos P., Davis K.L., Altshuler L.L., Bartzokis G.. Myelination, oligodendrocytes, and serious mental illness. Glia. 2014; 62:1856–1877. [DOI] [PubMed] [Google Scholar]
- 50. Grade S., Bernardino L., Malva J.O.. Oligodendrogenesis from neural stem cells: perspectives for remyelinating strategies. Int. J. Dev. Neurosci. 2013; 31:692–700. [DOI] [PubMed] [Google Scholar]
- 51. Rivers L.E., Young K.M., Rizzi M., Jamen F., Psachoulia K., Wade A., Kessaris N., Richardson W.D.. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat. Neurosci. 2008; 11:1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sprinkle T.J. 2΄,3΄-cyclic nucleotide 3΄-phosphodiesterase, an oligodendrocyte-Schwann cell and myelin-associated enzyme of the nervous system. Crit. Rev. Neurobiol. 1989; 4:235–301. [PubMed] [Google Scholar]
- 53. Deber C.M., Reynolds S.J.. Central nervous system myelin: structure, function, and pathology. Clin. Biochem. 1991; 24:113–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prut L., Belzung C.. The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: a review. Eur. J. Pharmacol. 2003; 463:3–33. [DOI] [PubMed] [Google Scholar]
- 55. Alves R., Barbosa de Carvalho J.G., Benedito M.A.. High and low rearing subgroups of rats selected in the open field differ in the activity of K+-stimulated p-nitrophenylphosphatase in the hippocampus. Brain Res. 2005; 1058:178–182. [DOI] [PubMed] [Google Scholar]
- 56. Hamm R.J., Pike B.R., O'Dell D.M., Lyeth B.G., Jenkins L.W.. The rotarod test: an evaluation of its effectiveness in assessing motor deficits following traumatic brain injury. J. Neurotrauma. 1994; 11:187–196. [DOI] [PubMed] [Google Scholar]
- 57. Carter R.J., Morton J., Dunnett S.B.. Motor coordination and balance in rodents. Curr. Protoc. Neurosci. 2001; doi:10.1002/0471142301.ns0812s15. [DOI] [PubMed] [Google Scholar]
- 58. Crawley J.N. Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res. 1999; 835:18–26. [DOI] [PubMed] [Google Scholar]
- 59. Momin A., McNaughton P.A.. Regulation of firing frequency in nociceptive neurons by pro-inflammatory mediators. Exp. Brain Res. 2009; 196:45–52. [DOI] [PubMed] [Google Scholar]
- 60. Hock R., Furusawa T., Ueda T., Bustin M.. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007; 17:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rochman M., Postnikov Y., Correll S., Malicet C., Wincovitch S., Karpova T.S., McNally J.G., Wu X., Bubunenko N.A., Grigoryev S. et al. . The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol. Cell. 2009; 35:642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Furusawa T., Lim J.H., Catez F., Birger Y., Mackem S., Bustin M.. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol. Cell. Biol. 2006; 26:592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Magri L., Swiss V.A., Jablonska B., Lei L., Pedre X., Walsh M., Zhang W., Gallo V., Canoll P., Casaccia P.. E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. J. Neurosci. 2014; 34:1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Orrego M., Ponte I., Roque A., Buschati N., Mora X., Suau P.. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 2007; 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zlatanova J., Doenecke D.. Histone H1 zero: a major player in cell differentiation. FASEB J. 1994; 8:1260–1268. [DOI] [PubMed] [Google Scholar]
- 66. Zhou V.W., Goren A., Bernstein B.E.. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011; 12:7–18. [DOI] [PubMed] [Google Scholar]
- 67. Lane A.A., Chapuy B., Lin C.Y., Tivey T., Li H., Townsend E.C., van Bodegom D., Day T.A., Wu S.C., Liu H. et al. . Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet. 2014; 46:618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nguyen G.D., Gokhan S., Molero A.E., Yang S.M., Kim B.J., Skoultchi A.I., Mehler M.F.. The role of H1 linker histone subtypes in preserving the fidelity of elaboration of mesendodermal and neuroectodermal lineages during embryonic development. PLoS One. 2014; 9:e96858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J. Mol. Neurosci. 2008; 35:3–12. [DOI] [PubMed] [Google Scholar]
- 70. Nagao M., Lanjakornsiripan D., Itoh Y., Kishi Y., Ogata T., Gotoh Y.. High mobility group nucleosome-binding family proteins promote astrocyte differentiation of neural precursor cells. Stem Cells. 2014; 32:2983–2997. [DOI] [PubMed] [Google Scholar]
- 71. Abuhatzira L., Shamir A., Schones D.E., Schaffer A.A., Bustin M.. The chromatin-binding protein HMGN1 regulates the expression of methyl CpG-binding protein 2 (MECP2) and affects the behavior of mice. J. Biol. Chem. 2011; 286:42051–42062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Davies G., Armstrong N., Bis J.C., Bressler J., Chouraki V., Giddaluru S., Hofer E., Ibrahim-Verbaas C.A., Kirin M., Lahti J. et al. . Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Mol. Psychiatry. 2015; 20:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gonzalez-Romero R., Eirin-Lopez J.M., Ausio J.. Evolution of high mobility group nucleosome-binding proteins and its implications for vertebrate chromatin specialization. Mol. Biol. Evol. 2015; 32:121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zalc B. The acquisition of myelin: a success story. Novartis Found. Symp. 2006; 276:15–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.