Abstract
Multiple G-quadruplex units in the 3΄-terminal overhang of human telomeric DNA can associate and form multimeric structures. The specific targeting of such distinctive higher-order G-quadruplexes might be a promising strategy for developing selective anticancer agents with fewer side effects. However, thus far, only a few molecules were found to selectively bind to telomeric multimeric G-quadruplexes, and their effects on cancer cells were unknown. In this study, a new triaryl-substituted imidazole derivative called IZNP-1 was synthesized and found to specifically bind to and strongly stabilize telomeric multimeric G-quadruplexes through intercalating into the pocket between the two quadruplex units. The pocket size might affect the binding behavior of IZNP-1. Further cellular studies indicated that IZNP-1 could provoke cell cycle arrest, apoptosis and senescence in Siha cancer cells, mainly because of telomeric DNA damage and telomere dysfunction induced by the interactions of IZNP-1 with telomeric G-quadruplexes. Notably, IZNP-1 had no effect on the transcriptional levels of several common oncogenes that have the potential to form monomeric G-quadruplex structures in their promoter regions. Such behavior differed from that of traditional telomeric G-quadruplex ligands. Accordingly, this work provides new insights for the development of selective anticancer drugs targeting telomeric multimeric G-quadruplexes.
INTRODUCTION
Human telomeres, which are essential for chromosomal stability and genomic integrity, are composed of thousands of double-stranded TTAGGG repeats and feature a 3΄-terminal single-stranded overhang of ∼200 nucleotides (1–3). The telomere terminus is protected from degradation by a T-loop, which forms by strand invasion of the 3΄-terminal overhang into the duplex part of the telomere and is further stabilized by a six-subunit protein complex called shelterin (4,5). It is now widely accepted that telomere maintenance plays a vital role in tumorigenesis. Therefore, interfering with telomere maintenance is considered to be an optional strategy in anticancer therapy (6,7). The 3΄-terminal G-rich overhang has a high propensity to fold into four-stranded helical secondary structures known as G-quadruplexes (8,9). The stabilization of telomeric G-quadruplexes by small molecule ligands can alter the T-loop structure, causing its degradation through a DNA damage response pathway and the release of some of shelterin proteins from telomeres (10–13). These events lead to a DNA damage response, telomeric dysfunction and then an induction of tumor cell senescence and apoptosis. Therefore, the development of highly specific telomeric G-quadruplex ligands as new anticancer agents has captured extensive attention (14–16).
To date, a number of studies have been performed to seek candidate telomeric G-quadruplex ligands (17–24). Most of these studies have applied the monomeric G-quadruplex model formed by a short telomeric DNA sequence (usually 21−26 nt) to screen G-quadruplex ligands. However, the 3΄-terminal single-stranded overhang contains tens of TTAGGG repeats (∼200 nt). Accumulating evidence indicates that it forms several consecutive quadruplex units connected by TTA linkers (25–31). Such higher-order structures are called telomeric multimeric G-quadruplexes.
Notably, only telomeric DNA has the capacity to form multimeric G-quadruplexes in the human genome (32). Such distinctive structure enables the possibility of the design of small molecules able to discriminate telomeric G-quadruplexes from a large number of other G-quadruplexes with various biological functions (33,34). Thus, it is believed that small molecules that specifically target telomeric multimeric G-quadruplexes might be more promising anticancer agents with fewer side effects. However, very few small molecules that specifically bind to telomeric multimeric G-quadruplexes with discrimination against monomeric G-quadruplexes have been reported (32,35,36). Furthermore, the effects of such molecules on cancer cells are unknown because they have been scarcely evaluated. Therefore, searching for highly specific telomeric multimeric G-quadruplex ligands and subsequent investigating their anticancer activity could provide an important theoretical basis for cancer therapies, which is challenging but urgently needed.
We recently discovered a series of multiaryl-substituted imidazole derivatives that are effective G-quadruplex ligands (37–41). Further biophysical studies demonstrated that these compounds selectively interact with G-quadruplexes against duplex DNAs, indicating their potential as anticancer agents. In this study, we synthesized a new triaryl-substituted imidazole derivative (IZNP-1, Figure 1A) and found that it could be used as a highly specific ligand of telomeric multimeric G-quadruplexes. The detailed interactions of IZNP-1 with telomeric multimeric G-quadruplexes were investigated. To test its potential as an anticancer agent, we explored the effects of this new compound in terms of its ability to induce cell cycle arrest, apoptosis and senescence in cancer cells. Furthermore, we discussed whether the anticancer activity of IZNP-1 was actually mediated by stabilizing telomeric G-quadruplexes and then interfering with telomere maintenance. To prove the selectivity of IZNP-1 for telomeric multimeric G-quadruplexes in cells, we also studied its effects on the transcriptional levels of certain pro-oncogenes that might form monomeric G-quadruplexes at their promoters.
Figure 1.
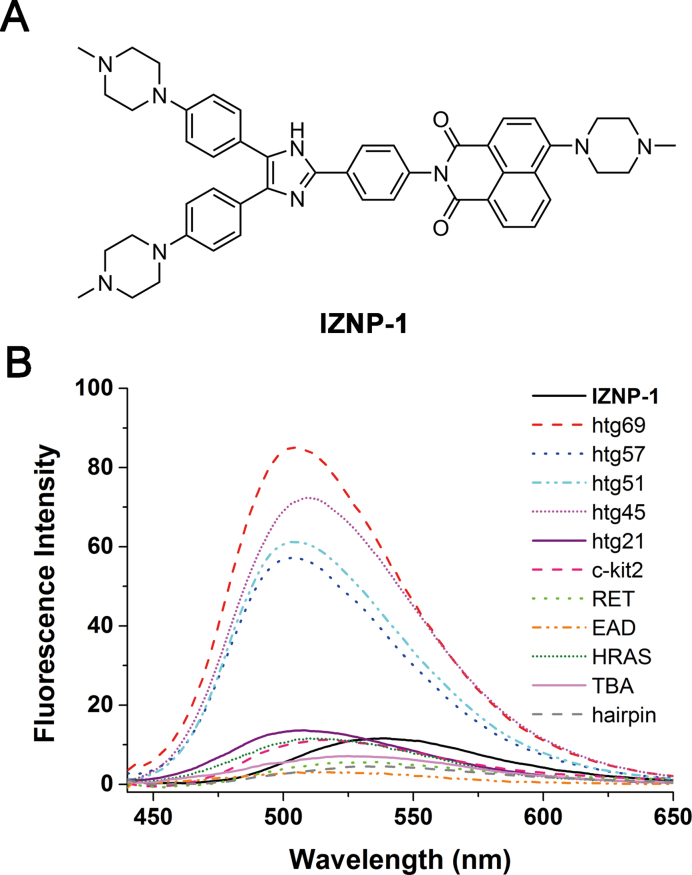
(A) The structure of IZNP-1. (B) The fluorescence spectra of IZNP-1 in the absence or presence of multimeric G-quadruplexes (htg45, htg51, htg57 and htg69), monomeric G-quadruplexes (htg21, c-kit2, RET, EAD, HRAS and TBA), and duplex DNA (hairpin). The concentrations of multimeric G-quadruplexes were set at 15 μM, and the concentrations of other DNA structure were set at 30 μM. All the experiments were conducted in 10 mM Tris-HCl buffer, 100 mM KCl, pH 7.2 at λex = 400 nm.
MATERIALS AND METHODS
Oligonucleotides and compound
All oligonucleotides (Supplementary Table S1) used in this study were purchased from Invitrogen (China) and Sangon (China). All oligonucleotides were dissolved in relevant buffers. Their concentrations were represented as single-stranded concentrations and determined from the absorbance at 260 nm using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, USA) based on their respective molar extinction coefficients. Stock solutions of IZNP-1 (10 mM) were dissolved in DMSO and stored at −80°C. Further dilutions of DNA samples and IZNP-1 to working concentrations were made with the relevant buffers immediately prior to use.
UV–Vis spectroscopic studies
UV–Vis spectroscopic studies were performed on a UV-2450 spectrophotometer (Shimadzu, Japan) using 1 cm path length quartz cuvettes. Solutions containing individual DNAs with the designated concentration in 10 mM Tris−HCl buffer (pH 7.2, 100 mM KCl) were prepared. Each solution was heated to 95°C for 5 min to remove any aggregates and then cooled slowly to 25°C. After overnight incubation at 4°C, 3 μM of IZNP-1 was added and allowed to equilibrate for at least 2 min. Afterward, the absorption spectra in the range of 300–600 nm were recorded. Absorption titration experiments were conducted by varying the DNA concentration but maintaining the IZNP-1 concentration at 3 μM. The sample solutions were prepared as above, and the absorption spectra in the range of 320–600 nm were recorded.
Fluorescence spectroscopic studies
Fluorescence studies were performed on a LS-55 luminescence spectrophotometer (Perkin-Elmer, USA). A quartz cuvette with a 2 mm × 10 mm path length was used for spectral recording. Solutions containing individual DNAs with the designated concentration in 10 mM Tris−HCl buffer (pH 7.2, 100 mM KCl) were prepared. Each solution was heated to 95°C for 5 min to remove any aggregates and then cooled slowly to 25°C. After overnight incubation at 4°C, 3 μM of IZNP-1 was added and allowed to equilibrate for at least 2 min. Fixing the excitation wavelength at 400 nm, the emission spectra in the range of 430–650 nm were collected at room temperature. The excitation slit and emission slit were both set at 10 nm. Fluorescence titration experiments were conducted by maintaining the IZNP-1 concentration at 3 μM but varying the DNA concentration. The sample solutions were prepared as above, and the fluorescence spectra in the range of 430–650 nm were recorded when excited at 400 nm.
2-Ap titration experiments
Solutions containing 2-Ap-labeled oligonucleotides with the designated concentration (1 μM) in 10 mM Tris−HCl buffer (pH 7.2, 100 mM KCl) were prepared. Each solution was heated to 95°C for 5 min to remove any aggregates and then cooled slowly to 25°C. Afterward, IZNP-1 was added to the solution. The final concentration of IZNP-1 was varied from 0 to 5 μM. After each addition of IZNP-1, the reaction was stirred and allowed to equilibrate for at least 2 min, and the fluorescence measurement was taken when excited at 305 nm.
Circular dichroism spectroscopic studies
CD studies were performed on a Chirascan circular dichroism spectrophotometer (Applied Photophysics, UK). A quartz cuvette with a 4 mm path length was used to record the spectra over a wavelength range of 230–330 nm with a 1 nm bandwidth, 1 nm step size and a time of 0.5 s per point. A CD melting assay was performed at a fixed concentration of G-quadruplex (2.5 μM), either with or without a fixed concentration (10 μM) of IZNP-1 in Tris−HCl buffer (10 mM, pH 7.2) with 100 mM KCl. The data were recorded at intervals of 5°C over the range 25–95°C with a heating rate of 1°C/min. A buffer baseline was collected in the same cuvette and was subtracted from the sample spectra. The final analysis of the data was conducted using Origin 9.0 (OriginLab Corp.).
Molecular modeling studies (38,39)
The telomeric multimeric G-quadruplex structure Hybrid 12 and monomeric structure Hybrid 1 were generated on the basis of their NMR structures by using Discovery Studio 2.5 and optimized with AMBER10. The structure of IZNP-1 was constructed and optimized with Gaussian 03 by using the HF/6-31G* basis set. Docking analyses were performed with the Schrodinger program. Simulations of molecular dynamics (MD) were performed by using the Sander module of the AMBER 10.0 program suite. The G-quadruplex and IZNP-1 complex was solvated in a rectangular box of TIP3P water molecules with solvent layers of 10 Å. The system was then heated from 0 to 300 K in a 100 ps simulation, followed by a 100 ps simulation to equilibrate the density of the system. A constant pressure MD simulation of 40 ns was subsequently performed in an NPT ensemble at 1 atm and 300 K. The output and trajectory files were saved every 0.1 and 1 ps for subsequent analysis, respectively. The MM/PBSA method implemented in the AMBER 10 suite was applied to calculate the binding free energies between the G-quadruplexes and the IZNP-1. A set of 4000 snapshots from the MD trajectories was collected to calculate the binding free energies. The average structure from the last 2 ns was visualized by using Discovery Studio 2.5 software package.
Short-term cell viability
Cells were seeded in 96-well plates (5.0 × 103 cells/well) and exposed to various concentrations of IZNP-1. After a 24-h treatment, 20 μl of 2.5 mg/ml methylthiazolyl tetrazolium (MTT) solution was added to each well, and the cells were further incubated for 4 h. The cells in each well were then treated with dimethyl sulfoxide (DMSO) (100 μl per well), and the optical density (OD) was recorded at 570 nm. All experiments were performed in parallel and in triplicate, and the IC50 values were derived from the curves of the mean OD values of the triplicate tests plotted against the drug concentration.
Long-term cell culture
Long-term proliferation experiments were conducted using Siha cells. The cells were grown in culture dishes at 2.0 × 105 per dish (60 mm) and were exposed to IZNP-1 at various subcytotoxic concentrations every four days. The cells in the control and drug-exposed dishes were counted, and the dishes were reseeded with 2.0 × 105 cells. The process was repeated until a growth platform appeared.
Senescence analysis
After completion of long-term cell culture, the growth medium was aspirated, and the cells were fixed in 2% formaldehyde/0.2% glutaraldehyde for 15 min at room temperature. The fixing solution was then removed, and the cells were gently washed with PBS twice. The cells were then stained using a β-Gal stain solution containing 1 mg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactoside and were incubated at 37°C overnight. The staining solution was then removed, and the cells were washed three times with PBS. The cells were viewed and photographed under an optical microscope.
Flow cytometry analysis
Cells treated with IZNP-1 were harvested and washed in PBS and fixed with 70% ethanol at 4°C overnight. Then, the cells were centrifuged and re-suspended in a staining solution (50 μg/ml propidium iodide (PI), 75 KU/ml RNase A in PBS) for 30 min at room temperature in the dark. The cells were analyzed by flow cytometry using an EPICS XL flow cytometer (Beckman Coulter, USA). For each analysis, 1.6 × 104 events were collected. The cell cycle distribution was analyzed using the EXPO32 ADC software.
Apoptosis analysis
Cells treated with IZNP-1 were harvested and washed in PBS. Then, they were centrifuged and re-suspended in Annexin-binding buffer. After that, the cells were incubated with Annexin V-FITC and PI for 15 min at room temperature in the dark and immediately analyzed by flow cytometry using an EPICS XL flow cytometer (Beckman Coulter, USA). For each analysis, 1.6 × 104 events were collected. The data are presented as bi-parametric dot plots showing PI red fluorescence against Annexin V-FITC green fluorescence.
Telomere length assay
Cells treated with IZNP-1 were incubated for 16 days. To measure the telomere length, genomic DNA was digested with Hinf1/Rsa1 restriction enzymes. The digested DNA fragments were separated on 0.8% agarose gel, transferred to a nylon membrane, and the transferred DNA was fixed on the wet blotting membrane by baking the membrane at 120°C for 20 min. The membrane was hybridized with a DIG-labeled hybridization probe for telomeric repeats, and incubated with anti-DIG-alkaline phosphatase. TRF was performed through chemiluminescence detection.
Immunofluorescence experiments
Cells grown on glass coverslips were fixed in 4% paraformaldehyde/PBS for 15 min and then permeabilized with 0.1% Triton-X 100/PBS at 37°C for 30 min. Then, the cells were finally blocked with 5% goat serum/PBS at 37°C for 3 h. Immunofluorescence was performed using standard methods, and the slides were incubated alternately with BG4 (40 ng/μl), anti-FLAG antibody (#2368, Cell Signaling Technology), γH2AX antibody (#9718, Cell Signaling Technology; ab26350, Abcam), TRF2 antibody (ab13579, Abcam), TRF1 antibody (ab1423, Abcam) or POT1 antibody (ab21382, Abcam) at 37°C for 3 h. The glass coverslips were washed six times with blocking buffer and were then incubated with the anti-rabbit Alexa 488-conjugated antibody (A21206, Life Technology), the anti-mouse Alexa 555-conjugated antibody (A21427, Life Technology), and 0.5 μg/ml of DAPI (Invitrogen) at 37°C for 3 h. The glass coverslips were again washed six times with blocking buffer, and then, digital images were recorded using a LSM710 microscope (Zeiss, GER) and analyzed with ZEN software. Fifty nuclei were counted in each group.
RT-PCR experiments
After a 12-h treatment with IZNP-1, the total RNA was extracted according to the protocol supplied by the Magen Company and eluted in DEPC-H2O with a final volume of 50 μl. The RNA was quantitated using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, USA). The total RNA was used as a template for reverse transcription using the following protocol: each 20 μl reaction contained 1 M-MLV buffer, 500 μmol of dNTP, 100 pmol of the oligo dT18 primer, 100 units of M-MLV reverse transcriptase, DEPC-H2O and 1 μg of total RNA. The mixtures were incubated at 42°C for 60 min for reverse transcription and then at 85°C for 5 min. Afterward, PCR was performed on a PCR apparatus. The total volume of the 20 μl RT-PCR reaction mixtures contained 10 μl of 2×HiFiTaq PCR StarMix (GenStar), 1 μl each of the forward and reverse primers (10 μM), 1 μl of cDNA and nuclease-free water to volume. The program used for all genes consisted of a denaturing cycle of 5 min at 95°C and 35 cycles of PCR (95°C for 30 s, 58°C for 30 s and 72°C for 40 s). The PCR product sizes were confirmed with agarose gel electrophoresis and Gel Red staining. The primers used in the RT-PCR were c-MYC A (5΄-TGGTGCTCCATGAGGAGACA-3΄), c-MYC S (5΄-GTGGCACCTCTTGAGGACCT-3΄), HRAS A (5΄-GTATCCAGGATGTCCAAC-3΄), HRAS S (5΄-GCTGATCCAGAACCATTT-3΄), c-KIT A (5΄-CACCGTGATGCCAGCTATTA-3΄), c-KIT S (5΄-CGTGGAAAAGAGAAAACAGTCA-3΄), VEGF A (5΄-CGGCTCACCGCCTCGGCTTG-3΄), VEGF S (5΄-TGCATTGGAGCCTTGCCTTG-3΄), BCL-2 A (5΄-GCCGGTTCAGGTACTCAGTC-3΄), BCL-2 S (5΄-GAGGATTGTGGCCTTCTTTG-3΄), β-actin A (5΄-GTTGCTATCCAGGCTGTGC-3΄), and β-actin S (5΄-GCATCCTGTCGGCAATGC-3΄).
RESULTS AND DISCUSSION
Synthesis of the triaryl-substituted imidazole IZNP-1
The synthesis of IZNP-1 is described in detail in the Supplementary Data. Its structure and purity were confirmed by 1H and 13C NMR spectrometry, HRMS spectrometry and HPLC analysis (Scheme S1 and Supplementary Figures S1–S4).
Spectroscopic studies of IZNP-1 interactions with nucleic acids
To test whether IZNP-1 could specifically recognize telomeric multimeric G-quadruplexes, we first studied the interactions of IZNP-1 with different nucleic acids using fluorescence spectroscopy, including four telomeric multimeric G-quadruplexes, six monomeric G-quadruplexes and one duplex DNA (Supplementary Table S1 and Supplementary Figure S5). As shown in Figure 1B, free IZNP-1 had weak fluorescence with maximum emission wavelength of 540 nm. However, the addition of the telomeric multimeric G-quadruplexes (htg45, htg51, htg57 and htg69) significantly enhanced the fluorescence of IZNP-1, while monomeric G-quadruplexes (htg21, c-kit2, RET, EAD, HRAS and TBA) and duplex DNA (hairpin) had very little effect on the level of fluorescence. Additionally, the Stokes’ shift was found to be as large as 110 nm in the presence of multimeric G-quadruplex htg45 (Supplementary Figure S6). These results suggested that IZNP-1 might selectively bind to telomeric multimeric G-quadruplexes because we found significant and positive correlations between the binding affinities and fluorescence enhancements of the triaryl-substituted imidazole derivatives when interacting with G-quadruplexes in previous studies (39,40).
To further understand the interactions of IZNP-1 with telomeric G-quadruplexes, fluorescence titration assays were performed. As shown in Figure 2A, IZNP-1 alone in buffer displayed a weak fluorescence emission at 540 nm. With the gradual addition of the multimeric G-quadruplex htg45, the emission peak shifted to 510 nm, which was accompanied by a significant fluorescence enhancement. In contrast, monomeric G-quadruplex htg21 also led to a blue shift, but the fluorescence signal displayed a very slight enhancement, even when the concentration of htg21 was set at a high level, which was probably because of a non-specific interaction between IZNP-1 and DNA. Additionally, fitting the titration data to Benesi–Hildebrand equation (42) revealed a 1:1 binding stoichiometry (IZNP-1/htg45), and the dissociation constants (KD) between IZNP-1 and htg45 was calculated to be 7.8 μM (Supplementary Figure S7). Otherwise, the binding stoichiometry and KD for monomeric G-quadruplex htg21 could not be determined because of weak interactions reflected in the negligible fluorescence enhancement upon the addition of up to 30 μM of DNA (Figure 2B). Similarly, all the KD values between IZNP-1 and other nucleic acids were obtained through fluorescence titrations and summarized in Supplementary Table S2. The KD values for multimeric G-quadruplexes htg57 and htg51 were determined as 13.3 μM and 11.0 μM, respectively, while no significant bindings were observed with regard to monomeric G-quadruplexes or duplex DNA. Collectively, these data revealed that IZNP-1 did bind to telomeric multimeric G-quadruplexes with a much stronger affinity.
Figure 2.
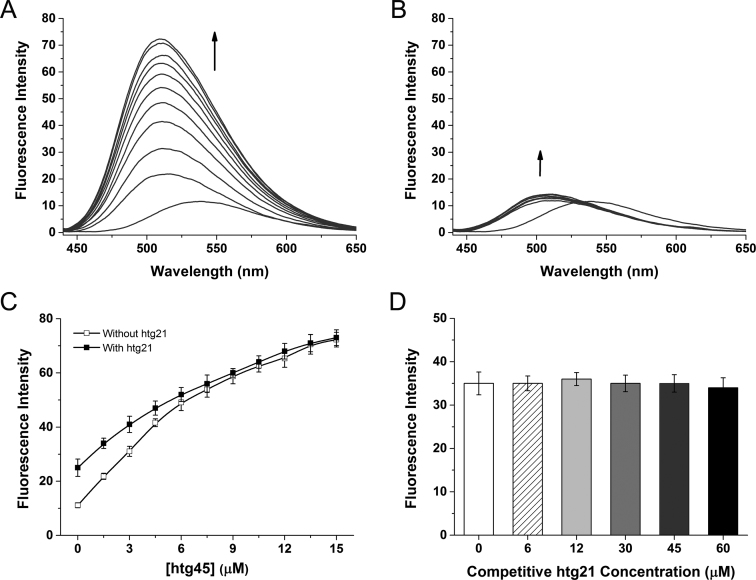
(A) The fluorescence spectra of 3 μM IZNP-1 with the stepwise addition of the multimeric G-quadruplex htg45 (arrow: 0–15 μM). (B) The fluorescence spectra of 3 μM IZNP-1 with the stepwise addition of the monomeric G-quadruplex htg21 (arrow: 0–30 μM). (C) The fluorescent titration curves of 3 μM IZNP-1 with the stepwise addition of htg45 in the absence and presence of 60 μM htg21. (D) The fluorescence intensities of 3 μM IZNP-1 with the stepwise addition of htg21 in Tris–HCl buffer containing 3 μM htg45. All the experiments were conducted in 10 mM Tris–HCl buffer, 100 mM KCl, pH 7.2 at λex = 400 nm.
The interactions of IZNP-1 with telomeric multimeric and monomeric G-quadruplexes were also explored by absorption titration assays. As shown in Supplementary Figure S8, with the addition of the multimeric G-quadruplex htg45, the absorption bands of IZNP-1 at both 323 and 400 nm continuously increased and then reached a relatively high level. However, the monomeric G-quadruplex htg21 had a little effect on the absorption spectrum of IZNP-1. This further demonstrated the high telomeric multimeric G-quadruplex binding selectivity of IZNP-1 against monomeric G-quadruplexes.
Competition titrations were performed to further confirm the selective interaction of IZNP-1 with telomeric multimeric G-quadruplexes compared to monomeric G-quadruplexes. As shown in Figure 2C, when gradually adding the multimeric G-quadruplex htg45 into a solution containing IZNP-1 and a high concentration of monomeric G-quadruplex htg21, the enhanced fluorescence emissions were practically identical to those in the experiment without a monomeric G-quadruplex competitor. In addition, in the presence of various amounts of the monomeric G-quadruplex htg21, the enhanced fluorescence of IZNP-1 with a constant concentration of the multimeric G-quadruplex htg45 was only slightly affected (Figure 2D). Thus, these competition experiments indicated that the presence of monomeric G-quadruplexes had little effect on the interactions between multimeric G-quadruplexes and IZNP-1, suggesting the much stronger binding affinity between IZNP-1 and telomeric multimeric G-quadruplexes. These results above confirmed the possibility of developing IZNP-1 as a specific telomeric multimeric G-quadruplex recognition ligand.
Melting studies of IZNP-1 interactions with nucleic acids
The ideal G-quadruplex ligand should possess two essential features: high G-quadruplex-recognition specificity and high G-quadruplex stabilization. Our experiments above demonstrated that IZNP-1 specifically recognized telomeric multimeric G-quadruplexes against monomeric G-quadruplexes and duplex DNAs. To test the ability of IZNP-1 to stabilize telomeric multimeric G-quadruplexes, the melting temperatures (Tm) of nucleic acids with and without IZNP-1 obtained through circular dichroism (CD) spectroscopic studies were compared. As shown in Table 1, IZNP-1 was highly effective at stabilizing multimeric G-quadruplexes, showing a ΔTm of 20 to 33°C. In contrast, the presence of IZNP-1 had a little effect on the Tm of the monomeric G-quadruplexes and the duplex DNA (Table 1 and Supplementary Table S3), confirming the above conclusion that a very weak interaction occurs between IZNP-1 and monomeric G-quadruplexes as well as duplex DNA. These results offered additional evidence that much stronger binding interactions exists between IZNP-1 and telomeric multimeric G-quadruplexes and further demonstrated that these interactions could increase the stability of telomeric multimeric G-quadruplexes.
Table 1. The G-quadruplex-stabilizing ability of IZNP-1a.
| G4 | T m (°C) under 100 mM KCl | ΔTm (°C) | |
|---|---|---|---|
| IZNP-1 (0 μM) | IZNP-1 (10 μM) | ||
| htg21 | 68 | 74 | 6 |
| htg45 | 63 | 86 | 23 |
| htg51 | 65 | 86 | 21 |
| htg57 | 64 | 84 | 20 |
| htg69 | 55 | 88 | 33 |
aMelting temperature (Tm) of G-quadruplex samples with and without IZNP-1 (incubating G-quadruplexes with IZNP-1 at room temperature) in 10 mM Tris-HCl buffer (100 mM KCl, pH 7.2) detected by circular dichroism spectroscopy.
Binding mode of IZNP-1 with telomeric multimeric G-quadruplexes
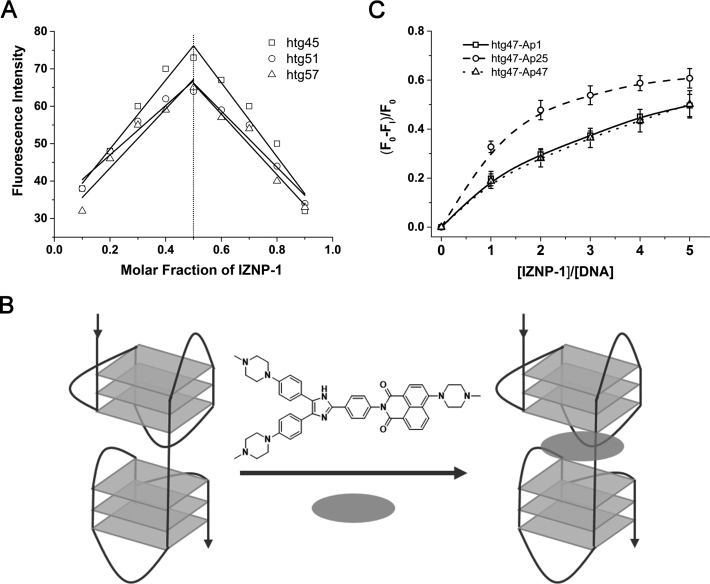
The results of the experiments described above suggested that IZNP-1 bound to telomeric multimeric and monomeric G-quadruplexes with dramatically different binding affinities. To understand such differences in behavior, we further investigated the binding modes of IZNP-1 to telomeric multimeric G-quadruplexes. It is already known that multimeric G-quadruplexes possess a particular quadruplex-quadruplex interface that is lacking in monomeric G-quadruplexes. Thus, we assumed that IZNP-1 might bind to telomeric multimeric G-quadruplexes by intercalating into the peculiar pockets between two adjacent G-quadruplex units. To test this, we first used Job plot analysis to determine the binding stoichiometry of IZNP-1 to multimeric G-quadruplexes. Three telomeric oligonucleotides were used, including htg45, htg51 and htg57. These sequences can fold into two G-quadruplex units. As shown in Figure 3A, an identical stoichiometry of 1:1 was observed. Furthermore, we analyzed another telomeric DNA oligonucleotide, htg69, which might fold into a multimeric G-quadruplex containing three G-quadruplex units. The Job plot analysis showed that IZNP-1 bound to htg69 with a stoichiometry ratio of nearly 2:1 (Supplementary Figure S9), which is mostly attributed to two IZNP-1 molecules intercalating into the two pockets of htg69. This result reinforced our hypothesis that IZNP-1 might bind to telomeric multimeric G-quadruplexes in an intercalative mode (Figure 3B). In contrast, no obvious inflection point was observed for the monomeric G-quadruplex htg21 (data not shown), suggesting a rather weak interaction between IZNP-1 and htg21.
Figure 3.
(A) The Job plot analysis for the binding stoichiometry of IZNP-1 to multimeric G-quadruplexes. The points of intersection of the best fit lines for the Job plots were near 0.5, showing a 1:1 stoichiometry of IZNP-1 binding to the G-quadruplexes. The excitation wavelength was set at 400 nm, and the emission was measured at 510 nm. (B) The proposed binding model of IZNP-1 to the multimeric G-quadruplex. (C) The plot of the normalized fluorescence intensity at 375 nm of 1 μM htg47 individually labeled with 2-Ap versus the binding ratio of [IZNP-1]/[htg47] at λex = 305 nm.
A modification of 2-aminopurine (2-Ap) in different loops has been widely used to estimate the binding mode of small molecules with G-quadruplexes (43). To gain more details on the interactions of IZNP-1 with telomeric multimeric G-quadruplexes, we performed fluorescence experiments using the multimeric G-quadruplex htg47 (two adenine bases were added at the 3΄- and 5΄-terminal of htg45) with 2-Ap substitutions at positions 1, 25 and 47, which are located at the two terminal G-quartets and the middle pocket (Supplementary Table S1). It was found that the fluorescence intensity of Ap25 in htg47 was significantly disturbed upon the addition of IZNP-1, compared with Ap1 and Ap47 (Figure 3C), indicating that IZNP-1 had close contact with this base, and further illustrated that IZNP-1 was inclined to bind to the pocket between the two quadruplex units in htg47. Such results again proved our hypothesis that IZNP-1 might intercalate into the quadruplex-quadruplex interfaces of multimeric G-quadruplexes (Figure 3B). In contrast, the fluorescence intensities of Ap1, Ap7, Ap13 and Ap19 in htg22 (one adenine base was added at the 5΄-terminal of htg21) were slightly affected by IZNP-1 (Supplementary Table S1 and Supplementary Figure S10), which proved that IZNP-1 might have a very weak interaction with htg22. These data were consistent with the fluorescence and absorption experimental results.
Effect of pocket size on the interaction between IZNP-1 and telomeric multimeric G-quadruplexes
As described above, IZNP-1 might intercalate into the pocket between two adjacent G-quadruplex units of telomeric multimeric G-quadruplexes. Because of this, we were curious as to whether the pocket size affected the interaction between IZNP-1 and multimeric G-quadruplexes. Thus, three mutant telomeric DNA sequences (htg43-1T, htg51-m and htg57-m) were designed to form multimeric G-quadruplexes with definite pocket sizes that differed from the native htg45 (Supplementary Table S1). In this case, htg43-1T, htg51-m and htg57-m might form two quadruplex units connected by a T linking loop, a TTATTATTA linking loop and a TTATTATTATTATTA linking loop, respectively. Therefore, multimeric G-quadruplexes with different pocket sizes were synthesized. Subsequently, we conducted fluorescence titration experiments using htg51-m and htg57-m. The fluorescence titration curves are shown in Supplementary Figure S11. When observing the binding of IZNP-1 to htg45, we observed that both the increased and the decreased pocket size might reduce the interaction between IZNP-1 and telomeric multimeric G-quadruplex, which was reflected by the decrease of the fluorescence level of the complex. Further calculation of the dissociation constant (KD) again revealed that the binding affinities actually decreased as the pocket size changed. The KD values for htg45 was 7.8 μM, while the KD for htg43-1T, htg51-m and htg57-m were determined as 20.5, 18.6 and 23.4 μM, respectively (Supplementary Figure S11). Such data indicated that the pocket formed by the two quadruplex units and the TTA linking loop in the multimeric G-quadruplex htg45 was most suitable for IZNP-1 binding, whereas a bigger or a smaller pocket might impede the formation of a tight complex of IZNP-1 and telomeric multimeric G-quadruplex.
Molecular modeling studies of IZNP-1 interactions with telomeric multimeric G-quadruplexes
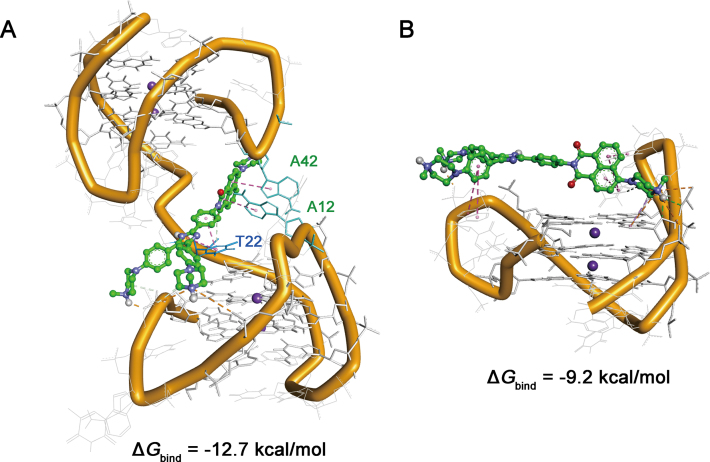
To gain more details on the interactions of IZNP-1 with telomeric multimeric G-quadruplexes, modeling studies were performed. According to our previous CD assays (Supplementary Figure S5), all the telomeric sequences used in this study formed into hybrid G-quadruplex topologies. We also observed that IZNP-1 did not affect the hybrid topology of multimeric telomeric G-quadruplex htg45 (Supplementary Figure S12). As is known, there are two known high-resolution NMR structures for the human telomeric sequence in K+ solution, Hybrid 1 and Hybrid 2 (44). Using these monomeric motifs as building blocks, we built a dimeric model Hybrid-12 (formed by a Hybrid 1 G-quadruplex at 5΄-end and a Hybrid 2 G-quadruplex at the 3΄-end) according to the previous study (27). Subsequently, we used the dimeric Hybrid 12 and the monomeric Hybrid 1 as the templates. Molecular models of IZNP-1 with Hybrid 12 and Hybrid 1 were first generated by docking studies and then examined by 40 ns molecular dynamics runs. The examined models and the estimated free energies of the binding of IZNP-1 in MM-PBSA calculations were shown in Figure 4. IZNP-1 intercalates into the pocket between two adjacent G-quadruplex units of Hybrid 12. In this case, the structure of IZNP-1 is twisted, and this reduced coplanarity cannot offer a robust stacking with the G-quartets at the interface. Actually, we observed that the electrostatic interactions with sugar-phosphate backbones and the π-π interactions with the bases of loops are the main interactions. In detail, as shown in Figure 4A and Supplementary Figure S13, the three outstretched physiologically positive 4-methylpiperazine groups can interact with the sugar-phosphate backbones of the two G-quadruplex units via electrostatic interactions. The triaryl-substituted imidazole moiety of IZNP-1 stacks on the thymine base of the loop bridging the two units (T22) via π–π interactions. Another part of IZNP-1, the 1,8-naphthalic anhydride moiety, can form stable stacking interactions with the two adenine bases of Hybrid 12 (A12 and A42), which further strengthens the binding affinity between IZNP-1 and Hybrid 12. Moreover, the intercalation of IZNP-1 into the pocket of Hybrid 12 induces the specific interactions between the two units, again stabilizing the dimeric structure. In contrast, IZNP-1 interacts with Hybrid 1 mainly via electrostatic interactions, and the whole structure cannot stack well on the G-quartets because the structure of IZNP-1 was twisted, resulting in the formation of a loose complex. In addition, IZNP-1 exhibited stronger binding with Hybrid 12 (−12.7 kcal/mol) than with Hybrid 1 (−9.2 kcal/mol). These findings were also consistent with the trends observed in absorption titrations, fluorescence titrations and CD melting assays, which may further strengthen the reliability of the models.
Figure 4.
(A) Averaged structures obtained through molecular dynamics (MD) simulations. (A) Complex model of IZNP-1 with the dimeric model Hybrid 12. (B) Complex model of IZNP-1 with the monomeric model Hybrid 1. IZNP-1 is shown using green sticks, and adenine and thymine bases stacked by IZNP-1 are shown using cyan sticks.
Effects of IZNP-1 on tumor cell proliferation
The above results support that IZNP-1 had a high binding affinity specific towards telomeric multimeric G-quadruplexes and possesses a remarkable multimeric G-quadruplex-stabilizing ability. IZNP-1 may accordingly have the potential to act as a favorable anticancer agent by disturbing telomere maintenance. In addition, the anticancer activities of previously reported telomeric multimeric G-quadruplex ligands have never been reported. Therefore, we made a biological evaluation of the effect of IZNP-1 on cancer cells.
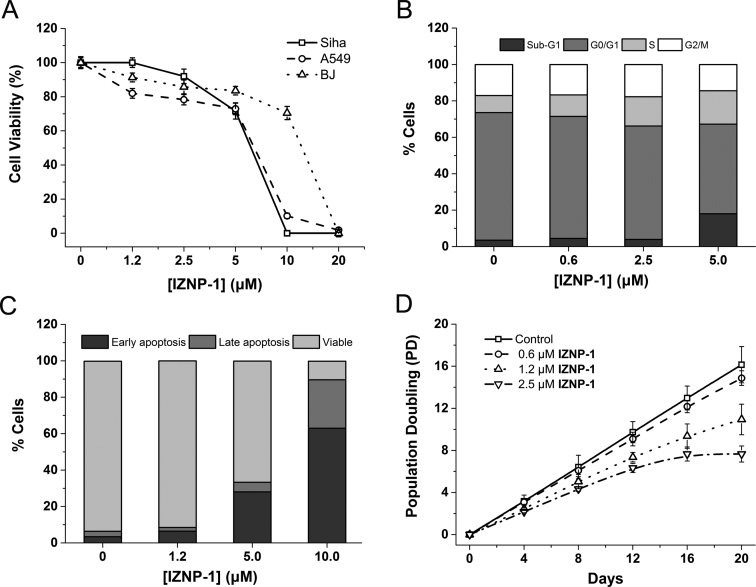
A short-term (24 h) cell viability assay was performed to assess the effects of IZNP-1 on cell proliferation in Siha, A549 and BJ cells. As shown in Figure 5A, IZNP-1 caused a significant dose-dependent cytotoxic effect on Siha and A549 cancer cells with IC50 values of 5.4 and 6.5 μM, respectively. In contrast, IZNP-1 induced growth inhibition in normal BJ fibroblasts with an IC50 of 13.3 μM, suggesting IZNP-1 was somehow more sensitive to cancer cells than normal cells.
Figure 5.
(A) The cell growth inhibition curves of tumor cells (Siha and A549 cells) and normal cells (BJ fibroblasts) after a 24-h treatment with IZNP-1. The data were reported as the percentage of growing cells in respect to that of the untreated cells. (B) The cell cycle analysis of Siha cells after 12-h treatment with IZNP-1. The cells were collected and stained with propidium iodide (PI). (C) An apoptosis evaluation of Siha cells after a 12-h treatment with IZNP-1. The cells were collected and stained with Annexin V–FITC and PI. (D) The proliferation curves of Siha cells treated with IZNP-1. The cells were counted and passaged at the indicated times, and the population doubling (PD) values were calculated from the cell amounts.
To further understand the short-term anti-proliferative activity of IZNP-1 on cancer cells, we explored whether IZNP-1 caused cell cycle arrest or apoptosis using flow cytometry assays. We first analyzed the percentage of cells in different phases of the cell cycle. As shown in Figure 5B and Supplementary Figure S14, after a 12-h treatment, IZNP-1 induced the apparent accumulation of cells in the S phase (increasing from 9.4% to 18.3%) and a sub-G1 peak (increasing from 3.5% to 18.1%) in a dose-dependent manner. Additionally, we also observed that IZNP-1 induced cellular apoptosis by Annexin V-FITC and PI staining assay. As shown in Figure 5C and Supplementary Figure S15, IZNP-1 induced apoptotic cell death in a dose-dependent manner. In Siha cells treated with 10 μM IZNP-1 for 12 h, the populations of apoptotic cells in the early stage and late stage were 26.6% and 63.0%. Otherwise, the non-treated cells had a negligible population of apoptotic cells. These effects of IZNP-1 on tumor cells were consistent with other reported telomeric G-quadruplex ligands. It is known that the accumulation of cells in the S phase is usually due to the induction of a DNA damage and repair pathway, and such events might result in cancer cell apoptosis (10,45). Thus, we proposed that IZNP-1, as a telomeric multimeric G-quadruplex ligand, might induce telomere DNA damage, thereby arresting the cell cycle in the S phase and causing cell apoptosis accordingly.
Telomeric G-quadruplex ligands might be capable of both directly causing telomere dysfunction (which leads to rapid anti-proliferation on cancer cells) and shortening telomeres during a relatively long period (which results in accelerated senescence) (16). Thus, the long-term cell viability assay (20 days) on Siha cells, which allowed a sufficient time for telomere shortening to occur, was further conducted using sub-cytotoxic concentrations of IZNP-1 (0.6, 1.2 and 2.5 μM). As shown in Figure 5D, after a 20-day treatment, growth arrest was observed in Siha cells. In addition, the long-term treatment of Siha cells with IZNP-1 led to apparent senescence, with larger cell size, a vacuolated cytoplasm, and β-galactosidase activity (Supplementary Figure S16). The percentage of SA-β-gal-positive cells reached 74.6% in Siha cells. Parallel, to investigate whether IZNP-1 could shorten telomeres, the telomere length was evaluated using the telomeric restriction fragment (TRF) length assay. As shown in Supplementary Figure S17, significant telomere shortening was observed after IZNP-1 treatment. These results demonstrated that IZNP-1 might induce telomere shortening, then cell senescence, and finally, the inhibition of the proliferation of cancer cells.
Telomeric DNA damage and telomere dysfunction induced by IZNP-1
Telomeres can form a T-loop structure, with the 3΄-G-rich strand invading the duplex telomeric repeats, which protects chromosome ends from being recognized and repaired as double strand breaks (DSB) and from triggering DNA damage responses (10). A protein complex consistent of six proteins (called shelterin) is responsible for the formation of the T-loop (5). A wealth of data indicates that telomeric G-quadruplex ligands, such as RHSP4, BRACO-19 and telomestatin, might interfere with telomere replication and disturb telomere capping (10–16), generating a telomere dysfunction in which the telomere architecture is disrupted (T-loop degradation), and chromosome ends are no longer properly protected. Without protection, telomeric ends would be rapidly recognized as damaged DNA and such an event would activate the DSB-mediated DNA damage response pathway (6,10,45). Moreover, uncapped telomeres can suffer degradation, inappropriate recombination, and end to end fusions and that telomere dysfunction can have an effect similar to that of DNA damage in eliciting cell cycle arrest, apoptosis and senescence (10,12).
To investigate whether IZNP-1 was able to induce DNA damage and whether the DNA damage was localized to telomeres, double immunofluorescence experiments were performed in Siha cells. Herein, BRACO-19 was used as a positive control. As shown in Figure 6A and B, a significant increase in the γH2AX foci (a hallmark of DNA double strand break response) was observed after treatment with IZNP-1 for 12 h, indicating that IZNP-1 actually induced DNA double-strand breaks. Besides, BRACO-19 induced a similar extent of DNA damage compared to IZNP-1. Furthermore, confocal microscopy revealed that most of the γH2AX foci induced by IZNP-1 co-localized with the TRF2 protein (about 62%), which formed the so-called telomere dysfunction-induced foci (TIF). Additionally, about 39% of the γH2AX foci induced by BRACO-19 co-localized with TRF2 protein, and this co-localization was much less than that of IZNP-1 (Figure 6A and C), demonstrating that IZNP-1 was more specific than BRACO-19. This was consistent with the fact that BRACO-19 targeted not only telomeric G-quadruplexes but also other G-quadruplexes in the genome (14,46,47). Such results suggested that IZNP-1 triggered DNA damage specifically located at telomeres.
Figure 6.
(A) Representative immunofluorescence images of γH2AX (green) and TRF2 (red) foci in Siha cells treated with or without 2.5 μM IZNP-1 or BRACO-19 for 12 h. The nuclei were stained with DAPI (blue), and typical co-localization foci are indicated by white arrows. (B) The quantification of γH2AX foci number per nucleus. (C) The quantification of TIF (telomere dysfunction-induced foci) number per nucleus. (D) Representative immunofluorescence images of BG4 (green) and TRF2 (red) foci in Siha cells treated with or without 2.5 μM IZNP-1 for 12 h. The nuclei were stained with DAPI (blue), and typical co-localization foci are indicated by white arrows. (E) The quantification of BG4 foci number per nucleus. (F) The quantification of co-localization of BG4 and TRF2 per nucleus. In all experiments, 50 nuclei were counted in each group. ***P < 0.001 compared with the controls. NS, not significant compared with the controls.
Furthermore, we investigated the effects of IZNP-1 on the dissociation of telomere-binding proteins and telomere uncapping. Our double immunofluorescence experiments revealed that 12-h treatment with IZNP-1 significantly delocalized POT1 and TRF1 (but not TRF2) from the nucleus to the cytoplasm (Supplementary Figure S18A and S18B). These events led to telomere uncapping, characterized by the formation of polynucleated nuclei (Supplementary Figure S18C). In part, the resulting dysfunctional telomere ultimately provoked cell cycle arrest, apoptosis and senescence. Besides, we also demonstrated that IZNP-1 did not show obvious binding to telomeric double-stranded DNA or alter the interaction between TRF1 and telomeric double-stranded DNA, suggesting that the delocalization of TRF1 was resulted from the T-loop degradation (Supplementary Figure S19). Therefore, we proposed that IZNP-1, by binding to and stabilizing G-quadruplex DNA at telomeres, impairs fork progression and/or telomere processing, and causes telomere uncapping, resulting in telomere dysfunctions and activation of DNA damage response pathway.
Effects of IZNP-1 on telomeric G-quadruplex in cellular
Whether IZNP-1 could stabilize endogenous telomeric G-quadruplexes was further investigated using the BG4 antibody, which was used for quantitative visualization of DNA G-quadruplexes in human cells (48). As shown in Figure 6D and E, 12-h treatment of IZNP-1 induced a significant increase of BG4 foci in the nucleus, indicating that IZNP-1 could increase the amount of G-quadruplexes in cellular. Moreover, to track whether the compound stabilized telomeric G-quadruplexes, double immunofluorescence experiments were also conducted to stain both BG4 and TRF2. As shown in Figure 6D and F, the co-localization of BG4 and TRF2 protein was considerably increased by IZNP-1 compared with that of the control cells, suggesting the inducement of endogenous telomeric G-quadruplex structures in cells by IZNP-1. Furthermore, to investigate whether the telomeric DNA damage was induced by ligand binding to and stabilizing telomeric G-quadruplexes, we again performed double immunofluorescence experiments to stain both BG4 and γH2AX in Siha cells. As shown in Supplementary Figure S20, IZNP-1 treatment significantly induced both BG4 and γH2AX foci, and most of the induced γH2AX foci co-localized with BG4 foci, suggesting that IZNP-1 could bind to and stabilize telomeric G-quadruplexes and then trigger DNA damage. Overall, we might conclude that through stabilizing telomeric G-quadruplexes, IZNP-1 induced DNA damage, telomere dysfunction and telomere shortening, and then led to cell cycle arrest, apoptosis and senescence.
However, we observed that the number of IZNP-1-induced BG4 foci was greater than the expected on the basis of telomere binding alone, and the extent of co-localization with TRF2 is also low (38%), suggestive of other targets. Considering that TERRA RNA might fold into multimeric G-quadruplexes, we thus examined IZNP-1 binding to TERRA RNA G-quadruplexes. As shown in Supplementary Figure S21, fluorescence titrations and CD melting assays demonstrated that IZNP-1 actually bound to and then stabilized TERRA multimeric G-quadruplexes, but with a reduced binding affinities (KD = 22.2 μM) compared to interacting with telomeric multimeric DNA G-quadruplexes. Therefore, we proposed that IZNP-1 might stabilize TERRA multimeric G-quadruplexes in cells and subsequently induce the BG4 foci. To provide more evidence, we further carried out double immunofluorescence experiments to stain both BG4 and TRF2 in Siha cells treated with IZNP-1 after RNase A digestion. The results were shown in Supplementary Figure S22. After RNase A treatment, a considerable portion of the IZNP-1-induced BG4 foci disappeared, whereas the co-localization foci of BG4 and TRF2 remained similar to that of the control, thus increasing the extent of co-localization with TRF2 (from 38% to 50%). Such data revealed that IZNP-1 might recognize and stabilize TERRA multimeric G-quadruplexes in cells.
Notably, excluding some background BG4 foci, there's still a small portion of IZNP-1-induced BG4 foci that did not belong to telomeric or TERRA G-quadruplexes. Indeed, a recent study that used a G4 ChIP–seq protocol employing BG4 revealed that an altered cellular state would cause a concomitant shift in the G-quadruplex profile in cells (49). We speculated that, to some extent, IZNP-1 altered the cell status and then might change the G-quadruplex landscape in cells. Thus, some of the undefined IZNP-1-induced BG4 foci might result from this reason. Besides, a number of noncanonical long loop and bulged G-quadruplex structures exist in the human genome (34) and we supposed that some unknown structures might resemble telomeric multimeric G-quadruplexes and possibly be recognized by IZNP-1. Apart from these uncertain events, IZNP-1 still exhibited favorable selectivity to telomeric multimeric G-quadruplexes.
Effects of IZNP-1 on the transcriptional levels of several common oncogenes
Monomeric G-quadruplex structures have been found in the promoter regions of many oncogenes, such as c-MYC, HRAS, c-KIT, KRAS, VEGF, BCL-2, etc., and ligands targeting these structures might repress transcription and also exhibit anti-proliferative activities (7,50). As IZNP-1 showed high selectivity for multimeric G-quadruplexes over monomeric ones, we thus evaluated its ability to down-regulate the transcription of several common oncogenes, including c-MYC, HRAS, c-KIT, VEGF and BCL-2, of which the promoters contained putative monomeric G-quadruplex structures. We determined the mRNA levels of these five genes in Siha cells treated for 12 h with IZNP-1. The results are shown in Supplementary Figure S23. IZNP-1 did not repress the transcription of c-MYC, HRAS, c-KIT, VEGF and BCL-2 at any level, even at a concentration of 10 μM. These data provided additional evidence for our conclusion that IZNP-1 inhibited cancer cell growth possibly by targeting telomeric multimeric G-quadruplexes. Notably, such behavior discriminates IZNP-1 from other reported telomeric G-quadruplex ligands that also showed interactions with promoter G-quadruplexes (51). Thus, this selective ligand has the potential to act as a lead compound or a chemical intervention tool in the research of telomeric G-quadruplexes.
CONCLUSION
Targeting G-quadruplexes has emerged as an appealing opportunity for drug intervention in anticancer therapy. The binding and stabilization of telomeric G-quadruplexes by small molecules are able to induce telomere dysfunction, thereby leading to cancer cell growth inhibition. The long G-rich single-stranded overhang of telomeric DNA has the potential to fold into higher-order multimeric G-quadruplexes. Such distinctive structures are different from that of a large number of monomeric G-quadruplexes in the human genome. Thus, small molecules that interact specifically with telomeric multimeric G-quadruplexes could be developed as promising anticancer agents with minimal side effects. However, to date, only a few ligands that possess multimeric G-quadruplex-targeting specificity have been reported. We have a great interest in searching for such ligands. In this study, a new triaryl-substituted imidazole derivative called IZNP-1 was synthesized, and its multimeric G-quadruplex recognition specificity was investigated. IZNP-1 had an excellent ability to discriminate between telomeric multimeric and monomeric G-quadruplexes. The interaction studies found that IZNP-1 selectively bound to telomeric multimeric G-quadruplexes with a much stronger affinity than it did with monomeric G-quadruplexes. Furthermore, the Job plot analysis, the 2-Ap titration assays and the further molecular modeling studies demonstrated that IZNP-1 might interact with telomeric multimeric G-quadruplexes by intercalating into the pocket between the two quadruplex units and forming a tight complex. Such a distinguished binding mode makes IZNP-1 a highly specific ligand for telomeric multimeric G-quadruplexes rather than other structures. In addition, an increase or a decrease in the pocket size was adverse for the intercalative mode.
IZNP-1 specifically bound to and effectively stabilized telomeric multimeric G-quadruplexes, making it a good candidate for cancer therapy. Hence, we evaluated the effects of IZNP-1 on tumor cells, demonstrating that it could provoke cell cycle arrest, apoptosis and senescence, which might be ascribed to the telomeric DNA damage and the telomere dysfunction induced by IZNP-1 stabilizing multimeric telomeric G-quadruplexes. Notably, this ligand had no effect on the transcriptional levels of several common oncogenes that have the potential to form G-quadruplexes in their promoters, suggesting that IZNP-1 had the potential to serve as a drug lead compound as well as a selective chemical tool for the study of telomeric G-quadruplex functions. To the best of our knowledge, this is the first report uncovering the telomere-related anticancer activity of a telomeric multimeric G-quadruplex ligand. This work also provides new insights for the development of novel anticancer drugs targeting telomeric multimeric G-quadruplexes.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81330077, 91213302, 21272291]; Guangdong Natural Science Funds for Distinguished Young Scholars [2015A030306004]; Guangdong Provincial Key Laboratory of Construction Foundation [2011A060901014]. Funding for open access charge: National Natural Science Foundation of China; Guangdong Natural Science Funds for Distinguished Young Scholar.
Conflict of interest statement. None declared.
REFERENCES
- 1. Neidle S., Parkinson G.N.. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003; 13:275–283. [DOI] [PubMed] [Google Scholar]
- 2. O'Sullivan R.J., Karlseder J.. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010; 11:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Lange T. How telomeres solve the end-protection problem. Science. 2009; 326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004; 5:323–329. [DOI] [PubMed] [Google Scholar]
- 5. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes. Dev. 2005; 19:2100–2110. [DOI] [PubMed] [Google Scholar]
- 6. Neidle S., Parkinson G.. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug. Discov. 2002; 1:383–393. [DOI] [PubMed] [Google Scholar]
- 7. Patel D.J., Phan A.T., Kuryavyi V.. Human telomere, oncogenic promoter and 5΄-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007; 35:7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaires J.B. Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010; 277:1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010; 277:1107–1117. [DOI] [PubMed] [Google Scholar]
- 10. Rizzo A., Salvati E., Porru M., D'Angelo C., Stevens M.F., D'Incalci M., Leonetti C., Gilson E., Zupi G., Biroccio A.. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 2009; 37:5353–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gomez D., O'Donohue M.F., Wenner T., Douarre C., Macadre J., Koebel P., Giraud-Panis M.J., Kaplan H., Kolkes A., Shin-ya K. et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006; 66:6908–6912. [DOI] [PubMed] [Google Scholar]
- 12. Sfeir A., de Lange T.. Removal of shelterin reveals the telomere end-protection problem. Science. 2012; 336:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan Z., Tang J., Kan Z.-Y., Hao Y.-H.. Telomere G-quadruplex as a potential target to accelerate telomere shortening by expanding the incomplete end-replication of telomere DNA. Curr. Top. Med. Chem. 2015; 15:1940–1946. [DOI] [PubMed] [Google Scholar]
- 14. Burger A.M., Dai F., Schultes C.M., Reszka A.P., Moore M.J., Double J.A., Neidle S.. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005; 65:1489–1496. [DOI] [PubMed] [Google Scholar]
- 15. Tauchi T., Shin-ya K., Sashida G., Sumi M., Okabe S., Ohyashiki J.H., Ohyashiki K.. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene. 2006; 25:5719–5725. [DOI] [PubMed] [Google Scholar]
- 16. Phatak P., Cookson J.C., Dai F., Smith V., Gartenhaus R.B., Stevens M.F., Burger A.M.. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br. J. Cancer. 2007; 96:1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010; 277:1118–1125. [DOI] [PubMed] [Google Scholar]
- 18. Cosconati S., Rizzo A., Trotta R., Pagano B., Iachettini S., De Tito S., Lauri I., Fotticchia I., Giustiniano M., Marinelli L. et al. Shooting for selective druglike G-quadruplex binders: evidence for telomeric DNA damage and tumor cell death. J. Med. Chem. 2012; 55:9785–9792. [DOI] [PubMed] [Google Scholar]
- 19. Di Leva F.S., Zizza P., Cingolani C., D'Angelo C., Pagano B., Amato J., Salvati E., Sissi C., Pinato O., Marinelli L. et al. Exploring the chemical space of G-quadruplex binders: discovery of a novel chemotype targeting the human telomeric sequence. J. Med. Chem. 2013; 56:9646–9654. [DOI] [PubMed] [Google Scholar]
- 20. Micco M., Collie G.W., Dale A.G., Ohnmacht S.A., Pazitna I., Gunaratnam M., Reszka A.P., Neidle S.. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013; 56:2959–2974. [DOI] [PubMed] [Google Scholar]
- 21. Maji B., Kumar K., Kaulage M., Muniyappa K., Bhattacharya S.. Design and synthesis of new benzimidazole-carbazole conjugates for the stabilization of human telomeric DNA, telomerase inhibition, and their selective action on cancer cells. J. Med. Chem. 2014; 57:6973–6988. [DOI] [PubMed] [Google Scholar]
- 22. Xiong Y.-X., Su H.-F., Lv P., Ma Y., Wang S.-K., Miao H., Liu H.-Y., Tan J.-H., Ou T.-M., Gu L.-Q. et al. A newly identified berberine derivative induces cancer cell senescence by stabilizing endogenous G-quadruplexes and sparking a DNA damage response at the telomere region. Oncotarget. 2015; 6:35625–35635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shinohara K., Sannohe Y., Kaieda S., Tanaka K., Osuga H., Tahara H., Xu Y., Kawase T., Bando T., Sugiyama H.. A chiral wedge molecule inhibits telomerase activity. J. Am. Chem. Soc. 2010; 132:3778–3782. [DOI] [PubMed] [Google Scholar]
- 24. Cummaro A., Fotticchia I., Franceschin M., Giancola C., Petraccone L.. Binding properties of human telomeric quadruplex multimers: a new route for drug design. Biochimie. 2011; 93:1392–1400. [DOI] [PubMed] [Google Scholar]
- 25. Dai J., Carver M., Yang D.. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008; 90:1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haider S., Parkinson G.N., Neidle S.. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008; 95:296–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petraccone L., Trent J.O., Chaires J.B.. The tail of the telomere. J. Am. Chem. Soc. 2008; 130:16530–16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu Y., Ishizuka T., Kurabayashi K., Komiyama M.. Consecutive formation of G-quadruplexes in human telomeric-overhang DNA: a protective capping structure for telomere ends. Angew. Chem., Int. Ed. 2009; 48:7833–7836. [DOI] [PubMed] [Google Scholar]
- 29. Petraccone L., Spink C., Trent J.O., Garbett N.C., Mekmaysy C.S., Giancola C., Chaires J.B.. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011; 133:20951–20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu H., Gu X., Nakano S., Miyoshi D., Sugimoto N.. Beads-on-a-string structure of long telomeric DNAs under molecular crowding conditions. J. Am. Chem. Soc. 2012; 134:20060–20069. [DOI] [PubMed] [Google Scholar]
- 31. Abraham Punnoose J., Cui Y., Koirala D., Yangyuoru P.M., Ghimire C., Shrestha P., Mao H.. Interaction of G-quadruplexes in the full-length 3΄ human telomeric overhang. J. Am. Chem. Soc. 2014; 136:18062–18069. [DOI] [PubMed] [Google Scholar]
- 32. Huang X.-X., Zhu L.-N., Wu B., Huo Y.-F., Duan N.-N., Kong D.-M.. Two cationic porphyrin isomers showing different multimeric G-quadruplex recognition specificity against monomeric G-quadruplexes. Nucleic Acids Res. 2014; 42:8719–8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Q., Liu Y.-C., Kong D.-M., Guo D.-S.. Tetraphenylethene derivatives with different numbers of positively charged side arms have different multimeric G-quadruplex recognition specificity. Chem. - Eur. J. 2015; 21:13253–13260. [DOI] [PubMed] [Google Scholar]
- 36. Zhao C., Wu L., Ren J., Xu Y., Qu X.. Targeting human telomeric higher-order DNA: dimeric G-quadruplex units serve as preferred binding site. J. Am. Chem. Soc. 2013; 135:18786–18789. [DOI] [PubMed] [Google Scholar]
- 37. Chen S.-B., Tan J.-H., Ou T.-M., Huang S.-L., An L.-K., Luo H.-B., Li D., Gu L.-Q., Huang Z.-S.. Pharmacophore-based discovery of triaryl-substituted imidazole as new telomeric G-quadruplex ligand. Bioorg. Med. Chem. Lett. 2011; 21:1004–1009. [DOI] [PubMed] [Google Scholar]
- 38. Chen S.-B., Wu W.-B., Hu M.-H., Ou T.-M., Gu L.-Q., Tan J.-H., Huang Z.-S.. Discovery of a new fluorescent light-up probe specific to parallel G-quadruplexes. Chem. Commun. 2014; 50:12173–12176. [DOI] [PubMed] [Google Scholar]
- 39. Hu M.-H., Chen S.-B., Guo R.-J., Ou T.-M., Huang Z.-S., Tan J.-H.. Development of a highly sensitive fluorescent light-up probe for G-quadruplexes. Analyst. 2015; 140:4616–4625. [DOI] [PubMed] [Google Scholar]
- 40. Hu M.-H., Chen X., Chen S.-B., Ou T.-M., Yao M., Gu L.-Q., Huang Z.-S., Tan J.H.. A new application of click chemistry in situ: development of fluorescent probe for specific G-quadruplex topology. Sci. Rep. 2015; 5:17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu M.-H., Chen S.-B., Wang Y.-Q., Zeng Y.-M., Ou T.-M., Li D., Gu L.-Q., Huang Z.-S., Tan J.-H.. Accurate high-throughput identification of parallel G-quadruplex topology by a new tetraaryl-substituted imidazole. Biosens. Bioelectron. 2016; 83:77–84. [DOI] [PubMed] [Google Scholar]
- 42. Benesi H.A., Hildebrand J.H.. A spectrophotometric investigation of the interaction of iodine with aromatic hydrocarbons. J. Am. Chem. Soc. 1949; 71:2703–2707. [Google Scholar]
- 43. Barbieri C.M., Srinivasan A.R., Rzuczek S.G., Rice J.E., LaVoie E.J., Pilch D.S.. Defining the mode, energetics and specificity with which a macrocyclic hexaoxazole binds to human telomeric G-quadruplex DNA. Nucleic Acids Res. 2007; 35:3272–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Klement K., Goodarzi A.A.. DNA double strand break responses and chromatin alterations within the aging cell. Exp. Cell Res. 2014; 329:42–52. [DOI] [PubMed] [Google Scholar]
- 46. Perrone R., Butovskaya E., Daelemans D., Palu G., Pannecouque C., Richter S.N.. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014; 69:3248–3258. [DOI] [PubMed] [Google Scholar]
- 47. Tippana R., Hwang H., Opresko P.L., Bohr V.A., Myong S.. Single-molecule imaging reveals a common mechanism shared by G-quadruplex–resolving helicases. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8448–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 50. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug. Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lemarteleur T., Gomez D., Paterski R., Mandine E., Mailliet P., Riou J.F.. Stabilization of the c-myc gene promoter quadruplex by specific ligands' inhibitors of telomerase. Biochem. Biophys. Res. Commun. 2004; 323:802–808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.