Abstract
AID deaminates C to U in either strand of Ig genes, exclusively producing C:G/G:C to T:A/A:T transition mutations if U is left unrepaired. Error-prone processing by UNG2 or mismatch repair diversifies mutation, predominantly at C:G or A:T base pairs, respectively. Here, we show that transversions at C:G base pairs occur by two distinct processing pathways that are dictated by sequence context. Within and near AGCT mutation hotspots, transversion mutation at C:G was driven by UNG2 without requirement for mismatch repair. Deaminations in AGCT were refractive both to processing by UNG2 and to high-fidelity base excision repair (BER) downstream of UNG2, regardless of mismatch repair activity. We propose that AGCT sequences resist faithful BER because they bind BER-inhibitory protein(s) and/or because hemi-deaminated AGCT motifs innately form a BER-resistant DNA structure. Distal to AGCT sequences, transversions at G were largely co-dependent on UNG2 and mismatch repair. We propose that AGCT-distal transversions are produced when apyrimidinic sites are exposed in mismatch excision patches, because completion of mismatch repair would require bypass of these sites.
INTRODUCTION
During adaptive immune responses, the affinity of antigen-specific antibodies increases over a time frame of a few days to weeks. Mutations are introduced into the Ig genes of activated B cells proliferating in germinal centres (1) by the DNA editing enzyme AID (activation-induced deaminase, gene Aicda) (2,3). AID deaminates C to U in targeted genes, producing a U:G mismatch founder mutation (4). AID has a preference to deaminate C within 5΄-WRCH-3΄, on either DNA strand (4,5). Ig genes concentrate this motif in the hypervariable regions of Ig V gene segments, and have undergone selection against it in most of the framework regions. As a consequence, mutation hotspots are concentrated in the hypervariable regions (6). A palindromic iteration of WRCH: AGCT, is the most favoured hotspot and AGCT motifs are highly abundant in Ig S regions (7–11).
If deaminations are not repaired, one daughter cell will inherit a U:A base pair in place of the original C:G base pair (a phase 1A mutation) and the other daughter will be un-mutated. DNA repair enzymes can correct U:G mismatches, but error-prone processing also diversifies mutation (reviewed in (12)). Specifically, the U can be excised by the uracil-specific enzyme UNG2 (uracil N-glycosylase), creating an apyrimidinic (AP) site (12). Conventionally, AP-endonucleases (APE1 or 2) can nick 5΄ to AP sites to facilitate repair synthesis by polymerase β, restoring the original base pair (13). Loss of UNG2 activity reduces the frequency of transversion mutations at C:G base pairs by >80% (14,15). It is frequently proposed that some AP-sites produced by the sequential action of AID and UNG2 are replicated before they are processed further by BER enzymes, requiring bypass of the AP sites by translesion DNA polymerases (phase 1B mutation) (12,16–19). Alternatively, the AP-lyase activity of the MRN complex may induce error-prone BER (20). Loss of UNG2 activity can also increase the frequency of C:G to T:A transition mutations, presumably as a result of increased uracil replication. This suggests that high fidelity uracil BER does occur in hypermutating B cells (9,11,19,21).
U:G mismatches are recognized by the mismatch binding protein MutSα: a heterodimer of MSH2 and MSH6. MutSα-deficiency substantially reduces the frequency of Ig mutations at A:T base pairs and also reduces transversion mutations at C:G base pairs. This has produced a consensus view that mismatch repair (MMR) plays a dominant role in AID-induced mutation at A:T base pairs, and a minor role in generating transversion mutations at C:G base pairs (12,16). Canonical MMR occurs post-replication: the binding of MutSα to a mismatch induces MutSα translocation, the formation of a MutSα/MutLα complex, and the recruitment of Exonuclease I (ExoI). Exo I excises a large single strand patch, starting from the nearest available 3΄-end (22,23). The excision patch is then in-filled by DNA polymerases δ or ε (24). In contrast, Ig A:T mutation exhibits little or no dependence on MutLα (see (12)), but still requires Exo I (25). It additionally requires PCNA capable of ubiquitination at K164 (26,27) and the translesion DNA polymerase η (pol η, (28)). K164Ub-PCNA/pol η is presumed to introduce mutations within Exo I excision patches and preferentially at A:T base pairs (phase II mutation, (16)), perhaps as a consequence of pol η's inherent infidelity (29), and/or as a consequence of low dNTP levels in G1-phase cells (30,31). It's not clear how nicks are created to enable Exo I entry during AID-induced MMR. UNG2 and SMUG1 induce at least some of the nicks via APE, but are semi-redundant with other nick generators; perhaps TDG, MBD4, MutLα or OGG1 (32–36).
Genetic ablation of MutSα did not increase the frequency of C:G to T:A transition mutations in hypermutating B cells in several studies (37–39), leading to the suggestion that faithful mismatch repair is poorly recruited to AID-induced founder mutations (21). However, it's possible that faithful MMR has been under-estimated because the most proliferated germinal center B cells are preferentially lost in MMR-deficient mice (37,38). Ablation of both UNG2 and MutSα results in a mutation spectrum consisting entirely of C:G to T:A transitions, on both strands. (9,19,40–41). This suggests that although SMUG1 and MBD4 do excise AID-induced uracils in MMR-competent cells (32,36,42), processing by these N-glycosylases does not occur in the absence of MutSα, or is non-mutagenic in V-regions when it does occur.
Thousands of AP sites are produced daily in normal cells independently of AID (43), but do not cause wholesale point mutation. The factors rendering AID-induced uracil excision mutagenic remain unclear. Here, we exploit mutation datasets collated using the SWHEL transduction/adoptive transfer model (see (30)) to analyse hypermutation at day 6 in a model immune response, prior to the narrowing of the response to a limited number of B cell dynasties. We find that both UNG2 and MMR repair many AID-induced uracils, especially in the top strand. Excluding C:G base pairs within and near AGCT hotspots—wherein UNG2 is mutagenic regardless of MMR activity—we find that interaction between UNG2 and MMR is required for up to 90% of transversions at C:G base pairs. Our data demonstrate that UNG2-mediated mutation occurs by distinct MMR-independent or MMR-dependent pathways, which are dictated by local sequence context.
MATERIALS AND METHODS
Mice
Male C57Bl/6 host mice were purchased from Animal Resources Centre (Canning Vale, Western Australia) and were used in experiments when 8–16 weeks old. SWHEL, Msh2ko/ko and Rag1ko/ko mice—all C57Bl/6 background (30,44) were interbred and maintained under SPF conditions in the Centenary Institute Animal Facility. Mouse experiments were approved and monitored by the University of Sydney Animal Ethics committee in accordance with the New South Wales Animal Research Act (1985 No. 123).
Retroviral transduction and adoptive transfer
pMiG-based retroviruses expressing EGFP and ugi-EGFP fusion protein have been described (19). cDNAs encoding mouse UNG2 (CCDS19560) or SMUG1 (CCDS27898) were cloned immediately 5΄ to the internal ribosome entry site (IRES) of the pMiG retroviral vector (45). cDNAs encoding UNG2-EGFP or SMUG1-EGFP fusion proteins were cloned in place of the IRES and EGFP sequences in the pMiG vector. Retroviral supernatants were produced using calcium phosphate-mediated transient transfection of Plat-E packaging cells (46). Primary SWHEL splenocytes were purified by density gradient separation on Histopaque 1083 (Sigma-Aldrich) and activated overnight by culture with recombinant CD40L as described (30), transduced by ‘spinfection’ (1100 g, 45 min, 20°C) with retrovirus supernatants in the presence of 4 μg/ml Polybrene, washed, then incubated for a further 2 days in activating medium. GFP+ve cells were purified by flow cytometric sorting (BD FACSAria IIU or Influx sorter), mixed with freshly HEL-conjugated sheep red blood cells (prepared as described (30)) and injected via a tail vein into 8–12 week old male C57Bl/6 hosts that had been primed i.p. with 108 non-conjugated SRBC in PBS seven days earlier. Each host received ≤104 GFP+ve cells, along with 108 HEL-SRBC in a bolus of 0.25 ml culture medium. Retroviral experiments were performed in accordance with a permit from the Office of the Gene Technology Regulator (Australia), overseen by the Royal Prince Alfred Hospital Institute Biosafety Committee (Sydney).
Measurement of SWHEL VDJ-mutation in single cells
Six days after adoptive transfer, individual transduced SWHEL B cells were recovered from the spleens of host mice into 96-well PCR plates (Bio-Rad) as GFP+ve cells that bound HEL conjugated to Alexa Fluor 647 (30). Following cell lysis and protein digestion, the VDJH-region of the single SWHEL allele present in each well was amplified by nested PCR, as described (30). PCR products from ≤60 wells per host in which amplification was successful were Sanger sequenced by Macrogen (South Korea). Mutations where secondary peaks formed <30% of the signal were confirmed using Sequencher software (version 5.1, Gene Codes Corporation). Processed sequences were sorted into phylogenetic trees (using neighbor joining and uncorrected ‘p’) with MacVector software (version 12.7.5 MacVector Inc) to check for clonal dynasties, then collated for a window spanning nucleotides 17–539 (counting from the translation start ATG codon as bases 1–3) using custom Microsoft Excel spreadsheets (version 15.23 for Mac, Microsoft Corporation). Statistical analyses of mutation data exported from Excel were performed using Prism software (version 7.0a for Mac, GraphPad Software).
RESULTS
Production of large datasets using the SWHEL adoptive transfer model
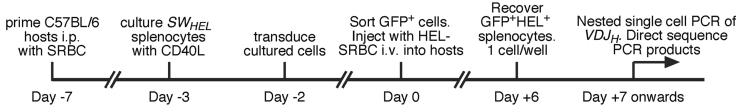
SWHEL mice carry the HyHEL10 VDJH3-rearrangement targeted to the IgH locus plus the HyHEL10 Igk as a low copy number randomly-integrated transgene, in the C57BL/6 strain. This enforces high affinity for hen egg lysozyme (HEL) upon B cells—as long as receptor editing has not occurred at the IgH locus (47). Editing is blocked in Rag1ko/koSWHEL mice, so all B cells are specific for HEL in this line (44). We blocked UNG2 activity in Rag1ko/koSWHEL (hereon simply called SWHEL) B cells via retroviral expression of the uracil glycosylase inhibitor (ugi) protein from phage (tagged with GFP), which blocks 98% of UNG activity (48). Retroviral expression of GFP alone served as a control. MutSα-deficient B cells were created by crossing Msh2ko alleles (49) into SWHEL mice, and Msh2ko/koSWHEL cells were transduced to express either GFP or ugi-GFP (19). We used our established procedure (30) to analyse hypermutation in transduced adoptive SWHEL cells (Figure 1). Briefly, C57BL/6 host mice were primed i.p. with 108 sheep red blood cells (SRBC) on day −7, to activate SRBC-specific T-cell help. On day −3, naïve donor SWHEL splenocytes were placed into culture with recombinant soluble CD40L. On day −2, these cells were transduced with retroviruses derived from pMiG (45). On day 0, GFP+ cells were purified and ≤104 GFP+ cells plus 108 HEL-conjugated SRBC were injected i.v. into each SRBC-primed host. By day +6, almost all adoptive SWHEL cells are in germinal centres (50) and comprise 0.1–0.5% of total splenocytes (data not shown). On day +6, individual HEL-binding GFP+ cells were recovered from host spleens into 96-well PCR plates, and the SWHEL VDJH allele present in each well was amplified by nested single cell PCR from genomic DNA, usually with ∼70% efficiency (30).
Figure 1.
Timeline for SWHEL transduction/adoptive transfer experiments.
Most of the mutations we collate are in exons (Supplementary Figure S1A), and subject therefore to Ag-selection, which has potential to bias the mutation spectrum towards rare mutations that favour antigen-binding. However, the HyHEL10 mAb from which SWHEL mice were derived has very high affinity for HEL (51), so Ag-selection is likely only to be against deleterious mutations, rather than for affinity enhancing mutations, especially given that adoptive SWHEL cells enter germinal centers only three days prior to our harvesting of SHM data (50). In support of this assertion, the distribution of mutations across all twelve possible point mutations in the adoptive SWHEL model is comparable to non-selected datasets derived from JH–CH intron sequences and from passenger Ig transgenes (based on comparison of Tables in (19) and (12)). Furthermore, as long as no more than fifty sequences are collated per host mouse, clonal dynasties are rare (30).
From now on, we will refer to cells expressing ugi-GFP or GFP as ‘UNG–’ or ‘UNG+’ cells, respectively, and refer to Msh2ko/ko or Msh2wt/wt cells as ‘M–’ or ‘M+’ cells, respectively. We collated mutations in UNG+M+, UNG–M+, UNG+M– or UNG–M–SWHEL cells, 6 days after antigen encounter, using 10 or 11 host mice per dataset (Supplementary Figure S1A, Table 1, Supplementary Table S1). As indicated in Table 1 (column s), sub-sets of the data collated here have been published for other purposes (19). Mutation frequencies were acceptably reproducible between independent hosts (Figure 2). The same hotspots were preferentially targeted in multiple mice within each dataset (Supplementary Figure S1B), demonstrating that hotspots arose as a result of repeat targeting, and were not an artifact of clonal dynasties. This indicated that our model could be used to quantitatively analyze antibody hypermutation.
Table 1. Summary of datasets and estimates of uracil processing as indicated by drops in transition mutations at C or G relative to the UNG−M– ‘baseline’, plus estimates of the relative contributions of UNG2 and MMR to other point mutations.
| a | b | c | d | e | f | g | h | i | j | k | l | m | n | o | p | q | r | s |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutations per 523bp sequence window: | Processing of baseline U | |||||||||||||||||
| No. of | G | C | drop in baseline Ts | U converted to Tv | contribution to: | |||||||||||||
| Transduced cells | Transduced protein | Shorthand | Hosts | Seq's | Mut'ns | Tv | Ts | Tv | Ts | A | T | G | C | G | C | A | T | published hosts (19) |
| Msh2ko/koSWHEL | ugi-GFP | UNG–M– | 10 | 368 | 999 | 0.02 | 0.97 | 0.008 | 1.70 | 0.005 | 0.008 | double-deficient baseline | 3 | |||||
| Msh2ko/koSWHEL | GFP | UNG+M– | 11 | 514 | 527 | 0.12 | 0.35 | 0.027 | 0.50 | 0.03 | 0.00 | 64% | 71% | 10% | 1.1% | 4% | -2% | 3 |
| SWHEL | ugi-GFP | UNG–M+ | 11 | 478 | 948 | 0.03 | 0.68 | 0.015 | 0.56 | 0.54 | 0.16 | 30% | 67% | 0.3% | 0.4% | 86% | 75% | 4 |
| SWHEL | GFP | UNG+M+ | 11 | 539 | 867 | 0.15 | 0.31 | 0.082 | 0.23 | 0.62 | 0.21 | 68% | 86% | 14% | 4.3% | 100% | 100% | 6 |
| PCR backgrounda | no AID | 2 | 375 | 3 | 0 | 0.005 | 0 | 0.003 | 0 | 0 | 1 | |||||||
aPCR background from sequencing CD4+B220–SWHELRag1+/+ cells.
Figure 2.
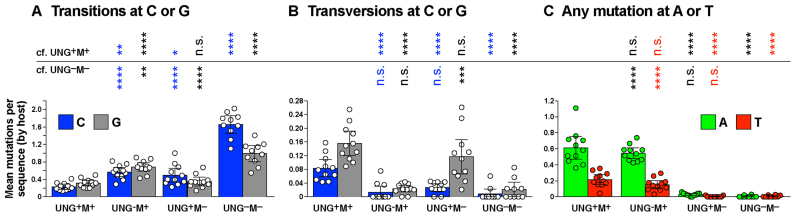
Mutations in the top strand of the SWHEL VDJH sequence window were averaged per host mouse (dots), with the overall mean of hosts (± 95% confidence interval) indicated by histograms. Significant differences (according to Tukey's ordinary one-way ANOVA) compared to (‘cf.’) the UNG+M+ or UNG–M– datasets are indicated: ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; n.s., not significant. Mutations are divided into three classes: (A) transition mutations at C or G; (B) transversion mutations at C or G; (C) any mutation at A or T.
Deficiency for UNG2 or MMR increases transition mutation at C:G base pairs
Deficiency for UNG2 activity (in UNG–M+ cells) or for Mutsα (in UNG+M– cells) increased transitions at C:G 2.3-fold or 1.6-fold, respectively, relative to UNG+M+ cells (Table 1). Reduced UNG activity increased transitions at C (p = 0.002) and at G (p < 0.0001) almost equally, while Msh2-knockout significantly increased transitions at C (p = 0.019), but not at G (Figure 2A). In double-deficient UNG–M– cells, transitions at C:G were increased 5-fold overall, relative to UNG+M+ cells (Table 1), with mutations at C increasing 2.3-fold more than mutations at G (Figure 2A). These transition increases in UNG–M– cells were highly significant, even in comparison to the UNG–M+ and UNG+M– datasets (Figure 2A). The data are consistent with MMR and UNG2 (especially UNG2) driving more faithful than mutagenic repair at deamination sites, but it is also possible that MMR and/or UNG2 curtail ongoing deamination by AID, such that deficiency for UNG2 or MMR gives AID greater leeway to deaminate cytosines in the first place. In other studies, mutation at C was also increased more than at G by double-deficiency for UNG2 and MutSα, but in two of those studies (9,40) the bias towards increased mutation at C was not as high as in this or another study (41). This may simply reflect experimental variability, which is better controlled in our study (because n ≥ 10 in each of our datasets) than in previous studies, or it may be due to differences in compensatory mechanisms induced by Ung-knockout versus ugi over-expression.
UNG2 and MMR semi-redundantly excise uracils
Transversion mutation at C:G (mostly phase 1B mutations (12)) and mutations at A:T (phase II mutations (12)) were almost eliminated in UNG–M– cells (Figure 2B and C, Table 1, Supplementary Table S1, Supplementary Figure S1A), as expected (40). The residual non-phase 1A mutations (16/999 mutations from 368 cells, Table 1) could be be due to a combination of PCR error (3 mutations per 375 cells—see Table 1), residual UNG2 activity (excess ugi leaves 2% of UNG activity intact (48)), and possibly the activity of other N-glycosylases such as SMUG1, TDG or MBD4. Thus, transition mutation at C:G in UNG–M– cells can be presumed to closely approximate the baseline deamination signature of AID in the absence of significant mutagenic uracil processing (52), and the residual phase 1B and phase II mutations we detected in UNG–M– cells can be considered little more than background noise.
We estimated the fraction of deaminations processed by UNG2 or MutSα by comparing C:G transition mutation in single-deficient UNG+M– cells or UNG–M+ cells to double-deficient UNG–M– cells. In UNG+M– cells, transitions were reduced by 71% at C and by 64% at G, relative to the UNG–M– baseline (Table 1). This suggests that UNG2 excises about two-thirds of deaminations in the absence of MMR, with 1.1-fold bias toward the non-template (upper) strand. Of course, dATP might be incorporated opposite deamination sites—indeed, dATP preferentially incorporates opposite AP sites in vitro (53,54). We presume, therefore, we have estimated a minimum amount of MMR-independent processing by UNG2. In UNG−M+ cells, transitions were reduced by 67% at C and by 30% at G, relative to the UNG–M– baseline (Table 1). Presumably this is also a minimum estimate of UNG2-independent processing by MMR in the absence of UNG2 activity, because Exo I excision of the G-bearing strand instead of the U-bearing strand of deamination sites would be indistinguishable from ignorant replication. MMR thus appeared to be much more biased towards excising upper strand deaminations than UNG2.
In UNG+M+ cells, transitions were reduced by 86% at C and by 68% at G, relative to the UNG–M– baseline; both of which are only a little greater than the uracil excision rate apparent in UNG+M–cells (Table 1). This result is consistent with incomplete redundancy between UNG2 and MMR for access to AID-induced uracils.
Collaboration between UNG2 and MMR to diversify mutation
As expected from the Neuberger model (12), transversion mutation at C:G was largely UNG2-dependent (because it was reduced by 85% in UNG−M+ cells, relative to UNG+M+ cells) and mutation at A:T was largely MSH2-dependent (because it was reduced by 96% in UNG+M– cells, relative to UNG+M+ cells; Table 1, Figure 2B–C, Supplementary Figure S1A). Some groups reported a similarly high dependence of A/T mutation on MMR (38,55), while others reported dependencies nearer 50% (37,39). The reason for this variability remains unknown, but it is unlikely to be due to variations in antigen selection, and might be due to variation between mouse strains.
Transversion mutation at C was substantially co-dependent on UNG and MMR. It was reduced by 84% in UNG–M+ cells (p < 0.0001) and by 68% in UNG+M– cells, relative to UNG+M+ cells (p < 0.0001, Figure 2B). Transversion mutation at G was reduced by 85% in UNG–M+ cells (p < 0.0001), but only by 25% in UNG+M– cells (p = 0.25, Figure 2B), so it was much less dependent on MMR. Mutation at A:T base pairs also appeared to be somewhat UNG2/MMR co-dependent, because the frequencies of A and T mutations in UNG+M– cells and UNG–M+ cells summed to values lower than their frequencies in UNG+M+ cells (see columns q-r in Table 1). Co-dependence of some phase 1B and phase II mutations on MMR and BER is already well recognised (12,32–33,35,56–57).
Processing of U:G base pairs by MMR or UNG2 appears to be dictated by local sequence context
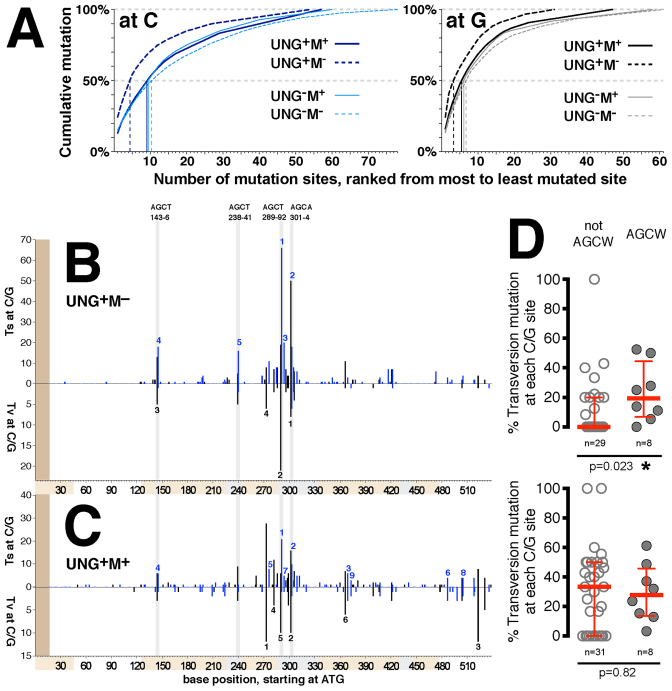
We were particularly interested to know whether local sequence context influenced the processing of deaminations. MMR-deficiency increases focusing of mutation on AID hotspots (25,37–39,55,58–59), but whether this occurs by altered AID-targeting (59) and/or by uneven uracil repair downstream of AID (28,33) is unclear. We wanted to quantify focusing in an unbiased manner. First, we sorted C or G mutation sites in each dataset from most to least mutated sites (Supplementary Figure S1B). We then calculated cumulative mutation at C or at G accordingly. The UNG+M– dataset produced outlying cumulative mutation plots for both C and G (Figure 3A). In the UNG+M– dataset, five C mutation sites or four G mutation sites (numbered in Figure 3B) accounted for ≥50% of mutation at C or at G, respectively (see 50% intercepts marked in Figure 3A). This was about half the number of mutation sites required in the other datasets. Seven of these nine most prominent UNG+M– hotspots occurred within AGCT or AGCA motifs (AGCW motifs, Figure 3B), in which the C on both strands conforms to the WRCH hotspot consensus. In other words, mutation focusing at C and at G in UNG+M– cells was about 2-fold greater than in the other treatment groups, and focussed towards palindromic hotspots (AGCT) or hotspots that are ‘quasi-palindromic’ from AID's viewpoint (AGCA). A direct role for MSH6 in AID-targeting (59) may contribute to mutation focusing in UNG+M– cells, but it is difficult for this role to explain the relaxed focusing evident in UNG–M– cells (see Figure 3A). Interference from MMR influences the fidelity of UNG2-mediated repair in vitro (33). This supports the proposal of Delbos et al. that mutation focusses to motifs that become hyper-resistant to faithful UNG2-mediated repair in MMR-deficient cells (28).
Figure 3.
Mutation focusing. (A) Mean mutations per mutation site ranked from most to least mutated (see Supplementary Figure S1B) were transformed into plots for cumulative mutation at C (blue, left) or at G (black, right) in the top strand. The x-axis indicates rank position (leftmost rank = most mutated C or G site), while the y-axis indicates cumulative mutation at C or at G. 50% intercepts indicate the least number of mutation sites required to account for ≥50% of all mutation at C or at G for each dataset. (B and C) Skylines of (above the x-axis) total transition or (below the x-axis, with a different y-scale) transversion mutations at (blue) C or (black) G in (B) UNG+M– cells or (C) UNG+M+ cells. Mutations were not collated in the most 5΄ region shaded brown. Grey stripes identify AGCW motifs. Beige and gray boxes at bottom indicate exons and CDRs (according to http://www.imgt.org), respectively. Top-ranked mutation sites that together accounted for ≥50% mutation at C or at G are indicated by integers above or below the x-axes, respectively. (D) Percentage of mutations that were transversions at each C or G site in the (top) UNG+M– or (bottom) UNG+M+ datasets, restricted to sites that acquired >7 mutations in the UNG–M– dataset, and partitioned into those lying (open symbols) outside or (closed symbols) inside an AGCW motif. Medians and interquartile ranges are indicated in red. p-values from Mann–Whitney tests are indicated.
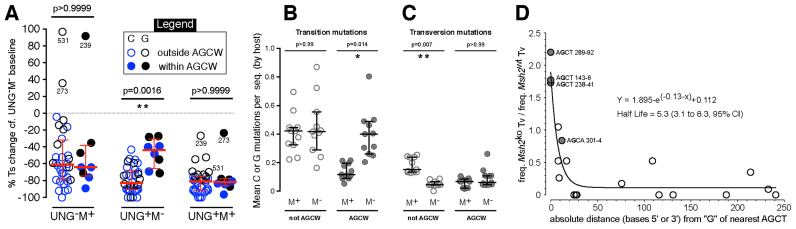
To test the proposal of Delbos et al., we measured changes in C or G transition frequency on a per-site basis in UNG–M+ cells and UNG+M– cells, compared to the UNG–M– baseline. This approach was based on the presumption that most C or G transitions are phase 1A mutations. To exclude rarely deaminated sites, only sites where >7 mutations were present in the UNG–M– dataset were considered. This arbitrary cut-off still encompassed 75% or 74% of mutation at C or G, respectively, in the UNG–M– dataset. Both MMR or UNG2 reduced baseline C and G transition at all sites considered, except for G531, G239 and G273 in the UNG–M+ dataset (Figure 4A). G239 and G273 were also noticeable outliers in the UNG+M+ dataset (Figure 4A). These outliers might be sites where MSH6 enhances AID-targeting, as proposed by Scharff's group (59). In MMR-competent UNG–M+ and UNG+M+ cells, baseline transitions were reduced outside and within AGCW sites almost equally (Figure 4A). However, in UNG+M– cells baseline transitions were reduced at sites outside AGCW almost twice as much as within AGCW motifs (p = 0.0016, Figure 4A). To confirm that these findings were consistent across replicate hosts, we plotted C:G transition frequencies outside or within AGCW motifs, for each host of UNG+M+ or UNG+M– cells (Figure 4B). This confirmed that C:G transition frequencies outside AGCW sites were comparable between UNG+M+ and UNG+M– cells, but within AGCW motifs were significantly higher in UNG+M– cells than in UNG+M+ cells (p = 0.014).
Figure 4.
Processing of deaminations outside versus within AGCW motifs. (A) The change in transition mutation in the top strand at (blue) C or (black) G in each dataset, relative to the baseline UNG–M– dataset, for every site that was mutated more than seven times in the UNG–M– dataset. Medians and interquartile ranges are indicated in red. Significant differences between (solid symbols) AGCW and (open symbols) non-AGCW sites are indicated by Kruskal-Wallis p-values. (B and C) Mean frequencies per sequence of (B) transition mutations or (C) transversion mutations at C:G base pairs lying (open symbols) outside or (closed symbols) inside AGCW sites, for individual hosts of (M+) UNG+M+ or (M–) UNG+M– cells. Bars indicate medians and inter-quartile ranges of hosts means. Significant differences between UNG+M+ cells and UNG+M– cells are indicated by Kruskall-Wallis p-values. (D) The MMR-dependence of normalised transversion mutation at G (y-axis) upon distance from ‘G’ in the nearest AGCT motif (x-axis). The best-fitting exponential decay curve (Prism software) is overlaid.
Thus, the AGCW sites upon which hypermutation became most focused in UNG+M– cells were sites that appeared to be especially resistant to MMR-independent processing by UNG2, as proposed by the groups of Weill and Ramiro (11,28). Nonetheless, the site-dependent changes in C:G transition mutations revealed in Figure 4A could be due to changes in targeting by AID, rather than changes in uracil-processing. If this were so, then the ratio between mutagenic and repair outcomes at AGCW versus non-AGCW deamination sites would not be expected to vary. We therefore determined if the frequency of mutagenic outcomes from deamination within AGCW versus outside AGCW varied.
Outside AGCW motifs, transversion mutation at C:G base pairs requires interaction between UNG2 and MMR
In the mutation skyline of UNG+M– cells shown in Figure 3B, transversion mutations at C/G were rare outside hotspots. We therefore calculated the percentage of mutations that were transversions at each C or G mutation site for the UNG+M– and UNG+M+ datasets (Figure 3D). Analysis was restricted to sites that acquired >7 mutations in the UNG–M– dataset. The percentages of mutations at C:G that were transversions were comparable within versus outside AGCW sites in UNG+M+ cells (p = 0.82, Figure 3D bottom). In contrast, they formed a reduced proportion of mutations at C:G outside AGCW sites, compared to inside AGCW sites in UNG+M– cells (p = 0.023, Figure 3D top). To confirm consistency across biological replicates, we plotted the frequencies of transversion mutations at C:G that lay outside or inside AGCW motifs, for each host of UNG+M+ or UNG+M– cells (Figure 4C). Transversion mutation at C:G outside AGCW sites was markedly reduced in UNG+M– compared to UNG+M+ cells (p = 0.007), but this was not the case within AGCW motifs (Figure 4C). This suggested that MMR was dispensable for UNG2-dependent mutation within AGCW motifs. Everywhere else, UNG2-dependent mutation was largely co-dependent on MMR. We noticed that most of the transversion mutations that remained outside AGCT motifs in our UNG+M– dataset were close to an AGCT site, including the transversions at AGCA (examine Figure 3B). This raised the possibility that proximity specifically to AGCT motifs, rather than AGCW motifs, might be the major factor governing transversion rate in UNG+M– cells. To normalize confounding influences, we plotted the ratio of transversion mutation in UNG+M– cells versus UNG+M+ cells as a function of absolute distance (i.e. 5΄ or 3΄) from the nearest AGCT motif. We only performed this analysis for transversions at G, because transversions at C were rare in all data sets (see Table 1). The plot produced was a good fit to exponential decay of MMR-independent G transversion mutation with distance from the nearest AGCT motif (Figure 4D; R square = 0.88; absolute sum of squares = 0.98), with a half-life of 5.3 bases (95% confidence interval = 3.1–8.3 bases), towards an asymptote of ∼0.1. In other words, repair by UNG2 was ∼90% error-free in M– cells, except within a roughly five base window either side of AGCT motifs. Within this window, MMR was redundant for UNG2-dependent mutagenesis.
UNG2 over-expression suppresses phase 1A and phase II mutations, but has little effect on phase 1B mutation
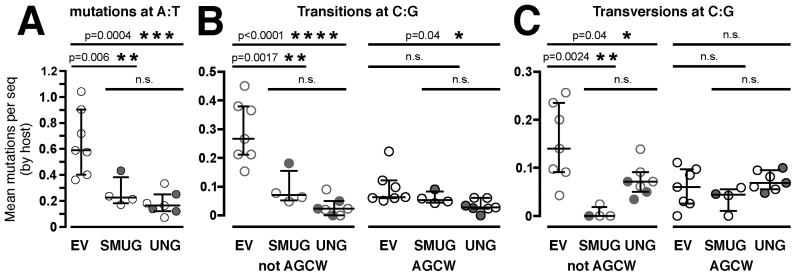
As a direct test for resistance of deaminated AGCW motifs to faithful processing by uracil N-glycosylases, we used the adoptive SWHEL model to over-express SMUG1 or UNG2 in MMR-proficient B cells and measured the impact on mutation within and outside AGCW hotspots. SMUG1 over-expression substantially suppressed mutation at A:T base pairs (p = 0.006, Figure 5A). Outside AGCW motifs, it also suppressed C/G transition mutation (p = 0.0017, Figure 5B) and almost ablated C/G transversion mutation (p = 0.0024, Figure 5C). However, SMUG1 over-expression had no significant impact on C/G mutation within AGCW motifs (Figure 5B and C). This suggested that SMUG1 was largely unable to process deaminations within AGCW motifs, while outside AGCW motifs, SMUG1 induced non-mutagenic BER almost exclusively or prevented deamination outside AGCW in the first place. The overall reduction in hypermutation induced by SMUG1 over-expression (Figure 5 and Supplementary Figure S2) is consistent with a previous report using human SMUG1-transgenic mice (42). Like SMUG1, ectopic UNG2 substantially suppressed mutation at A:T and C/G transition mutation outside AGCW motifs (p < 0.0001, Figure 5A and B), and had no impact upon C/G transversion mutation within AGCW motifs (Figure 5C). In addition, UNG2 over-expression slightly reduced transition mutation within AGCW motifs (p = 0.04, Figure 5B) and slightly reduced transversion mutation outside AGCW motifs (p = 0.04, Figure 5C).
Figure 5.
Uracil N-glycosylase over-expression suppresses mutation outside AGCW motifs. Mean frequencies of (A) mutations at A:T, (B) transitions at C:G or (C) transversions at C:G per sequence for each host mouse that received adoptive SWHEL B cells transduced with (SMUG) SMUG1-expressing vector, (UNG) UNG2-expressing vector or (EV) empty pMiG vector. Open symbols: co-expression of native protein and GFP. Closed symbols: expression of SMUG1-GFP or UNG2-GFP fusion proteins. Bars indicate median ± inter-quartile range of the host means. Holm–Sidak's p-values are shown.
Regardless of any impact SMUG1 or UNG2 over-expression may have on deamination rates, Figure 5 indicates firstly that most Ig C and G transitions arise from replication of unrepaired uracils, and secondly that deaminations within AGCW sites are resistant to faithful repair by UNG2 or SMUG1, even when these N-glycosylases are over-expressed. Furthermore, the virtual absence of transversion mutations at C:G outside AGCW motifs in SMUG1 over-expressing cells (Figure 5C) suggests that, unlike UNG2, SMUG1 cannot collaborate with MMR to produce AGCT-distal phase 1B mutations.
DISCUSSION
Incomplete redundancy between UNG2 and MMR to process AID-induced uracils
In contrast to early studies using memory or Peyer's patch B cells, loss of either UNG2 or MutSα activity increased V-region transition mutation at C:G bases in our day 6 germinal center B cells, especially at C (Figure 2A). The increases could be due to increased ignorance of deaminations, and/or to these factors curtailing deamination by AID. In theory, excision of template strand uracils by UNG2 could curtail iterative deamination, because AP-sites block transcription in yeast (60) and might therefore be expected to alter recruitment of AID by transcription complexes. This could explain why C and G mutation in UNG–M– cells were less focused than in wild-type UNG+M+ cells (Figure 3A). Nonetheless, we suspect the presence of AP sites does not have a major effect on AID-recruitment, firstly because mutation focusing in UNG–M– cells was only slightly more relaxed than in UNG+M+ cells (Figure 3A), and secondly because transversion mutation at C:G was only marginally reduced in cells over-expressing UNG2 (Figure 5C).
If we accept that UNG2 or MutSα induce only minor curtailment of deamination, Table 1 presents evidence that Mutsα is largely, but not completely redundant to UNG2 for excision of AID-induced uracils. Furthermore, the ability of ectopic UNG2 and SMUG1 to suppress MMR-dependent mutation (Figure 5A) suggests that excess uracil N-glycosylase activity out-competes MMR to process U:G base pairs. Recent data collated from day 8 anti-NP cells implied that MMR is completely redundant to UNG2 for processing U:G base pairs, because UNG2, MMR or combined UNG2/MMR activities reduced transitions at C:G by 81%, 64% or 78%, respectively, relative to Ung−/−Msh2−/− cells (our calculations from Supplemental Data in (9)). The deviations from our MMR data might be explained by target sequence composition, which has a profound impact on both AID-recruitment and processing downstream of AID ((8–9,11), this paper), or by the higher risk of sampling error in the NP study (due to fewer replicates per genotype) compared to this study, which has used the highest number of biological replicates per dataset to date. It is also possible that DNA repair-deficiency in TFH cells impinges upon B cell hypermutation when Ung−/− or Msh2−/− mice are immunised. This complication would not occur in our datasets, because only the adoptive B cells carried DNA repair defects.
A role for uracil BER in phase II mutations?
Despite the ability of elevated uracil N-glysolysase activity to inhibit MMR-induced mutation (Figure 5A), loss of UNG2 activity did not increase MMR's ability to produce mutations at A:T base pairs in our study (see Table 1), nor in any other studies. Some have concluded from this that UNG2 and MMR do not compete to process deaminations (61). However, it is also consistent with a fraction of phase II mutation being UNG2 and MMR co-dependent. Any potential co-dependence cannot be explained by recruitment of MMR to AP sites, because MutSα does not recognize BER intermediates (62). However, entry by Exo I at BER-induced nicks (32–35) does provide a plausible collaboration mechanism. Suppression of A:T mutation by ectopic uracil N-glycosylases (Figure 5A) might seem to rule this idea out. However, excess uracil N-glycosylase probably deprives MutSα of U:G substrate, suppressing activation of MMR. UNG/SMUG1-double knockout and UNG/APE2-double knockout significantly reduced mutation at A:T base pairs (32,35). This is strong evidence of a major role for uracil BER in A:T mutation. UNG2/SMUG1-double knockout, but not UNG/APE2-double knockout, also reduced the strand bias of A:T mutation (32,35). AID and UNG2 seem to have 1.75-fold and 1.1-fold bias, respectively, towards activity in the top strand in our datasets (see Table 1). The reduction in transitions we induced with ectopic SMUG1 was 1.2-fold biased towards C over G (data not shown). Assuming that AP sites can be accessed by AP-endonucleases in each strand equally, we calculate that combined UNG2 and SMUG1 activity should produce 1.9 to 2.2-fold more nicks in the top strand than in the template strand of mutating SWHEL alleles. This could bias Exo I entry into the top strand, should a nearby deamination site recruit MMR. Alternatively, AP-endonucleases may simply have better access to AP sites in the non-transcribed DNA strand. Either way, much of the strand bias of MMR-mediated uracil processing and of A:T mutation (see Table 1 and Figure 6) can be explained if BER is a major mediator of Exo I entry during AID-induced MMR. Semi-redundant nick generation by uracil BER and MutLα, and perhaps oxidation-induced BER (34) potentially explain why MutLα-knockout has only marginal impact on antibody hypermutation (57). Nick generation via MutLα might account for the markedly reduced strand bias of A:T mutation in Ung−/−Smug1−/− mice (32), because there is no reason to assume that MutLα can distinguish between the non-deaminated and deaminated strands of hypermutating Ig genes.
Figure 6.
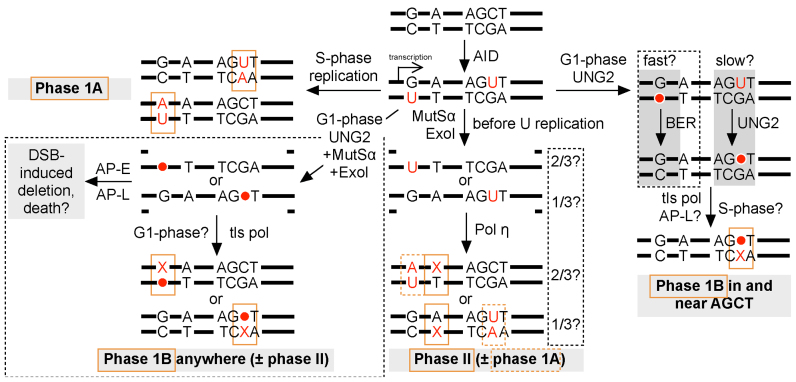
A revised hypermutation model based on (12,16,19–20). Dashed boxes enclose refinements added by this paper. The class of mutation (phase 1A, phase 1B or phase II) produced by each pathway is indicated by orange boxes. (•) AP site; (AP-E) AP-endonuclease; (AP-L) AP-lyase; (Pol η) DNA polymerase η; (tls pol) translesion polymerases; (Phase 1A) transition at C or G; (Phase 1B) transition or transversion at C or G; (Phase II) mutation at A or T.
Two distinct pathways for generating phase 1B mutations, dictated by proximity to AGCT motifs
Deaminations in AGCW motifs appeared more resistant to excision by UNG2 than deaminations at other sites (Figure 4A). In fact, the increase in C:G transition mutation induced by Msh2-knockout (relative to wild-type) was confined to these sites (Figure 4B). A similar phenomenon was noted in sequences 3΄ to JH4 (28). It's possible that targeting of AGCW motifs by AID increases when MMR is lost from UNG-competent cells because MSH6 can directly influence targeting by AID (see (59)). However, increased mutation-focusing in UNG+M– cells cannot be fully explained by abilities of MSH6 or MMR to influence AID targeting, because mutation was not similarly focussed in our UNG–M–cells (Figure 3A). Relaxed targeting was also apparent in Ung−/−Msh2−/− cells (see (28,40)). Mutagenic processing was particularly focused to AGCW sites in UNG+M– cells (Figure 3D). Thus, AGCW sites in UNG+M– cells were resistant both to processing by UNG2 and to conventional BER steps downstream of UNG2, diverting to mutagenic outcomes at a higher rate than other sites in UNG+M– cells. Even when UNG2 was over-expressed, C:G mutation rates within AGCW motifs were barely affected (Figure 5B and C). A similar phenomenon was observed in synthetic target genes in 3T3 cells expressing ectopic AID (11). Taken together, our results suggest that mutation-focusing previously described in UNG2-competent Msh2−/−, Msh6−/−, and Exo1−/− B cells (25,37–39,55,58) is more a consequence of altered uracil processing by UNG2 than of altered uracil production by AID.
We uncovered a mathematical relationship between distance from AGCT motifs and the MMR-dependence of transversion mutation at C:G (Figure 4D). We can predict from Figure 4D that the dependence of transversion mutation on MMR should decrease as the density of AGCT sequences in a target sequence increases. We can use the data of Chen et al.—who compared hypermutation in a conventional VDJH knock-in allele with hypermutation in an AGCT-rich knock-in allele (9)—to test this prediction. Ung-knockout reduced C:G transversion mutation by 85% and 89% (relative to Ung+/+Msh2+/+ cells) in Chen et al.'s conventional and AGCT-rich target alleles, respectively. These reductions compare to the 85% drop observed in our UNG–M+SWHEL cells. Msh2-knockout decreased C:G transversion mutation (relative to wild-type cells) in their conventional allele and our SWHEL allele by 44% and 40%, respectively. These reductions are also comparable, but contrast with the 29% reduction Msh2-knockout caused in Chen et al.'s AGCT-rich allele (our calculations, using Supplemental Data in (9)). This contrast is what we would predict from Figure 4D.
Based on our findings, we propose a refinement of the Neuberger hypermutation model (Figure 6), wherein AID-induced phase 1B mutation occurs by two distinct mechanisms: (1) In the near vicinity of AGCT motifs, phase 1B mutation depends on UNG2 and does not require MMR; (2) distal to AGCT motifs, phase 1B mutation is ∼90% co-dependent on UNG2 and MutSα. We propose that these distal mutations involve exposure of AP sites in Exo I excision patches. Critical to our model is the point that MMR could recruit ExoI to excise either the U-bearing or the G-bearing strand of U:G base pairs (albeit with a bias apparently towards ExoI entry into the top strand; see Discussion earlier), regardless of which strand was deaminated in the U:G base pair actually bound by MutSα. This creates the potential for uracils or AP-sites to be exposed in single-stranded MMR excision patches. If ExoI exposed AP-sites created by UNG2, or if UNG2 excised uracils after their exposure in ExoI excision patches, nicking by an AP-endonuclease or AP- lyase (20,35) would create a problematic double-strand break – likely leading to a deletion and/or cell death if nicking occurred in G1-phase (see Figure 6). AP-sites exposed within G1-phase ExoI excision patches would therefore require lesion bypass using a translesion polymerase to be resolved, potentially causing a phase 1B mutation (Figure 6). In the absence of MMR, or when MMR's access to U:G base pairs is over-whelmed by uracil N-glycosylase over-expression, AGCT-distal uracils appear to be processed efficiently by UNG2, leading almost exclusively to faithful BER. We noticed that SMUG1 over-expression almost ablated transversions at C:G outside AGCW sites (Figure 5C). SMUG1 has been reported to bind the AP sites it creates more persistently than UNG2 (63). Bearing this in mind, Figure 6 can explain why excess SMUG1 prevents phase 1B mutation distal to AGCT motifs: if SMUG1, rather than UNG2, created an AP site within an ExoI excision patch, persistent binding of SMUG1 to its product AP site would block excision patch in-fill and force the creation of a double-strand break once an AP-endonuclease or AP-lyase was recruited.
Deaminations within AGCT sequences appeared to be more resistant to processing by UNG2 and were also resistant to faithful BER steps downstream of UNG2 regardless of MMR activity (see Figures 3D and 4). Since transversion mutation at C:G depends on G1-phase UNG2 regardless of proximity to AGCT motifs (19), we propose that slow G1-phase BER within and near AGCT motifs leads to the persistence of G1-phase AP sites into S-phase, where their entry into replication forks induces mutagenic lesion bypass (Figure 6), as we proposed before (19), and/or induces mutagenic processing by the MRN complex (20). In contrast, bypass of AP-sites exposed in Exo I excision patches is likely to occur entirely within G1-phase – without any need for AP sites to persist into S-phase (Figure 6). It's possible, therefore, that MMR-independent versus MMR-dependent phase 1B mutation occur in distinct cell cycle phases. Our model (Figure 6) posits that the same fraction of deaminations within AGCT motifs will undergo BER-induced lesion bypass regardless of MMR activity; in MMR-competent cells they will undergo lesion bypass by either of the phase 1B pathways shown in Figure 6, while in MMR-deficient cells, they will undergo lesion bypass via the classic Neuberger pathway (i.e. ‘slow BER’) only.
Why are deaminated AGCT motifs resistant to uracil-BER?
Why do deaminated AGCT motifs appear to be resistant to uracil BER? Option A: In vivo, deaminated AGCT sequences might innately form a structure inaccessible to UNG2 and other BER enzymes. Option B: AID or co-factors (e.g. 14-3-3 proteins, Spt5, RPA or RNA pol II) might preferentially accumulate at AGCT sites in hypermutating B cells (7,28,64–68), thus reducing access by UNG2 and subsequently by AP-endonucleases or AP-lyases. Since AGCT sites form an AID hotspot in both strands, it's conceivable that AID or co-factors continue to be recruited to the opposite strand even after one strand has been deaminated or processed by UNG2. This would enhance the likelihood that deaminations or AP sites created in AGCT motifs in G1-phase persisted into S-phase. Deaminated AGCT motifs were not more resistant to processing by MMR than other deamination sites (Figure 4A). This can be explained by the potential for ExoI to excise uracil(s) distal to the site of initial MutSα-binding, regardless of whether MutSα ever bound to the uracil(s) excised by ExoI. Figure 4D implies that the foot-print of BER-resistance at AGCT sites is about 10.6 bases (i.e. 2X the Figure 4D half-life, which represents one half of a bell curve footprint). This is consistent with the size of the DNA binding grooves in AID homologues APOBEC2 (69) and APOBEC3G (70) (Xiaojiang Chen, personal communication). However, it is also consistent with the size of the single-stranded DNA bubble formed by RNA pol II (∼11 bases), which may be particularly prone to stalling at AGCT motifs in Ig genes independently of AID (64,68)].
CONCLUSION
In this study, we have refined the Neuberger model of antibody hypermutation, having provided evidence that UNG2-induced phase 1B mutation distal to AGCT hot-spots is MMR-dependent. Furthermore, our refined model potentially explains the previously enigmatic strand bias of phase II mutation. Our refinements are necessary to encompass the remarkable complexity of antibody hypermutation that has emerged since the Neuberger DNA deamination model was first developed.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the provision of cell sorting facilities and expertise by Sydney Cytometry (Centenary Institute), and the provision of mouse care and breeding by the Centenary Institute Animal Facility. All work was carried out with approval from and oversight by the University of Sydney and Royal Prince Alfred Hospital (RPAH) Animal Ethics/Welfare Committees and by the RPAH Institute Biosafety Committee, in accordance with NSW and Federal Australian legislation.
Footnotes
Present addresses:
George Sharbeen, Lowy Cancer Research Centre, Prince of Wales Clinical School, UNSW Sydney, NSW 2052, Australia.
K.K. Edwin Lau, Haematology Coagulation, Westmead Hospital, Westmead NSW 2145, Australia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council (NHMRC) Project Grants [1067891, 1051820 and 1012291 to C.J.] and a Postgraduate Scholarship [to E.L.]; Australian Postgraduate Awards [to E.T. and G.S.]; Cancer Institute NSW Scholarship [to G.S.]. Funding for open access charge: Centenary Institute Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gearhart P.J. The effect of somatic mutation on antibody affinity. Ann. N. Y. Acad. Sci. 1983; 418:171–176. [DOI] [PubMed] [Google Scholar]
- 2. Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T.. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000; 102:553–563. [DOI] [PubMed] [Google Scholar]
- 3. Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A. et al. . Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 2000; 102:565–575. [DOI] [PubMed] [Google Scholar]
- 4. Wang M., Rada C., Neuberger M.S.. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J. Exp. Med. 2010; 207:141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rogozin I.B., Diaz M.. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 2004; 172:3382–3384. [DOI] [PubMed] [Google Scholar]
- 6. Jolly C.J., Wagner S.D., Rada C., Klix N., Milstein C., Neuberger M.S.. The targeting of somatic hypermutation. Semin. Immunol. 1996; 8:159–168. [DOI] [PubMed] [Google Scholar]
- 7. Xu Z., Fulop Z., Wu G., Pone E.J., Zhang J., Mai T., Thomas L.M., Al-Qahtani A., White C.A., Park S.R. et al. . 14-3-3 adaptor proteins recruit AID to 5΄-AGCT-3΄-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 2010; 17:1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei L., Chahwan R., Wang S., Wang X., Pham P.T., Goodman M.F., Bergman A., Scharff M.D., MacCarthy T.. Overlapping hotspots in CDRs are critical sites for V region diversification. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E728–E737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Z., Eder M.D., Elos M.T., Viboolsittiseri S.S., Chen X., Wang J.H.. Interplay between target sequences and repair pathways determines distinct outcomes of AID-initiated lesions. J. Immunol. 2016; 196:2335–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yeap L.-S., Hwang J.K., Du Z., Meyers R.M., Meng F.-L., Jakubauskaitė A., Liu M., Mani V., Neuberg D., Kepler T.B. et al. . Sequence-intrinsic mechanisms that target AID mutational outcomes on antibody genes. Cell. 2015; 163:1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pérez-Durán P., Belver L., de Yébenes V.G., Delgado P., Pisano D.G., Ramiro A.R.. UNG shapes the specificity of AID-induced somatic hypermutation. J. Exp. Med. 2012; 209:1379–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di Noia J.M., Neuberger M.S.. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007; 76:1–22. [DOI] [PubMed] [Google Scholar]
- 13. Dianov G.L., Hubscher U.. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013; 41:3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Di Noia J., Neuberger M.S.. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002; 419:43–48. [DOI] [PubMed] [Google Scholar]
- 15. Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S.. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002; 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 16. Weill J.-C., Reynaud C.-A.. DNA polymerases in adaptive immunity. Nat. Rev. Immunol. 2008; 8:302–312. [DOI] [PubMed] [Google Scholar]
- 17. Daly J., Bebenek K., Watt D.L., Richter K., Jiang C., Zhao M.-L., Ray M., McGregor W.G., Kunkel T.A., Diaz M.. Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase zeta activity. J. Immunol. 2012; 188:5528–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kano C., Hanaoka F., Wang J.Y.. Analysis of mice deficient in both REV1 catalytic activity and POLH reveals an unexpected role for POLH in the generation of C to G and G to C transversions during Ig gene hypermutation. Int. Immunol. 2012; 24:169–174. [DOI] [PubMed] [Google Scholar]
- 19. Sharbeen G., Yee C.W., Smith A.L., Jolly C.J.. Ectopic restriction of DNA repair reveals that UNG2 excises AID-induced uracils predominantly or exclusively during G1 phase. J. Exp. Med. 2012; 209:965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Larson E.D., Cummings W.J., Bednarski D.W., Maizels N.. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Mol. Cell. 2005; 20:367–375. [DOI] [PubMed] [Google Scholar]
- 21. Saribasak H., Gearhart P.J.. Does DNA repair occur during somatic hypermutation. Semin. Immunol. 2012; 24:287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kadyrov F.A., Dzantiev L., Constantin N., Modrich P.. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006; 126:297–308. [DOI] [PubMed] [Google Scholar]
- 23. Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008; 18:85–98. [DOI] [PubMed] [Google Scholar]
- 24. Pena-Diaz J., Jiricny J.. Mammalian mismatch repair: error-free or error-prone. Trends Biochem. Sci. 2012; 37:206–214. [DOI] [PubMed] [Google Scholar]
- 25. Bardwell P.D., Woo C.J., Wei K., Li Z., Martin A., Sack S.Z., Parris T., Edelmann W., Scharff M.D.. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 2004; 5:224–229. [DOI] [PubMed] [Google Scholar]
- 26. Langerak P., Nygren A.O., Krijger P.H., van den Berk P.C., Jacobs H.. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J. Exp. Med. 2007; 204:1989–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roa S., Avdievich E., Peled J.U., Maccarthy T., Werling U., Kuang F.L., Kan R., Zhao C., Bergman A., Cohen P.E. et al. . Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:16248–16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Delbos F., Aoufouchi S., Faili A., Weill J.C., Reynaud C.A.. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2007; 204:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pavlov Y.I., Rogozin I.B., Galkin A.P., Aksenova A.Y., Hanaoka F., Rada C., Kunkel T.A.. Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase eta during copying of a mouse immunoglobulin kappa light chain transgene. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:9954–9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharbeen G., Cook A.J., Lau K.K., Raftery J., Yee C.W., Jolly C.J.. Incorporation of dUTP does not mediate mutation of A:T base pairs in Ig genes in vivo. Nucleic Acids Res. 2010; 38:8120–8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peòa-Diaz J., Bregenhorn S., Ghodgaonkar M., Follonier C., Artola-Bor·n M., Castor D., Lopes M., Sartori A.A., Jiricny J.. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell. 2012; 47:669–680. [DOI] [PubMed] [Google Scholar]
- 32. Dingler F.A., Kemmerich K., Neuberger M.S., Rada C.. Uracil excision by endogenous SMUG1 glycosylase promotes efficient Ig class switching and impacts on A:T substitutions during somatic mutation. Eur. J. Immunol. 2014; 44:1925–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schanz S., Castor D., Fischer F., Jiricny J.. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5593–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zlatanou A., Despras E., Braz-Petta T., Boubakour-Azzouz I., Pouvelle C., Stewart G.S., Nakajima S., Yasui A., Ishchenko A.A., Kannouche P.L.. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol. Cell. 2011; 43:649–662. [DOI] [PubMed] [Google Scholar]
- 35. Stavnezer J., Linehan E.K., Thompson M.R., Habboub G., Ucher A.J., Kadungure T., Tsuchimoto D., Nakabeppu Y., Schrader C.E.. Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9217–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grigera F., Wuerffel R., Kenter A.L.. MBD4 facilitates immunoglobulin class switch recombination. Mol. Cell. Biol. 2016; doi:10.1128/MCB00316-16 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rada C., Ehrenstein M.R., Neuberger M.S., Milstein C.. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998; 9:135–141. [DOI] [PubMed] [Google Scholar]
- 38. Frey S., Bertocci B., Delbos F., Quint L., Weill J.C., Reynaud C.A.. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998; 9:127–134. [DOI] [PubMed] [Google Scholar]
- 39. Jacobs H., Fukita Y., van der Horst G.T., de Boer J., Weeda G., Essers J., de Wind N., Engelward B.P., Samson L., Verbeek S. et al. . Hypermutation of immunoglobulin genes in memory B cells of DNA repair- deficient mice. J. Exp. Med. 1998; 187:1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rada C., Di Noia J.M., Neuberger M.S.. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 2004; 16:163–171. [DOI] [PubMed] [Google Scholar]
- 41. Shen H.M., Tanaka A., Bozek G., Nicolae D., Storb U.. Somatic hypermutation and class switch recombination in Msh6(-/-)Ung(-/-) double-knockout mice. J. Immunol. 2006; 177:5386–5392. [DOI] [PubMed] [Google Scholar]
- 42. Di Noia J.M., Rada C., Neuberger M.S.. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 2006; 25:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakamura J., Swenberg J.A.. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999; 59:2522–2526. [PubMed] [Google Scholar]
- 44. Cook A.J.L., Oganesian L., Harumal P., Basten A., Brink R., Jolly C.J.. Reduced switching in SCID B cells is associated with altered somatic mutation of recombined S regions. J. Immunol. 2003; 171:6556–6564. [DOI] [PubMed] [Google Scholar]
- 45. Refaeli Y., Van Parijs L., Alexander S.I., Abbas A.K.. Interferon gamma is required for activation-induced death of T lymphocytes. J. Exp. Med. 2002; 196:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morita S., Kojima T., Kitamura T.. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000; 7:1063–1066. [DOI] [PubMed] [Google Scholar]
- 47. Phan T.G., Amesbury M., Gardam S., Crosbie J., Hasbold J., Hodgkin P.D., Basten A., Brink R.. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. J. Exp. Med. 2003; 197:845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mol C.D., Arvai A.S., Sanderson R.J., Slupphaug G., Kavli B., Krokan H.E., Mosbaugh D.W., Tainer J.A.. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995; 82:701–708. [DOI] [PubMed] [Google Scholar]
- 49. de Wind N., Dekker M., Berns A., Radman M., de Riele H.. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination and predisposition to cancer. Cell. 1995; 82:321–330. [DOI] [PubMed] [Google Scholar]
- 50. Phan T.G., Gardam S., Basten A., Brink R.. Altered migration, recruitment, and somatic hypermutation in the early response of marginal zone B cells to T cell-dependent antigen. J. Immunol. 2005; 174:4567–4578. [DOI] [PubMed] [Google Scholar]
- 51. Chan T.D., Wood K., Hermes J.R., Butt D., Jolly C.J., Basten A., Brink R.. Elimination of germinal-center-derived self-reactive B cells is governed by the location and concentration of self-antigen. Immunity. 2012; 37:893–904. [DOI] [PubMed] [Google Scholar]
- 52. Xue K., Rada C., Neuberger M.S.. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2-/- ung-/- mice. J. Exp. Med. 2006; 203:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shibutani S., Takeshita M., Grollman A.P.. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the "A rule. J. Biol. Chem. 1997; 272:13916–13922. [DOI] [PubMed] [Google Scholar]
- 54. Simonelli V., Narciso L., Dogliotti E., Fortini P.. Base excision repair intermediates are mutagenic in mammalian cells. Nucleic Acids Res. 2005; 33:4404–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Phung Q.H., Winter D.B., Cranston A., Tarone R.E., Bohr V.A., Fishel R., Gearhart P.J.. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J. Exp. Med. 1998; 187:1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Frieder D., Larijani M., Collins C., Shulman M., Martin A.. The concerted action of Msh2 and UNG stimulates somatic hypermutation at A. T base pairs. Mol. Cell. Biol. 2009; 29:5148–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zivojnovic M., Delbos F., Girelli Zubani G., Jule A., Alcais A., Weill J.C., Reynaud C.A., Storck S.. Somatic hypermutation at A/T-rich oligonucleotide substrates shows different strand polarities in Ung-deficient or -proficient backgrounds. Mol. Cell. Biol. 2014; 34:2176–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wiesendanger M., Kneitz B., Edelmann W., Scharff M.D.. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 2000; 191:579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li Z., Zhao C., Iglesias-Ussel M.D., Polonskaya Z., Zhuang M., Yang G., Luo Z., Edelmann W., Scharff M.D.. The mismatch repair protein Msh6 influences the in vivo AID targeting to the Ig locus. Immunity. 2006; 24:393–403. [DOI] [PubMed] [Google Scholar]
- 60. Kim N., Jinks-Robertson S.. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 2010; 30:3206–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Krijger P., Storb U., Jacobs H.. Fugmann S, Diaz M, Papavasiliou N. DNA Deamination and the Immune System: AID in Health and Disease. 2010; World Scientific; 97–126. [Google Scholar]
- 62. Wilson T.M., Vaisman A., Martomo S.A., Sullivan P., Lan L., Hanaoka F., Yasui A., Woodgate R., Gearhart P.J.. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J. Exp. Med. 2005; 201:637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pettersen H.S., Sundheim O., Gilljam K.M., Slupphaug G., Krokan H.E., Kavli B.. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007; 35:3879–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ronai D., Iglesias-Ussel M.D., Fan M., Li Z., Martin A., Scharff M.D.. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J. Exp. Med. 2007; 204:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V. et al. . Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010; 143:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Basu U., Meng F.-L., Keim C., Grinstein V., Pefanis E., Eccleston J., Zhang T., Myers D., Wasserman C.R., Wesemann D.R. et al. . The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011; 144:353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yamane A., Resch W., Kuo N., Kuchen S., Li Z., Sun H.W., Robbiani D.F., McBride K., Nussenzweig M.C., Casellas R.. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 2011; 12:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Maul R.W., Cao Z., Venkataraman L., Giorgetti C.A., Press J.L., Denizot Y., Du H., Sen R., Gearhart P.J.. Spt5 accumulation at variable genes distinguishes somatic hypermutation in germinal center B cells from ex vivo-activated cells. J. Exp. Med. 2014; 211:2297–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prochnow C., Bransteitter R., Klein M.G., Goodman M.F., Chen X.S.. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007; 445:447–451. [DOI] [PubMed] [Google Scholar]
- 70. Holden L.G., Prochnow C., Chang Y.P., Bransteitter R., Chelico L., Sen U., Stevens R.C., Goodman M.F., Chen X.S.. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008; 456:121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.