Abstract
The adenovirus L4-22K protein both activates and suppresses transcription from the adenovirus major late promoter (MLP) by binding to DNA elements located downstream of the MLP transcriptional start site: the so-called DE element (positive) and the R1 region (negative). Here we show that L4-22K preferentially binds to the RNA form of the R1 region, both to the double-stranded RNA and the single-stranded RNA of the same polarity as the nascent MLP transcript. Further, L4-22K binds to a 5΄-CAAA-3΄ motif in the single-stranded RNA, which is identical to the sequence motif characterized for L4-22K DNA binding. L4-22K binding to single-stranded RNA results in an enhancement of U1 snRNA recruitment to the major late first leader 5΄ splice site. This increase in U1 snRNA binding results in a suppression of MLP transcription and a concurrent stimulation of major late first intron splicing.
INTRODUCTION
Most adenovirus late proteins are translated from mRNAs originating from the major late transcription unit (MLTU, Figure 1A). The MLTU produces >20 alternatively spliced mRNAs that are grouped into five families, L1 to L5, where each family shares the same 3΄ end polyadenylation site (reviewed in (1)). The MLTU is under the transcriptional control of the adenovirus major late promoter (MLP). The MLP is active at all stages of a virus infection. At early times of infection the MLP has an activity comparable to the other early viral promoters. However, at late times of infection the activity of the MLP increases dramatically. The core MLP contains an initiator element, a consensus TATA box, and two cis-acting upstream elements (UPE and CAAT box, Figure 1B), which bind cellular transcription factors regulating MLP activity (reviewed in (2)). However, the late-specific increase in MLP activity requires the so-called downstream element (DE) (Figure 1B) (3–5), which binds two adenovirus-induced transcription factors, DEF-A and DEF-B. The DEF-B factor has been suggested to be a homodimer of the adenoviral IVa2 protein (6,7) whereas the core factor of DEF-A appears to be the L4-22K protein (8). The DE element is essential for maximal activation of the MLP at late times of infection (3–5,8,9). L4-22K stimulates MLP transcription by binding to two 5΄-TTTG-3΄ motifs located in the DE element (8). Since L4-22K is part of the MLTU it stimulates its own expression in a feed forward type of mechanism.
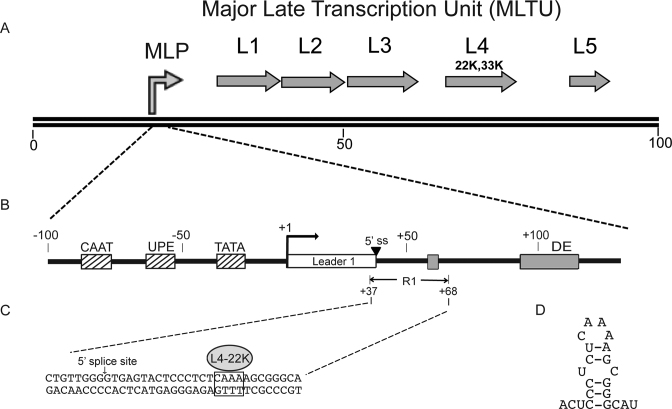
Figure 1.
(A) Schematic drawing depicting the organization of the major late transcription unit. Arrows show the relative position of the five families of late mRNAs with the genomic position of the L4-22K and L4-33K marked. (B) Schematic diagram of the MLP showing the position of upstream transcription factor-binding sites (hatched boxes), the first tripartite leader exon (open box), the major late first leader 5΄ splice site (5΄ss), and the L4-22K binding sites in the first leader intron (gray boxes) in relation to the MLP transcription start site (+1). (C) Nucleotide sequence expansion of the R1 region with the L4-22K binding site indicated. (D) Potential hairpin structure of the sense-strand R1 RNA sequence encompassing the L4-22K binding site.
We recently demonstrated that L4-22K also binds to an additional low affinity site in the MLTU first intron, the so-called R1 region (Figure 1B) (10). Binding of L4-22K to R1 has a suppressive effect on MLP activity by stimulating the recruitment of the cellular transcription factor Sp1 to the DNA at a site overlapping the major late first leader 5΄ splice site.
L4-22K has a closely related partner protein called L4-33K. Both proteins have been shown to serve important functions as regulators of the temporal early to late shift in adenoviral late gene expression (reviewed in (11)). The L4-22K and L4-33K proteins share the 105 amino-terminal amino acids but have unique carboxy-terminal ends. L4-33K is translated from a spliced variant of the L4-22K mRNA where an intron interrupting the L4-22K protein coding sequence has been removed, resulting in a frame shift and a unique carboxy-terminus in the L4-33K protein-coding sequence (reviewed in (11)). L4-33K is a nuclear phosphoprotein that suppresses transcription from the L4P promoter (12), promotes cytoplasmic accumulation of late viral mRNAs and, at least in vitro, activates splicing of MLTU mRNAs with a weak 3΄ splice site context (13,14). L4-22K appears to be a multifunctional protein that also enhances adenoviral late mRNA abundance by post-transcriptional mechanisms (reviewed in ref. 11).
Here, we have further characterized the activity of L4-22K as a regulator of MLP transcription. In contrast to previous studies (8,10,12,15–17), which have focused on the DNA-binding activity of L4-22K we show that L4-22K preferentially binds to double-stranded RNA (dsRNA) and single-stranded RNA (ssRNA) of the same polarity as the nascent MLTU RNA (sense-strand RNA). This binding is sequence-specific. L4-22K binds to the 5΄-CAAA-3΄ motif located 15 nucleotides downstream of the major late first leader 5΄ splice site and stimulates recruitment of U1 snRNA to the 5΄ splice site containing RNA. The increase in U1 snRNA binding has dual effects. Firstly, it results in a suppression of MLP transcription and, secondly, a stimulation of major late first intron splicing.
MATERIALS AND METHODS
Plasmids and recombinant protein purification
Reporter plasmid pGL4-MLP-Luc (referred to as MLP) and plasmid pGL4-MLP-ΔDE-Luc, lacking the DE element, contains the MLP sequence fused to the firefly luciferase coding sequence (8,10). Plasmid pGL4-MLP-5ssM-Luc contains four point mutations inactivating the major late first leader 5΄ splice site (GGCGTCTAT) (18) Plasmid pGL4-MLP-Luc-MR4 contains three point mutations changing the 5΄-CAAA-3΄ motif into 5΄-CGCG-3΄. Plasmid pGL4-MLP-12S 5΄ss-Luc, (referred to as MLP12S) contains nine nucleotides spanning the E1A 12S 5΄ splice site (AGGGTGAGG) replacing the major late first leader 5΄ splice site and pGL4-MLP-12S5΄ssM-Luc (referred to as MLP12S-5΄ssM) contains two point mutations in the E1A 12S 5΄ splice site (positions +5G+6G mutated to +5A+6T) that can be suppressed by cotransfection of plasmid pUC-U1-4u containing a compensatory base change in U1 snRNA (see ref. (19) for details). All plasmids were generated by standard PCR cloning strategies. Plasmid pBSAd1 encoding the AdML pre-mRNA transcript was previously described (20). Plasmid pBSAd1-MR4 contains point mutations changing the 5΄-CAAA-3΄ motif into 5΄-CGCG-3΄. Plasmid pcDNA3-L4-22K encodes the HAdV-5 L4-22K protein coding sequence fused to a carboxy-terminal Flag epitope tag (8,10). Plasmid pET-24a-L4-22K produces the L4-22K protein with a carboxy-terminal His-tag (8). The His-tagged L4-22K protein was expressed in Escherichia coli (BL21(DE3), Novagen) and purified on a HisTrap HP column (GE Healthcare).
Transcript synthesis
Radiolabeled single-stranded wild type and mutant R1 RNAs and AdML pre-mRNAs were generated by in vitro run-off transcription in the presence of 62.5 μCi [α-32P]CTP (3000 Ci/mmol; PerkinElmer), 1 mM cap analog (m7G-(5΄)ppp(5΄)G; Amersham Biosciences), 500 μM ATP, 50 μM GTP, 50 μM CTP, 500 μM UTP, 12 mM DTT, 5 U RNase inhibitor (Applied Biosystems) and 25 U T7 RNA polymerase in the supplied T7 RNA polymerase buffer (New England Biolabs). Reactions were incubated at 37°C for 3 h, followed by 0.5 U DNase I treatment and gel purification. The wild type and mutant R1 RNAs were transcribed from synthetic oligonucleotide pairs encoding the R1 region (+1 to +68: Figure 1). The wild type and mutant AdML transcripts were generated from a PCR product of plasmid pBSAd1 or pBSAd1-MR4 using primer pair pBSAd1:1U40 and pBSAd1:371L22 as described previously (21). The non-radioactive single-stranded wild type and mutant R1 RNAs were generated by TranscriptAid™ T7 High Yield transcription Kit (Fermentas).
Transient transfection and RNA isolation
Subconfluent monolayers of HEK293 cells were seeded in 60-mm plates and cultured in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 100 units/ml penicillin–streptomycin (Invitrogen) and 10% newborn calf serum (Saveen Werner). Cells in each plate were transfected with a total of 2.0 μg of DNA (1.5 μg reporter plasmid and 0.1 or 0.2 μg activator plasmid pcDNA3-L4-22K) using the Turbofect transfection reagent (Thermo Scientific). Cells were lysed by IsoB/NP40 buffer treatment for cytoplasmic RNA extraction 24 h post-transfection (13). The RNA was purified by phenol/chloroform/isoamylalcohol (25:24:1) extraction followed by ethanol precipitation.
Primer extension
Five μg cytoplasmic RNA isolated from transfected cells was hybridized with 0.1 pmol 5΄-end labeled primer (5΄- GAGTATAGACGCCAACAGCTGGCCCTCGCAGACAGCGATGCG-3΄) in 1.3 × Superscript reaction buffer (Promega) for 10 min at 70°C. The sample was slowly cooled down to 37°C and 200 U of M-MLV Reverse Transcriptase (Promega) added. The mixture was incubated for 1 h at 37°C in final reaction conditions of 50 mM Tris–HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2 and 10 mM DTT and 1 mM dNTP (22). The reaction was terminated by addition of 1 μl 5 M NaOH followed by a 20 min incubation at 65°C. After ethanol precipitation, the cDNA was resolved on a 15% denaturing polyacrylamide gel.
U1 snRNA depletion
Oligonucleotide-directed RNase H depletion of U1 snRNA in uninfected HeLa nuclear extracts (HeLa–NE) was performed as previously described (23). The oligonucleotides used were 5΄C (5΄-TTCAGGTAAGTACTCA-3΄) directed against the 5΄-end of the U1 snRNA (+2 to +11) and R5S (5΄-TAGATCAGACGAGATA-3΄) directed towards the 5S rRNA (+32 to +45) (24). In brief, 50 μM of oligonucleotides were incubated with 0.5 mg of HeLa–NE, 1.5 mM ATP, 5 mM creatine phosphate, 2 mM MgCl2 and 10 U RNase H at 30°C for 1 h followed by two 15 min treatments with 1 U DNase I.
In vitro run-off transcription
The template DNA for in vitro transcription was generated by NcoI cleavage of plasmid MLP. 0.1 μg linearized template DNA was mixed with 0.2 or 0.5 μg of recombinant L4-22K protein and 100 μg of U1 snRNA- or 5S rRNA-depleted HeLa–NE and pre-incubated at room temperature for 15 min in a buffer containing 60 mM KCl, 7.5 mM MgCl2, 0.12 mM EDTA, 0.3 mM DTT, 12 mM Hepes–KOH, pH 7.9 and 12% glycerol (22.5 μl in total). The transcription reaction was initiated by addition of 3.5 μl nucleotide mix (final concentrations 0.5 mM ATP, GTP, UTP and 0.05 mM CTP and 10 μCi of [α-32P]CTP, 3000 Ci/mmol; PerkinElmer) and continued at 30°C for 1.5 h (8). The reaction was terminated by diluting mixture to 200 μl in a buffer containing 40 μg proteinase K, 150 mM NaCl, 100 mM Tris-HCl, pH 7.5, 12.5 mM EDTA, pH 8.0, 1% SDS and 10 μg yeast tRNA followed by a further incubation for 40 min at 65°C. After phenol extraction and ethanol precipitation the transcription products were resolved on an 8% denaturing polyacrylamide gel.
In vitro RNA splicing
In a standard splicing reaction mixture 0.02 pmol radiolabeled AdML or AdML-MR4 transcripts were incubated in 70 μg HeLa–NE with or without 0.4 or 0.5 μg of recombinant L4-22K protein in a buffer containing 4.2 mM MgCl2, 33 mM creatine phosphate, 3.3 mM ATP, 33% polyvinyl alcohol at 30°C for 90 min in a total volume of 15 μl (3,21). Reactions were terminated by proteinase K treatment as described in the in vitro run-off transcription section. After phenol extraction and ethanol precipitation splicing products were resolved on an 8% denaturing polyacrylamide gel.
Gel shift assays
The DNA and RNA gel shift assays were done under the same experimental conditions. In brief, the 5΄-end labeled DNA probe or the [α-32P]CTP-radiolabeled RNA probe were incubated with or without the recombinant L4-22K protein and increasing amounts of competitor DNA or RNA in a buffer containing 1.25 mM MgCl2, 1 mM EDTA, 10 mM Hepes–KOH, pH 7.9, 2.5% Ficoll and 24 ng poly(dI-dC) (DNA gel shift assays only) (15 μl total) for 30 min at room temperature (see Figure legends for details). For RNA gel shift assays (Figure 5B–D) the [α-32P]CTP-radiolabeled RNA probe was incubated in HeLa–NE for 1 h at 30°C, followed by a 15 μl salt treatment (2 M NaCl, 40 mM Tris pH 7.6 and 20% glycerol) for 10 min at 30°C. The resulting complexes were resolved on a 6% native polyacrylamide gel.
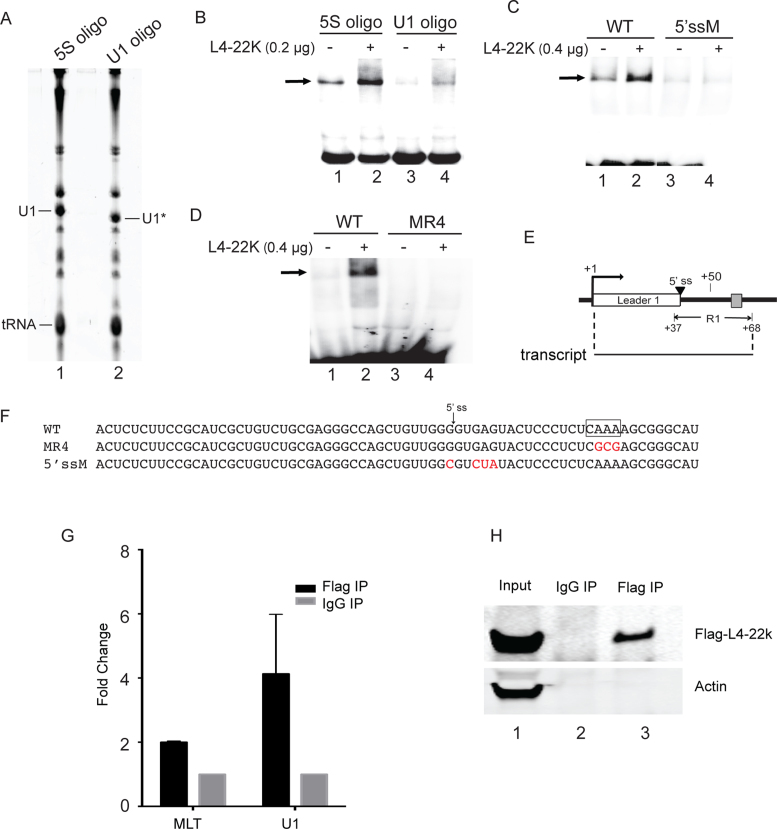
Figure 5.
L4-22K recruits U1 snRNP to the major late leader 1 exon. (A) RNA from HeLa–NE pretreated with RNase H and a complementary oligonucleotide directed against the 5΄-end of U1 snRNA (U1 oligo) or the 5S rRNA (5S oligo) was separated on an 8% denaturing polyacrylamide gel. The position of the full length (U1) and shortened (U1*) U1 snRNA are indicated. (B) 0.2 μg recombinant L4-22K protein was incubated with 0.4 pmol radiolabeled sense-strand major late first leader-R1 ssRNA (depicted in panel E) and 1.5 μg U1 snRNA-depleted HeLa–NE (U1 oligo) or 5S rRNA-depleted HeLa–NE (5S oligo) in a gel shift assay. (C and D) 0.4 μg recombinant L4-22K protein was incubated with 2.6 μg HeLa–NE and 0.4 pmol radiolabeled sense-strand major late first leader-R1 ssRNA (WT in panel C and D), the sense-strand major late first leader-R1 5΄splice site mutant ssRNA (5΄ssM in panel C) or the sense-strand major late first leader-R1 5΄-CAAA-3΄ motif mutant ssRNA (MR4 in panel D). ‘–’ denotes no recombinant L4-22K protein added. (E) Schematic drawing depicting the structure of the major late first leader-R1 ssRNA used in the gel shift assays. (F) The sequence of the R1 wild type, mutant MR4 and the 5΄ splice site mutant sense-strand ssRNAs used as probes in the gel shift assays. (G) HEK293 cells were co-transfected with a Flag-L4-22K expressing plasmid and an MLP reporter plasmid lacking the DE element. Following a 36 hours incubation extracts were UV crosslinked and immunoprecipitated with an anti-Flag or anti-IgG antibodies, RNA was extracted and quantitated by RT-qPCR. Results are shown as the fold change of the major late first leader-R1 ssRNA (MLT) or U1 snRNA in Flag-IP versus IgG-IP. (H) Immunoblotting of the same samples used in panel G using antibodies directed against Flag-L4-22K and Actin. The experiments shown in panels B and D have been conducted three times and in panel C four times. The data shown in panel G is the result from three independent experiments with error bars indicating the standard deviation.
35S-methionine/cysteine metabolic labeling of infected cells
HEK293 cells were infected with HAdV-5 at 10 FFU/cell. Conditions for pulse labeling of cells, extract preparation and immunoblotting were as previously described (25). The following primary antibodies were used: anti-Actin (1:3000, SantaCurz), anti-HAdV-5 (1:5000, Abcam) and anti-Flag (1:1000, Sigma).
RNA immunoprecipitation and RT-qPCR
HEK293 cells (∼107 cells) were transfected with 10 μg of plasmid pGL4-MLP-ΔDE-Luc and 5 μg activator plasmid pcDNA3-L4-22K using the Jetprime transfection reagent (Polyplus). At 36 h post-transfection cells were washed once in 1× PBS, UV crosslinked at 250 mJ/cm2, followed by cell lysis using RIPA buffer (150 mM NaCl, 50 mM HEPES [pH 7.4], 0.5% sodium deoxycholate, and 0.1% SDS supplemented with 2 U DNAseI) with sonication for 5 cycles (30 s each). Cell lysates were incubated with the Flag-M2 antibody (Sigma) or IgG (Santa Cruz) (2 μg antibody/1 ml lysate) coupled with Dynabeads Protein G overnight at 4°C. The beads were washed three times with RIPA buffer. Finally RNA extraction was preformed with proteinase K digestion, phenol extraction and ethanol precipitation as described above. The cDNA synthesis was preformed as described in (26). QPCR was preformed on Applied Biosystems 7900 system (Life Technologies) using HOT FIREPol® EvaGreen® qPCR Supermix (Solis BioDyne) with primer pairs specific for U1 snRNA (Forward- CCATGATCACGAAGGTGGTTT, Reverse- ATGCAGTCGAGTTTCCCACAT) and the major late Transcript (Forward-ACTCTCTTCCGCATCGCTGT, Reverse-CGGGCCAGGTGAATATCAAATC). Relative enrichment was calculated by subtracting raw Ct value of Flag-Ip from IgG-IP ((Ct Flag-IP) – (Ct IgG-IP)) and using the comparative Ct method (2−DCt).
RESULTS
L4-22K binds preferentially to the sense strand R1 single stranded DNA and RNA
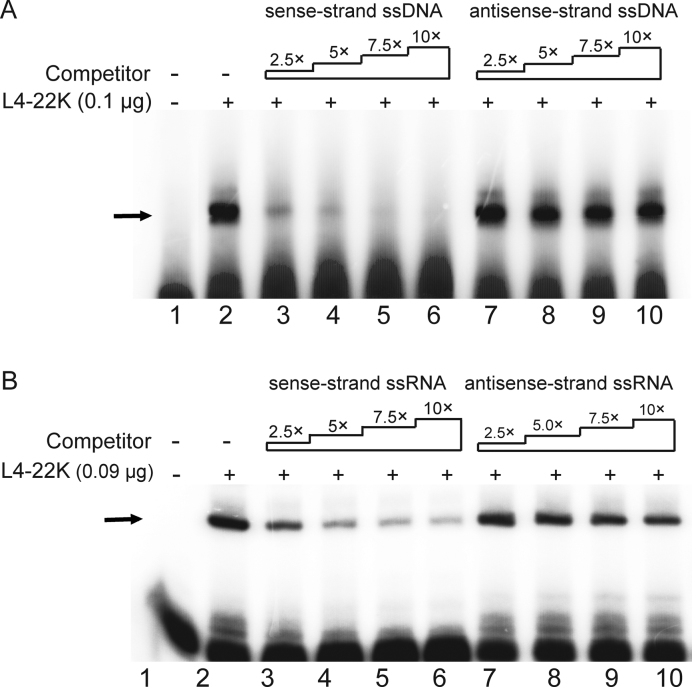
We have previously shown that L4-22K binds to both the DE element and the R1 region in vitro (8,10). To determine whether L4-22K also has a capacity to bind to single stranded (ss) nucleic acids we incubated the recombinant L4-22K protein with the 5΄-end labeled R1 dsDNA probe in the presence of increasing amounts of the unlabeled sense- or antisense-strand of R1 ssDNA as a competitor. As shown in Figure 2A, the L4-22K binding to the dsDNA probe was efficiently competed by the sense-strand ssDNA (lanes 3–6), but not by the antisense-strand ssDNA (lanes 7–10), suggesting that L4-22K has a specific affinity for the R1 sense-strand ssDNA.
Figure 2.
L4-22K binds preferentially to the sense strand R1 ssDNA and ssRNA. (A) The 5΄-end labeled double stranded R1 ssDNA probe (0.4 pmol) was incubated with 0.1 μg of recombinant L4-22K protein (lanes 2–10), together with a 2.5, 5, 7.5 or 10-fold excess of an unlabeled single strand sense (lanes 3–6) or antisense (lanes 7–10) R1 ssDNA competitor. (B) The radiolabeled R1 dsRNA probe (0.4 pmol) was incubated with 0.09 μg of recombinant L4-22K protein (lanes 2–10), together with a 2.5-, 5-, 7.5- or 10-fold excess of the unlabeled sense (lanes 3–6) or antisense (lanes 7–10) R1 ssRNA competitor. ‘–’ denotes no recombinant L4-22K protein added. The experiments shown in panels A and B have been conducted three times.
The finding that L4-22K binds to the R1 ssDNA made it interesting to investigate whether L4-22K also binds to the sense-strand of the R1 ssRNA. Such an observation could suggest that L4-22K might control gene expression by targeting the nascent major late transcript. To test this possibility we tested the RNA binding capacity of L4-22K. As shown in Figure 2B, L4-22K also binds to the double-stranded R1 RNA (R1 dsRNA) (lane 2). To study the binding affinity of L4-22K to the sense- and antisense-strand R1 ssRNA, L4-22K binding to the radiolabeled R1 dsRNA was competed with increasing amounts of the unlabeled sense- or antisense-strand R1 RNA. As shown in Figure 2B, L4-22K binding was competed efficiently by the sense-strand R1 RNA (lanes 3–6), whereas the antisense-strand was inefficient as a binding competitor (lanes 7–10). Taken together, these results suggest that L4-22K binds to both the R1 RNA and R1 DNA. Further, the results strongly suggest that L4-22K has a higher binding affinity to the sense-strand of both the R1 RNA and R1 DNA.
The relative binding affinity of L4-22K is higher for RNA compared to DNA
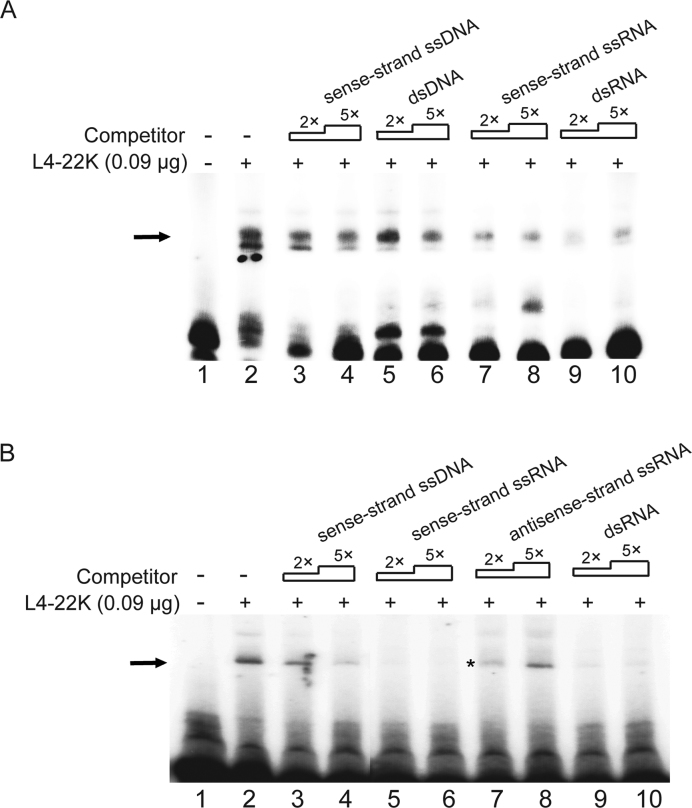
Previous studies have focused on the dsDNA binding capacity of L4-22K (8,10,12,15–17). Our finding that L4-22K binds to the sense-strand R1 RNA made it important to investigate whether the relative affinity of L4-22K was higher for DNA or RNA. For this experiment we used the competitive binding assay described above. We used conditions where L4-22K binding to the radiolabeled sense-strand R1 ssRNA (Figure 3A) or sense-strand R1 ssDNA (Figure 3B) was competed with the unlabeled RNA and DNA competitors described in Figure 3. As expected from the previous result (Figure 2B), the R1 ssRNA and R1 dsRNA efficiently competed the binding of L4-22K to the sense-strand R1 ssRNA (Figure 3A, compare lane 2 with lanes 7, 8 and 9, 10). In contrast, the R1 ssDNA or R1 dsDNA were less effective as competitive binding partners (Figure 3A, compare lane 2 with lanes 3–6), suggesting that, in fact, L4-22K prefers to bind to RNA. This hypothesis was further tested by competing L4-22K binding to the 5΄-end labeled sense-strand R1 ssDNA. As shown in Figure 3B, the sense-strand R1 ssRNA (lanes 5 and 6) and the R1 dsRNA (lanes 9 and 10) were much more effective as competitors compared to the same sequence as a DNA version (lanes 3 and 4). Importantly, the high competitive activity of R1 RNA was not unspecific since the antisense-strand R1 ssRNA did not compete efficiently for L4-22K binding (lane 8). Collectively, these results strongly suggest that L4-22K preferentially binds to the R1 RNA with a high preference for the sense-strand R1 ssRNA. Thus, as a binding protein to the R1 region L4-22K is not primarily a DNA binding protein but instead an RNA binding protein.
Figure 3.
L4-22K possesses higher binding affinity to R1 RNA compared to R1 DNA. (A) The radiolabeled sense-strand R1 ssRNA probe (0.4 pmol) was incubated with 0.09 μg of recombinant L4-22K protein (lanes 2–10), together with a 2- or 5-fold excess of an unlabeled R1 DNA competitor (sense-strand ssDNA, lanes 3–4 or dsDNA, lanes 5–6), or an R1 RNA competitor (sense-strand ssRNA, lanes 9–10 or dsRNA, lanes 9–10). (B) The 5΄-end labeled sense-strand R1 ssDNA probe (0.4 pmol) was incubated with 0.09 μg of recombinant L4-22K protein (lanes 2–10), together with a 2- or 5-fold excess of an unlabeled single strand sense R1 DNA (lane 3–4), single strand sense (lane 5–6), antisense (lane 7–8) R1 ssRNA or R1 dsRNA (lane 9–10) competitor. The asterisk was added to point out that some of the sample was lost during loading on lane 7. ‘–’ denotes no recombinant L4-22K protein added. The experiments shown in panels A and B have been repeated three times.
L4-22K binds to the 5΄-CAAA-3΄ motif in the sense-strand R1 RNA
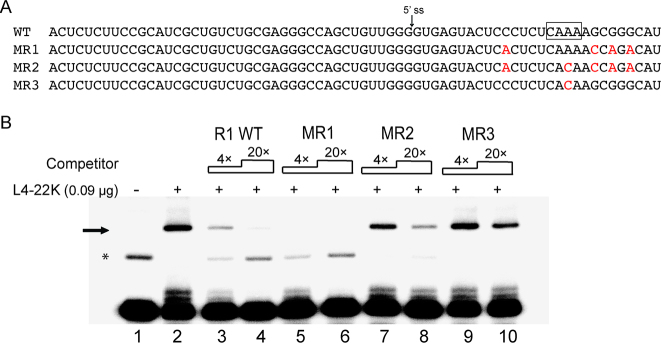
L4-22K was previously shown to bind to the distal part of the R1 DNA (Figure 1C) (10) covering the 5΄-TTTG-3΄ motif that has been proposed as the consensus L4-22K binding site (16). The observation here that L4-22K has an affinity for the sense-strand R1 ssRNA and ssDNA suggests that L4-22K might interact with the 5΄-CAAA-3΄ sequence in the previously identified consensus binding site (i.e. the reverse complement sequence). To study the specific L4-22K binding site within the R1 ssRNA, several sense-strand R1 ssRNA mutants were constructed (Figure 4A) and tested as competitors for L4-22K binding to the wild type sense-strand R1 ssRNA probe. As shown in Figure 4B, L4-22K binding to the radiolabeled sense-strand R1 ssRNA was efficiently reduced by an excess of the unlabeled sense-strand wild type R1 (lanes 3 and 4) and mutant MR1 (lanes 5 and 6) containing four point mutations disrupting the potential hairpin structure of the sense-strand R1 RNA (Figure 1D). In contrast, the binding was only partially reduced by the unlabeled mutant MR2 RNA (lanes 7 and 8) containing the same four point mutations as mutant MR1 plus an A to C point mutation in the 5΄-CAAA-3΄ motif (Figure 4A). In fact, mutant MR3, containing the single A to C point mutation in the 5΄-CAAA-3΄ motif was the least effective competitor of L4-22K binding to the sense-strand R1 ssRNA probe (lanes 9 and 10). Collectively, these results suggest that L4-22K binds to the same sequence motif in the R1 ssRNA as in the R1 DNA (Figure 1C) (10). The results further suggest that the potential hairpin formed around the L4-22K binding site (Figure 1D) is not essential for L4-22K binding to the R1 ssRNA (mutant MR1, Figure 4B, lanes 5 and 6).
Figure 4.
L4-22K binds to the 5΄-CAAA-3΄ motif in the sense strand R1 ssRNA. (A) The sequence of the R1 wild type, the three mutant (MR1-MR3) sense-strand ssRNAs used as competitors in the gel shift assay (B). The predicted L4-22K binding site is shown boxed in the R1 wild type sequence and the mutations introduced are shown in red letters. (B) The 5΄-end labeled sense-strand R1 ssRNA (0.4 pmol) was incubated with 0.09 μg recombinant L4-22K protein (lanes 2–10), together with a 4 or 20-fold excess of the unlabeled R1 wild type (lane 3 and 4), MR1 (lane 5 and 6), MR2 (lane 7 and 8) or MR3 (lane 9 and 10) sense-strand competitor RNAs. The asterisk indicates the position of a potential slower migrating secondary structure variant of the R1 probe (see text for discussion). ‘–’ denotes no recombinant L4-22K protein added. The experiment has been conducted three times.
L4-22K stimulates U1 snRNA binding to the major late first leader 5΄ splice site
The observation here that L4-22K binds to the sense-strand R1 ssRNA (Figure 4) at a site covering the 5΄-CAAA-3΄ motif (Figure 1C) suggested the possibility that L4-22K might act as a splicing regulator to control MLTU gene expression at a post-transcriptional level.
At an early step of the spliceosome assembly process, U1 snRNP is recruited to the 5΄ splice site of a pre-mRNA via a base pairing between the U1 snRNA and the 5΄ splice site (reviewed in ref. 27). To study whether L4-22K stimulated binding of U1 snRNP to the major late first leader 5΄ splice site we used a mobility shift assay where the recombinant L4-22K was incubated together with the radiolabeled first leader-R1 transcript (nucleotide +1 to +68, Figure 5E) in HeLa–NE. The contribution of U1 snRNP to complex formation was tested after oligonucleotide-directed RNase H depletion of U1 snRNA in the HeLa–NE (Figure 5A, compare U1 and U1*). As shown in Figure 5B, formation of the slower migrating species was much reduced in the U1 snRNA depleted HeLa–NE (compare lanes 1 and 3), suggesting that the shifted band contains U1 snRNP. Further, addition of L4-22K did not enhance this complex formation in U1 snRNA depleted HeLa–NE (lane 4) to the same level as seen in the control depleted HeLa–NE (lane 2). A mutation destroying the major late first leader 5΄splice site (Figure 5F) resulted in a complex formation that was not enhanced by L4-22K addition (Figure 5C, compare lanes 3 and 4). Importantly, the complex formation on the transcript where the L4-22K binding motif was mutated (MR4, Figure 5F), was not enhanced in the presence of the L4-22K protein (Figure 5D). Taken together these experiments suggest that L4-22K stimulates U1 snRNA binding to the functional 5΄ splice site on the R1 ssRNA and further that efficient L4-22K binding to the ssRNA is required for this enhancer activity.
To further confirm that L4-22K, the major late first leader region containing RNA and U1 snRNA forms a complex we used a cotransfection assay where an L4-22K expressing plasmid and a reporter construct lacking the DE element (pGL4-MLP-ΔDE-Luc) were cotransfected into HEK293 cells. Since the DE element, which is a characterized L4-22K dsDNA-binding site (8,16), is located less than 30 bp from the R1 region this experiment was done with a reporter plasmid lacking the DE element. Cells were UV crosslinked at 36 hours post-transfection and the L4-22K associated RNAs enriched by immunoprecipitation with a Flag antibody recognizing the epitope tagged L4-22K protein and the major late first intron RNA and U1 snRNA detected by RT-qPCR. The specificity of the immunoprecipitation was confirmed by western blotting using antibodies detecting Flag-L4-22K and Actin (Figure 5H). As shown in Figure 5G, L4-22K transfection resulted in an enrichment of the major late first intron RNA and U1 snRNA, which is consistent with the results shown in Figure 5B–D suggesting that L4-22K forms a specific complex with the major late first leader RNA and U1 snRNA.
L4-22K enhances major late first intron splicing in vitro
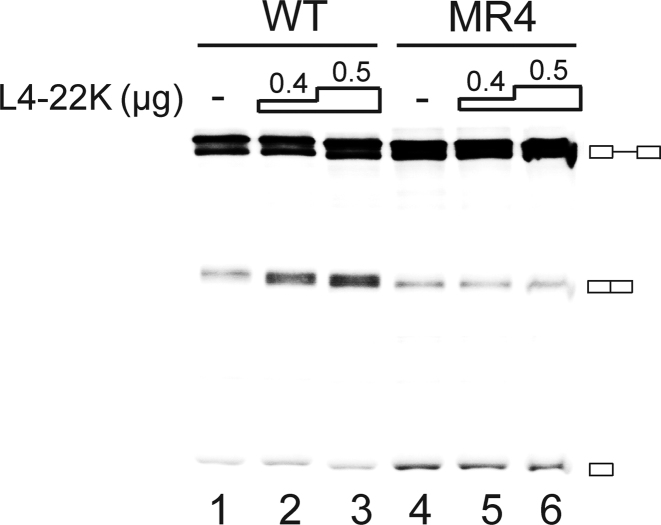
Since U1 snRNP is an essential splicing factor required for the commitment of a pre-mRNA for splicing we tested whether the L4-22K-mediated enhancement of U1 snRNA binding to the first leader-R1 RNA had a positive effect on major late first intron splicing. For this experiment the recombinant L4-22K was incubated with the radiolabeled AdML transcript in HeLa–NE. In this reporter transcript the first leader 5΄ splice site has been brought in close proximity to its natural second leader 3΄ splice site by a large deletion taking out ∼75% of the major late first intron (28). As shown in Figure 6, addition of the recombinant L4-22K protein resulted in a dose dependent increase in the accumulation of the spliced product with the wild type radiolabeled AdML transcript (lanes 1–3) but not with the AdML-MR4 mutant (lanes 4–6) containing point mutations in the 5΄-CAAA-3΄ motif (Figure 5F), which impair efficient L4-22K binding. The results suggest that L4-22K can function as a splicing enhancer protein stimulating major late first intron splicing.
Figure 6.
L4-22K stimulates major late first intron splicing. 0.4 or 0.5 μg of the recombinant L4-22K protein was incubated with 70 μg HeLa–NE in reactions supplemented with 0.02 pmol radiolabeled wild type (WT) or the 5΄-CAAA-3΄ motif mutant (MR4, Figure 5F) AdML pre-mRNA. Splicing products were resolved on an 8% denaturing polyacrylamide gel. ‘–’ denotes no recombinant L4-22K protein added. The position of the pre-mRNA, the spliced product and the first exon intermediate is graphically shown at the right. The experiment has been conducted three times.
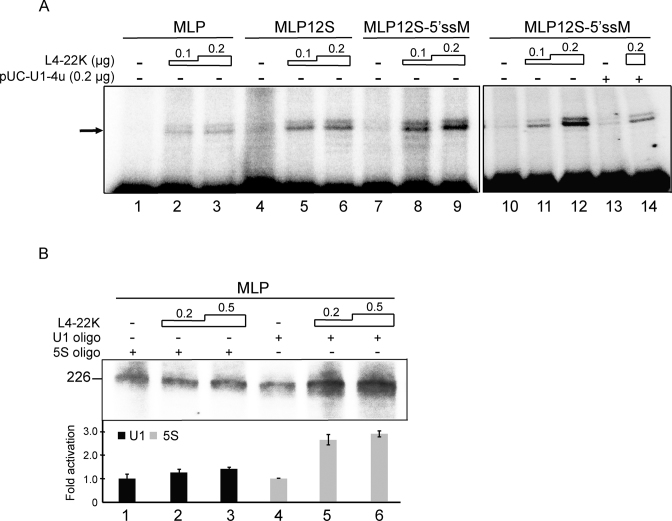
U1 snRNA interaction with the major late first leader 5΄ splice site has an inhibitory effect on MLP transcription
We recently demonstrated that a mutation at a site covering the major late first leader 5΄ splice site resulted in an increase in L4-22K-mediated activation of the MLP (10). The inhibitory effect of the 5΄ splice site region on transcription correlated with an L4-22K-mediated enhanced recruitment of Sp1 to the DNA at a site overlapping the major late first leader 5΄ splice site (10). To test whether the enhanced U1 snRNA binding to the sense-strand first leader-R1 RNA (Figure 5E) had an effect on MLP RNA accumulation we constructed chimeric substrates where the natural first leader 5΄ splice site was substituted with a nine nucleotide sequence covering the wild type E1A 12S 5΄ splice site or a 12S 5΄ splice site mutant disrupting a functional U1 snRNA interaction (19). HEK293 cells were cotransfected with the reporter plasmids with or without increasing amounts of an L4-22K expressing plasmid. Total RNA was prepared 24 h post-transfection and gene expression assayed by primer extension. As shown in Figure 7A, replacing the first leader 5΄ splice site with the E1A 12S 5΄ splice site (MLP12S) resulted in a reporter that, like the wild type MLP reporter, was enhanced by L4-22K plasmid cotransfection (lanes 1–6). A mutation inactivating the 12S 5΄ splice site (MLP12S-5΄ssM) resulted in a further 2-fold increase in L4-22K-mediated activation of RNA accumulation (lanes 7–9), indicating that also the functional 12S 5΄ splice site has an inhibitory effect on L4-22K-mediated activation of MLP RNA accumulation. Interestingly, the inhibitory effect of the mutated 12S 5΄ splice site was restored by cotransfection of a modified U1 snRNA containing a compensatory mutation restoring U1 snRNA/5΄ splice site interaction (lanes 10–14). This result suggests that, in addition to Sp1 binding to the DNA, an interaction between U1 snRNA and the 5΄ splice site contributes to the inhibitory effect of the first leader 5΄ splice site region on L4-22K-mediated activation of MLP RNA accumulation in vivo.
Figure 7.
U1 snRNP interaction with the major late first leader 5΄ splice site has a suppressive effect on L4-22K-activated MLP transcription. (A) 5 μg total RNA from HEK293 cells cotransfected with 1.5 μg of luciferase reporter plasmid MLP, MLP12S or MLP12S-5΄ssM and activator plasmid pcDNA3-L4-22K (0.1 or 0.2 μg) was assayed by the primer extension technique. Cells were additionally cotransfected with 0.2 μg of pUC-U1-4u plasmid (lanes 13 and 14). ‘–’, denotes no L4-22K activator plasmid DNA added. The arrow points to the full-length primer extension product. (B) In vitro run-off transcription, 0.1 μg MLP template DNA linearized by NcoI cleavage was incubated with the recombinant L4-22K protein (0.2 or 0.5 μg) in 5S rRNA (lanes 1–3) or U1 snRNA depleted HeLa–NE (lanes 4–6). The MLP full-length transcript is 226 nucleotides in length. ‘–’ denotes no recombinant L4-22K protein added. The experiments shown in panels A and B have been conducted three and four times, respectively.
To more directly demonstrate that U1 snRNA has a negative effect on MLP transcription we used an in vitro run-off transcription assay. In this experiment, we used the same strategy as described in Figure 5 to functionally inactivate U1 snRNA in HeLa–NE by oligonucleotide-directed RNase H depletion. As shown in Figure 7B, decapitating the capacity of U1 snRNA to recognize the major late 5΄ splice site had no detectable effect on basal MLP transcription (compare lanes 1 and 4). However, a functional inactivation of U1 snRNA resulted in a significant increase in the capacity of L4-22K to activate MLP transcription (compare lanes 1–3 and 4–6). Collectively, these results suggest that the L4-22K-mediated increase in U1 snRNP recruitment to the major late first leader 5΄splice site has a negative effect on MLP transcription both in vivo and in vitro.
L4-22K overexpression during a HAdV-5 infection suppresses late viral protein synthesis
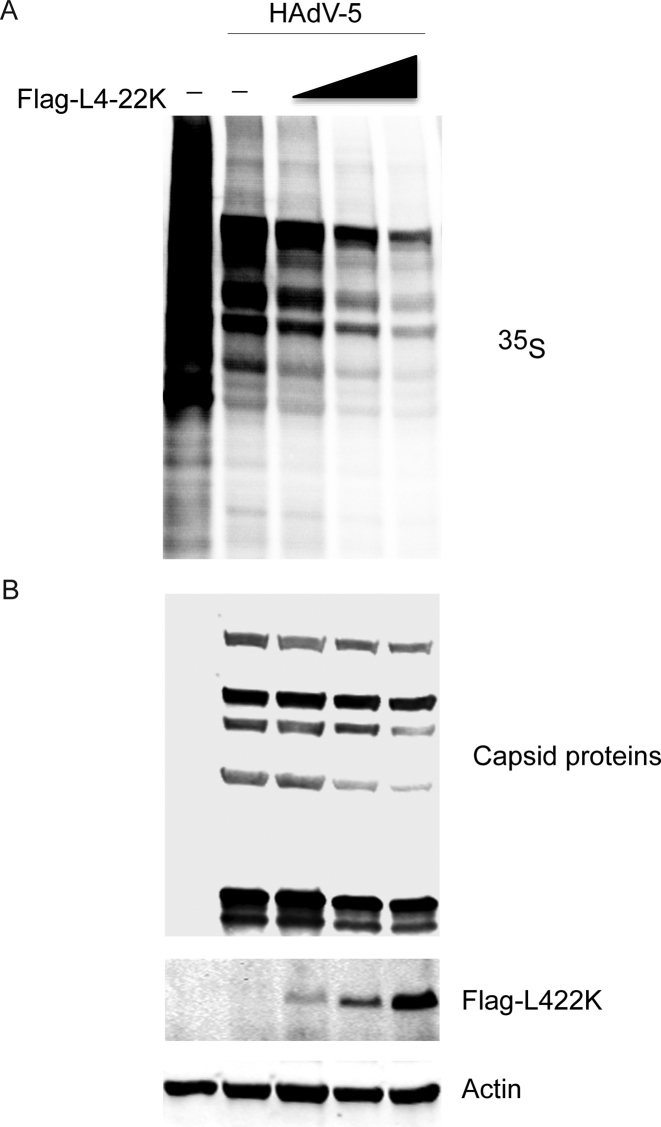
An expected consequence of the inhibitory effect of L4-22K on MLP transcription is that overexpression of this protein during a HAdV-5 infection should suppress late viral protein synthesis. This hypothesis was tested by transfection of HEK293 cells with increasing amounts of plasmid pcDNA3-L4-22K followed by a HAdV-5 infection 24 hours post-transfection. At 20 h post-infection, cells were pulse-labeled with 35S-methionine/cysteine mix for 2 h and proteins separated on a SDS-PAGE. HAdV-5 new late proteins synthesis was visualized by autoradiography (Figure 8A) and steady-state accumulation by western blot analysis (Figure 8B), respectively. The results from both assays show that the viral late proteins synthesis rate and steady-state levels were significantly reduced as a consequence of the L4-22K protein overexpression (Figure 8B).
Figure 8.
L4-22K overexpression suppresses HAdV-5 capsid protein synthesis and accumulation in vivo. HEK293 cells were transfected with increasing concentration of pcDNA3-Flag-L4-22K, while total amount of DNA was kept constant in all transfected sample with an empty plasmid. Twenty four hours later, cells were HAdV-5- (10 FFU/cell) or mock-infected for another 18 h followed by 35S-methionine/cysteine pulse labeling for 2 h and total proteins were extracted. (A) Rate of protein synthesis was visualized by autoradiography, accumulation of hexon late viral protein is demonstrated by arrow. (B) Immunoblotting from the same samples against capsid proteins, Flag-L4-22K and Actin.
DISCUSSION
The L4-22K protein serves two essential functions during a lytic infection. First, the protein functions in viral DNA encapsidation by binding to several sites in the DNA packaging domain (16). Second, the protein contributes to the control of the temporal early to late switch in viral gene expression by participating in the control of the regulated expression of the full repertoire of late viral mRNAs (9,17). In the context of a virus infection, L4-22K stimulates the accumulation of a selected set of late genes (9). The results from virus infections have been complicated by the observation that the L4-22K protein appears to function at multiple steps in viral gene expression. For example, L4-22K stimulates protein V and penton base expression by enhancing accumulation of the spliced V and penton base mRNAs, suggesting an effect on splicing/stability/nuclear export (9). In contrast, L4-22K enhancement of hexon protein accumulation has been reported to occur without a change in hexon mRNA abundance, suggesting an effect on translation and/or protein stability. Whereas L4-22K is important for the timely expression of the late viral genes, the protein also suppresses early viral gene expression (17). Interestingly, the L4-22K protein has also been suggested to directly stimulate L4-33K mRNA splicing (29). Collectively, these observations have suggested that L4-22K might play a role in viral gene expression in a complex way, and potentially at multiple transcriptional and post-transcriptional steps.
These types of observations make it important to dissect the basic function(s) of the protein. Here, we used transient transfection experiments and in vitro approaches to study some aspects of L4-22K function. To use a reductionistic experimental system has the advantage that different activities can be dissected and ascribed to the protein. However, this approach also has its limitations because in the context of a virus infection other viral proteins with redundant, overlapping or competing functions may overshadow some of the effects observed in our reductionistic approach. Hence, both types of experimental approaches need to be done.
Here, we show that L4-22K binds to both the RNA and DNA form of the R1 region (Figures 2 and 3). Surprisingly, L4-22K binds preferentially to the R1 RNA, not the R1 DNA. L4-22K binding to ssRNA is sequence-specific with the same motif characterized for DNA binding (10,16) functioning as the binding site on ssRNA (Figure 4). Our detailed binding studies have revealed that the L4-22K protein has a higher affinity towards the sense-strand R1 ssRNA compared to the ssDNA counterpart (Figure 3A). This unexpected result would suggest the presence of a potential RNA-binding domain (RBD) in the L4-22K protein. Although L4-22K does not encompass an annotated RBD it is possible that it contains a novel RNA binding surface allowing specific interaction with the 5΄-CAAA-3΄ ssRNA motif. Hence, the further studies are needed to elucidate L4-22K structural constraints controlling its interactions with RNA. It is also curious that L4-22K can bind to both ds and ss RNA and DNA. How a single protein can bind to both ss and ds nucleic acid remains to be established. Potentially the small protein contains separate binding domains for ss and ds nucleic acids, or alternatively the protein has an intrinsically disordered region (reviewed in (30)) that can adopt alternative structures depending on the type of nucleic acid that is presented. Additionally L4-22K has been shown to form high-order structures (31), which also might contribute to this unusual nucleic acid binding properties.
Also, our results suggest that L4-22K enhances U1 snRNA binding to the major late first leader 5΄ splice site both in vitro (Figure 5B–D) and in transient transfection assays in vivo (Figure 5G), which in turn results in a suppression of MLP transcription (Figure 7). In fact, our data is compatible with the model that L4-22K makes a physical complex containing U1 snRNA and the major late first leader region RNA (Figure 5G). Previous studies have suggested that U1 snRNA binding to promoter-proximal introns or 5΄ splice sites might stimulate transcription in vivo (32,33). This was interpreted to indicate that U1 snRNP binding to promoter-proximal 5΄ splice sites via a feedback mechanism enhances transcription initiation. However, subsequent studies have suggested an alternative explanation that a functional U1 snRNP is needed to suppress the usage of cryptic polyadenylation sites during the elongation phase (34,35). Thus, a number of studies have shown that U1 snRNP, in addition to its function in RNA splicing, also has the capacity to inhibit 3΄ end processing by blocking the polyA polymerase and/or cleavage factors (reviewed in ref. 36). A key question for the future is to explain how U1 snRNA binding to the major late first leader 5΄splice site exerts its negative effects on MLP transcription.
Taken together our results appear to suggest a two-step model for how the L4-22K protein may inhibit MLP transcription. Thus, L4-22K binding to R1 DNA has a negative effect on MLP transcription by recruiting Sp1 to a site covering the 5΄ splice site region (10) whereas L4-22K binding to the nascent MLTU transcript stimulates recruitment of U1 snRNA to the major late first leader 5΄splice site, a binding that has suppressive effects on MLP transcription (Figure 7).
Our data further suggests that L4-22K-mediated enhancement of U1 snRNA binding to the major late first leader 5΄ splice site (Figure 5) stimulates first intron splicing (Figure 6). L4-22K is most likely not a general splicing factor activating multiple adenovirus pre-mRNAs. Although L4-22K has previously been shown to enhance L4-33K splicing (29) we have previously shown that L4-22K does not have a stimulatory effect on L1-IIIa pre-mRNA splicing (8), under conditions where the L4-33K protein functions as an efficient splicing enhancer protein (13). Our results indicate that L4-22K may function as a splicing enhancer protein on a limited number of transcripts, which we predict would have a L4-22K binding motif in close proximity to a 5΄ splice site.
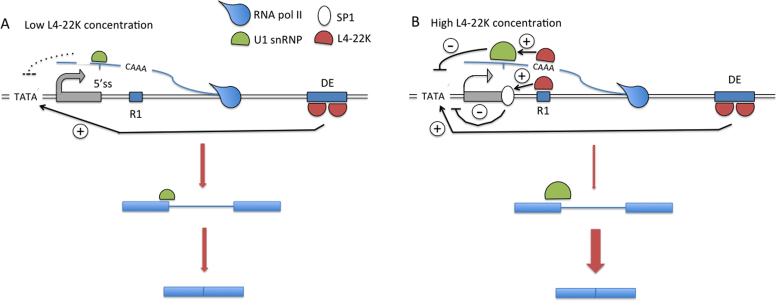
Combining our results suggest a model where the L4-22K protein might have a feedback regulatory effect on MLP gene expression (Figure 9). This type of self-limiting mechanism might be important for the virus to control late viral protein expression during a lytic or persistent infection. Thus, at low concentrations L4-22K binds with high affinity to the DE element and stimulates MLP transcription. Since L4-22K itself is expressed from the MLTU this enhanced transcription results in an increase in L4-22K protein expression. As the concentration increases the protein starts to bind to the R1 region, both at the DNA and ssRNA level. Binding to this low affinity site results in Sp1 recruitment to the DNA and as a consequence a suppression of transcription counteracting the DE-mediated activation of MLP transcription (10). Also, L4-22K binding to the 5΄-CAAA-3΄ motif on the nascent RNA stimulates binding of U1 snRNP to the major late first leader 5΄ splice site, which results in a further suppression of transcription and concomitantly an increased efficiency of major late first intron splicing. Interestingly, the two L4-22K binding sites in the DE element are inverted compared to the site in R1, suggesting that L4-22K would not be expected to bind to the nascent MLTU transcript at the position of the DE element. It is noteworthy that our virus infection experiment (Figure 8) underscores the importance of the L4-22K protein concentration as a factor controlling efficient viral late protein synthesis and accumulation in HAdV-5 infected cells, creating a possible feedback loop to control the activity of the MLP.
Figure 9.
A model describing a potential mechanism by which the L4-22K protein has both a positive and negative effect on MLP activity. (A) At low concentrations the L4-22K protein binds to its high affinity site, the DE element, and activates MLP transcription. (B) At high concentrations L4-22K additionally associates to its low affinity R1 binding site, both at the DNA and nascent MLTU transcript levels, which results in a reduction of MLP transcription and a concomitant enhancement of major late first intron splicing. See Discussion for further details.
The results presented here provide new pieces of evidence that help explain how the L4-22K protein might participate in the fine-tuning of MLTU gene expression. However, much future work will be needed to unravel how the interactions between L4-22K, U1 snRNP, Sp1 and additional, yet to be discovered, factors operate in the control of MLP transcription.
ACKNOWLEDGEMENTS
We thank the members of the adenovirus group for constructive criticism.
FUNDING
Swedish Cancer Society [120678 and 130410]; Swedish Research Council through a grant to the Uppsala RNA Research Centre [2006-5038-36531-16]. Funding for open access charge: The Swedish Cancer Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Akusjärvi G. Temporal regulation of adenovirus major late alternative RNA splicing. Front. Biosci. 2008; 13:5006–5015. [DOI] [PubMed] [Google Scholar]
- 2.Young C.S. The structure and function of the adenovirus major late promoter. Curr. Top. Microbiol. Immunol. 2003; 272:213–249. [DOI] [PubMed] [Google Scholar]
- 3.Mansour S.L., Grodzicker T., Tjian R.. Downstream sequences affect transcription initiation from the adenovirus major late promoter. Mol. Cell. Biol. 1986; 6:2684–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansen-Durr P., Boeuf H., Kedinger C.. Replication-induced stimulation of the major late promoter of adenovirus is correlated to the binding of a factor to sequences in the first intron. Nucleic Acids Res. 1988; 16:3771–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leong K., Lee W., Berk A.J.. High-level transcription from the adenovirus major late promoter requires downstream binding sites for late-phase-specific factors. J. Virol. 1990; 64:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tribouley C., Lutz P., Staub A., Kedinger C.. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 1994; 68:4450–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz P., Kedinger C.. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J. Virol. 1996; 70:1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backström E., Kaufmann K.B., Lan X., Akusjärvi G.. Adenovirus L4-22K stimulates major late transcription by a mechanism requiring the intragenic late-specific transcription factor-binding site. Virus Res. 2010; 151:220–228. [DOI] [PubMed] [Google Scholar]
- 9.Morris S.J., Leppard K.N.. Adenovirus serotype 5 L4-22K and L4-33K proteins have distinct functions in regulating late gene expression. J. Virol. 2009; 83:3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan S., Östberg S., Punga T., Akusjärvi G.. A suppressive effect of Sp1 recruitment to the first leader 5΄ splice site region on L4-22K-mediated activation of the adenovirus major late promoter. Virus Res. 2015; 210:133–140. [DOI] [PubMed] [Google Scholar]
- 11.Biasiotto R., Akusjärvi G.. Regulation of human adenovirus alternative RNA splicing by the adenoviral L4-33K and L4-22K proteins. Int. J. Mol. Sci. 2015; 16:2893–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright J., Atwan Z., Morris S.J., Leppard K.N.. The human adenovirus type 5 L4 promoter is negatively regulated by TFII-I and L4-33K. J. Virol. 2015; 89:7053–7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Törmanen H., Backström E., Carlsson A., Akusjärvi G.. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J. Biol. Chem. 2006; 281:36510–36517. [DOI] [PubMed] [Google Scholar]
- 14.Törmanen Persson H., Aksaas A.K., Kvissel A.K., Punga T., Engström A., Skalhegg B.S., Akusjärvi G.. Two cellular protein kinases, DNA-PK and PKA, phosphorylate the adenoviral L4-33K protein and have opposite effects on L1 alternative RNA splicing. PLoS One. 2012; 7:e31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewing S.G., Byrd S.A., Christensen J.B., Tyler R.E., Imperiale M.J.. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J. Virol. 2007; 81:12450–12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostapchuk P., Anderson M.E., Chandrasekhar S., Hearing P.. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 2006; 80:6973–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu K., Orozco D., Hearing P.. The adenovirus L4-22K protein is multifunctional and is an integral component of crucial aspects of infection. J. Virol. 2012; 86:10474–10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreivi J.P., Zerivitz K., Akusjärvi G.. A U1 snRNA binding site improves the efficiency of in vitro pre-mRNA splicing. Nucleic Acids Res. 1991; 19:6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang Y., Weiner A.M.. A compensatory base change in U1 snRNA suppresses a 5΄ splice site mutation. Cell. 1986; 46:827–835. [DOI] [PubMed] [Google Scholar]
- 20.Konarska M.M., Sharp P.A.. Association of U2, U4, U5, and U6 small nuclear ribonucleoproteins in a spliceosome-type complex in absence of precursor RNA. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:5459–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutzelberger M., Backstrom E., Akusjarvi G.. Substrate-dependent differences in U2AF requirement for splicing in adenovirus-infected cell extracts. J. Biol. Chem. 2005; 280:25478–25484. [DOI] [PubMed] [Google Scholar]
- 22.Xu N., Gkountela S., Saeed K., Akusjärvi G.. The 5΄-end heterogeneity of adenovirus virus-associated RNAI contributes to the asymmetric guide strand incorporation into the RNA-induced silencing complex. Nucleic Acids Res. 2009; 37:6950–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins S.J., Hames B.D.. RNA Processing: A Practical Approach. 1994; Oxford University Press. [Google Scholar]
- 24.Black D.L., Chabot B., Steitz J.A.. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985; 42:737–750. [DOI] [PubMed] [Google Scholar]
- 25.Kamel W., Segerman B., Punga T., Akusjärvi G.. Small RNA sequence analysis of adenovirus VA RNA-derived miRNAs reveals an unexpected serotype-specific difference in structure and abundance. PLoS One. 2014; 9:e105746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamel W., Segerman B., Öberg D., Punga T., Akusjärvi G.. The adenovirus VA RNA-derived miRNAs are not essential for lytic virus growth in tissue culture cells. Nucleic Acids Res. 2013; 41:4802–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Will C.L., Luhrmann R.. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001; 13:290–301. [DOI] [PubMed] [Google Scholar]
- 28.Konarska M.M., Sharp P.A.. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987; 49:763–774. [DOI] [PubMed] [Google Scholar]
- 29.Guimet D., Hearing P.. The adenovirus L4-22K protein has distinct functions in the posttranscriptional regulation of gene expression and encapsidation of the viral genome. J. Virol. 2013; 87:7688–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calabretta S., Richard S.. Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem. Sci. 2015; 40:662–672. [DOI] [PubMed] [Google Scholar]
- 31.Yang T.C., Maluf N.K.. Self-association of the adenoviral L4-22K protein. Biochemistry. 2010; 49:9830–9838. [DOI] [PubMed] [Google Scholar]
- 32.Furger A., O'Sullivan J.M., Binnie A., Lee B.A., Proudfoot N.J.. Promoter proximal splice sites enhance transcription. Genes Dev. 2002; 16:2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damgaard C.K., Kahns S., Lykke-Andersen S., Nielsen A.L., Jensen T.H., Kjems J.. A 5΄ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell. 2008; 29:271–278. [DOI] [PubMed] [Google Scholar]
- 34.Kaida D., Berg M.G., Younis I., Kasim M., Singh L.N., Wan L., Dreyfuss G.. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010; 468:664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almada A.E., Wu X., Kriz A.J., Burge C.B., Sharp P.A.. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013; 499:360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaida D. The reciprocal regulation between splicing and 3΄-end processing. Wiley Interdiscipl. Rev. RNA. 2016; 7:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]