Abstract
Polyadenylation is thought to be involved in the degradation and quality control of bacterial RNAs but relatively few examples have been investigated. We used a combination of 5΄-tagRACE and RNA-seq to analyze the total RNA content from a wild-type strain and from a poly(A)polymerase deleted mutant. A total of 178 transcripts were either up- or down-regulated in the mutant when compared to the wild-type strain. Poly(A)polymerase up-regulates the expression of all genes related to the FliA regulon and several previously unknown transcripts, including numerous transporters. Notable down-regulation of genes in the expression of antigen 43 and components of the type 1 fimbriae was detected. The major consequence of the absence of poly(A)polymerase was the accumulation of numerous sRNAs, antisense transcripts, REP sequences and RNA fragments resulting from the processing of entire transcripts. A new algorithm to analyze the position and composition of post-transcriptional modifications based on the sequence of unencoded 3΄-ends, was developed to identify polyadenylated molecules. Overall our results shed new light on the broad spectrum of action of polyadenylation on gene expression and demonstrate the importance of poly(A) dependent degradation to remove structured RNA fragments.
INTRODUCTION
Polyadenylation is a ubiquitous post-transcriptional modification, which profoundly affects the activity and fate of RNAs (1). First discovered in prokaryotes, polyadenylation has been primarily studied in eukaryotes, where it contributes to the export of mRNAs to the cytoplasm and promotes mRNA stability and translation. Subsequently polyadenylation was also demonstrated to have an RNA destabilizing function that is conserved in bacteria, organelles and both the nuclei and cytoplasm of eukaryotes (2–5). In this paper we investigate for the first time the entire genome-wide polyadenylation landscape in Escherichia coli.
In E. coli the rate-limiting step in the degradation of mRNAs is an endoribonucleolytic cleavage, performed in the majority of cases, by RNase E, but also by RNase III for a small number of mRNAs. These cleavages generate small RNA fragments which are then degraded by the 3΄-5΄ exoribonucleases RNase II, PNPase and RNase R (4). In some cases the addition of poly(A) tails by poly(A)polymerase I (PAP I) to the 3΄ end of RNA fragments has been shown to facilitate the degradation of short folded RNAs by providing a toe-hold for the exoribonucleases (4). Poly(A)-assisted decay is also implicated in RNA quality control, as demonstrated for a defective tRNA precursor (6). In contrast to eukaryotes, the pcnB gene, encoding poly(A)polymerase, is not essential in E. coli (7). This allows to compare polyadenylation in wild-type and a pcnB-deficient strain, as will be done here.
Polyadenylation is not only a biological waste-disposal system to recycle non-functional RNA molecules, it has also been shown to play an integral role in the regulation, either positive or negative, of the expression of certain bacterial genes in E. coli (8–12). The gene for PAP I (pcnB) was initially identified because it controls plasmid copy number. PAP I is required for the destabilization of RNA I, CopA, Sok and RNA-OUT antisense RNAs encoded by plasmids, phages and transposons and as a consequence controls their function (13). In addition, poly(A)polymerase I, with PNPase, participates in the end repair process of tRNA, when tRNA nucleotidyltransferase is absent, to maintain functional tRNA levels (14). PNPase also has a template-independent 3΄-oligonucleotide polymerase activity, and it was reported to be responsible for the addition of short heteropolymeric tails detected in the absence of PAP I and of non-A residues inserted into poly(A)tails in the wild-type strain (15).
Previous reports on the degradation pathway of the rpsO mRNA have revealed that about 10% of these mRNA harbor short oligo(A) tails ranging from one to five A residues. Poly(A) tails were mapped to several positions within the rpsO mRNA, in addition to the 3΄ end of the primary transcript, corresponding to positions expected to be both single stranded or within in a 3΄ folded termini. This showed that any 3΄ RNA extremity can be tagged by a poly(A) tail and that poly(A)polymerase has little or no sequence specificity for the addition of oligo(A) tails (16). This is in agreement with the idea that poly(A) tails are added indiscriminately and that repetitive rounds of polyadenylation and exonucleolytic digestion progressively can remove secondary structures that otherwise would impair 3΄-5΄ degradation of RNA (16–19).
Polyadenylation affects the decay of a significant number of mRNAs and has also been shown to affect a few non-coding RNA. However, these previous studies aiming to identify PAP I targets were performed in the absence of exoribonucleases or when PAP I was overproduced (6,20) but not under physiological conditions. In this paper, we make use of innovations in RNA-seq (21–26) to compare the transcriptomes of a wild-type strain and a strain deleted for the pcnB gene. Our proposal was to make a catalogue of RNAs, which are polyadenylated. By analyzing the 3΄ extremities of these RNAs, we demonstrate the range of PAP I substrates and gain insight to the specificity of PAP I target selection. Inactivation of poly(A)polymerase induces the massive accumulation of RNA fragments containing stable secondary structures. We identify many transcripts which are up- or down-regulated in the pcnB mutant. This differential accumulation of functional transcripts brings further evidence of the complex role, either direct or indirect, of polyadenylation, in the control of gene expression in E. coli.
MATERIALS AND METHODS
Bacterial strains, growth conditions
Strains N3433 (HfrH lacZ43 λ relA1 spoT1 thi1 from D. Apirion) and its ΔpcnB KanR derivative, IBPC903 (8) were used in this work. Strains were grown in LB medium at 37°C.
RNA extraction, northern blot analysis, 5΄ RNA tagging and RNA seq
Total RNA was prepared from bacteria grown to an A650 = 0.35–0.4 using the hot-phenol procedure (27). Ten micrograms of total RNA were electrophoresed either on 1% agarose formaldehyde gel or 6% polyacrylamide gels containing 7 M urea and analyzed by Northern blotting (28,29). Templates for the synthesis of the RNA probes were obtained by PCR amplification using the pair of m and T7 oligonucleotides (Supplementary Table S1). RNA probes were synthesized by T7 RNA polymerase with (α-32P) UTP yielding uniformly labeled RNAs (30). Membranes were also probed for 5S rRNA with 5΄-labeled oligonucleotide (Supplementary Table S1). RNA levels were quantified by phosphorimagery.
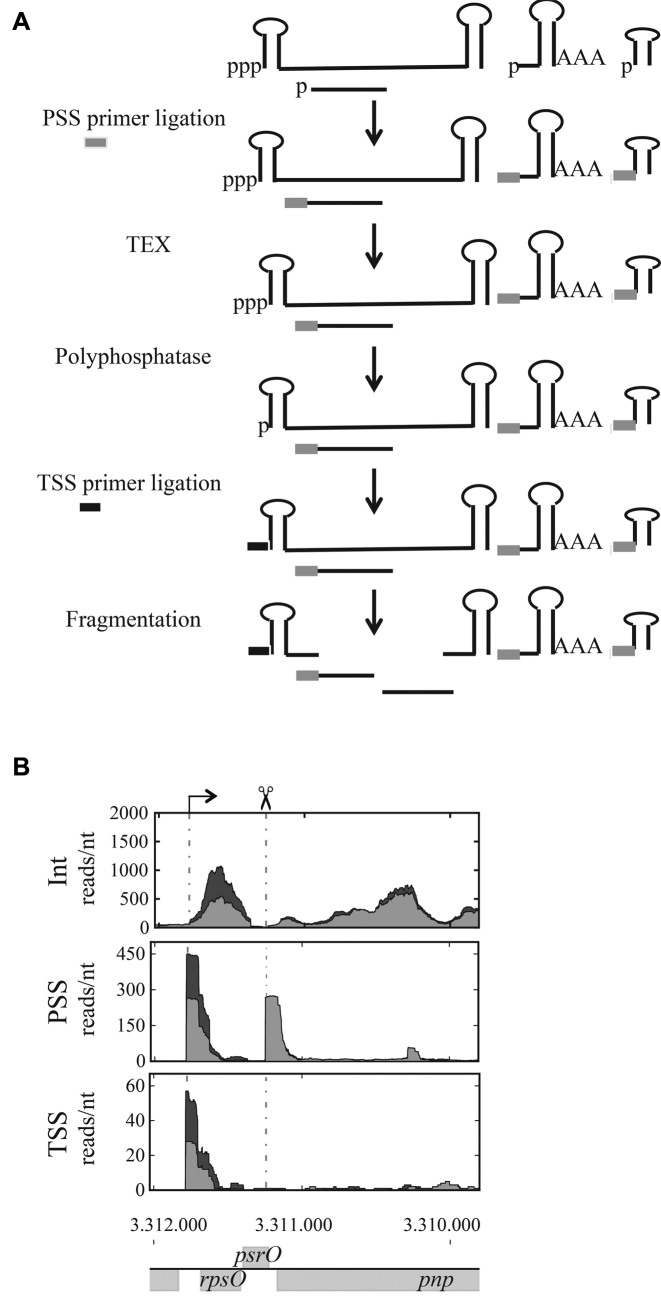
For RNA-seq analysis, after DNase RQ1 digestion (Promega), RNAs were re-extracted with phenol–chloroform and precipitated with ethanol. RNAs were prepared by using a modified version of the 5΄-tagRACE approach, including TEX treatment to remove untagged 5΄ mono-phosphates (26,31) (Figure 1A). Total RNA (40 μg) was ligated to the PSS RNA primer (Processing Start Site specific primer) (Supplementary Table S1) with T4 RNA ligase (Biolabs) in denaturing buffer (50 mM Hepes pH 7.5, 20 mM MgCl2, 3 mM DTT, 0.1 mM ATP, 10% DMSO, 10 μg/μl BSA) and precipitated with ethanol. Terminator™ 5΄-Phosphate-Dependent Exonuclease (TEX) was added to digest remaining RNA having a 5΄-monophosphate (Epicentre Biotechnologies). RNA primers and nucleotides were removed by gel filtration (Illustra MicroSpin G-50 Columns). γ and β-phosphates were sequentially removed from 5΄-triphosphorylated RNAs by RNA 5΄-polyphosphatase (Epicentre Biotechnologies). RNAs were re-extracted with phenol-chloroform and precipitated with ethanol. The TSS RNA primer (Transcription Start Site specific primer) (Supplementary Table S1) was then ligated and non-ligated primers removed by gel filtration. The PSS and TSS primers sequences were chosen as short sequences not found on E. coli chromosome similar enough to each other to avoid any sequence specific artifact during the ligations and without any secondary structure. RNAs were then fragmented according to the NEBnext Magnesium RNA fragmentation module and purified on an RNEasy column (Qiagen). Purified fragmented molecules were incubated with antarctic alkaline phosphatase then T4 polynucleotide kinase (NEB). Those molecules were next prepared according to the TruSeq Small RNA Library Prep kit replacing the last gel purification step by magnetic beads purification for a larger size selection.
Figure 1.
Scheme of the RNA preparation prior to the RNA-seq. (A) A hypothetical transcript and its degradation intermediates are illustrated. A mRNA with 5΄ triphosphate, stabilizing secondary structure near the 5΄-end and a terminator structure at the 3΄-end is shown. Three fragments with 5΄-monophosphate ends generated by endonucleolytic cleavages are also shown. Total RNA preparations from wt and pcnB mutant were incubated with RNA ligase and the PSS primer to tag 5΄-monophosphorylated RNAs. Excess PSS adaptors were eliminated and samples were incubated with TEX to remove untagged 5΄-monophosphorylated RNA molecules. RNA polyphosphatase was then used to eliminate γ and β-phosphates from primary transcripts, the TSS adaptor was ligated and TSS adaptor in excess eliminated. Total RNAs were then fragmented. Other extremities such as 5΄-hydroxyls generated by toxin cleavage, or RNA harboring a 5΄-NAD modification as a bacterial cap (66,67) will not have been tagged. (B) Differential expression profiles for the transcripts of the rpsO-pnp operon in the untagged (Int), PSS and TSS fractions from the wild-type (light grey) and pcnB deletion (dark grey). The arrow and the scissor refer to the rpsO transcription start and the RNase III upstream cleavage site, respectively. Expression level is indicated as reads/nt as a function of the gene's coordinates, which are shown under the RNA-seq profiles.
Directional sequencing was performed by the high throughput sequencing platform of IMAGIF (Centre de Recherche de Gif-www.imagif.cnrs.fr) on an Illumina HiSeq 2000, in a 2 × 100 bp run on three technical replicates for each condition.
RNA sequencing alignment, differential expression analysis and RNA secondary structure prediction
After demultiplexing, tag sorting and quality trimming, reads were mapped on the E. coli K12 genome (Genbank accession U00096.3) using BWA (v0.7.5a-r405) with default option and allowing multiple mismatches. Paired-end reads were merged and counted in each annotated region and inter-genic region with bedtools multicov (v2.19.0). Read counts were normalized by total number of reads in each condition and by their length (rpkm). Looking at the mean-log distribution, regions with at least 40 rpkm in one of the two strains are enough covered to be above the background noise and were taken into account for the differential expression analysis.
Mapped paired-end reads:
| Int | PSS | TSS | |
|---|---|---|---|
| wt | 33 418 029 | 5 201 903 | 363 327 |
| pcnB | 33 374 213 | 4 666 615 | 346 133 |
Detection level threshold of the 3΄ end extremity was determined as 2/3 of the value of the highest 5΄-monophosphate site density in each pcnB PSS upregulated region. A nucleotide n bp downstream of the PSS was considered as part of the fragment as long as the derivative 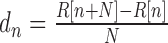 (with N = 10 and R = pile-up value) is <0.5.
(with N = 10 and R = pile-up value) is <0.5.
RNA extracted fragments were folded and minimum fold energy (mfe) determined using the RNAfold software v.2.1.9 (Vienna Package 2.0, (32)). Random sequences in the genome are picked up following the same normal length distribution as the extracted RNA fragments and were used as controls.
Validation of the experimental approach
An analysis of the RNA profiles in two regions, which have been previously studied in detail, were used to validate our method. Differential expression profiles from the RNAseq for the transcripts of the rpsO-pnp operon confirmed that the vast majority of the pnp transcripts harbored a PSS-tag at the downstream RNase III cleavage site (33), while the rpsO transcripts harbored either the TSS-tag, or the PSS-tag at the transcription start site (34) (Figure 1B and Supplementary Figure S1ABD). This indicates a conversion of the 5΄ terminus of the rpsO transcript from a triphosphate to a monophosphate (Supplementary Figure S1D), as has been shown previously (35). PAP I facilitates the degradation of GlmY and GlmZ sRNAs, which are required for substantial glmS expression. As a consequence the glmS mRNA is stabilized in the pcnB but very rapidly degraded in the wt, probably due to its low translation, although we cannot exclude that it is also polyadenylated (8–10) (Supplementary Table S2 and Figure S2).
Our attempts to validate the 5΄-tagging of the samples surprisingly revealed that mature 5S rRNA was resistant to TEX treatment, probably because of its recessed 5΄-monophosphate end (Supplementary Figure S1AC). Whilst being aware of this possible limitation to detect 5΄ extremity of transcripts, we decided to continue with this approach because our goal was to compare the levels of 5΄-tagged (and untagged, see below) RNAs in the wt and pcnB strains. After tagging the native 5΄ ends in the RNA preparations, subsequent RNA fragmentation before the sequencing allowed to get an overview of the entire transcriptomes. These fragmented RNA did not harbor any tag and were labeled Int for internal fragments. Transcripts, harboring the PSS or the TSS tags, were labeled PSS and TSS transcripts, respectively. Gene counts representative of RNA levels in the tagged (PSS and TSS) and untagged (Int) fractions from the wt and pcnB mutant are presented in Supplementary Table S2. Inspection of the mean-log plots distribution in tagged and untagged fractions, two cut-offs based on fold changes (FC) were determined; one medium (log2FC = 1.5) and one stringent (log2FC = 2.0), the stringent one being of higher confidence (Supplementary Figure S3). Transcripts which were either up- or down-regulated in the mutant when compared to the wild-type strain are presented in Table 1 and Supplementary Table S3.
Table 1. Differential expression analysis.
| coding | non-coding | |||
|---|---|---|---|---|
| Int | PSS | Int | PSS | |
| Up-high | 24 | 78 | 68 | 153 |
| Up-medium | 41 | 42 | 108 | 62 |
| unchanged | 2373 | 838 | 1679 | 1112 |
| Down-medium | 46 | 33 | 68 | 64 |
| Down-high | 46 | 26 | 35 | 52 |
The values indicate the number of coding and non coding regions up- and down-regulated in the pcnB mutant in the untagged (Int) and PSS fractions. Cut-off values for gene expression fold changes higher than log2FC = 2.0 determined up-high and up-down transcripts, while up-medium and down-medium transcripts present a FC determined as (1.5 < log2FC < 2).
Analysis of unmapped 3΄-ends
Demultiplexed raw reads were processed using PEAR (Paired-End reAd mergeR) with default options (36) in order to simultaneously merge mates of paired-end reads into single but longer reads and remove sequencing adapters. The merging process succeeded for 84–85% of raw reads leading to ∼52 million merged reads for each sample. Tags were removed using Flexbar v2.5 (37) and command line arguments “-be LEFT_TAIL -bt 2.5”.
Reads which included an RNA sequence, with a 3΄ tail that does not match the sequence on the coding strand of the DNA sequence, so called ‘unmapped’ 3΄ ends, were detected by an iterative aligning-and-trim procedure as follows: reads are aligned with Bowtie v1.0.0 (38), allowing only for perfect matches (‘-v 0’ command line option). For unaligned reads that are longer than 25 nt, one nucleotide is trimmed from the 3΄-end and memorized as the ‘tail’ of the read before a new mapping attempt is performed. At the end of this procedure, reads that still do not align are reconstructed and, for those longer than 25 nt, trimmed of one nucleotide at the 3΄-end and sent through a new series of iterative align-and-trim attempts. In total, 98% of merged reads were mapped this way in each sample.
For all reads that align after the removal of one or more nucleotides from their 3΄ end, we constructed four signals similar to ‘pile-ups’ but counting, instead of aligned reads, the number of unalignable nucleotides of each type at each given position (Supplementary Figure S4). Those four signals will be denoted further as SA, SC, SG and ST.
Finally, we construct an estimator defined as
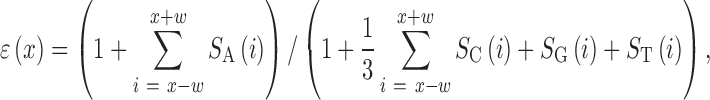 |
where SN(i) denotes the value of the corresponding signal at position i and w is a parameter that controls a tradeoff between noise rejection and accuracy, the value of which should be on the order of the expected size of a typical poly(A) tail. In this work, we set w = 5 and note that the analysis outcome does not vary much for w between 3 and 20.
RESULTS
Genome-wide localization of polyadenylation sites
In order to perform a genome-wide localization of polyadenylation sites and of other heterogeneous 3΄ tails in the wt strain and the pcnB mutant (15,39), we searched in the whole data set for reads partially mapping and ended with a 3΄ ‘tail’ not matching the sequence on the coding strand (Supplementary Figure S4).
We identified the nucleotides composition of 208 989 tails, clustering into 16 867 regions and calculated the fraction of Adenine in the tails using the estimator ε (Materials and Methods). Supplementary Figure S5A shows that there are many more A-rich unmapped tails in the wild-type strain. On the other hand RNAs with unmapped tails in the pcnB strain correspond to nucleotides added by PNPase (15,39) or could be the result of experimental artifacts. By choosing a cut-off, defining the most asymmetrical points in the wt strain (depicted by the dashed line in Supplementary Figure S5), we retrieved 878 unmapped 3΄-ends, which cluster into 138 regions (Supplementary Table S4). Twenty-eight candidates were eliminated by manual curation because they either fell into regions with very low expression levels and/or were very low frequency amongst all the 3΄ ends detected in that region. Of 110 remaining sites, polyadenylation of only three of them; lpp, rmf and GlmY, has been previously reported (9,12,40).
Newly identified polyadenylated molecules
mRNAs
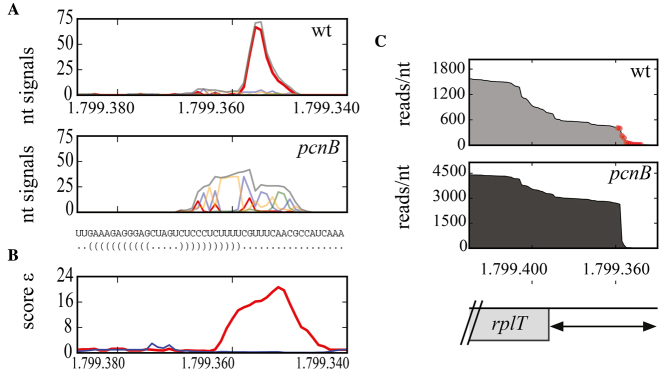
A striking new example of a polyadenylated RNA is the rplT mRNA, encoding the L20 ribosomal protein (Supplementary Table S4). The rplT RNA levels were considerably higher in the pcnB mutant in both the PSS and Int fractions (Supplementary Table S2). Figure 2A and B shows that the composition of unmapped 3΄ ends and the tail pile-up signals, ε, obtained with the estimator, are very different in the wt and the pcnB strain. The unmapped 3΄ ends detected in the wt strain were essentially A residues located downstream of the Rho-independent terminator and were coincident with a massive reduction in transcript abundance at this position (Figure 2B and C). The 3΄ non-template extensions detected in the pcnB mutant did not show a preference for any nucleotide (Figure 2A). They were located throughout the terminator region and corresponded to only a small percentage of the total rplT RNA. The vast majority of rplT transcripts terminated abruptly at the terminator. This difference between the rplT RNA and polyadenylation profiles in the wt and the pcnB strain illustrates how the PAP I assists the exoribonucleases to achieve the degradation of RNA harboring a terminator. Analogous results were obtained for frr, hdeB and rpmF RNA (Supplementary Figure S6).
Figure 2.
Polyadenylation sites at the 3΄ end of rplT. (A) Signals from unmapped tails detected in the 3΄-UTR of the rplT transcript in the wt and the pcnB strain are presented with A residues in red, C in blue, G in green, U in orange and total in grey. Sequence and predicted RNA structure obtained with RNAfold (ViennaRNA package 2) of the 3΄ end of rplT containing the Rho independent terminator are indicated. (B) Tail pile-up signals ε obtained with the estimator, representing the number of residues and their A content in the unmapped sequences in the wt (red) and pcnB (blue) strains (see Materials and Methods). (C) Expression profile of 3΄ end of rplT in the Int fraction in both strains. Post-transcriptionally A-added nucleotides detected as unmapped 3΄ additions are indicated in red dots in the wt strain.
non-coding RNAs
REP sequences represent a major class of polyadenylated molecules (14/110) in the wt strain which are more abundant in the pcnB strain (Supplementary Tables S2 and S4). tRNA precursors corresponded to the second most represented class of polyadenylated transcripts (5/110), with poly(A) located downstream of the 3΄-end of the mature tRNA and, in the case of aspV, downstream of a strong secondary structure (Supplementary Figure S6). Analogous results were obtained for asnV, leuZ, metY and serT (Supplementary Table S4). We also detected poly(A) tails on sRNAs (4/110) which are terminated by Rho-dependent (SibD) and Rho-independent terminators (IstR and GlmY) (41,42), indicating that Rho-dependent terminators are also poly(A) targets (Supplementary Table S4). No poly(A) tails were detected at the internal polyadenylation site of GlmY (9,10) but rather at the extremity of its Rho-independent terminator, which correlates with the down-regulation of the full length sRNA.
These examples illustrate another proposed function of poly(A)polymerase, which is to participate to the maturation of functional tRNA molecules (6,43). Altogether unmapped A sequences were found downstream of the terminators of mRNA and sRNAs, after the stable secondary structures of REP sequences and at the 3΄ extremities of tRNA precursors. We designate these RNAs as direct targets of PAP I.
Other sites of polyadenylation have been mapped in strains deficient in exonucleases (16). Since these sites are not associated with unmapped poly(A) sequences in the wt strain, we suspect that they are normally too rapidly degraded by exonucleases in the wt strain, to be detected in our analysis.
Small structured RNA molecules accumulate in the pcnB mutant
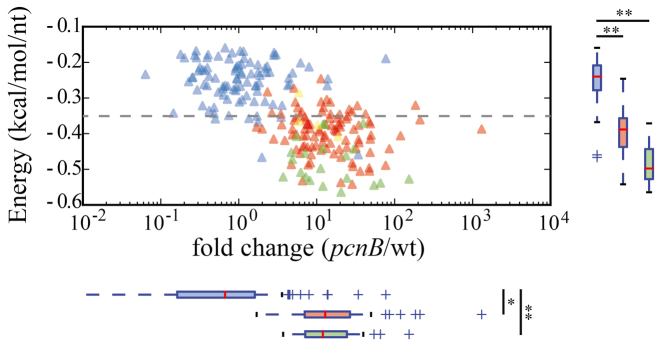
In order to further investigate the involvement of PAP I in the degradation of RNAs, we next analyzed PSS tagged molecules, which are more abundant in the pcnB strain. Their sequences were extracted, the resulting fragments were folded in silico and the minimum free energy of folding (mfe) of these molecules was calculated (Materials and Methods). This pool of fragments includes 102 short RNA fragments from full-length transcripts (including ptsG, pheS, malT and fur fragments), 21 REP sequences and 6 sRNAs, whose lengths range from 75 to 342 nt. They have stronger secondary structures than random sequences, and their level of accumulation are significantly higher than folding energy of random sequences (Figure 3). This is particularly true for REP sequences which, as shown above, are major targets of polyadenylation, thus confirming the direct role of PAP I in the destabilization of these stable RNA fragments. Most of these fragments had a mfe below 0.35 kcal/mol/nt (Figure 3). This value could represent the limiting energy above which an RNA structure requires addition of poly(A) for degradation. These small structured RNAs are thus likely to be direct targets for PAP I, even though poly(A) tails were not found on all of them using the estimator.
Figure 3.
Genome wide analysis of misregulated transcripts. The most abundant RNA fragments detected in the PSS fraction (> 40 rpkm) accumulated in the mutant relative to the wt strain (log2 FC > 1.5) were selected. The folding energy of these 129 RNAs was normalized relative to their length (normalized mfe) and presented as a function of the relative accumulation between the two strains (FC). Fragments derived from CDS are shown by red triangles and from REP sequences by green triangles. Randomly selected sequences (see Materials and Methods) were also folded and plotted as a function of their FC (blue triangles). Distributions of FC and normalized energy of the various populations are presented at the bottom and the right side of the graph respectively, with the same colors. Significantly different distributions from a random selection are indicated with one star (P-value < 0.05) or two stars (P-value < 10−6).
Poly(A)polymerase deficiency increases the level of specific regions of transcripts
Next we wanted to determine whether poly(A)polymerase also impacts the level of functional/full size transcripts (both mRNAs or non-coding RNA). We compared the FC values of RNA in the wt and the pcnB mutant in the Int fraction versus the FC values in the PSS fraction (Supplementary Figure S7). The majority of RNAs upregulated in the pcnB mutant shows a higher fold change in the PSS fraction indicating that it was predominately endonucleolytically generated RNA fragments which were stabilized in the absence of PAP I, and they may also represent direct polyadenylation targets. The few RNAs upregulated in the pcnB mutant, which have similar FC in the Int and the PSS fractions could correspond to indirect targets, such as glmS, which is regulated by the sRNA GlmY, itself a direct poly(A) target (9).
We found 65 RNAs containing REP sequences which were upregulated in the PSS fraction in the pcnB mutant, but were mostly unchanged in the Int fraction (Supplementary Table S2). One explanation could be that the stability of long REP-containing transcripts depends upon endoribonucleolytic cleavage, which permits rapid exonucleolytic degradation of the non-REP part. Alternatively short REP elements may be more efficiently polyadenylated than long REP containing transcripts. In either case, REP containing RNA fragments produced from transcripts by processing are less efficiently degraded in the pcnB mutant than the transcripts from which they originate.
Other short fragments derived from longer transcripts also accumulated massively in the pcnB strain and are presumably the result of specific endonucleolytic cleavages generating fragments which are normally subject to poly(A) dependent degradation (Supplementary Figure S8). For example, a fragment containing the fur Shine–Dalgarno sequence and start codon, accumulated in the pcnB mutant. fur expression is downregulated by RyhB sRNA. In our growth conditions RyhB sRNA exhibited similar levels in the wt and the pcnB mutant. This suggests that the cleavage, which generates the fragment detected in the pcnB mutant, should be independent of RyhB control. Other examples, notably in ptsG, pheM and malT, are presented in Supplementary Figure S8.
Full size functional RNAs upregulated in the PAP I mutant
In addition to the direct effects on small RNA fragments, Poly(A) Polymerase also affects the abundance of functional, full length transcripts, of both sRNAs and mRNA.
Thirteen sRNAs, were present in higher amounts in the pcnB strain, and were detected in the PSS and Int fractions (Supplementary Table S2). In the case of SroH and SraF both full-length transcripts and processed forms accumulated in the pcnB mutant (Supplementary Figure S9 and Table S2) and poly(A) sequences were detected at the 3΄ extremity of the SroH primary transcript (Supplementary Table S3). In the case of RyjA (SraL) (Supplementary Figure S9) which is also polyadenylated (Supplementary Table S4) (44,45) only the full-length transcript accumulated in the mutant. This indicates that the degradation of the primary transcript was initiated by polyadenylation.
The flu transcript (encoding the surface antigen, Ag43) is an example demonstrating the stabilization of whole mRNA in the pcnB mutant (Supplementary Tables S2, S3 and Figure S9GH). Its expression is controlled by antitermination of the upstream IsrC RNA (46). However, the stabilized flu mRNA did not contain IsrC, indicating that isrC-flu transcript had been processed and the leader region degraded. Although Ag43 expression is controlled by OxyR (repression) and Dam (activation) (47), there was no substantial variation in the levels of the oxyR or dam transcripts upon pcnB inactivation (Supplementary Table S2). At the moment we cannot distinguish between a direct or indirect effect of pcnB on flu levels.
Poly(A)polymerase affects the levels of some toxin–antitoxin pairs
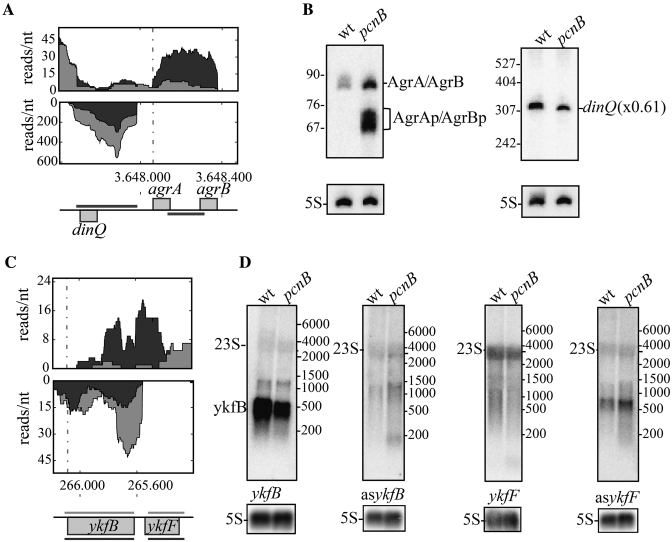
The dinQ-agr locus shows the classical features of a Type I toxin/antitoxin (TA) system. It encodes AgrA and AgrB sRNAs transcribed in the opposite direction to dinQ with their own promoters and terminators (48). Both were upregulated in the mutant and AgrB was polyadenylated in the wt (Supplementary Table S2 and Figure S6). Shorter AgrA/B forms were also detected in the pcnB strain, revealing that these RNAs were processed (Figure 4AB). Formation of the dinQ RNA/AgrB complex inhibits the endonucleolytic cleavage that normally produces the translatable dinQ mRNA (48). The dinQ transcript was less abundant in the pcnB mutant, probably as a consequence of the higher levels of AgrA and AgrB in the same strain (Figure 4AB). SibD and IstR, two other polyadenylated Type I antitoxins sRNA, were more abundant in the mutant strain as well as their corresponding ibsD and tisB toxin RNA (Supplementary Tables S2 and S4). The chromosome of E. coli encodes for five Hok-like toxins with an associated Sok antisense RNA, among which only SokB displayed a 3-fold higher level in the pcnB mutant (Supplementary Table S2). Two other antisense sRNAs, sibA and sibB were abundant and downregulated in the mutant by 2- and 2.5-fold, respectively. They are transcribed antisense to the ibsA and ibsB genes, encoding short toxic proteins (49) whose transcripts were nearly undetectable (Supplementary Table S2). These examples demonstrate that polyadenylation, by modulating the levels of some RNA antitoxins, participates in the control of Type I TA systems.
Figure 4.
Antisense transcripts and anti-toxins are stabilized in the pcnB mutant Int fraction. (A and C) Expression profile in the Int fraction in the wild-type strain (light grey) and the pcnB mutant (dark grey) as described in Figure 1B. (B and D) Northern blot validation of the RNA-seq data using single stranded RNA probes shown with a light grey line for the mRNA or the sRNAs and a dark grey line for the antisense RNA. Probing for AgrA and AgrB was performed with a agrAB probe already described which does not allow to distinguish the two very similar sRNAs (48). Radioactivity corresponding to transcript was quantified and normalized relative to 5S RNA used as loading control. The ratio of RNA levels of specific transcripts between the pcnB mutant and the wt is given in parentheses, when possible.
Antisense transcripts are only minimally affected by pcnB inactivation
Our RNAseq analysis detected transcripts on the opposite strand of open reading frames in the Int fraction (Supplementary Table S2). Some of these antisense transcripts have already been described; e.g. antisense to sRNAs MicA or ArcZ, in a RNase III deficient strain (23,50) and to mRNA, e.g. aseutB and asholE (24). We also detected two new antisense transcripts; aslpp and asdeoA transcripts (Supplementary Table S2). However, pcnB inactivation had no impact on their levels or sizes (data not shown). Our analysis also detected antisense transcripts in the ykfBK operon from the CP4-6 prophage (Supplementary Table S2). It did not reveal any ykfF transcripts, while an asykfF RNA about 500 nt in length was detected in the wild-type strain which was slightly, more abundant in the pcnB mutant. In contrast, ykfB transcript was more abundant in the wt than in the mutant and an asykfB about 200 nt long was only detected in the mutant (Figure 4CD). It was recently shown that the yafY gene located upstream of ykfF contains a binding site for the FliA sigma factor that drives significant expression of ykfB (51). The increased expression of fliA in the wt strain compared to the pcnB could be responsible for the higher level of ykfB mRNA in the wt strain.
RNAs downregulated in the PAP I mutant
Transcripts downregulated in the pcnB strain are expected to be indirect targets of the destabilizing function of PAP I, since no transcripts are known to be stabilized by poly(A) addition in E. coli. We previously showed that polyadenylation positively controls the expression of FliC, FliA and FlhD (11). The RNAseq analysis showed that this down-regulation in the pcnB strain extends to the whole FliA regulon, controlling flagella biosynthesis (Supplementary Table S2, Figure S10).
TnaC is a regulatory peptide encoded by the leader of the tnaAB operon, which codes tryptophanase (52,53). tnaC, tnaCA and tnaCAB transcripts were all down-regulated in the pcnB mutant (Supplementary Figure S11A and B, Supplementary Tables S2 and S3). Global investigation in enterohemorrhagic E. coli (EHEC) showed that expression of the analogous tnaABL operon was upregulated when the GlmY and GlmZ sRNAs were inactivated (54). Thus, the higher levels of these two sRNAs in the pcnB mutant could explain the lower level of tnaCAB transcripts in this strain (9).
A transcript the size of srlAEBD was down-regulated in the pcnB mutant (Supplementary Figure S11C and D, Supplementary Tables S2 and S3). pcnB inactivation also decreased by 3-fold the level of the downstream srlM (gutM) activator, with a smaller effect on the downstream srlR repressor (Supplementary Tables S2 and S3). This modification of the balance between the activator and the repressor may explain the down-regulation of transcripts from this operon in the mutant.
The fhuACDB operon, which encodes components of the ferrichrome transport system, was down-regulated in the mutant but with fhuC and fhuD more strongly affected (Supplementary Tables S2 and S3 and Figure S11E). Northern analysis, using a probe specific for the 5΄ part of the fhuA gene, revealed a smear of degraded RNA both in the wt and the pcnB mutant (Supplementary Figure S11F). These RNA fragments were produced by a cleavage in the 3΄-extremity of fhuA mRNA as revealed by probing the 3΄-part of the messenger (Supplementary Figure S11G). This cleavage initiated the 3΄-5΄ degradation of the 5΄ part of the fhuA message and generated the smaller 3΄ RNA fragment of about 700 nt, detected only in the mutant. This cleavage is independent of RNase E or RNase III in spite of the stabilization of the fhuA transcript when RNase E is inactivated (Supplementary Figure S12). Surprisingly, the PAP I-dependent down-regulation of the full-length transcript remained dominant in the absence of both RNase III and RNase E activity. Hfq inactivation had no impact on the level or the cleavage of the transcript in the wt or the pcnB, which would rule out a role for a Hfq-dependent sRNA (Supplementary Figure S12). Another hypothesis, that PAP I may modulate the level of a transcription regulator, also seems unlikely, since the 3΄-part of fhuA mRNA was still detectable at a high level in the pcnB mutant. Fur is an inhibitor of fhuA transcription (55), which may contribute to the pcnB+ mediated accumulation of fhuA since fur mRNA increased in the pcnB strain (Supplementary Figure S8H band c). Altogether, this shows that poly(A) polymerase can exert complex and indirect effects that have major consequences on gene expression in E. coli.
DISCUSSION
The destabilizing activity of 3΄ poly(A) addition is involved in RNA degradation and quality control found in almost all organisms. While the predominant function of poly(A) tails is to facilitate degradation of short structured RNA decay intermediates, Poly(A)polymerase also impacts gene expression (2,4). In this study, we identify new RNAs whose expression level changes in the pcnB strain (Supplementary Tables S2 and S3) and RNAs that harbor a poly(A)tail (Supplementary Table S4), revealing the landscape of RNA polyadenylation at the transcriptome level. This work provides extensive new evidence that PAP I controls gene expression both positively and negatively in E. coli.
The major targets of poly(A) addition are short structured RNAs and it is therefore not surprising that structured REP containing RNA fragments are abundant poly(A) targets. Interestingly REP RNA fragments are poly(A) targets but not REP containing transcripts, indeed when part of long functional mRNAs, REP have little effect on poly(A) sensitivity of the mRNA. We show that both Rho-dependent and Rho-independent terminated RNAs are PAP I targets. Several new sRNAs as exemplified by regulatory RNAs, Type I antitoxin RNAs of TA systems and tRNA precursors were also modulated by poly(A)polymerase.
Our experiments also give invaluable information on 3΄-5΄ degradation mechanisms of individual RNAs. In the wt strain, using a rather stringent criteria, we identified only 110 poly(A) sites out of a total of 16 867 non-encoded 3΄ ends. Their detection means that at these sites oligo(A) removal is slower than its addition. The repartition of poly(A) tails at the 3΄ end of Rho-independent terminators together with a reduction in transcript abundance in the wt strain reveals the difficulty of 3΄-5΄ exoribonucleases to overcome these secondary structures and the need to add A residues to degrade these molecules (Figure 2, Supplementary Figure S6). In contrast in the pcnB mutant the vast majority of 3΄ non-encoded ends are spread over this secondary structure and most, if not all, transcripts have their 3΄ end at the terminator. This led to the conclusion that the unmapped tails detected in the pcnB mutant have no role in RNA decay. The nucleotide content of these 3΄ end tails does not present any preference for A, C, G or U residues and does not support a previous finding of residual polyadenylation in the mutant (39). Moreover there is no evidence for any uridylation of E. coli RNAs, that has been reported to participate in the control of RNA stability in various eukaryotes (56,57). Whether the heterologous tails detected in our RNAseq analysis are synthesized by PNPase (15,58) or correspond to experimental noise is unknown.
Three classes of RNA molecules were individually quantified in our analysis; 5΄ tri- (TSS), 5΄ mono-phosphorylated (PSS) fragments and the bulk RNA after fragmentation (Int). Very few transcripts with a 5΄-triphosphate with the exception of sRNAs, were affected by inactivation of PAP I (Supplementary Table S2). This may be due to the conversion of the 5΄-terminus of primary transcript from a triphosphate to a monophosphate by RppH (35) (Figure 1B and Supplementary Figure S1D) or to the presence of a stable hairpin at the 5΄ end, making them inaccessible to RNA polyphosphatase and/or to 5΄-ligation (Supplementary Figure S1C). Either scenario would lead to an underestimation of the number of TSS tagged RNA molecules. As a consequence we cannot draw any conclusion on a difference in the polyadenylation between primary and processed targets. The recovery of all kinds of transcripts after fragmentation as untagged RNAs reveals additional full-length transcripts misregulated in the pcnB strain, as exemplified by srlAEBD or tnaA(C) mRNAs. Numerous RNAs harboring a PSS tag were also affected. They should correspond to indirect PAP I targets, via an effect of polyadenylation on a regulator or a sigma factor (e.g. FliA, see below). When upregulated in the pcnB mutant, they mostly corresponded to degradation products generated by an endoribonuclease and are direct PAP I targets. This is exemplified by REP-containing transcripts and transcripts with Rho-independent terminators. One possible explanation is that degradation of translated (and unstructured) transcripts mainly depends on endoribonucleolytic cleavages that generate fragments, which are further degraded by exonucleases without the help of poly(A) tails, while the structured 3΄ terminators and REP sequences require PAP I to facilitate their exonucleolytic decay as previously proposed (17,59). However, when the initial cleavage is inhibited or inefficient, these transcripts are polyadenylated at their 3΄ ends and degraded through the poly(A)dependent exonucleolytic pathway (18).
A recent paper proposes a new function for a REP containing RNA in the organization of the E. coli chromosome by connecting chromosomal domains through the histone like protein HU (60). The determinant role of PAP I in the degradation of REP containing RNAs may impact such regulatory network. Finally, the many RNA fragments, which are mostly small and highly structured and sometimes harbor a Rho-independent terminator as exemplified by the ptsG 3΄-extremity pose questions on their possible biological functions. We hypothesize that some of them may interact with other RNA molecules and therefore regulate gene expression as reported for the 3΄-part of the gltL transcript (61).
One other notable outcome of this work is the generalization of our previous observation, that FliC is down-regulated in the absence of polyadenylation (11), to an effect on the whole FliA regulon (Supplementary Figure S10 and Table 2). The levels of the vast majority of the transcripts of the Class 2 flagella operon fliLMNOPQR, and Class 3 operons that encode products required in late flagellar assembly, chemotaxis and cyclic-di-GMP regulation of motility (51) were decreased in the pcnB mutant. In agreement, the FliA promoters located inside flhC, yjdA and yafY genes also drive increased transcription of the downstream motA, yjcZ and ykfB genes in the wild-type strain (51). The flhDC operon that encodes the master regulator of the flagellar gene cascade acting upstream of FliA is targeted by numerous regulators. For example, the 5΄-untranslated region of the flhDC mRNA is targeted for repression by ArcZ, OmrA, OmrB and OxyS sRNAs and for activation by McaS (62,63). Our observation that McaS sRNA is three times more abundant in the wild-type strain than in the pcnB mutant could be one clue to the mechanism of how PAP I enhances expression of FlhDC (11). However we cannot exclude the involvement of another regulator in the poly(A)dependent control of fliA transcription.
Table 2. Functional categories of genes differentially expressed in the pcnB mutant.
| up | down | |
|---|---|---|
| membrane protein | 7/7 | 9/12 |
| prophage | 1/4 | 2 |
| fimbriae | 1/5 | 0 |
| RNA metabolism | 2/2 | 1 |
| ammonium metabolism | 3/1 | 1/4 |
| miscellaneous | 10/22 | 9/10 |
| flagella | 0 | 11/15 |
| catabolism | 0 | 2/5 |
Up- and down-regulated transcripts in the pcnB strain in the Int fraction classified by functional categories according to the Gene Ontology database. The first number represents the number of regulated transcripts with a log2FC > 2 and the second, regulated transcripts with 1.5 < log2FC < 2.
tar, tap, trg encode methyl-accepting chemotaxis proteins that together with cheA and cheY products, are the signaling intermediates, which allow the bacteria to sense the environmental stimuli leading to chemotaxis (64). This permits the bacteria to change either the rotational direction or speed of flagella rotation and causes tumbling of the cell. Our observation that all these transcripts are less abundant in the pcnB mutant explains the decreased motility of the mutant on soft agar (11). Another finding of this work concerns the misregulation of numerous membrane proteins in the absence of PAP I (Table 2). Membrane proteins make up 22% and 27% of up- and down-regulated transcripts, respectively, which is in agreement with the increased sensitivity of the pcnB mutant to cell envelope stress (11). The fact that many membrane proteins are regulated by sRNAs, together with the major effects of poly(A)polymerase on the degradation of numerous regulatory RNAs (Table 3), provides an important clue on how polyadenylation may impact on membrane protein expression. This is exemplified by SroH sRNA whose function is still unknown, but the sroH-deleted mutant is sensitive to cell envelope stress (65).
Table 3. Summary of differential expression analysis.
| Expression detected only in pcnB | Expression detected only in wt | Top 5 upregulated | log2 fold-change | Top 5 down-regulated | log2 fold-change |
|---|---|---|---|---|---|
| sdsR | rydC | yjiY | 4.76 | grcA | -3.98 |
| micF | rprA | ryjA | 3.84 | pcnB | -3.96 |
| ohsC | rdlD | tsgA | 3.74 | nanC | -3.57 |
| arrS | agrB | 3.73 | ydhL | -3.33 | |
| sokA | glmS | 3.25 | fhuD | -3.32 |
Genes the expression of which was detected in only one of the strain together with the five most up-regulated and down-regulated genes in pcnB deficient mutant (positive log2 fold-change indicates a higher expression in the pcnB strain).
Finally we also show that poly(A)polymerase is part of the fine-tuning of Type I TA systems, by modulating the level of antitoxin RNAs (AgrB, SibA, SibB, SokB RNAs). We suspect that this function is underestimated due to the low level of expression of these RNAs in our experimental conditions.
This study has enlarged the vision we had on the role of polyadenylation, which is not only a way to eliminate molecular waste but which contributes to more complex regulatory network acting post-transcriptionally to control gene expression in bacteria.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to Jackie Plumbridge and Antonin Marchais for discussions and critical reading of the manuscript. This work has benefited from the facilities and expertise of the high throughput sequencing platform of IMAGIF (Centre de Recherche de Gif- www.imagif.cnrs.fr).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique [UPR9073 now UMR8261]; University Paris-Diderot and Agence Nationale de la Recherche [asSUPYCO, ANR-12-BSV6-0007-03 to E.H.]; ‘Initiative d'Excellence’ program from the French State [‘DYNAMO’, ANR-11-LABX-0011 to E.H.]; Swedish Science Council [621-2012-2982 to E.A.]; Academy of Finland through Finland Distinguished Professor Programme and the Center of Excellence COIN [to E.A.]. Funding for open access charge: CNRS [UPR9073 now UMR8261].
Conflict of interest statement. None declared.
REFERENCES
- 1.Dreyfus M., Régnier P.. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002; 111:611–613. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J.T. RNA turnover: Unexpected consequences of being tailed. Curr. Biol. 2005; 15:R635–R638. [DOI] [PubMed] [Google Scholar]
- 3.Lange H., Sement F.M., Canaday J., Gagliardi D.. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant. Sci. 2009; 14:497–504. [DOI] [PubMed] [Google Scholar]
- 4.Régnier P., Hajnsdorf E.. Poly(A)-assisted RNA decay and modulators of RNA stability. Prog. Mol. Biol. Transl. Sci. 2009; 85:137–185. [DOI] [PubMed] [Google Scholar]
- 5.Slomovic S., Fremder E., Staals R.H., Pruijn G.J., Schuster G.. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:7407–7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maes A., Gracia C., Hajnsdorf E., Régnier P.. Search for poly(A) polymerase targets in E. coli reveals its implication in surveillance of Glu tRNA processing and degradation of stable RNAs. Mol. Microbiol. 2012; 83:436–451. [DOI] [PubMed] [Google Scholar]
- 7.Cao G.-J., Sarkar N.. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl Acad. Sci. U.S.A. 1992; 89:10380–10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joanny G., Le Derout J., Bréchemier-Baey D., Labas V., Vinh J., Régnier P., Hajnsdorf E.. Polyadenylation of a functional mRNA controls gene expression in E. coli. Nucleic Acids Res. 2007; 35:2494–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichenbach B., Maes A., Kalamorz F., Hajnsdorf E., Görke B.. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008; 36:2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban J.H., Vogel J.. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008; 6:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maes A., Gracia C., Brechemier D., Hamman P., Chatre E., Lemelle L., Bertin P.N., Hajnsdorf E.. Role of polyadenylation in regulation of the flagella cascade and motility in Escherichia coli. Biochimie. 2013; 95:410–418. [DOI] [PubMed] [Google Scholar]
- 12.Aiso T., Yoshida H., Wada A., Ohki R.. Modulation of mRNA stability participates in stationary-phase-specific expression of ribosome modulation factor. J. Bacteriol. 2005; 187:1951–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Régnier P., Marujo P.E.. Lapointe J, Brakier-Gingras L. Translation Mechanisms. 2002; 184–196. [Google Scholar]
- 14.Reuven N.B., Zhou Z., Deutscher M.P.. Functional overlap of tRNA nucleotidyltransferase, poly(A)polymearse I and polynucleotide phosphorylase. J. Biol. Chem. 1997; 272:33255–33259. [DOI] [PubMed] [Google Scholar]
- 15.Mohanty B.K., Kushner S.R.. Polynucleotide phosphorylase functions both as a 3΄->5΄ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 2000; 97:11966–11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haugel-Nielsen J., Hajnsdorf E., Régnier P.. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996; 15:3144–3152. [PMC free article] [PubMed] [Google Scholar]
- 17.Hajnsdorf E., Braun F., Haugel-Nielsen J., Régnier P.. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 1995; 92:3973–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marujo P.E., Braun F., Haugel-Nielsen J., Le Derout J., Arraiano C.M., Régnier P.. Inactivation of the decay pathway initiated at an internal site by RNase E promotes poly(A)-dependent degradation of the rpsO mRNA in Escherichia coli. Mol. Microbiol. 2003; 50:1283–1294. [DOI] [PubMed] [Google Scholar]
- 19.Le Derout J., Folichon M., Briani F., Dehò G., Régnier P., Hajnsdorf E.. Hfq affects the length and the frequency of short oligo(A) tails at the 3. end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 2003; 31:4017–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohanty B.K., Kushner S.R.. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006; 34:5695–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero D.A., Hasan A.H., Lin Y.F., Kime L., Ruiz-Larrabeiti O., Urem M., Bucca G., Mamanova L., Laing E.E., van Wezel G.P. et al. A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing. Mol. Microbiol. 2014; 94:963–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinhara A., Matsui M., Hiraoka K., Nomura W., Hirano R., Nakahigashi K., Tomita M., Mori H., Kanai A.. Deep sequencing reveals as-yet-undiscovered small RNAs in Escherichia coli. BMC Genomics. 2011; 12:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lybecker M., Zimmermann B., Bilusic I., Tukhtubaeva N., Schroeder R.. The double-stranded transcriptome of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:3134–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomason M.K., Bischler T., Eisenbart S.K., Forstner K.U., Zhang A., Herbig A., Nieselt K., Sharma C.M., Storz G.. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 2015; 197:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghavan R., Sloan D.B., Ochman H.. Antisense transcription is pervasive but rarely conserved in enteric bacteria. mBio. 2012; 3, doi:10.1128/mBio.00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Innocenti N., Golumbeanu M., D'Heroue L.A., Lacoux C., Bonnin R.A., Kennedy S.P., Wessner F., Serror P., Bouloc P., Repoila F. et al. Whole-genome mapping of 5΄ RNA ends in bacteria by tagged sequencing: a comprehensive view in Enterococcus faecalis. RNA. 2015; 21:1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun F., Hajnsdorf E., Régnier P.. Polynucleotide phosphorylase is required for the rapid degradation of the RNase E-processed rpsO mRNA of Escherichia coli devoid of its 3΄ hairpin. Mol. Microbiol. 1996; 19:997–1005. [DOI] [PubMed] [Google Scholar]
- 28.Hajnsdorf E., Régnier P.. E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J. Mol. Biol. 1999; 286:1033–1043. [DOI] [PubMed] [Google Scholar]
- 29.Hajnsdorf E., Steier O., Coscoy L., Teysset L., Régnier P.. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams-rnb-pnp mutant. EMBO J. 1994; 13:3368–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajnsdorf E., Régnier P.. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A)polymerase I. Proc. Natl Acad. Sc. U.S.A. 2000; 97:1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouquier d'Herouel A., Wessner F., Halpern D., Ly-Vu J., Kennedy S.P., Serror P., Aurell E., Repoila F.. A simple and efficient method to search for selected primary transcripts: non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res. 2011; 39:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Portier C., Dondon L., Grunberg-Manago M., Régnier P.. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5΄ end. EMBO J. 1987; 6:2165–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portier C., Régnier P.. Expression of the rpsO and p. np genes: Structural analysis of a DNA fragment carrying their control regions. Nucleic Acids Res. 1984; 112:6091–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deana A., Celesnik H., Belasco J.G.. The bacterial enzyme RppH triggers messenger RNA degradation by 5΄ pyrophosphate removal. Nature. 2008; 451:355–358. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J., Kobert K., Flouri T., Stamatakis A.. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014; 30:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodt M., Roehr J.T., Ahmed R., Dieterich C.. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology. 2012; 1:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalapos M.P., Cao G.J., Kushner S.R., Sarkar N.. Identification of a second poly(A) polymerase in Escherichia coli. Biochem. Biophys. Res. Commun. 1994; 198:459–465. [DOI] [PubMed] [Google Scholar]
- 40.Cao G.-J., Sarkar N.. Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc. Natl Acad. Sci. U.S.A. 1992; 89:7546–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters J.M., Mooney R.A., Kuan P.F., Rowland J.L., Keles S., Landick R.. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:15406–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogel J., Argaman L., Wagner E.G., Altuvia S.. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004; 14:2271–2276. [DOI] [PubMed] [Google Scholar]
- 43.Li Z., Pandit S., Deutscher M.P.. Polyadenylation of stable RNA precursors in vivo. Proc. Natl Acad. Sci. U.S.A. 1998; 95:12158–12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G.H., Margalit H., Altuvia S.. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001; 11:941–950. [DOI] [PubMed] [Google Scholar]
- 45.Wassarman K.M., Repoila F., Rosenow C., Storz G., Gottesman S.. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001; 15:1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wallecha A., Oreh H., van der Woude M.W., deHaseth P.L.. Control of gene expression at a bacterial leader RNA, the agn43 gene encoding outer membrane protein Ag43 of Escherichia coli. J. Bacteriol. 2014; 196:2728–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haagmans W., van der Woude M.. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 2000; 35:877–887. [DOI] [PubMed] [Google Scholar]
- 48.Weel-Sneve R., Kristiansen K.I., Odsbu I., Dalhus B., Booth J., Rognes T., Skarstad K., Bjoras M.. Single transmembrane peptide DinQ modulates membrane-dependent activities. PLoS Genet. 2013; 9:e1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fozo E.M., Kawano M., Fontaine F., Kaya Y., Mendieta K.S., Jones K.L., Ocampo A., Rudd K.E., Storz G.. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008; 70:1076–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udekwu K.I. Transcriptional and post-transcriptional regulation of the Escherichia coli luxS mRNA; involvement of the sRNA MicA. PLoS One. 2010; 5:e13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fitzgerald D.M., Bonocora R.P., Wade J.T.. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 2014; 10:e1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanofsky C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA. 2007; 13:1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li G., Young K.D.. A cAMP-independent carbohydrate-driven mechanism inhibits tnaA expression and TnaA enzyme activity in Escherichia coli. Microbiology. 2014; 160:2079–2088. [DOI] [PubMed] [Google Scholar]
- 54.Gruber C.C., Sperandio V.. Global Analysis of Posttranscriptional Regulation by GlmY and GlmZ in Enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2015; 83:1286–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Z., Lewis K.A., Shultzaberger R.K., Lyakhov I.G., Zheng M., Doan B., Storz G., Schneider T.D.. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 2007; 35:6762–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilusz C.J., Wilusz J.. New ways to meet your (3΄) end oligouridylation as a step on the path to destruction. Genes Dev. 2008; 22:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sement F.M., Ferrier E., Zuber H., Merret R., Alioua M., Deragon J.M., Bousquet-Antonelli C., Lange H., Gagliardi D.. Uridylation prevents 3΄ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 2013; 41:7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slomovic S., Portnoy V., Yehudai-Resheff S., Bronshtein E., Schuster G.. Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases. Biochim. Biophys. Acta. 2008; 1779:247–255. [DOI] [PubMed] [Google Scholar]
- 59.Khemici V., Carpousis A.J.. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 2004; 51:777–790. [DOI] [PubMed] [Google Scholar]
- 60.Qian Z., Macvanin M., Dimitriadis K.E., He X., Zhurkin V., Adhya S.. A new noncoding RNA arranges bacterial chromosome organization. mBio. 2015; 6, doi:10.1128/mBio.00998-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyakoshi M., Chao Y., Vogel J.. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015; 34:1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Lay N., Gottesman S.. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012; 86:524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomason M.K., Fontaine F., De Lay N., Storz G.. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 2012; 84:17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hazelbauer G.L. Bacterial chemotaxis: the early years of molecular studies. Annu. Rev. Microbiol. 2012; 66:285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hobbs E.C., Astarita J.L., Storz G.. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J. Bacteriol. 2010; 192:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cahova H., Winz M.L., Hofer K., Nubel G., Jaschke A.. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015; 519:374–377. [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi Y., Inouye M.. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 2009; 85:467–500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.