Abstract
During primed CRISPR adaptation spacers are preferentially selected from DNA recognized by CRISPR interference machinery, which in the case of Type I CRISPR–Cas systems consists of CRISPR RNA (crRNA) bound effector Cascade complex that locates complementary targets, and Cas3 executor nuclease/helicase. A complex of Cas1 and Cas2 proteins is capable of inserting new spacers in the CRISPR array. Here, we show that in Escherichia coli cells undergoing primed adaptation, spacer-sized fragments of foreign DNA are associated with Cas1. Based on sensitivity to digestion with nucleases, the associated DNA is not in a standard double-stranded state. Spacer-sized fragments are cut from one strand of foreign DNA in Cas1- and Cas3-dependent manner. These fragments are generated from much longer S1-nuclease sensitive fragments of foreign DNA that require Cas3 for their production. We propose that in the course of CRISPR interference Cas3 generates fragments of foreign DNA that are recognized by the Cas1–Cas2 adaptation complex, which excises spacer-sized fragments and channels them for insertion into CRISPR array.
INTRODUCTION
The CRISPR–Cas (Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated genes) adaptive immunity systems of prokaryotes confer protection against mobile genetic elements such as bacteriophages and plasmids (1–3). A CRISPR–Cas system is composed of two essential parts: a set of cas genes and a CRISPR array. CRISPR arrays consist of short repeats separated by unique ‘spacer’ sequences, some of which are derived from invader DNA (4). The CRISPR–Cas systems can be classified into two classes, six types and multiple subtypes (4,5). Despite this variety, all CRISPR–Cas systems share a common mechanism of action. Once a CRISPR array is transcribed, its transcript is processed into small crRNAs (each containing a spacer sequence and flanking repeat sequences) that are bound by Cas proteins. The resulting effector complex then recognizes ‘protospacers’—target sequences complementary to crRNA spacer (1,6). In CRISPR–Cas systems that solely target DNA (Types I, II and V) protospacer recognition is accompanied by localized DNA melting and formation of an R-loop containing an RNA–DNA heteroduplex between crRNA spacer and one strand of protospacer DNA. The other, non-target protospacer strand is displaced and remains single-stranded (7–9). The DNA bound by the effector complex is cleaved and, eventually, destroyed, either by the protein component of the effector complex (Type II and V systems) (10,11) or by a separate executor nuclease/helicase Cas3 (Type I systems) (12–14). The entire sequence of events is referred to as ‘CRISPR interference’ and is responsible for the protective function of CRISPR–Cas systems.
New spacers are introduced into CRISPR arrays during a process termed ‘CRISPR adaptation’ (15). The acquisition of new spacers predominantly occurs at promoter-proximal side of CRISPR array and requires at least one repeat and a fragment of the upstream leader region (16,17). Spacer acquisition also requires Cas1 and Cas2, the most conserved protein components of all CRISPR–Cas systems (18). An addition of spacer also leads to the appearance of a new repeat copy.
For Type I CRISPR–Cas systems, two modes of adaptation have been described. Naïve adaptation requires just the Cas1 and Cas2 proteins. While biased towards incorporation of spacers from extrachromosomal DNA (17,19,20), it is relatively inefficient. A much more efficient ‘primed adaptation’ requires all components of the CRISPR–Cas system and a crRNA whose spacer matches, partially or fully, a protospacer in foreign DNA. Priming specifically increases acquisition of spacers located in cis with the protospacer recognized by the effector complex. In the case of the Escherichia coli Type I-E system priming leads to preferential acquisition of spacers from the non-target strand (21–23). In contrast, naïve adaptation by this system proceeds without a strand bias (17).
Recently, important details of molecular mechanisms of spacer incorporation into CRISPR array by Type I Cas1 and Cas2 were revealed. It was shown that the two proteins form a complex that introduces single-stranded breaks on both sides of the leader-proximal CRISPR repeat. Intermediates of spacer incorporation at the sites of Cas1–Cas2 generated nicks were detected in vivo and in vitro (24,25). Similar intermediates are known for transposase-mediated reactions suggesting that spacer acquisition and transposon integration reactions are mechanistically similar (26,27). The Cas1–Cas2 complex was crystallized bound to partially double-stranded splayed DNA fragments that may correspond to physiologically relevant fragments of foreign DNA on their way of becoming spacers (28,29).
In general, spacers must be selected for their subsequent functionality in CRISPR interference and to avoid autoimmunity (30,31). Efficient interference requires, in addition to a match between crRNA spacer and target protospacer, the presence of PAM (protospacer-associated motif) (6,32,33). In E. coli, consensus interference-proficient AAG PAM is also preferentially recognized during adaptation (17).
Little is known about the earliest stages of CRISPR adaptation during which spacers are selected and mechanisms responsible for preferential selection of new spacers from one strand of target DNA during primed adaptation are still obscure. In this work, we studied DNA association of the Cas1 protein during primed CRISPR adaptation by the E. coli type I-E system. We show that Cas1 is associated with protospacer-sized non-double-stranded fragments of foreign DNA. These fragments are excised from longer non-double-stranded fragments of foreign DNA that are generated by Cas3. Our results suggest an intimate mechanistic link between CRISPR interference and primed adaptation and unite both parts of the CRISPR response.
MATERIALS AND METHODS
Strains and plasmids
Escherichia coli KD263 (K-12 F+, lacUV5-cas3 araBp8-cse1, CRISPR I: repeat-spacer g8-repeat, ΔCRISPR II) has been described (34). Escherichia coli KD454 is a derivative of KD263 carrying deletion of cas3 gene, it was obtained by recombineering (35). Escherichia coli AM7-7 is a derivative of KD263 carrying deletion of ihfA gene, it was obtained by P1-mediated transduction (36). Escherichia coli BW40297 has been described (21).
Plasmids pG8 and pG8mut have been described previously (21). Plasmid pG8mut_CCG is pG8mut derivative containing CCG PAM instead of AAG PAM in front of hot protospacer 1 (GTGCTCATCATTGGAAAACGTTCTTCGGGGCGA). The mutation was introduced by standard site-directed mutagenesis protocol with primers HS1_CCG for and HS1_CCG rev (primer sequences are available in Supplementary Table S1).
Antibody preparation and purification
A pET28-based expression plasmid for co-overproduction of N-terminalally 6-His-tagged Cas1 and untagged Cas2 was constructed by amplifying an E. coli genomic fragment containing cas1 and cas2 with appropriate primers and cloning under the inducible T7 RNAP promoter. Plasmid-borne cas genes were expressed in E. coli BL21 (DE3) strain in LB medium containing 30 μg/ml kanamycin. Cells were grown at 37°C until OD600 reached 0.6 followed by induction with 1 mM isopropyl 1-thio-β-d-galactopyranoside and further growth for 2 h. Cells were harvested by centrifugation for 20 min at 5000 × g at 4°C and frozen at −80°C. Cell pellets were resuspended in buffer A (20 mM Tris pH 8, 0.5 M NaCl) containing 1 mg/ml lysozyme. Cells were disrupted by sonication and cells lysate was clarified by centrifugation at 16 000 × g for 1 h and filtering through a 0.45 μm filter. The extract was loaded on a 1 ml Chelating HP column (GE Healthcare) loaded with Ni2+ and equilibrated with buffer A. The column was washed with buffer A containing 20 mM and 50 mM imidazole and bound proteins were eluted with 300 mM imidazole in buffer A. A gel showing material in eluted fractions is shown in Supplementary Figure S1A. Fractions 6 and 7 were pooled and used to immunize rats. Antisera were tested by Western blotting against material used for immunization and further purified on an affinity column containing recombinant Cas1 (purified as described above from cells expressing hexahistidine-tagged Cas1 only from a pET28 plasmid) immobilized on cyanogen bromide-activated sepharose (Sigma-Aldrich) according to manufacturer instructions. The reactivity of the antibody (1:5000 dilution) on a western blot against proteins from whole cell extracts of induced and uninduced KD263 E. coli cells is shown in Supplementary Figure S1B). While the final purified antibody preparation was reactive against Cas1, it pulled-down Cas2 from whole-cells extracts of induced cells (Supplementary Figure S1C).
In vivo induction of CRISPR–Cas system
Escherichia coli KD263, AM7-7, KD454 or BW40297 were transformed with pG8mut, pG8mut_CCG or pT7blue (Novagen) plasmids. Transformants were selected on LB agar plates containing 100 μg/ml ampicillin. Individual transformants were grown in liquid medium and induced as described (37). Three hours post-induction aliquots of induced and uninduced cultures were processed for ChIP or total DNA purification.
Chromatin immunoprecipitation and real-time PCR quantification
Ten milliliters of induced and uninduced cultures were harvested 3 hours post-induction. Chromatin immunoprecipitation procedure with purified antibody was performed as described with minimal modifications (38). In short, formaldehyde was added to cultures to final concentration 1% and incubated for 20 min at RT with rotation. Three biological replicates or every immunoprecipitation experiment were performed. The reaction was quenched by adding glycine (0.5 M final concentration) and incubated under same condition for 5 min. Twenty milliliters of cross-linked cells were pelleted by centrifugation and washed three times with TBS (pH 7.5). One milliliter of lysis buffer (10 mM Tris (pH 8.0), 20% sucrose, 50 mM NaCl, 10 mM EDTA, 20 mg/ml lysozyme and 0.1 mg/ml RNaseA) was added and samples were incubated at 37°C for 30 min. After adding of 4 ml of IP buffer (50 mM HEPES–KOH (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS and 1 mM PMSF) the samples were subjected to sonication on Vibra-Cell VCX130 machine (Sonics) at 80% power for 5 min yielding in DNA fragments of 200–300 bp length. This and later steps were performed on ice. After centrifugation, 800 μl of supernatant was preincubated with 20 μl of Protein A/G Sepharose beads (Thermoscientific) to pull down unspecific interactors with the resin and unbound fraction was combined with 30 μl of BSA-blocked Protein A/G Sepharose and 7 μl of anti-Cas1 antibody and incubated overnight on a rotary platform. Standard washing with IP buffer, high salt IP buffer, wash buffer and TE buffer and elution steps were performed as described (38). Immunoprecipitated samples and sheared input samples DNA were de-cross linked in 0.5× elution buffer containing 0.8 mg/ml Proteinase K at 42°C for 2 h followed by 65°C for 6 h. DNA was precipitated with glycogen and dissolved in 20 μl of MilliQ water. A typical yield of DNA yield was 40–60 ng. Each qPCR reaction was carried out in triplicate (technical repeats) in a 20 μl reaction volume with 0.8 units of HS Taq DNA polymerase (Evrogen) and 0.01 μl of Syto13 intercalating dye (LifeTechnology) using DTlite4 (DNA-Technology) amplifier. For each reaction, melting curves were analyzed to ensure amplicon quality and exclude primer dimer formation during amplification. Amplicons from qPCR reactions were cloned and, for each amplicon, several randomly chosen recombinant plasmids sequenced. In each case the cloned inserts size and sequence matched the expectation (Supplementary Table S2). Enrichment ratio ΔΔCt = ΔCt ind (mean Ct IP – mean Ct input) − ΔCt unind(mean Ct IP – mean Ct input) was determined. To convert ΔΔCt values to relative differences in amplicon concentrations a 2−ΔΔCt value was determined. 10 μl aliquots of ChIP material were treated with 5 units of TaiI/FaiI restriction endonucleases (ThermoScientific) following manufacturer's instructions. After precipitation, qPCR was conducted as described above. The fold enrichment between treated and untreated DNA was next calculated as 2−(ΔΔCt[treated] − ΔΔCt[untreated]).
Total DNA purification and analysis
Total DNA was prepared from cells collected from 2 ml of induced or uninduced cell cultures using Genomic DNA Purification Kit (ThermoScientific) following manufacturer's instructions and adding glycogen (ThermoScientific) during precipitation steps to promote recovery of short and singe-stranded DNA fragments. Total DNA (∼5 μg) was dissolved in 25 μl of deionized water. 10 μl aliquots were treated with 5 units of S1 nuclease (ThermoScientific) following manufacturer's instructions. After precipitation, qPCR was conducted as described above with three biological replicates performed. Normalization has been performed as follows: ΔCt = mean Ct sample – mean Ct gyr, where the latter term is obtained for amplification of a gyrA gene fragment.
Spacer acquisition analysis
Aliquots of induced and uninduced total DNA samples were subjected to PCR with primers P4518 and P4581 annealing at both sides of the CRISPR array. The results were analyzed by agarose gel electrophoresis. To analyze the pattern of newly acquired spacers the PCR product corresponding to expanded CRISPR array was gel purified with QIAquick Gel Extraction Kit (QIAGEN) and sequenced using Miseq Illumina in pair-end 250-bp long reads mode according to manufacturer's protocols. Analysis was performed as described earlier (39). To compare pattern of spacer choice specificity between different experiments, the Pearson coefficient, which is a measure of the linear dependence between two variables was used. A Pearson coefficient of 1 indicates total positive linear correlation, 0—no linear correlation and −1—total negative linear correlation.
Primer extension analysis
Oligonucleotides HS1 for pr/ext, HS1 rev pr/ext, HS2 for pr/ext or HS2 rev pr/ext were radiolabeled with γ-[32P]ATP at the 5΄ end with T4 PNK. Extension reactions (10 μl) were performed with 200 ng of total DNA as a template using 40 thermal cycles of 15 s at 95°C, 30 s at 50°C and 30 s at 72°C. The reactions contained 1 pmol of labeled primer, 0.2 mM dNTP, 1 μl of 10× buffer, 5 units of Taq polymerase. As a marker, sequencing reactions with the same primer were set up on the purified pG8mut plasmid as template using the Thermo Sequenase Cycle Sequencing Kit (Affymetrix) following the manufacturer's instructions. The products were separated by denaturing (urea) 6% polyacrylamide gel and visualized using a Phosphorimager.
Generation of model substrates
20 pmol of oligonucleotides HS1_full/cmp for and HS1_full/cmp rev (or HS1_part/cmp for and HS1_part/cmp rev) were subjected to annealing (5 min at 95°C followed by slow temperature reduction in 1× annealing buffer (20 mM Tris–HCl (pH 7.5), 50 mM NaCl, 20 mM MgCl2) in 100 μl reactions. The annealed substrate was precipitated with glycogen (ThermoScientific) and used in downstream experiments. 210-bp long model substrate was prepared by PCR with appropriate primer pairs using pG8mut plasmid as a template for amplification. PCR-products were purified with GeneJET PCR Purification Kit (ThermoScientific). 1 pmole of model fragments was used to S1 or TaiI/FaiI treatment.
RESULTS
Cas1 is associated with CRISPR array during primed adaptation
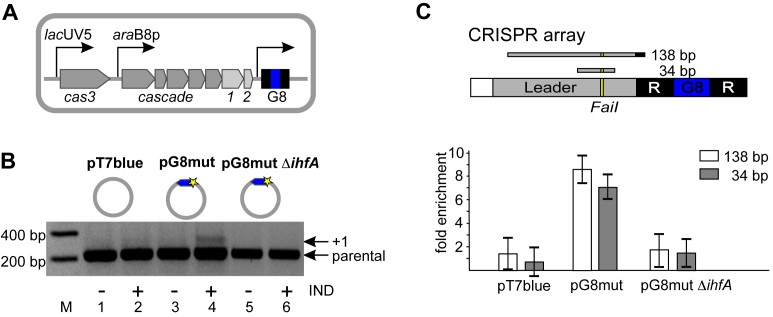
Escherichia coli strain KD263 cells are capable of inducible cas gene expression and contain an engineered CRISPR array with a single spacer named G8 (Figure 1A). Uninduced KD263 were transformed with a pT7blue-based plasmid pG8mut harboring a C to T substitution in the first position of the G8 protospacer (21) or control empty pT7blue vector. By introducing a mismatch with crRNA, the C1T substitution decreases CRISPR interference but strongly stimulates primed adaptation (21). Transformed cells were grown without antibiotic required for plasmid maintenance but in the presence of inducers of cas genes expression. As expected, expansion of CRISPR array in induced cultures harboring the protospacer plasmid but not the vector control was detected (Figure 1B). Cultures analyzed in Figure 1B were also subjected to formaldehyde crosslinking followed by immunoprecipitation with polyclonal antibody raised against Cas1 (see Materials and Methods). The precipitated material was subjected to qPCR with a pair of primers that amplified a 138 bp fragment spanning the CRISPR leader and a portion of the first repeat or a shorter, 34 bp, leader fragment (Figure 1C). For both DNA fragments, the fold enrichment between induced and uninduced cultures harboring each plasmid was determined. There was no enrichment of leader DNA in antibody-associated fraction from induced cells harboring pT7blue. In contrast, at least 8-fold enrichment in cells undergoing primed adaptation was observed (Figure 1C, Supplementary Figure S2A). Since the amount of Cas1 in both induced cultures was similar (Supplementary Figure S2B, compare lanes 2 and 4), we conclude that Cas1 associates with the CRISPR array leader only in cells undergoing primed adaptation.
Figure 1.
Cas1 is associated with CRISPR array leader in cells undergoing primed adaptation. (A) The KD263 Escherichia coli cells containing inducible cas genes and a single CRISPR array containing G8 spacer (blue) are schematically shown. (B) KD263 cells were transformed with pT7blue vector or pG8mut plasmid containing a G8 protospacer (blue) with a mismatch mutation (yellow star). The KD263 derivative strain carrying a deletion of the ihfA gene was transformed with pG8mut plasmid. Cells were grown with or without induction (‘+/−’, correspondingly) and PCR with oligonucleotide primers annealing upstream and downstream of CRISPR array was performed. ‘Parental’ marks a PCR amplification product corresponding to unexpanded, parental CRISPR array. The ‘+1’ band corresponds to CRISPR array expanded by one spacer-repeat unit. (C) The KD263 CRISPR array and upstream leader region is schematically shown at the top. 138 and 34 bp DNA products of amplification are shown. The FaiI recognition site is indicated. Below, results of Cas1 ChIP analysis with primer pairs amplifying each of the two leader fragments and cells shown in panel B are presented. Results from two additional biological replicates are shown in Supplementary Figure S2A. Here and in other figures showing qPCR data fold enrichment values correspond to ratios of relative concentrations of each fragment subjected to amplification in induced and uninduced cells containing indicated plasmids. Mean values and standard deviations in triple technical replicates are presented.
It has recently been reported that IHF, an architectural DNA binding protein that interacts with the AT-rich leader, is required for spacer acquisition (40). Indeed, the KD263 cells harboring the disrupted ihfA gene and transformed with pG8mut did not acquire new spacers (Figure 1B). There was also no enrichment of leader DNA in Cas1-antibody-associated fraction from these cells (Figure 1C).
Cas1 is preferentially associated with protospacers efficiently acquired from target DNA
Amplified DNA corresponding to extended CRISPR array in cells harboring the pG8mut plasmid (Figure 1B, lane 4) was subjected to high-throughput sequencing. Most newly acquired spacers corresponded to protospacers with consensus PAMs and matched the non-targeted strand of the plasmid. Certain plasmid protospacers behaved as hot spots and were preferentially used as a source of spacers (Figure 2A, the height of bars corresponding to individual protospacers reflects the frequency of occurrence of the corresponding spacer in expanded arrays). One ‘hot’ protospacer (HS1, 99 244 reads, Figure 2A) and one ‘cold’ protospacer (CS, 25 reads, Figure 2A) were chosen for further analysis. When the HS1 consensus AAG PAM was changed to CCG (plasmid pG8mut_CCG, Figure 2B) no spacers corresponding to HS1 were acquired, while efficiency of other protospacers use was unaffected (Pearson correlation co-efficient of 0.95, P-value < 2.2e−16). The material precipitated with Cas1-specific antibody from induced and uninduced KD263 cultures harboring pG8mut, pG8mut_CCG, and pT7blue was analyzed by qPCR with primer pairs that amplified 33 nucleotide-long HS1 or CS protospacers (Figure 2C). Strong (∼16-fold) enrichment for HS1 in induced cells carrying pG8mut but not pG8mut_CCG or pT7blue was observed. In contrast, the enrichment level of CS in induced cells carrying pG8mut was insignificant and similar to that in pG8mut_CCG or pT7blue carrying cells.
Figure 2.
Cas1 is associated with hot protospacers in target DNA during primed adaptation. (A, B) Spacers acquired by KD263 cells transformed with pG8mut (A) and pG8mut_CCG (B) plasmids. The priming G8 protospacer is shown as a blue arrow with an asterisk indicating mutation introducing a mismatch with G8 crRNA spacer. The heights of the bars emanating from circles representing each plasmid indicate the relative efficiency of protospacer use (number of times reads with matching acquired spacers were observed). CS and HS1 are, correspondingly, cold and hot protospacers. In A, the bar indicating the efficiency of spacer acquisition from the HS1 protospacer is highlighted in green. No spacers from HS1 were acquired from pG8mut_CCG (B). (C) Results of Cas1 ChIP analysis with primer pairs amplifying 33 bp amplicons corresponding to HS1 and CS are presented. Results from two additional biological replicates are shown in Supplementary Figure S3A. (D) The HS1 protospacer (green) and the flanking plasmid DNA sequences are shown. The AAG PAM is highlighted with a red box. The TaiI recognition site is underlined. Below, primers used to amplify HS1 containing fragments of different lengths are schematically shown. (E) Results of Cas1 ChIP analysis with primer pairs amplifying HS1 amplicons of different lengths (marked as in panel D) are presented. Results from two additional biological replicates are shown in Supplementary Figure S3A. (F) Results of PCR amplification (40 cycles) of pre- and post-immunoprecipitation material from KD263 cells transformed with pG8mut plasmid. Primers complementary to HS1 and a region downstream of CRISPR array were used to amplify incorporated HS1 spacer (‘HS1’, right-hand side). Amplification of 138-bp leader fragment (‘Leader’, left hand-side) with appropriate primers was performed as a positive amplification control. (G) Results of Cas1 ChIP analysis with primer pairs amplifying 33 bp DNA fragment corresponding to HS1 in KD263 and derivative strain with deletion of the ihfA gene are presented.
Cas1-associated DNA was also probed with primer pairs amplifying HS1-containing plasmid fragments of longer lengths (Figure 2D and E). The result showed that fold enrichment decreased when primer pairs amplifying 47-bp DNA fragments extended at either side of the HS1 protospacer were used. A longer, 61-bp amplicon that contained HS1 in its center was not enriched in the Cas1-associated fraction. A similar result was obtained with another hot protospacer, HS2 (Supplementary Figure S3B–D). The observed length dependence of Cas1-associated material suggests that target DNA fragments preferentially associated with Cas1 are protospacer (or spacer)-sized.
To exclude a possibility that preferential amplification of protospacer-sized Cas1-associated DNA is due to amplification of spacers that have been already acquired into the CRISPR array of cells transformed with pG8mut, we monitored incorporation of HS1 spacer in CRISPR array using an appropriate specific primer and another primer annealing downstream of the CRISPR array. The expected chromosomal amplification product was readily detectable in input material before precipitation with Cas1 antibodies but was absent even after 40 amplification cycle of DNA associated with Cas1 (Figure 2F). The same result was obtained with HS2 (Supplementary Figure S3E).
KD263 cells lacking functional IHF and unable to acquire new spacers were also tested. While these cells did not contain Cas1-associated leader DNA (Figure 1C), the level of enrichment for HS1 was even higher (∼24-fold versus ∼16-fold) than in KD263 cells with functional IHF (Figure 2G).
Together, these results establish that Cas1 is associated with protospacers from target DNA rather than with newly incorporated spacers in the CRISPR array. The Cas1-associated fragments are short and are thus no longer part of the original plasmid from which spacers are selected from. The enrichment of Cas1-associated protospacer DNA is correlated with the efficiency of protospacer use during adaptation.
Recently, data have been presented suggesting that Cas1 and Cas2 stimulate Cas3 recruitment to the priming site (41). We tested whether the priming G8 protospacer in the pG8mut plasmid is associated with Cas1 but found no enrichment (Supplementary Figure S3F).
Cas1-associated target protospacers are not in a standard double-stranded form
To assess the state of Cas1-associated target DNA we made use of the presence of TaiI restriction endonuclease recognition site in the HS1 protospacer (Figure 2D). After reversal of cross-linking, Cas1-associated DNA was treated with TaiI followed by amplification. Such treatment had no effect on the enrichment of the 33-nucleotide HS1 fragment (Figure 3A). Likewise, treatment with FaiI had no effect on enrichment of a 33-nucleotide DNA fragment for HS2, one of the top 20 most used protospacers that contains an internal FaiI site (Supplementary Figure S4A). In contrast, FaiI treatment abolished enrichment of a Cas1-associated 34-nucleotide leader amplicon (Figure 3A), an expected result for a double-stranded chromosomal DNA. Fully double-stranded model 33-bp HS1 or HS2 substrates were also efficiently destroyed by, respectively, TaiI or FaiI, while corresponding single-stranded DNA oligonucleotides of the same size were fully resistant (Figure 3B, Supplementary Figure S4B). As an additional control, we digested input DNA (after reversal of cross-linking but before Cas1 precipitation) with TaiI and then determined the effect of this digestion on a randomly selected 33-bp amplicon of E. coli genomic DNA containing a TaiI site. The TaiI treatment led to disappearance of the amplicon (Figure 3B). Together, these data show that target DNA fragments associated with Cas1 during primed adaptation are resistant to restriction endonuclease digestion and are therefore not present in a standard double-stranded form.
Figure 3.
Cas1-associated protospacer DNA is resistant to restriction endonuclease digestion. (A) KD263 cells transformed with pG8mut were processed for Cas1 ChIP analysis. After immunoprecipitation aliquots of Cas1-associated DNA were treated with FaiI or TaiI restriction endonucleases. qPCR analysis with a primer pairs amplifying the HS1 protospacer (33 nucleotide) or 34-nucleotide CRISPR array leader fragment was performed. The ratios of fold enrichment values between TaiI-(for HS1) or FaiI- (for leader) treated (‘+R’) and corresponding untreated (‘−R’) samples are presented. (B) Model single-stranded (ss) or double-stranded (ds) HS1 substrates were treated with TaiI followed by qPCR analysis. ‘ds*’ is a partially double stranded HS1-based substrate whose structure corresponds to nucleic acid fragments bound to Cas1–Cas2 in available structures (25,29). For bars labeled ‘ds’, ‘ss’, and ‘ds*’ qPCR analysis was performed with a primer pair amplifying HS1 protospacer. For bar labeled ‘host’ total E. coli DNA was subjected to TaiI treatment and primer pair amplifying a 33 bp genomic amplicon containing an internal TaiI site was performed. The ratios of apparent concentrations (calculated from Ct values) with and without TaiI treatment are presented. Mean values and standard deviations for triple replicates are shown.
Recently, a structure of the E. coli Cas1–Cas2 adaptation complex bound to a partially double-stranded model substrate was determined (28,29). Similar substrates based on the HS1 protospacer were fully sensitive to TaiI digestion (Figure 3B). Thus, Cas1-associated fragments from cells undergoing primed adaptation are different from substrates bound to the adaptation complex in published structures.
Target DNA in cells undergoing CRISPR interference or primed adaptation is sensitive to single-strand specific nuclease digestion
To assess the state of plasmid DNA in cells undergoing primed adaptation, total DNA was prepared from induced and uninduced KD263 cells carrying the pG8mut plasmid. As controls DNA was also prepared from cultures of KD454 (a KD263 derivative lacking cas3 and unable to undergo CRISPR interference) and BW40297 (no functional cas1, able to undergo CRISPR interference) (Figure 4A) transformed with pG8mut. The KD454 and BW40297 cells were incapable of primed adaptation, as expected (Supplementary Figure S5A). After treating total DNA prepared from induced an uninduced cultures with S1 nuclease, qPCR was performed with a primer pair amplifying a 210 nt pG8mut fragment containing the HS1 protospacer. A fragment of this length is fully resistant to S1 treatment if double-stranded (Supplementary Figure S5B). Indeed, S1 treatment of DNA from uninduced cultures had no effect on qPCR signal (Figure 4B). In contrast, S1 treatment of DNA from induced KD263 cultures increased the qPCR threshold cycle (Ct) value by at least three cycles (equivalent to ∼8-fold decrease in template DNA), suggesting that a significant portion of plasmid DNA in cells undergoing primed adaptation is present in single-stranded form. An increase of Ct value was also observed when total DNA from cells harboring pG8mut_CCG was treated with S1 prior to qPCR (Supplementary Figure S5C). The S1 treatment had no effect on amplification of a 138 bp fragment of CRISPR leader (Figure 4B), an expected result for double-stranded chromosomal DNA. S1 treatment of total DNA from KD454 cells also had no effect on the Ct value of 210 nt pG8mut amplicon (Figure 4B). In contrast, BW40297 cells behaved like KD263 (Figure 4B). It therefore follows that extended-length S1 sensitive fragments of target DNA require functional Cas3 and do not require catalytically active Cas1.
Figure 4.
CRISPR interference leads to appearance of S1 nuclease sensitive target DNA in vivo. (A) Three E. coli strains used are shown. KD263 is capable of both CRISPR interference and adaptation. KD454 is incapable of CRISPR interference. BW40297 is capable of interference but not adaptation. (B) Total DNA was prepared from cells shown in A transformed with pG8mut and grown in the presence or in the absence of cas gene expression inducers. Half of each sample was treated with S1 nuclease. qPCR with primer pairs amplifying a 210 nucleotide plasmid fragment or 138 nucleotide CRISPR leader fragment was performed. For each sample, Ct values are presented. For KD263 and BW40297 samples, carrying pG8mut plasmid, higher Ct values observed for plasmid amplicon in the presence of inducers are due to CRISPR interference.
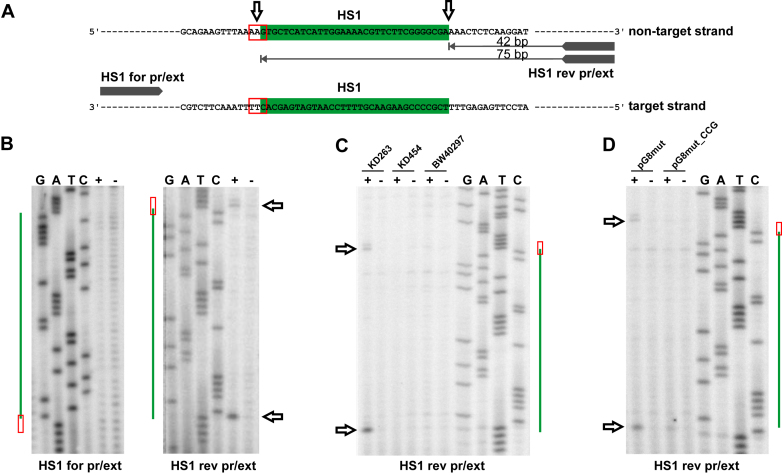
Protospacer-sized fragments are excised from one strand of target DNA
As shown in previous sections, the Cas1 is associated with short non-double-stranded DNA fragments of target DNA. S1 sensitive fragments generated by Cas3 nuclease are considerably longer. To determine which strand of target DNA protospacer-sized fragments originate from, total DNA from KD263 cells carrying the pG8mut plasmid was subjected to primer extension analysis with primers annealing upstream and downstream of the HS1 protospacer (Figure 5A). No primer extension products were detected with a primer annealing upstream of HS1. In contrast, two distinct primer extension products at the boundaries of the HS1 protospacer (and including the last G of the AAG PAM) were detected with the downstream primer (anneals 42 bp away from HS1 to a strand targeted by the priming protospacer) (Figure 5B). A similar result was obtained for HS2 protospacer (Supplementary Figure S6). The primer extension products at protospacer boundaries were only detected in the presence of functional Cas3 and Cas1 (Figure 5C). When primer extension reaction was conducted with DNA prepared from cultures harboring pG8mut_CCG, no cleavage at PAM was detected (Figure 5D). Interestingly, primer extension product corresponding to the downstream cleavage was not strongly affected. We propose that primer extension products mark the boundaries of protospacers excised by Cas1 from DNA intermediates generated by Cas3 and channeled for incorporation into the CRISPR array.
Figure 5.
Target DNA is nicked at both sides of protospacer in cells undergoing primed adaptation. (A) A fragment of plasmid DNA containing the HS1 protospacer (green) is shown and positions of annealing of primers used for primer extension reactions are indicated. For HS1 rev pr/ext primer, leftward-oriented arrows indicate primer extension products detected. (B) Results of primer extension experiment using total DNA purified from KD263 cells transformed with pG8mut with (‘+’) and without (‘−’) cas gene induction. At the side of each autoradiogram, the position of the HS1 protospacer (green) and PAM (red box, see also А) is shown. Horizontal arrows indicate the positions of primer extension products migration on the gel. (C) As in B but using DNA prepared from indicated cells for primer extension with the HS1 rev pr/ext primer. (D) Primer extension reactions were performed with DNA prepared from KD263 cells transformed with pG8mut or pG8mut_CCG plasmids.
DISCUSSION
The process of CRISPR adaptation must consist of multiple steps. A protospacer in foreign DNA with a functional PAM must be selected, a protospacer-sized fragment of foreign DNA must be generated, and, finally, the reaction of spacer incorporation in the leader-proximal end of the CRISPR array must occur. Recently, significant progress in late events of the spacer adaptation pathway has been achieved (42,43). In contrast, the early events of the pathway remain poorly understood. During primed adaptation, protospacers located in cis to the priming protospacer bound by the effector complex must be selectively recognized and a strand bias in spacer acquisition must be maintained somehow. Here, we present evidence that in E. coli cells undergoing primed adaptation, Cas1 is associated with protospacer-sized fragments of plasmid DNA. These fragments are not in the standard double-stranded DNA form as they are resistant to restriction endonuclease digestion. The abundance of the Cas1-associated fragments is correlated with efficiency of protospacer use as spacer donors. We propose that these fragments correspond to in vivo intermediates of the CRISPR adaptation pathway on their way to be incorporated in the array. Cas1 can also be detected on the CRISPR array, but only at conditions of ongoing CRISPR adaptation.
The adaptation complex Cas1–Cas2 can either (i) itself generate protospacer-sized DNA fragments from plasmids containing priming protospacers; (ii) rely on upstream interference machinery, specifically, the Cas3 nuclease/helicase, to generate fragments ready for incorporation into the CRISPR array or (iii) use the products of target DNA degradation by the interference machinery to generate the adaptation substrates. Our data are consistent with the latter scenario. In the presence of active Cas3, significant portion of plasmid DNA carrying the priming protospacer is present as extended (at least 200 nt) fragments that are sensitive to S1 nuclease digestion. These fragments must be generated by Cas3 after the Cascade effector complex recognizes the priming protospacer. The abundance of these fragments does not depend on the frequency of use of protospacers that they contain, however, their presence is required for primed adaptation. We propose that affinity of the Cas1–Cas2 complex to protospacers carried on fragments generated by Cas3 is a major determinant of protospacer ‘hotness’ during primed adaptation.
Earlier data suggested that effector complex interactions with fully matching protospacers lead to CRISPR interference while interactions with partially matching protospacers that abolish interference lead to primed adaptation (21,44–47). Accordingly, it was postulated that two structurally distinct types of complexes, (i) capable of Cas3 recruitment and interference and (ii) capable of Cas3, Cas1 and Cas2 recruitment and adaptation, are formed on, respectively, fully matching and partially matching protospacers (41,48). Recent experiments show, however, that Cascade effector complex interaction with mismatched priming protospacer targets causes interference, albeit at rates slower than those seen for mismatched targets (49). Moreover, when the rates of degradation of matched and mismatched protospacer-carrying DNA are made equal, the former is actually much more efficient in promoting primed adaptation (37). The apparent lack of adaptation with matched targets could thus be a trivial consequence of their rapid destruction, which also eliminates Cas3 degradation products from which spacers are selected by Cas1–Cas2.
Primer extension analysis reveals nicks in the non-target strand of plasmid DNA that should produce protospacer-sized DNA fragments. We hypothesize, that these fragments are the same as those associated with Cas1 during the ChIP experiments. Detection of two primer extension products at each side of the protospacer in our experiments indicates that the first cut is introduced at the PAM (which is distal from the primer) followed by the second cut at another end of the protospacer. This is an expected scenario for Cas1–Cas2 must first recognize a PAM (AAG in case of primed adaptation in E. coli type I-E system (22,34) and then use a ruler-like mechanism to introduce another cut further downstream. The efficiency of selection or spacers from sequences associated with AAG PAMs varies by several orders or magnitude, with some protospacers behaving as hot spots (22). The reasons for such preferential use are not known but clearly, cannot be determined by PAM alone. It has been suggested that additional sequences at the other end of the protospacer can contribute to adaptation efficiency (50). Our analysis of a hot spot with mutated PAM is consistent with this notion, since the downstream cut is maintained in the hot protospacer with PAM mutation, though spacer acquisition is abolished by the mutation.
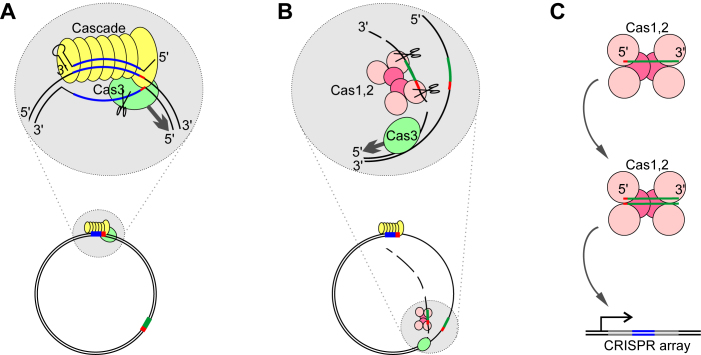
A mechanism of spacer acquisition that is consistent with our data is presented in Figure 6. It is based on known properties of the effector complex (binding to mismatched protospacers and R-loop formation), preferential cleavage by the E. coli Cas3 of the non-targeted strand in the R-loop and its 3΄-5΄ helicase activity, and the data obtained in this work. The model posits that Cas3 processively unwinds target DNA moving along the non-targeted strand in the 3΄ to 5΄ direction. The Cas3 nuclease then generates extended-length S1 nuclease sensitive fragments from the non-target strand, from which Cas1–Cas2 excise protospacers. These fragments are then channeled for integration into CRISPR array. The major features of the model are consistent with recent results obtained in reconstituted in vitro primed adaptation system that showed that Cas3 generated partially single-stranded fragments fuel primed adaption (51), though in this work the S1-sensitivity of Cas3 generated fragments was not assessed.
Figure 6.
A model of primed adaptation by the E. coli Type I-E CRISPR–Cas system. (A) The priming protospacer is recognized by the effector complex, leading to R-loop formation. (B) Cas3 processively unwinds and degrades the DNA strand non-targeted by the priming complex leading to formation of extended single-stranded fragments. The Cas1–Cas2 complex excises protospacers from fragments generated by Cas3. The Cas1–Cas2 complex may (as suggested by the in vitro findings of Redding et al., 2015) or may not associate with Cas3. (C) The Cas1–Cas2 complex, first bound to non-double-stranded protospacers, is converted to adaptation-proficient complex with fully or partially double-stranded protospacers that are inserted in the CRISPR array.
Published data convincingly show that in vitro Cas1–Cas2 use fully or partially double stranded substrates for incorporation in CRISPR array (25,29,51). To make our data consistent with these observations, one has to postulate that fragments of the target strand must reassociate with Cas1–Cas2 bound single-stranded protospacer DNA or that a special mechanism that creates the second strand of Cas1–Cas2 bound protospacers must exist. It has been suggested that the RecBCD nuclease-helicase is responsible for generating material for spacer acquisition during naïve adaptation (20). Available data indicate that RecBCD generated fragments are also single-stranded (52). Thus, whatever the mechanism responsible for generation of intermediates used for incorporation into CRISPR array is, it should be operational both for naïve and primed adaption.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sofia Medvedeva for advising in high throughput data analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [NIGMS RO1 10407] and Russian Science Foundation [14-14-00988] to K.S. and Russian Foundation for Basic Research grant 16-04-00767 to E.S. Funding for open access charge: Skolkovo Institute of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Barrangou R., Fremaux C., Deveau H., Richards M., Moineau S., Romero D.A., Horvath P., Barrangou R., Fremaux C., Deveau H. et al. . CRISPR provides aquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 2. Brouns S.J.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J.H., Snijders A.P.L., Dickman M.J., Makarova K.S., Koonin E. V., van der Oost J.. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008; 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marraffini L.A., Sontheimer E.J.. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008; 322:1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J.J., Charpentier E., Haft D.H. et al. . An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K. et al. . Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell. 2015; 60:385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deveau H., Barrangou R., Garneau J.E., Labonte J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S.. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008; 190:1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul Ü., Wurm R., Wagner R. et al. . Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011; 18:529–536. [DOI] [PubMed] [Google Scholar]
- 8. Szczelkun M.D., Tikhomirova M.S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P., Siksnys V., Seidel R.. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9798–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wiedenheft B., Lander G.C., Zhou K., Jore M.M., Brouns S.J.J., van der Oost J., Doudna J.A., Nogales E.. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011; 477:486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gasiunas G., Barrangou R., Horvath P., Siksnys V.. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA–guided DNA endonuclease in adaptive baceterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Westra E.R., van Erp P.B., Kunne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K. et al. . CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012; 46:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mulepati S., Bailey S.. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 2013; 288:22184–22192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sinkunas T., Gasiunas G., Waghmare S.P., Dickman M.J., Barrangou R., Horvath P., Siksnys V.. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013; 32:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van der Oost J., Jore M.M., Westra E.R., Lundgren M., Brouns S.J.. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009; 34:401–407. [DOI] [PubMed] [Google Scholar]
- 16. Wei Y., Chesne M.T., Terns R.M., Terns M.P.. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 2015; 43:1749–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yosef I., Goren M.G., Qimron U.. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012; 40:5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koonin E. V, Makarova K.S.. CRISPR–Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013; 10:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Díez-Villaseñor C., Guzmán N.M., Almendros C., García-Martínez J., Mojica F.J.M.. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR–Cas I-E variants of Escherichia coli. RNA Biol. 2013; 10:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R.. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015; 520:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E.. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012; 3:945. [DOI] [PubMed] [Google Scholar]
- 22. Savitskaya E., Semenova E., Dedkov V., Metlitskaya A., Severinov K.. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013; 10:716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Swarts D.C., Mosterd C., van Passel M.W.J., Brouns S.J.J.. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012; 7:e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arslan Z., Hermanns V., Wurm R., Wagner R., Pul Ü.. Detection and characterization of spacer integration intermediates in type I-E CRISPR–Cas system. Nucleic Acids Res. 2014; 42:7884–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nuñez J.K., Lee A.S.Y., Engelman A., Doudna J.A.. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature. 2015; 519:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mizuuchi K., Adzuma K.. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991; 66:129–140. [DOI] [PubMed] [Google Scholar]
- 27. Rollie C., Schneider S., Brinkmann A.S., Bolt E.L., White M.F.. Intrinsic sequence specificity of the integrase directs new spacer acquisition. eLife. 2015; 4, doi: 10.7554/eLife.08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nuñez J.K., Harrington L.B., Kranzusch P.J., Engelman A.N., Doudna J.A.. Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature. 2015; 527:535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J., Li J., Zhao H., Wang M., Yin M., Wang Y., Wang J., Li J., Zhao H., Sheng G. et al. . Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR–Cas systems. Cell. 2015; 163:840–853. [DOI] [PubMed] [Google Scholar]
- 30. Stern A., Keren L., Wurtzel O., Amitai G., Sorek R.. Self-targeting by CRISPR: Gene regulation or autoimmunity?. Trends Genet. 2010; 26:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vercoe R.B., Chang J.T., Dy R.L., Taylor C., Gristwood T., Clulow J.S., Richter C., Przybilski R., Pitman A.R., Fineran P.C.. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013; 9:e1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mojica F.J., Diez-Villansenor C., Garcia-Martinez J., Almendros C.. Short motif sequences determin the targets of the prokaryotic CRISPR defence system. Microbiology. 2009; 155:733–740. [DOI] [PubMed] [Google Scholar]
- 33. Horvath P., Romero D.A., Coute-Monvoisin A.C., Richards M., Deveau H., Moineau S., Boyaval P., Fremaux C., Barrangou R.. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008; 190:1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shmakov S., Savitskaya E., Semenova E., Logacheva M.D., Datsenko K. A., Severinov K.. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res. 2014; 42:5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2, doi:10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Semenova E., Savitskaya E., Musharova O., Strotskaya A., Vorontsova D., Datsenko K., Logacheva M., Severinov K.. Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I-E CRISPR–Cas interfering complex. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7626–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kahramanoglou C., Seshasayee A.S.N., Prieto A.I., Ibberson D., Schmidt S., Zimmermann J., Benes V., Fraser G.M., Luscombe N.M.. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011; 39:2073–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Semenova E., Kuznedelov K., Datsenko K. A., Boudry P.M., Savitskaya E.E., Medvedeva S., Beloglazova N., Logacheva M., Yakunin A.F., Severinov K.. The Cas6e ribonuclease is not required for interference and adaptation by the E. coli type I-E CRISPR–Cas system. Nucleic Acids Res. 2015; 43:6049–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nunez J.K., Bai L., Harrington L.B., Hinder T.L., Doudna J.A.. CRISPR immunological memory requires a host factor for specificity. Mol. Cell. 2016; 62:824–833. [DOI] [PubMed] [Google Scholar]
- 41. Redding S., Sternberg S.H., Wiedenheft B., Jennifer A., Greene E.C., Redding S., Sternberg S.H., Marshall M., Gibb B., Bhat P. et al. . Surveillance and processing of foreign DNA by the Escherichia coli CRISPR–Cas system. Cell. 2015; 163:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amitai G., Sorek R.. CRISPR–Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016; 14:67–76. [DOI] [PubMed] [Google Scholar]
- 43. Sternberg S.H., Richter H., Charpentier E., Qimron U.. Adaptation in CRISPR–Cas systems. Mol. Cell. 2016; 61:797–808. [DOI] [PubMed] [Google Scholar]
- 44. Fineran P.C., Gerritzen M.J.H., Suarez-Diez M., Kunne T., Boekhorst J., van Hijum S.A.F.T., Staals R.H.J., Brouns S.J.J.. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E1629–E1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li M., Wang R., Zhao D., Xiang H.. Adaptation of the Haloarcula hispanica CRISPR–Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014; 42:2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Richter C., Dy R.L., McKenzie R.E., Watson B.N.J., Taylor C., Chang J.T., McNeil M.B., Staals R.H.J., Fineran P.C.. Priming in the Type I-F CRISPR–Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014; 42:8516–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J.J., Severinov K.. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blosser T.R., Loeff L., Westra E.R., Vlot M., Künne T., Sobota M., Dekker C., Brouns S.J.J., Joo C.. Two distinct DNA binding modes guide dual roles of a CRISPR–Cas protein complex. Mol. Cell. 2015; 58:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xue C., Seetharam A.S., Musharova O., Severinov K., Brouns S.J.J., Severin A.J., Sashital D.G.. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015; 43:10831–10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yosef I., Shitrit D., Goren M.G., Burstein D., Pupko T., Qimron U.. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc. Natl. Acad. Scie. U.S.A. 2013; 110:14396–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kunne T., Kieper S.N., Bannenberg J.W., Vogel A.I., Miellet W.R., Klein M., Depken M., Suarez-Diez M., Brouns S.J.. Cas3-derived target DNA degradation fragmnents fuel primed CRISPR adaptation. Mol. Cell. 2016; 63:852–864. [DOI] [PubMed] [Google Scholar]
- 52. Dillingham M.S., Kowalczykowski S.C.. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008; 72:642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.