Abstract
DNA methylation plays a critical role in the regulation and maintenance of cell-type specific transcriptional programs. Targeted epigenome editing is an emerging technology to specifically regulate cellular gene expression in order to modulate cell phenotypes or dissect the epigenetic mechanisms involved in their control. In this work, we employed a DNA methyltransferase Dnmt3a–Dnmt3L construct fused to the nuclease-inactivated dCas9 programmable targeting domain to introduce DNA methylation into the human genome specifically at the EpCAM, CXCR4 and TFRC gene promoters. We show that targeting of these loci with single gRNAs leads to efficient and widespread methylation of the promoters. Multiplexing of several guide RNAs does not increase the efficiency of methylation. Peaks of targeted methylation were observed around 25 bp upstream and 40 bp downstream of the PAM site, while 20–30 bp of the binding site itself are protected against methylation. Potent methylation is dependent on the multimerization of Dnmt3a/Dnmt3L complexes on the DNA. Furthermore, the introduced methylation causes transcriptional repression of the targeted genes. These new programmable epigenetic editors allow unprecedented control of the DNA methylation status in cells and will lead to further advances in the understanding of epigenetic signaling.
INTRODUCTION
Epigenetic modifications control cellular gene expression profiles and maintain the cell's differentiation state (1,2). DNA methylation has emerged as a key mechanism governing cellular reprogramming processes, such as cell differentiation, cellular senescence and disease (2). Aberrations in DNA methylation patterns caused by mutations or mis-regulation of the DNA methylation machinery are commonly found in cancer cells and contribute to carcinogenesis (2,3). In mammalian cells, DNA methylation is abundantly found in the context of CpG dinucleotides and methylation of CpG-rich promoter regions is correlated with their transcriptional repression (1,4). DNA methylation patterns are established by the de novo DNA methyltransferases Dnmt3a and Dnmt3b, the activity of which is enhanced by interaction with the Dnmt3L protein (5,6). Methylation patterns are maintained by the hemi-methylation specific DNA methyltransferase Dnmt1. However, it is becoming increasingly clear that the maintenance and regulation of DNA methylation is much more dynamic than initially anticipated (1,7,8).
To date, most DNA methylation studies have been descriptive in nature and although they provide important statistical and correlative information about the distribution of the DNA methylation mark in diverse cells and tissues, they do not allow the study of the direct mechanistic principles governing the establishment of methylated or unmethylated states at native genomic loci and their epigenetic consequences. Synthetic epigenetics approaches on the other hand provide means to precisely introduce or remove chromatin marks in the genome (9–11), therefore allowing the study of causal interferences regarding the presence or absence of an epigenetic mark. These tools rely on a sequence specific delivery of chromatin editors to genomic target loci, thus forcing a change of the epigenetic state in the selected region. To date, most of the targeted epigenetic tools have relied on programmable C2H2 zinc fingers or TALE arrays (reviewed in (9,11,12)), which have the disadvantage of the need to re-design and re-construct the targeting domain for each novel target sequence. The emergence of the bacterial Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) (13) system and the nuclease-inactivated CRISPR variant (dCas9) as a programmable genome targeting technology has opened new possibilities to target epigenetic effectors (14–16).
In this work, we show that targeting of an engineered Dnmt3a–Dnmt3L single-chain DNA methyltransferase (Dnmt3a3L) fused to the nuclease-inactivated dCas9 leads to efficient and widespread DNA methylation of CpG islands located within targeted promoters (up to 1200 bp). Targeted methylation with the Dnmt3a3L fusion protein is ∼4–5 times stronger than the methylation achieved through targeting of Dnmt3a alone. Peaks of methylation are observed around 25 bp upstream and 40 bp downstream of the PAM site, while 20–30 bp of the dCas9 binding site itself are protected against methylation. In general, targeting with single guide RNAs is sufficient for methylation and multiplexing does not increase its efficiency. In addition, using this system we illustrate that the multimerization of the Dnmt3a3L protein on DNA contributes to introduction and spreading of DNA methylation within the targeted genomic region.
MATERIALS AND METHODS
Generation of Cas9-Dnmt3a–Dnmt3L –SC fusion construct
The M-SPn-Cas9-VP64 plasmid (Addgene plasmid #48674) (17) was used as the base for the introduction of the Dnmt3a–Dnmt3L single-chain construct (18). The vector backbone was amplified using vector specific primers listed in Supplementary Data. The murine Dnmt3a catalytic domain was amplified from ZNF-Dnmt3a CD plasmid and Dnmt3a CD fused to the Dnmt3L C-terminal domain from the ZNF-Dnmt3a3L plasmids (18) with D3a_Cas9_f, D3a_Cas9_r and D3a3L_Cas9_f, D3a3L_Cas9_r primers using Q5® High-Fidelity DNA Polymerase (NEB). The insert and the vector backbone were joined using Gibson assembly (NEB). Proper sequence of the resulting clones was confirmed with Sanger sequencing. The plasmid maps are depicted in Supplementary Data).
Design of the guide RNAs for the EPCAM, CXCR4 and TFRC promoters
The sequences of the promoter regions (containing the CpG islands) of human EpCAM, CXCR4 and TFRC genes were extracted using UCSC genome browser. Potential guide RNA targeting sequences specific for these promoters were identified using the E-CRISP server (19). For EpCAM, a selection of twelve unique guide RNA binding sites was chosen to cover the region with 100–200 bp in between the gRNAs (shown in Supplementary Data). For CXCR4 and TFRC, four and ten gRNAs locations were selected, respectively. Each of the guide RNA constructs was generated as a separate gRNA plasmid. The gRNA plasmids were synthesized using overlapping ssDNA oligonucleotides which were cloned into the empty gRNA plasmid (Addgene plasmid # 41824) (20) using Gibson Assembly® Master Mix (NEB) following the manufacturer protocol. All gRNA constructs were validated with Sanger sequencing. Potential off-target binding sites for the gRNAs were predicted using CRISPOR (http://crispor.tefor.net/crispor.py). Four highest scoring off-target sites were selected based on the MIT or CFD scores and primers for bisulfite sequencing were designed directly surrounding these regions (primer sequences are provided in Supplementary Data).
Cell culture, transfections, MACS selection
The human ovary adenocarcinoma cell line, SKOV-3 (a generous gift of Dr Marianne Rots) and HEK293 cells were maintained at 37°C with 5% CO2 in DMEM media supplemented with 10% FCS (Sigma-Aldrich), 1× penicillin/streptomycin and 8 mM glutamine. For SKOV-3 cells transfection, the cells were trypsinized and seeded in six-well plates or T25 flasks at 40% density. The following day, the cells were immersed in the transfection cocktail composed of 5% of a modified pVenus-NLS plasmid (derived from Addgene #27794 plasmid) (21), 5% of the pLNGFR plasmid (Miltenyi Biotec), 70–88% of individual or equimolar mixture of pooled gRNA plasmids and 2–20% of SPn-Cas9-Dnmt3a3L-SC plasmid (% of total DNA, 2 or 4 μg for six wells or T25s, respectively) containing 6 or 12 μg PEI MAX MW 40 000 (Polysciences). The cells were washed with Dulbecco's PBS and immersed in normal cell media 14 h post-transfection. Subsequently, the medium was exchanged every two days. Magnetic activated cell sorting (MACS) was performed following the manufacturer's instructions at fifth day post-transfection and the sorted cells were frozen for further analysis. The percentage of the Venus positive cells before and after MACS selection was quantified using fluorescence microscopy (EVOS) and FACS analysis (BD FACS Calibur). Approximately 10 000 cells were analyzed for each sample. HEK293 cells were maintained as described above and transfected using a similar protocol, with the exception that 10% mVenus C1 and no pLNGFR were used.
Bisulfite sequencing – DNA methylation analysis
Genomic DNA from frozen cells was extracted using QIAamp DNA Mini Kit (Qiagen) and bisulfite converted using EZ DNA Methylation-Lightning™ Kit (Zymo Research) following manufacturer's instructions. The bisulfite treated DNA was used for PCR amplification using the amplicon specific primers (listed in Supplementary Data) and HotStarTaq DNA Polymerase (Qiagen), subsequently purified and sub-cloned using StrataClone PCR Cloning Kit (Agilent Technologies). Single positive colonies were picked and sequenced with standard Sanger sequencing using vector specific primers.
NGS library preparation and high-throughput sequencing
For Illumina library preparation, genomic DNA was isolated from each experimental samples using QIAamp DNA Mini Kit (Qiagen), bisulfite converted and the specific amplified regions of the targeted promoters were cleaned up with SPRI beads (Agencourt AMPure XP, Beckman-Coulter). For each experimental sample, the separate amplicons covering the different investigated CpG islands were mixed in an equimolar ratio, end repaired and A-tailed using the components of the SureSelect (Agilent Technologies) library preparation kit. Subsequently, the mixed amplicons belonging to one experimental sample were ligated to unique TruSeq HT double indexed adapters and cleaned up with SPRI beads. The uniquely indexed DNA samples were pooled and PCR amplified (eight amplification cycles) using the Illumina specific primers, SPRI purified and quantified with NEBNext Library quantification kit for Illumina (NEB). Sequencing was performed on a MiSeq machine with 2×300 PE run at the Microbiome Core Facility (UNC School of Medicine, Chapel Hill, USA).
Bisulfite sequencing analysis
The high-throughput sequencing results were demultiplexed using Qiime package, subsequently quality filtered and adapter trimmed with Trim Galore (v0.4.1, using default parameters—Phred score: 20; Paired, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), mapped to the human genome (GRCh38) with Bismark (v0.14.4; options –non_directional) and further visualized with SeqMonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). The methylation levels were determined for each CpG site within the regions of interest with SeqMonk using the ‘Difference quantification’ function (as a percent of methylated CpGs calls over total calls for each site) and retrieved using the ‘Annotated probe report’ function. Further visualization and analysis was performed in MS Excel. In the figures, average methylation per CpG site is provided or average methylation of a region calculated as the mean of average methylation of all CpG sites present in the reported region. At least a 1000-fold coverage of the CpG sites was achieved. The methylation profiles of the sub-cloned amplicons were analyzed with BISMA (22) using default parameters. The BISMA results are directly shown in Supplementary Data.
RT-qPCR
RT-qPCR was performed on total RNA extracted from frozen untransfected and transfected HEK293 and SKOV-3 cells. For this, total RNA from frozen cells was extracted with RNeasy Mini Plus Kit (Qiagen) for HEK293 cells or Ambion PureLink RNA Mini kit (Life Technologies) for SKOV-3 cells and reverse transcribed (using oligo-dT18-primers (NEB)) with M-MuLV Reverse Transcriptase (NEB) while protected by RNasin plus (Promega). Quantitative PCR was performed with SsoFast EvaGreen Supermix on a CFX96 Connect Real-Time detection system (both from Bio-Rad) with ACTB as a reference gene in at least two biological replicates (each with three technical repeats) and analyzed using the ΔΔCt method. All the primers used for quantification are listed in the Supplementary Data.
RESULTS
Design of the targeted methylation study
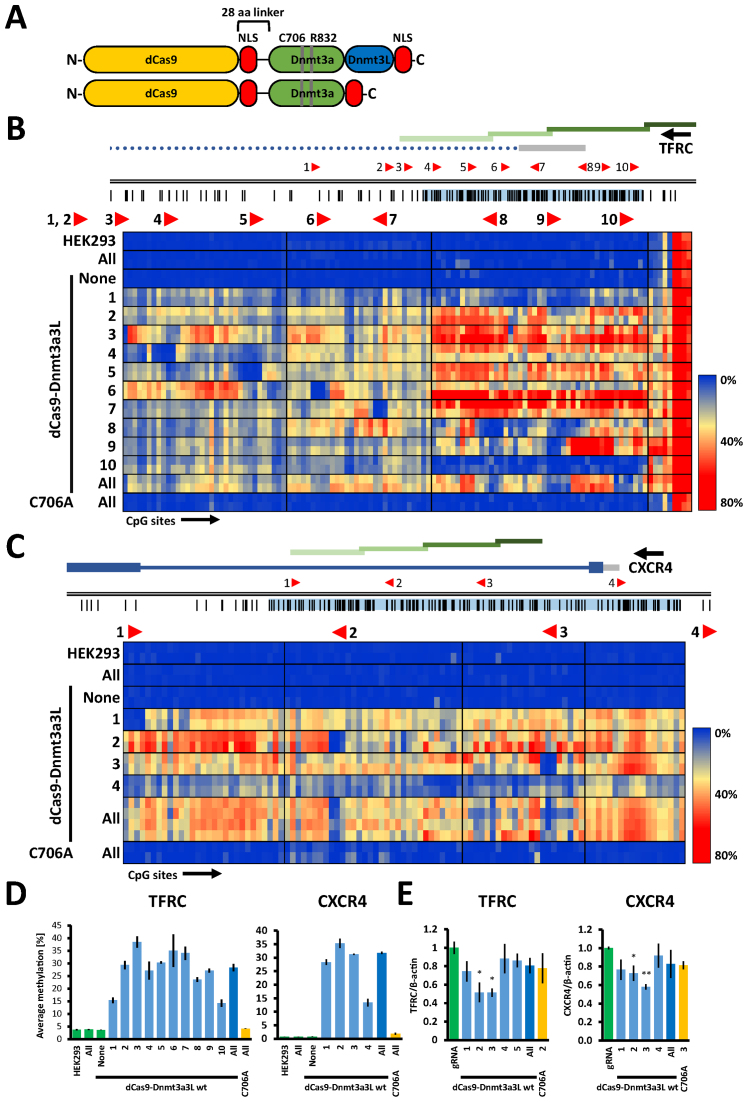
To take advantage of the targeting flexibility of the CRISPR/dCas9 system, we fused the catalytic C-terminal domain of the mouse Dnmt3a DNA methyltransferase (MTase) or the Dnmt3a–Dnmt3L C-terminal domain single-chain construct (18) to the nuclease-inactivated dCas9 (23,24) (Figure 1A). These fusion proteins were employed to introduce DNA methylation at the unmethylated CpG islands within the gene promoters of epithelial cell adhesion molecule (EpCAM), transferrin receptor protein 1 (TFRC, also known as CD71) and C-X-C chemokine receptor type 4 (CXCR4). In order to achieve this, we tiled the CpG islands of the target promoters with 12, 10 and 4 gRNAs binding sites, respectively, spaced 100–400 bp apart (Figures 1B, C and 2A). Next, we transiently co-transfected the dCas9-DNA MTase fusions along with single or pooled gRNA constructs into HEK293 cells (TFRC and CXCR4) or SKOV-3 cells (EpCAM). We obtained typical transfection efficiencies of around ∼80% for HEK293 and 30–40% for SKOV-3 cells, for which MACS selection was performed to enrich the transfected cell population to ∼80% (Supplementary Data). Both dCas9-methyltransferase fusions were efficiently expressed in the cells as illustrated by Western blotting (Supplementary Data). The transfected cells were cultured for 5 days, harvested and the methylation levels in the selected promoters were determined by Sanger or next-generation targeted bisulfite sequencing.
Figure 1.
The dCas9–Dnmt3a3L fusion protein deposits DNA methylation at endogenous human TFRC and CXCR4 promoters in HEK293 cells. (A) Schematic structure of the dCas9–Dnmt3a3L and Dnmt3a CD fusion proteins. The catalytically inactive dCas9 gene was fused to an engineered Dnmt3a–Dnmt3L C-terminal domain single-chain construct or Dnmt3a C-terminal domain with a 28 amino acid linker (containing NLS peptide). An additional NLS was added at the C-termini of both constructs. The residues that were mutated in this study within the catalytic domain of Dnmt3a in both Dnmt3a CD and Dnmt3a3L proteins are indicated (C706A inactivates the methyltransferase and R832E disrupts the multimerization of Dnmt3a). Not drawn to scale. (B) Genomic region of the human TFRC promoter that was targeted with the dCas9 fusions (Chr. 3: 196081310–196082794). Regions analyzed by bisulfite sequencing are indicated with green bars. The location and directionality of the Cas9 target sites are shown by red arrow heads. The black vertical lines indicate CpG sites. The gray bar denotes the 5΄ UTR and the dotted line represents an intron. The heat map shows average methylation per CpG site. Rows represent separate HEK293 co-transfection experiments where targeted methylation was induced with single or pooled gRNAs targeting the annotated region, columns represent separate CpG sites in the investigated region. Direction of the transcription is indicated with black arrow above the gene name. (C) Targeted methylation of the CXCR4 promoter in HEK293 cells (Chr.2: 136116618–136117805) illustrated as described in (B). The heat map shows acquisition of DNA methylation in the CXCR4 promoter region when targeting the dCas9–Dnmt3a3L with single or pooled gRNAs. (D) Average DNA methylation over all CpG sites within the analyzed regions in the HEK293 cells that were co-transfected with single or pools of gRNA. The analyzed TFRC and CXCR4 regions comprise 118 and 101 CpG sites respectively. The error bars denote STDEV from methylation averages measured in two biological samples. Color of the bars denotes experimental sets: green—control experiments, light blue—targeting of the dCas9 fusions with single gRNAs, dark blue—targeting with multiple gRNAs and yellow corresponds to targeting of C706A catalytic mutant. (E) Relative mRNA expression of TFRC and CXCR4 in the cells co-transfected with dCas9–Dnmt3a3L and single or pools of gRNAs as determined by RT-qPCR (error bars represent SEM from at least two biological replicates performed in technical triplicate). *P < 0.05, **P < 0.0001, unpaired t-test relative to ‘gRNA only’ control experiment.
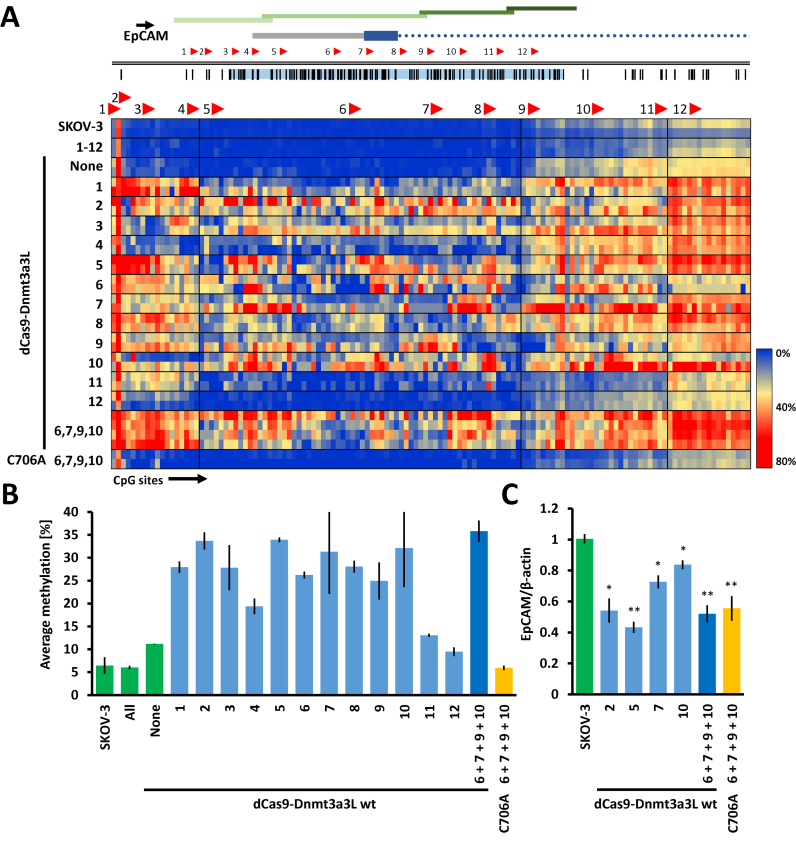
Figure 2.
Targeted methylation of the endogenous human EpCAM promoter in SKOV-3 cells with dCas9–Dnmt3a3L. (A) The dCas9–Dnmt3a3L fusion protein was targeted either to separate or multiple sites within the CpG island in the EpCAM promoter (Chr. 2: 47368894–47370157). Methylation was investigated in four consecutive amplicons covering the CpG island (indicated with green bars). For details cf. the legend to Fig. 1B. The heat map shows targeted DNA methylation within the EpCAM promoter CpG island when the dCas9–Dnmt3a3L fusion protein was targeted to single or multiple locations within the island. (B) Average methylation level observed over the whole analyzed EpCAM promoter region containing 131 CpG sites. The guide RNAs that were used in the targeting experiments are indicated. Error bars represent SEM of average methylation from two biological replicates. Color of the bars denotes experimental sets: green—control experiments, light blue—targeting of the dCas9 fusions with single gRNAs, dark blue—targeting with multiple gRNAs and yellow corresponds to targeting of C706A catalytic mutant. (C) Relative EpCAM mRNA expression levels measured with RT-qPCR. The numbers below the bars denote gRNAs that were used to target the dCas9–Dnmt3a3L. The error-bars denote SEM from at least two biological repeats performed in technical triplicate). *P < 0.05, **P < 0.0001, unpaired t-test relative to ‘SKOV-3’ control experiment.
Control experiments showed that expression of gRNAs alone did not cause any changes in the DNA methylation at the target promoters (Figures 1B, C, 2A and Supplementary Data). Similarly, no significant increase in DNA methylation was observed in the TFRC or CXCR4 promoters after expression of the untargeted dCas9–Dnmt3a3L construct. At the EpCAM promoter, a slight increase in methylation from 6.4 ± 1.8% in untreated SKOV-3 cells to 11.2 ± 0.1% after treatment (for details refer to Supplementary Data) was detected predominantly within the EpCAM gene body (Figure 2A and B).
Targeting the dCas9–Dnmt3a3L with single gRNAs leads to efficient genomic methylation
Next, we targeted the dCas9–Dnmt3a3L fusion construct to defined single sites within the analyzed promoters using 10 different gRNAs for TFRC (Figure 1B), 4 gRNAs for CXCR4 (Figure 1C) and 12 gRNAs for EpCAM (Figure 2A). In contrast to the control experiments, co-transfection of the gRNAs and dCas9–Dnmt3a3L plasmids caused introduction of robust methylation at the targeted promoters with most of the gRNAs. The strongest methylation of the TFRC promoter was observed with gRNATFRC 3, 6 and 7, which resulted in 38.5 ± 2.2%, 35.1 ± 6.5% and 34.0 ± 2.6% average methylation, respectively (Supplementary Data). The average methylation introduced by most of the gRNATFRC ranged between 23.6 ± 1.9% and 30.3 ± 0.5%. Co-transfection of dCas9–Dnmt3a3L with gRNATFRC 1, which targets a site located ∼340 bp upstream of the amplicons used for bisulfite sequencing, resulted in 15.3 ± 1.1% average methylation (Figure 1B and D). gRNATFRC 10, which is located at the boundary of the CpG island caused only 14.2 ± 1.4% methylation. Similarly, targeting the CXCR4 promoter with gRNAsCXCR4 1, 2 or 3 resulted in efficient methylation of the CpG island, reaching 28.3 ± 1.0%, 35.3 ± 1.8% and 31.2 ± 0.3% average methylation, respectively. gRNACXCR4 4, which binds to a site ∼370 bp upstream of the bisulfite amplicons, caused introduction of 13.3 ± 1.5% methylation within the analyzed amplicons. In the EpCAM promoter, targeting the dCas9–Dnmt3a3L construct with the individual gRNAsEpCAM 1–3 or 5–10 triggered methylation between 25% (for gRNAEpCAM 9) to 33.7% (for gRNAEpCAM 2). gRNAEpCAM 11 and 12, which are targeting the boundary of the CpG island, caused only weak methylation, comparable to the methylation increase observed with untargeted dCas9–Dnmt3a3L. Interestingly, targeted methylation with gRNAEpCAM 4 in the EpCAM promoter was less efficient than with other neighboring gRNAs, which might be explained by the fact that the gRNA binding site overlaps with the annotated TSS, where the transcriptional machinery could prevent stable binding of the dCas9 fusion.
In summary, we conclude that single gRNAs co-transfected with dCas9–Dnmt3a3L can cause robust DNA methylation. Strikingly, although the deposited methylation was most prominent on the 3΄ side next to the dCas9 targeted sites, where based on modeling the fusion partner is presented (reaching up to 60.5 ± 1% for gRNATFRC 3 or 49.1 ± 2.7% for gRNACXCR4 2), the DNA methylation signal was propagated over the entire promoter in most cases (Figures 1B, C, 2A and Supplementary Data).
Our data show that the efficiency of dCas9 mediated targeted methylation is comparable to previous targeting systems. For example, targeting the EpCAM promoter in SKOV-3 cells Nunna et al. observed introduction of 29% methylation in transient transfection experiment, whereas in stably transfected cells 46–48% DNA methylation was detected (albeit within a shorter analyzed region) (25).
Targeting multiple sites does not increase the DNA methylation levels
Subsequently, we tested whether co-targeting of the loci with multiple gRNAs could further increase the methylation levels. However, targeting with all ten gRNAs to the TFRC promoter did not result in increased methylation efficiency (28.3 ± 1.5%) when compared to single gRNA targeting experiments (e.g. gRNATFRC 3—38.5 ± 2.2%). Similarly, no significant enhancement of targeted methylation was achieved at the CXCR4 promoter after co-targeting with all four gRNAsCXCR4 (31.8 ± 0.4%), nor at the EpCAM promoter co-targeted with gRNAEpCAM 6, 7, 9 and 10 (35.8 ± 2.3%) (Figures 1D, 2B). This may be caused by a less efficient expression of gRNA transfected as pools or by a competition of gRNAs for dCas9-DNA MTase proteins, which finally prevents an increase in efficiency. Future work will show if improved systems can be developed to overcome these barriers. Importantly, the dCas9-fusions targeting a catalytic mutant Dnmt3a3L C706A, which carries an amino acid exchange in the catalytic center, could not trigger DNA methylation (Figures 1B, C, D and 2A, B), showing that the catalytic activity of the targeted DNMT is necessary for efficient DNA methylation.
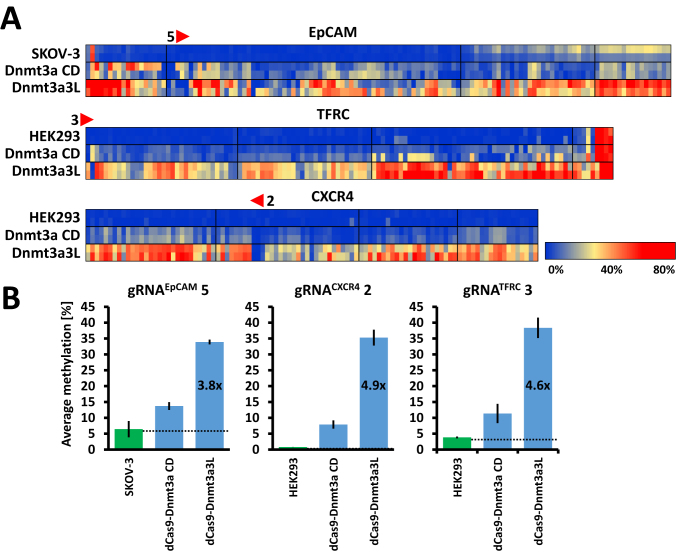
Dnmt3a3L causes more efficient methylation at the target promoters than Dnmt3a catalytic domain
In order to directly compare the efficiency of targeted DNA methylation introduced by the dCas9–Dnmt3a-CD and dCas9–Dnmt3a3L we targeted both MTase fusions in parallel experiments to three separate loci. For this, we have chosen the single gRNAs targeting EpCAM, CXCR4 and TFRC promoters that showed the highest efficiency with dCas9–Dnmt3a3L.The results indicate that Dnmt3a3L outperformed Dnmt3a CD (despite similar expression levels in the transfected cells (Supplementary Data)) in all three experiments, with 3.8× (gRNAEpCAM 5), 4.9× (gRNACXCR4 2) and 4.6× (gRNATFRC 3) higher methylation efficiency (Figure 3A and B).
Figure 3.
Comparison of targeted DNA methylation efficiencies imposed by dCas9–Dnmt3a CD and dCas9–Dnmt3a3L. (A) Heatmap representation of CpG methylation averages (per CpG site) imposed by the dCas9–Dnmt3a CD and dCas9–Dnmt3a3L which were targeted to promoters using single gRNAs (as indicated). (B) Comparison of average methylation (calculated as mean methylation of all CpG sites within the analyzed amplicons). The fold increase in methylation efficiency was calculated after subtracting the background methylation level already present in the untreated cells.
Distribution of DNA methylation levels and spreading of methylation
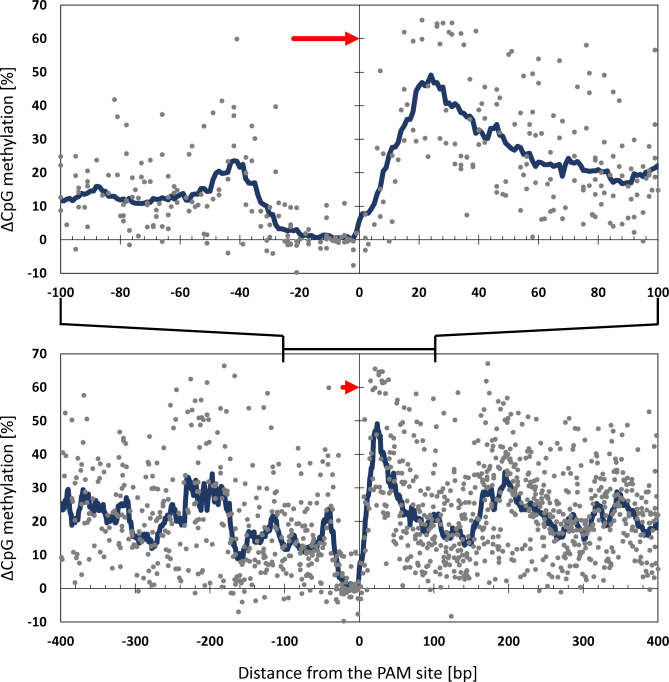
Comparison of all data shows highest methylation right upstream of the PAM site (around +25 bp). A second (weaker) peak of methylation is observed downstream of the PAM site (around –40 bp). These maxima may represent the footprint of the bound dCas9-DNA MTase fusion protein and indicate the nearest free DNA available for binding of the methyltransferase. The fact that DNA methylation is not introduced directly at the dCas9 binding sites suggests that dCas9 occupancy protects the overlapping CpG sites from methylation by the DNA methyltransferase (Figure 4). The profiles also present interesting additional peaks of methylation ∼200 bp upstream and downstream of the PAM site. This finding may suggest that the region in between is partially blocked by nucleosomes and the bound MTase most efficiently reaches the next available linker DNA region. Similar spreading of DNA methylation was also observed in other targeted methylation studies using zinc fingers (26,27) and TALE effectors (28).
Figure 4.
Relative distribution of targeted DNA methylation. Scatter plot showing the average gain of DNA methylation at all CpG sites observed at the EpCAM, TFRC and CXCR4 promoters with single guide RNAs targeting dCas9–Dnmt3a3L. The location of the CpG sites relative to the targeted dCas9 sites is indicated on the X axis. Each dot represents the average methylation gain (Y axis) of a specific CpG site in one of the targeting experiment corrected for the DNA methylation present at that site in untreated cells. The blue line represents the mean methylation level measured in a sliding window of 15 bp. The guide RNA binding site and orientation are depicted as a red arrow. Upper and lower panels show regions between –100 bp to 100 bp and –400 bp to 400 bp from the PAM site, respectively.
Specificity of targeted DNA methylation
Having shown the efficient methylation of the target sites, the question appeared if off-target DNA methylation is introduced at additional genomic locations. To check this, we have predicted off-target gRNA binding sites for four of our most efficient gRNAs and analyzed the methylation status of these off-target loci and determined methylation at unrelated CpG islands and using untargeted dCas9-MTase constructs. Out of four analyzed off-targets, two showed a mild methylation increase (9.2% and 11.1% increase of methylation at the off-targets compared to 25.6% and 34.7% at the targets using gRNATFRC 2 and 3, respectively). The other two regions were already fully methylated in the untreated cells hence non-informative (Supplementary Data). Moreover, untargeted dCas9–Dnmt3a3L did not cause methylation of the TFRC, CXCR4 or EpCAM promoters, nor four additional unrelated CpG islands which do not contain sequences resembling the gRNA target sites (Supplementary Data).
Targeted DNA methylation causes gene repression
DNA methylation of CpG rich promoters is associated with repression of gene expression; therefore, we investigated whether the introduced DNA methylation is capable of silencing expression of the associated genes. Indeed, when targeting the TFRC promoter with gRNATFRC 2 or 3 that caused the most efficient DNA methylation, we observe roughly 50% repression of TFRC expression. Similarly, targeting the CXCR4 promoter with gRNACXCR4 3 also decreased the gene expression to 58 ± 3% (Figures 1E and 2C). When interpreting these numbers it must be kept in mind, that transient experiments were conducted with transfection yields around 80%. When targeting the inactive Dnmt3a3L (C706A R832E) mutant with the same gRNAs, we also observed a smaller decrease in the expression of the target genes to 78 ± 3% with gRNATFRC 2 and 81 ± 4% for gRNACXCR4 3. This can be explained by the binding of the dCas9 protein, which can inhibit transcriptional elongation by blocking the passage of the RNA polymerase (29), steric clashes with binding of transcription factors or components of the preinitiation complex. This suggests that the observed transcriptional silencing is caused by introduced DNA methylation, but partially also by the strong binding of dCas9 that interferes with transcription.
Mechanism of targeted DNA methylation
As described above, targeting of the dCas9–Dnmt3a3L construct resulted in robust methylation across the entire EpCAM, CXCR4 and TFRC promoters, with methylation observed as far as 1 kb away from the gRNA binding site. The cooperative multimerization of Dnmt3a and Dnmt3a/Dnmt3L complexes along the DNA has been reported in vitro with purified recombinant proteins (30,31) and this process was shown to contribute to the efficient methylation of DNA in vitro (32). To test if the multimerization of Dnmt3a has a role in targeted DNA methylation in cells, we targeted the Dnmt3a mutants which affect the catalytic center (C706A), or disrupt the polymerization interface (R832E) (31) or both (C706A R832E). It should be noted that the R832E mutation, in agreement with the role of multimerization for DNA methylation activity, decreases the catalytic activity of Dnmt3a by ∼2-fold in vitro (31). Next, we compared the methylation patterns of the investigated promoters upon targeting the wild type or mutant dCas9–Dnmt3a3L fusion proteins. The wild type dCas9–Dnmt3a3L caused strong and widespread methylation over the entire CpG island of the EpCAM promoter (Figure 5A) when co-targeted to four sites (gRNAEpCAM 6, 7, 9 and 10). The R832E mutant, however, introduced less methylation that was confined to the nearest vicinity of the targeted sites (between 10 and 50 bp away from the binding sites), while the C706A and the double C706A/R832E mutants did not methylate the locus at all (Figures 1, 2 and 5). Similar results were obtained in the analyses of the CXCR4 and TFRC promoters (Supplementary Data). Interestingly, with the dCas9–Dnmt3a3L R832E mutant we also observed weak methylation on remote well-defined sites (denoted with * in Supplementary Data). However, no efficient DNA methylation spreading was observed for the R832E mutant, showing that the lateral multimerization of the Dnmt3a/Dnmt3L complex contributes to the establishment of DNA methylation at the targeted sites and in larger regions of the genome. Moreover, the lack of strong introduced DNA methylation for the C706A mutant indicates that the fusion protein does not recruit the native Dnmt3a, Dnmt3b and Dnmt3L machinery, presumably due to the low expression levels of the endogenous Dnmt3 enzymes in the cell lines used here. Collectively, these data suggest that the multimerization of the Dnmt3a/Dnmt3L complexes on the DNA is a mechanism which allows efficient methylation and spreading of DNA methylation across a selected genomic region upon targeting of the Dnmt3a/3L complex, for example by a transcription factor (Figure 5B).
Figure 5.
Spreading of DNA methylation is dependent on Dnmt3a3L multimerization. (A) Heat map of DNA methylation deposited within the EpCAM promoter region in untransfected SKOV-3 cells and cells co-transfected with a pool of guide RNAs (EpCAM gRNAs 6, 7, 9 and 10) and the dCas9–Dnmt3a3L wild type, catalytically inactive mutant C706A, non-multimerizing R832E mutant or a C706A R832E double mutant. (B) Model illustrating the mechanism of DNA methylation setting and spreading. Binding of the effector domain of targeted dCas9–Dnmt3a3L can be achieved both in cis and in trans relative to the DNA strand bound by dCas9 (here shown for two different guide RNAs). This leads to the deposition of DNA methylation (black filled circles) in the direct vicinity of the bound site or on a DNA strand that comes into spatial proximity to the dCas9 bound site. The Dnmt3a3L dimers can multimerize along the DNA via the R832 interaction interface (top). The R832E mutant with a disrupted multimerization interface cannot form fibers, leading to more locally defined DNA methylation either directly next to the dCas9 binding site or further away via DNA looping (middle). In a native situation (bottom), a transcription factor (brown) can recruit native Dnmt3a (green) or in complex with Dnmt3L (blue) to a specific site; this in turn can serve as a nucleation point to elongate the formed fiber causing cooperative deposition of DNA methylation in a larger genomic region.
DISCUSSION
In this study, we show that the CRISPR/Cas9 programmable genome targeting system can be successfully employed to introduce DNA methylation at defined endogenous genomic loci, leading to transcriptional silencing. We further show that efficient methylation of an entire CpG island can be achieved by targeting the Dnmt3a3L fusion construct to a single site within the island. Notably, DNA methylation is most efficiently deposited directly next to dCas9 binding sites; nevertheless, we observe efficient DNA methylation across the entire CpG islands (up to 1000 bp away from the target site). We detected a mild methylation increase (9.2% and 11.1%) at two of the predicted, highest scoring off-target regions, which was significantly weaker than the methylation at the corresponding on-target sites for the used gRNAs (25.6% and 34.7%). In addition, untargeted dCas9–Dnmt3a3L did not cause methylation of the CXCR4, TFRC promoters nor of four unrelated CpG islands. We speculate that the observed weak off-target methylation at near-cognate binding sites could be attributed to transient dCas9 binding at these sites, thus allowing a time window for DNA methylation deposition. Highly specific Cas9 mutants developed recently (33,34) could be used to increase the specificity of targeted DNA methylation in cells. Alternatively, the expression level of the dCas9 fusion proteins could be reduced to increase the specificity of target occupancy.
Furthermore, our results indicate that the efficient methylation and spreading could be attributed to the multimerization of Dnmt3a/Dnmt3L complexes on the DNA, as illustrated by the fact that less methylation and no efficient spreading was observed when we targeted the non-multimerizing R832E mutant of Dnmt3a3L. This illustrates for the first time the role of Dnmt3a3L multimerization in the establishment and spreading of targeted de novo DNA methylation in cells. Mechanistically, this finding can be explained by a model considering a multistep pathway finally leading to efficient DNA methylation at the target site that is initiated by a diffusional encounter of the tethered MTase with a substrate DNA site. The ability to multimerize increases the DNA binding strength of Dnmt3a, which raises the probability that a transient DNA encounter leads to the formation of a stable Dnmt3a3L–DNA complex followed by DNA methylation. Notably, formation of a long Dnmt3a3L fiber that would cover the whole CpG island (∼1000 bp) is rather unlikely, as this would cause a larger distortion of the local chromatin structure, including displacement of nucleosomes. However, we envision a dynamic formation of shorter multimers that would cause local methylation of the bound DNA. Methylation of remote regions within the locus could be achieved through DNA looping and subsequent multimer formation and methylation at the distant site. As the propensity of a diffusional encounter of the tethered enzyme and DNA declines with the distance of the DNA site to the targeted region, efficient binding of the tethered MTase to remote DNA sites is even more dependent on the ability of Dnmt3a to multimerize on DNA.
We show here the targeted methylation of larger DNA regions by dCas–Dnmt3a3L. This approach mimics establishment of natural DNA methylation patterns and it increases the probability of causing a strong biological response (as for example changes in gene expression as shown here). Additionally, we observed that the CpG sites that are covered by the bound dCas9 protein are almost completely protected from DNA methylation, illustrating that inactivated Cas9 stays firmly bound to its target site. This explains why dCas9 alone can be used to out-compete and displace DNA binding proteins (such as transcription factors) to directly affect transcription, as observed by others (24). In a more general view, it suggests that targeting of dCas9 to sites bound by other protein factors can be applied in order to investigate their functions at a specific genomic site. The pattern of the targeted DNA methylation suggests that Dnmt3a preferentially methylates linker DNA, which is in agreement with earlier in vitro data and cellular methylation studies (35–39).
Targeting of our epigenetic editing tools based on DNA methyltransferase domains caused down-regulation of gene expression at three different promoters tested, indicating that it could be used as a universal tool for gene repression. Alternative tools, like CRISPRi or based on the targeted recruitment of transcriptional repressors (like for example KRAB domain) have been developed and successfully used to down-regulate gene expression as well (14,24,29). The possible advantage of targeted methylation is that it has the potential of causing durable effects. Future work will show which of these techniques finally will prove most useful for targeted gene regulation.
During preparation of this manuscript, dCas9 targeted DNA methylation was described by two other groups using dCas9 fused to the catalytic domain of Dnmt3a (40,41). Vojta and colleagues observed efficient targeted methylation occurring in a distance of ∼35 bp downstream of the dCas9 binding sites, which is consistent with our findings. However, in contrast to the dCas9–Dnmt3a3L targeting used here, co-targeting of multiple locations within the BACH2 promoter was required to achieve methylation of a wider region (40). Similarly, McDonald et al. reported successful targeted methylation using dCas9–Dnmt3a CD of the CDKN2A, Cdkn1a and ARF promoters (41).
We envision that our dCas9–Dnmt3a3L programmable DNA methylation writer will find numerous applications in establishing the functional significance of DNA methylation in gene repression and the control of the cellular differentiation state, as well as to dissect the principles of network responses to epigenetic changes in synthetic biology settings. Moreover, targeted DNA methylation offers the unique potential to correct epimutations in disease states.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: P.S., R.Z.J. and T.P.J. designed the experiments. P.S., R.Z.J., G.K. performed experiments. P.S., G.K., R.Z.J., T.C., F.K., A.J. and T.P.J. analyzed the data, P.S. and T.P.J. wrote the initial manuscript draft. A.J., W.R., R.R. and T.P.J. supervised research. All authors contributed to the editing of the manuscript and approved its final version.
Footnotes
Present address: Renata Z. Jurkowska, BioMed X Innovation Center, Im Neuenheimer Feld 583, D-69120 Heidelberg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by Baden-Württemberg Stiftung [95011370 to A.J. and T.P.J.]; European Foundation for the Study of Diabetes [84011359 to T.P.J.]; Deutsche Forschungsgemeinschaft [Je252/10 to A.J.]. This publication was supported by the Open Access Publishing Fund of the University of Stuttgart.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 2. Bergman Y., Cedar H.. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013; 20:274–281. [DOI] [PubMed] [Google Scholar]
- 3. Yang L., Rau R., Goodell M.A.. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015; 15:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weichenhan D., Plass C.. The evolving epigenome. Hum. Mol. Genet. 2013; 22:1–6. [DOI] [PubMed] [Google Scholar]
- 5. Chedin F., Lieber M.R., Hsieh C.-L.. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gowher H., Liebert K., Hermann A., Xu G., Jeltsch A.. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005; 280:13341–13348. [DOI] [PubMed] [Google Scholar]
- 7. Jeltsch A., Jurkowska R.Z.. New concepts in DNA methylation. Trends Biochem. Sci. 2014; 39:310–318. [DOI] [PubMed] [Google Scholar]
- 8. Lee H.J., Hore T.A., Reik W.. Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell. 2014; 14:710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jurkowski T.P., Ravichandran M., Stepper P.. Synthetic epigenetics—towards intelligent control of epigenetic states and cell identity. Clin. Epigenet. 2015; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keung A.J., Joung J.K., Khalil A.S., Collins J.J.. Chromatin regulation at the frontier of synthetic biology. Nat. Rev. Genet. 2015; 16:159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kungulovski G., Jeltsch A.. Epigenome editing: state of the art, concepts, and perspectives. Trends Genet. 2016; 32:101–113. [DOI] [PubMed] [Google Scholar]
- 12. de Groote M.L., Verschure P.J., Rots M.G.. Epigenetic editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012; 40:10596–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A Programmable Dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thakore P.I., D'Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., Gersbach C.A.. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015; 12:1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hilton I.B., D'Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A.. Epigenome editing by a CRISPR-Cas9 based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015; 33:510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kearns N.A., Pham H., Tabak B., Genga R.M., Silverstein N.J., Garber M., Maehr R.. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods. 2015; 12:401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M.. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013; 10:1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siddique A.N., Nunna S., Rajavelu A., Zhang Y., Jurkowska R.Z., Reinhardt R., Rots M.G., Ragozin S., Jurkowski T.P., Jeltsch A.. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a–Dnmt3L single-chain fusion protein with increased DNA methylation activity. J. Mol. Biol. 2013; 425:479–491. [DOI] [PubMed] [Google Scholar]
- 19. Heigwer F., Kerr G., Boutros M.. E-CRISP: fast CRISPR target site identification. Nat. Methods. 2014; 11:122–123. [DOI] [PubMed] [Google Scholar]
- 20. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koushik S. V, Chen H., Thaler C., Puhl H.L., Vogel S.S.. Cerulean, Venus, and VenusY67C FRET reference standards. Biophys. J. 2006; 91:L99–L101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rohde C., Zhang Y., Reinhardt R., Jeltsch A.. BISMA–fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010; 11:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K.. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013; 10:977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nunna S., Reinhardt R., Ragozin S., Jeltsch A.. Targeted methylation of the epithelial cell adhesion molecule (EpCAM) promoter to silence its expression in ovarian cancer cells. PLoS One. 2014; 9:e87703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stolzenburg S., Beltran A.S., Swift-Scanlan T., Rivenbark A.G., Rashwan R., Blancafort P.. Stable oncogenic silencing in vivo by programmable and targeted de novo DNA methylation in breast cancer. Oncogene. 2015; 34:5427–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li F., Papworth M., Minczuk M., Rohde C., Zhang Y., Ragozin S., Jeltsch A.. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007; 35:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bernstein D.L., Le Lay J.E., Ruano E.G., Kaestner K.H.. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J. Clin. Invest. 2015; 125:1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013; 154:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jurkowska R.Z., Anspach N., Urbanke C., Jia D., Reinhardt R., Nellen W., Cheng X., Jeltsch A.. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 2008; 36:6656–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rajavelu A., Jurkowska R.Z., Fritz J., Jeltsch A.. Function and disruption of DNA methyltransferase 3a cooperative DNA binding and nucleoprotein filament formation. Nucleic Acids Res. 2012; 40:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emperle M., Rajavelu A., Reinhardt R., Jurkowska R.Z., Jeltsch A.. Cooperative DNA binding and protein/DNA fiber formation increases the activity of the Dnmt3a DNA methyltransferase. J. Biol. Chem. 2014; 289:29602–29613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K.. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gowher H., Stockdale C.J., Goyal R., Ferreira H., Owen-Hughes T., Jeltsch A.. De novo methylation of nucleosomal DNA by the mammalian Dnmt1 and Dnmt3A DNA methyltransferases. Biochemistry. 2005; 44:9899–9904. [DOI] [PubMed] [Google Scholar]
- 36. Takeshima H., Suetake I., Tajima S.. Mouse Dnmt3a preferentially methylates linker DNA and is inhibited by histone H1. J. Mol. Biol. 2008; 383:810–821. [DOI] [PubMed] [Google Scholar]
- 37. Felle M., Joppien S., Németh A., Diermeier S., Thalhammer V., Dobner T., Kremmer E., Kappler R., Längst G.. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011; 39:8355–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morselli M., Pastor W.A., Montanini B., Nee K., Ferrari R., Fu K., Bonora G., Rubbi L., Clark A.T., Ottonello S. et al. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. Elife. 2015; 4:e06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schübeler D.. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015; 520:243–247. [DOI] [PubMed] [Google Scholar]
- 40. Vojta A., Dobrinić P., Tadić V., Bočkor L., Korać P., Julg B., Klasić M., Zoldoš V.. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016; 44:5615–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDonald J.I., Celik H., Rois L.E., Fishberger G., Fowler T., Rees R., Kramer A., Martens A., Edwards J.R., Challen G.A.. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol. Open. 2016; 5:866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.