Abstract
Communication between distantly spaced genomic regions is one of the key features of gene regulation in eukaryotes. Chromatin per se can stimulate efficient enhancer-promoter communication (EPC); however, the role of chromatin structure and dynamics in this process remains poorly understood. Here we show that nucleosome spacing and the presence of nucleosome-free DNA regions can modulate chromatin structure/dynamics and, in turn, affect the rate of EPC in vitro and in silico. Increasing the length of internucleosomal linker DNA from 25 to 60 bp results in more efficient EPC. The presence of longer nucleosome-free DNA regions can positively or negatively affect the rate of EPC, depending upon the length and location of the DNA region within the chromatin fiber. Thus the presence of histone-free DNA regions can differentially affect the efficiency of EPC, suggesting that gene regulation over a distance could be modulated by changes in the length of internucleosomal DNA spacers.
INTRODUCTION
Distant communication between DNA regulatory regions and their targets in cis is involved in the regulation of gene expression (see (1,2) for reviews). Activating DNA regions (enhancers) physically interact with their target promoters; these interactions are mediated by sequence-specific DNA-bound proteins and accompanied by the formation of chromatin loops (see (3,4) for reviews). Enhancers can efficiently activate transcription in cis over a wide range of distances, from hundreds to hundreds of thousands of base pairs (5).
Chromatin structure must be flexible and dynamic to allow for regulatory interactions between enhancers and promoters; higher-order chromatin structures within activated chromatin domains most likely reduce to the level of chromatin fibers (6,7). Although chromatin fibers are highly mobile structures (8,9), the impact of this mobility on the efficiency of enhancer-promoter communication (EPC) in cis has not yet been extensively studied. Previously it was shown that a chromatin fiber is a rigid polymer and the presence of histone-free DNA is likely required for efficient formation of chromatin loops (10,11). Thus, the flexibility of the chromatin regions localized between communicating enhancers and promoters and the presence of histone-free DNA regions are likely to be important factors that could dictate the overall efficiency of communication in chromatin. Indeed, enhancers are often associated with nearby nucleosome-depleted regions (NDRs). The appearance of NDRs near potentially active enhancers can activate them and contribute to cancer progression (12). Furthermore, irregular nucleosome positioning could lead to the formation of chromatin structures with dynamics different from chromatin fibers containing regularly positioned nucleosomes (13).
In addition to the presence of histone-free DNA, the overall structure and communication properties of chromatin fibers likely depend upon the length of internucleosomal linker DNA. The average nucleosome repeat length (NRL) varies across species, tissues, and cell-cycle states. Average NRL values range from ∼155–190 bp in transcriptionally active cells to ∼190–240 bp in mature, transcriptionally inactive states (14). The role of internucleosomal spacing in chromatin dynamics during regulation of gene expression has not been addressed experimentally (11).
In this work, we examine the role of chromatin structure/dynamics in the regulation of the rate of enhancer-promoter communication (EPC). Using a highly purified and efficient in vitro system that allows for quantitative analysis of the rate of distant communication in chromatin and complementary computer simulations of long, nucleosome-decorated DNA chains, we show that the presence of histone-free DNA regions has a considerable effect on the dynamics of the chromatin fiber.
MATERIALS AND METHODS
Proteins and plasmids
Proteins and protein complexes were purified as described (15). Plasmids pSW12 (601207×13 template), pEN12 (601172×13 template), pSW616 (601207×13-1CN), and pSW08 (601177×8) were constructed with the approach given in Supplementary Table S1. Plasmids pYP07 (601177×4), pYP08 (601177×7), pYP09 (601177×10) and pYP05 (601177×13) were described previously (16).
Chromatin assembly
H1/H5-depleted chicken erythrocyte donor chromatin was prepared as described (17). In vitro reconstitution of chromatin on linearized DNA templates was conducted at different mass ratios of donor chromatin DNA to template DNA (1:1.25, 1:1, 1:0.75) using continuous dialysis from 1 M to 10 mM NaCl to achieve different levels of chromatin assembly (17).
Analysis of nucleosome positioning and occupancy of nucleosome positioning sites (NPS)
The expected positioning of nucleosomes within the array was verified using a different version of the restriction enzyme sensitivity assay followed by primer extension (18). Chromatin was assembled on XhoI-linearized plasmids and digested with an excess of restriction enzymes AluI or ScaI. Purified DNA was subjected to primer extension with Taq DNA polymerase using a radioactively end-labeled primer, which anneals immediately upstream of the promoter (18). Homogeneity of chromatin arrays was evaluated by electrophoresis in 0.8% agarose gel containing 0.5× TBE (15,19,20).
Transcription
Conditions for in vitro transcription were optimized for maximal utilization of the chromatin templates. Transcription was conducted as described (15).
All templates were linearized, and single-round transcription assays were carried out in 40-μl aliquots in a transcription buffer (TB) containing 50 mM Tris–OAc (pH 8.0), 100 mM KOAc, 8 mM Mg(OAc)2, 27 mM NH4OAc, 0.7% PEG-8000 and 0.2 mM DTT at 1 nM DNA or chromatin concentrations and 10 nM core RNA polymerase, 300 nM σ54, 120 nM NtrC and 400 nM NtrB transcription factors. First, all components were mixed together in the TB buffer, in total volume of 40 μl; then the reaction mixture was incubated for 15 min at 37°C to form the closed initiation complex (RPC). Next, 5 μl of 40 mM ATP in 1× TB was added to the reaction to 4 mM final concentration, and the reaction was incubated at 37°C for 2 min to form the open initiation complex (RPO), which is competent to start transcription. Then a mixture of all four ribonucleotide-triphosphates (4 mM each) in 1× TB with 2.5 μCi of [α-32P]-GTP (3000 Ci/mmol) and 2 mg/ml heparin was added to the reaction to start transcription and to limit it to a single round. The reaction was continued at 37°C for 15 minutes before it was stopped by rapid addition of a phenol:chloroform mixture (1:1). Labeled RNA was purified and analyzed by denaturing PAGE. The gel was dried, exposed, and scanned as described above. The data were analyzed using the OptiQuant software.
Computational modeling
Numerical simulations of long-distance enhancer–promoter communication along nucleosome arrays of size and composition identical to the templates studied experimentally were carried out along the lines described recently (16,21). Collections of chromatin structures were generated for nucleosome-decorated DNA chains bearing intact histone proteins at the specified locations. The saved configurations were recorded at regular intervals during the simulation, typically following ∼200 successful configurational moves of the protein-free DNA linkers and histone tail charges. The linker DNA was treated as a series of base pairs subject to bending and twisting deformations and the nucleosomes as rigid bodies with electrostatic and configurational features consistent with high-resolution structural data, including the wedge-like shape of the histone protein core (22), the acidic patches on the upper and lower faces of the protein-DNA assembly (23), the roll-and-slide features of local nucleosomal DNA distortion (24), and the conformational variability of the histone tails (25,26). The changes in linker configuration were governed by a potential that ignores the sequence-dependent structural and deformational features of DNA. The chain was assumed to be a naturally straight, inextensible homopolymer capable of isotropic bending and independent fluctuations in twist of magnitudes consistent with the solution properties of mixed-sequence DNA (27).
The probabilities of communication were estimated from the distances between the centers of rigid NtrC and RNAP protein assemblies attached respectively to the enhancer and promoter binding sites on the DNA model. The predicted communication enhancement is the ratio of the contact probability determined for a nucleosome-bound DNA compared to that found for a nucleosome-free chain of the same length. The methodology takes detailed account of the electronic and spatial features of the regulatory proteins, the known shapes of the regulatory proteins, and the precise pathways of the associated DNA. The DNA linkers are subject to incremental, as opposed to random moves, and excluded volume, i.e. molecular overlap, is detected with software from a rigid-body simulator (OpenDE; www.ode.org). Average and representative individual structures were generated from the average and specific values of the rigid-body parameters (28,29) used to describe the orientations and displacement of successive base pairs along the simulated chains. See the Supplementary Materials for further details.
RESULTS
The experimental approach for analysis of EPC on saturated nucleosomal arrays
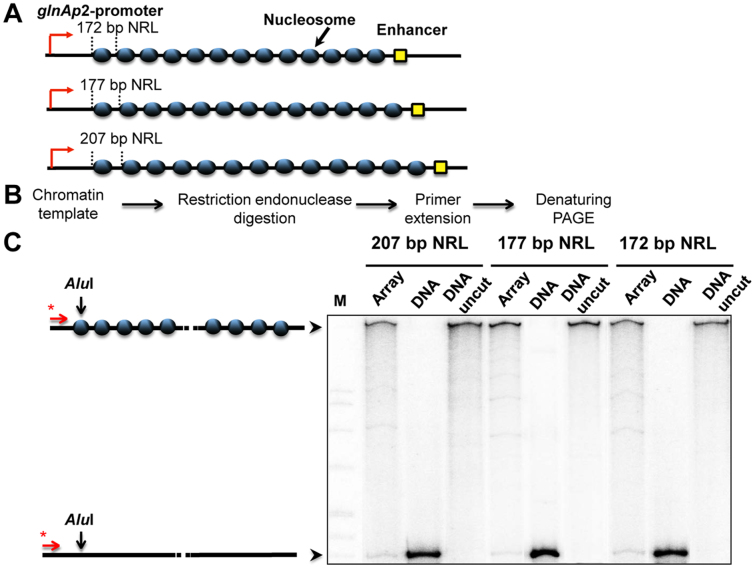
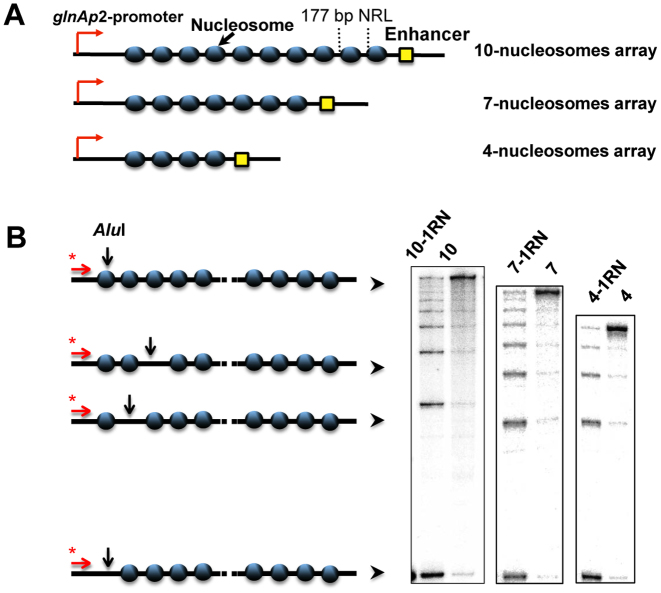
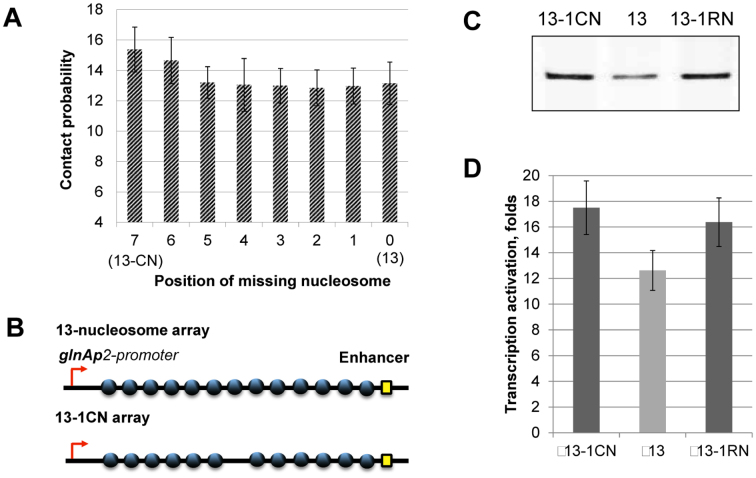
Arrays containing regularly spaced, precisely positioned nucleosomes capable of spontaneous formation of chromatin fibers were assembled using high-affinity histone-binding 601 DNA sequences that maintain precise nucleosome positioning in vitro and prevent nucleosome formation on the enhancer and promoter (30). DNA templates having different DNA spacers separating the 601 nucleosome-positioning sequences (NPS's, Figure 1A) were subjected to restriction digestion, primer extension, and denaturing PAGE to analyze the level of chromatin assembly (Figure 1B). The NPS's spaced by 30-bp DNA linkers yield 601177 arrays with 177-bp nucleosome repeat lengths (NRL). The length of the linker DNA between the NPS's on the designed arrays was varied from 25 up to 60 bp. The resulting 601172–207 nucleosomal arrays can form chromatin fibers without linker histones (31,32). Chromatin reconstitution was conducted on linearized DNA at different mass ratios of donor chromatin DNA to plasmid DNA by transfer of histone octamers from donor –H1 chromatin using dialysis from 1 M NaCl (18). Nucleosomes are formed predominantly on the high-affinity NPS's, but not on the enhancer and/or promoter sequences (Figure 1, Supplementary Figure S1). The extent of nucleosome saturations was determined by quantitation of radioactivity in the combined NFR cuts versus the radioactivity in completely cut DNA. Only less than 7% of templates contained one missing nucleosome. Saturated chromatin arrays (containing 4, 7 or 10 nucleosomes) and chromatin arrays missing a single nucleosome in random positions (10-RN, 7-RN, 4-RN) were obtained (Figure 2B).
Figure 1.
Analysis of nucleosome occupancy of 601 arrays having different nucleosome repeat lengths. (A) Schematic diagram of 13-nucleosome arrays having 172-, 177- and 207-bp nucleosomal repeat lengths (NRL). (B) Experimental approach: a restriction enzyme sensitivity assay. 601207×13, nucleosomal arrays were assembled on linearized plasmids at different ratios of histones to DNA and incubated in the presence of an excess of restriction enzyme AluI (see Materials and Methods). Purified DNA was subjected to primer extension followed by denaturing PAGE. (C) Saturated 13-nucleosomal arrays (207-, 177- and 172-bp NRL) were characterized using the restriction digestion sensitivity assay. Analysis of end-labeled DNA by denaturing PAGE (right). Note that chromatin assembly results in almost quantitative protection of the templates from AluI. M: MspI digest of plasmid pBR322 (left).
Figure 2.
Analysis of nucleosome occupancy of 601177 arrays. (A) Schematic diagram of the arrays containing 10, 7 or 4 nucleosome-positioning sequences. (B) Saturated nucleosomal arrays (10, 7, 4) and arrays missing a single nucleosome in random positions (10-1RN, 7-RN, 4-RN) were characterized using the restriction digestion sensitivity assay described in Figure 1. Analysis of end-labeled DNA by denaturing PAGE (right).
Using these nucleosomal arrays and the transcriptional assay, the rate of enhancer–promoter communication (EPC) was quantitatively measured as described by (16,18). The overall experimental approach for the transcriptional analysis of EPC in chromatin is outlined in Figure 3A. A template containing the NtrC-dependent enhancer, which strongly activates the Escherichia coli glnAp2 promoter, was used (18). The enhancer is activated by the NtrC protein complex, which is phosphorylated by the NtrB protein kinase (33). When phosphorylated, enhancer-bound NtrC interacts with the σ54-containing RNA polymerase holoenzyme and stimulates conversion of the inactive, closed complex to the open, functionally active initiation complex (34,35). During this direct enhancer–promoter interaction, the intervening DNA is transiently bent or looped out (36). Loop formation is a rate-limiting step in this process, as was shown previously (37,38). These experiments were conducted under assumption that the efficiency of transcription is proportional to the probability of looping, suggested by our previous experiments (38). The addition of labeled rNTPs with heparin allows for transcript synthesis, allows only a single round of transcription, and eliminates the nucleosomal barrier to transcribing RNA polymerase (RNAP). The amounts of transcript accumulated soon after the addition of NTPs are directly proportional to the rate of EPC; therefore the rate of EPC can be quantitatively measured using this approach (37). The rates of EPC in chromatin arrays were normalized to the rate of EPC on linear, histone-free DNA.
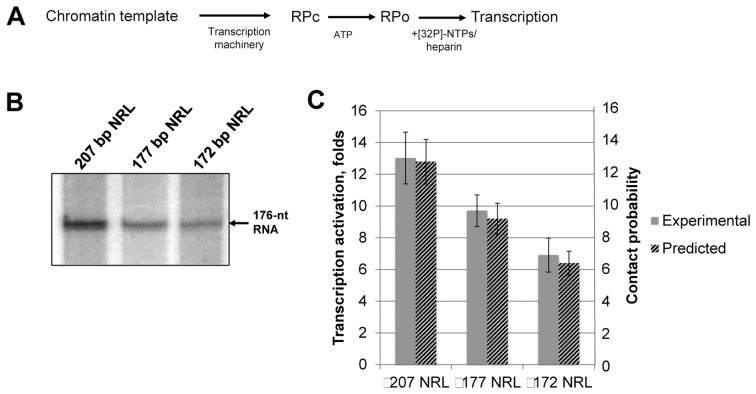
Figure 3.
Increase in the length of the internucleosomal linker DNA results in more efficient EPC in chromatin. (A) Experimental approach for analysis of the rate of EPC in chromatin. Closed initiation complexes of RNA polymerase formed on the promoter (RPC) were converted to the open initiation complexes (RPO) after addition of ATP for a limited time period, followed by addition of NTPs and heparin to limit transcription to a single round and to remove the nucleosomal barrier to transcription. The efficiency of transcription is directly proportional to the rate of EPC (16). (B) Transcription of 13-nucleosome arrays with 207-, 177- and 172-bp NRL. Analysis of labeled transcripts by denaturing PAGE. The rates of transcription activation (units on the left of the plot) were measured as described in Figure 3A. (C) Quantitative analysis of the 176-nt transcripts (gray bars) shown in Figure 3B. Error bars indicate standard deviations based on four independent measurements using two different reconstitutes (see Supplementary Table S2 for the results of t-test). Computational likelihood of long-range enhancer–promoter contacts (normalized to the values for histone-free DNA) are shown on the same plot. The contact probabilities (units on the right of the plot) correspond to the ratios of the computed probabilities that nucleosome-saturated and nucleosome-free DNA constructs of the specified composition adopt configurations that bring transcriptional proteins on the enhancer and promoter into close contact.
Increase of the length of internucleosomal linker DNA results in more efficient EPC in chromatin
Nucleosome spacing affects chromatin structure and dynamics (39) and is expected to affect the efficiency of chromatin looping and the rate of EPC. Our previous studies were conducted on arrays with a 177-bp nucleosomal repeat length (NRL): these arrays are well characterized (16), but are not typical in higher eukaryotes where the nucleosomal repeat is longer (185–205 bp) (40). In particular, active chromatin in mammalian cells is characterized by ∼200-bp NRL values. To evaluate the effect of DNA linker length on the rate of EPC in chromatin experimentally, templates having 207, 177 and 172 bp NRLs were constructed (Figure 1A). Saturated chromatin arrays were assembled (Figure 1C) and the rate of EPC was measured using the transcription assay (Figure 3A). Increasing the DNA linker length from 25 to 30 to 60 bp results in progressively increasing rates of EPC (Figure 3B and C). Although some of this effect could be rationalized by increased chromatin flexibility stemming from the presence of longer DNA linkers, the difference in the rates of communication between arrays with 172- and 177-bp spacing likely reflects very different chromatin structures (21,41). The correspondence between the experimentally obtained EPC rates and the predicted enhancement of NtrC-RNA polymerase contacts on simulated chromatin fibers (Figure 3C) lends support to this idea.
The computations reveal significant shortening of the distances between the ends of simulated 13-nucleosome arrays with 172-bp versus 177-bp repeats (∼40% closer on average; Supplementary Figure S2A). Moreover, the former construct is stiffened with respect to the latter in terms of the narrower range of end-to-end distances and is thus less capable of bringing transcriptional elements outside of the nucleosome-decorated fragment into direct contact. The differences in chain extension and deformability reflect the large-scale differences in average fiber structure associated with the change in spacing (Supplementary Figure S2B). The 5-bp increment in DNA linker length, roughly half the double-helical repeat, reorients the nucleosomes with respect to one another and with respect to the fiber axis. By contrast, the addition of a full helical turn to the DNA linkers introduces a modest enhancement in simulated chain deformations and little, if any, change in average nucleosome orientation. As reported previously (21), the interactions and effective stiffness of modeled arrays of regularly spaced nucleosomes exhibit an overall decreasing oscillatory pattern with linker length, with the oscillations spanning a period roughly equal to the DNA helical repeat. The cylindrical axes of the nucleosomes along the 172-bp construct run roughly perpendicular to the axis of the simulated fiber and those along the 177-bp construct in nearly the same direction as the fiber axis. In other words, the upper and lower faces of the nucleosomes in the former system are analogous to the risers and those in the latter system to the treads of a staircase. Furthermore, the face-to-face packing of nucleosomes in the two systems gives rise to different modes of global organization. The stacks of nucleosomes along the ‘average’ 172-bp fiber describe two helical pathways and those along the ‘average’ 177-bp fiber three (color-coded in Supplementary Figure S2B), so-called two-start and three-start models (double and triple helical pathways) of nucleosome packing (42,43). Although the structural regularity is distorted in individual configurations (thin curves in Supplementary Figure S2B), the large-scale differences in chain compaction and nucleosome packaging persist. The differences in nucleosome packaging underlie the simulated differences in long-range communication. The denser packing of nucleosomes in the 172-bp fiber limits the deformations of DNA compared to those in the more extended 177-bp construct. The added stiffness in the DNA, in turn, limits the deviations of the fiber as a whole from its ‘average’ global structure and makes it more difficult for proteins on the enhancer and promoter sites to come into direct contact (Figure 3C and Supplementary Figure S2B). The increase in nucleosome spacing from 177 to 207 bp has a much lesser effect on chain extension. The wider range of accessible structures broadens the distribution of end-to-end distances (Supplementary Figure S2A) and allows for greater frequency of communication between proteins at the enhancer and promoter sites. The simulated 207-bp construct, only ∼5% longer on average than its 177-bp counterpart, samples a broad range of spatial configurations. Individual configurations deviate widely from the average fiber structure, a three-start, triple helical pathway roughly twice the diameter of the average 177-bp fiber structure (Supplementary Figure S2B).
Our combined data point to the importance of DNA linker length in supporting efficient distant EPC in chromatin. The data indicate that both the structure of the chromatin fiber and its flexibility are essential for efficient long-range communication in chromatin.
Single, randomly positioned nucleosome-free gaps facilitate EPC on nucleosome arrays having more than 7 nucleosomes
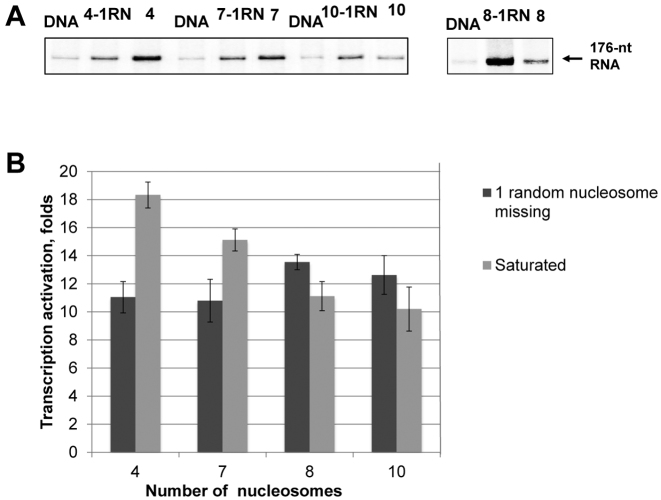
Chromatin fibers have irregular nucleosome positioning and nucleosome-depleted regions in vivo. How nucleosome-free regions affect chromatin dynamics and EPC is unknown. Previously we showed that formation of saturated chromatin arrays results in a decrease of the rate of EPC (16). Our experimental system allows for the assembly of chromatin arrays having different numbers of nucleosomes precisely positioned between the enhancer and promoter. Using different ratios of donor chromatin to DNA during chromatin assembly makes it possible to construct chromatin fibers either containing saturated nucleosomal arrays with a specified NRL or having nucleosome-free DNA regions at some positioning sites. Taking advantage of this approach, we prepared saturated chromatin arrays and arrays having a single, randomly positioned nucleosome-free gap (Figure 2). The effect of the nucleosome-free gap on EPC was analyzed using 601177×N arrays, where N is the number of nucleosomes in a saturated array (4, 7 or 10 Figure 2A), in combination with the transcription assay (Figure 3A). The effects of chromatin assembly on the rates of EPC in the different chromatin arrays missing one nucleosome were comparable: the rates were ∼11–12 times higher than the rates on corresponding histone-free DNA (Figure 4). At the same time, the assembly of saturated nucleosomal arrays revealed more variable effects of the gaps on the rates of EPC. In particular, the nucleosome-free gap had a negative effect on EPC in the 601177×4/601177×7 arrays and a positive effect in the 601177×10 array (Figure 4). The data suggest that the type of effect that a gap has on the rate of EPC depends upon the number of nucleosomes in the array. Therefore we considered the question: what is the maximal number of nucleosomes in an array that leads to inhibition of EPC by nucleosome-free regions? The presence of the gap leads to an increase in the rate of EPC on an 8-nucleosome array (Figure 4), suggesting that a 7-mer array is the longest array of nucleosomes spaced at 177-bp increments supporting a rate of EPC lower than in the absence of nucleosome-free gaps. Thus the effect of randomly localized nucleosome-free gaps present in the chromatin fiber on the rate of EPC depends on the number of nucleosomes in the array: the presence of gaps results in a decrease of the rates of EPC on shorter (≤7 nucleosomes) and an increase on longer (≥8 nucleosomes) arrays.
Figure 4.
Single, randomly positioned nucleosome-free gaps strongly affect EPC on 601177 arrays. (A) Transcription of saturated 601177×N nucleosomal arrays (N = 4, 7, 8 or 10 nucleosomes in the array) and the same arrays missing one nucleosome in random positions (-1RN). Analysis of labeled transcripts by denaturing PAGE. The rates of EPC were measured as described in Figure 3A. Removal of a single nucleosome from any position along the nucleosome arrays with more than seven nucleosomes leads to more efficient EPC. (B) Quantitative analysis of the specific transcripts shown in Figure 4A. Error bars indicate standard deviations based on four independent measurements using two different reconstitutes (see Supplementary Table S2 for the results of t-test).
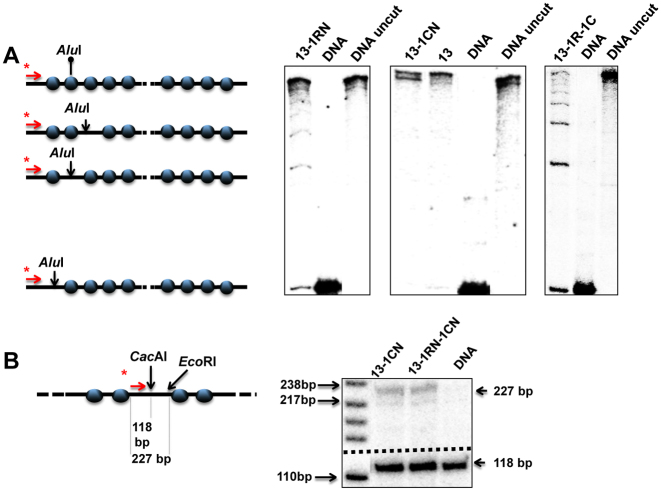
We next studied the effect of nucleosome-free gaps on EPC in 601207×13 arrays with nucleosome spacing characteristic of higher eukaryotes and either saturated or missing a single nucleosome (Figure 5A). Templates lacking a single nucleosome in the middle of the 601207×13 array (601207×(13-1CN)) or missing a single nucleosome in a random position (601207×(13-1RN)) were obtained (Figure 5A). The central high-affinity 601 nucleosome-positioning sequence in the 601207×(13-1CN) template was replaced by a DNA sequence (a fragment of the lambda bacteriophage genome) having low affinity to histones. Therefore, after reconstitution in the presence of an excess of competitor DNA nucleosomes did not form there. The absence of the central nucleosome in the 601207×(13-1CN) array was confirmed by the nearly full sensitivity of a unique CacAI site on the substituted sequence to restriction endonuclease digestion (Figure 5B).
Figure 5.
Characterization of nucleosomal arrays with different nucleosome occupancies. (A) Characterization of saturated 601207×13 nucleosomal arrays (13) and arrays missing a single nucleosome in random positions (13-1RN), a central nucleosome (13-1CN), or up to two nucleosomes (13-1RN-1CN). The templates were characterized using the restriction enzyme sensitivity assay (Figure 1B). Analysis of end-labeled DNA by denaturing PAGE (right). (B) The nucleosome-free region of the 13-1CN nucleosomal array is not protected by a nucleosome (left). Chromatin was digested with an excess of restriction enzyme CacAI. DNA was purified, digested by enzyme EcoRI, and subjected to primer extension. Analysis of end-labeled DNA by denaturing PAGE (right).
The effect of a single nucleosome-free gap on EPC could depend on its position within an array of nucleosomes. Computational simulations predict small, nearly identical positive effects on enhancer-promoter interactions by a single nucleosome-free gap at different locations along a 601207×13 nucleosomal array, with a slightly higher stimulating effect of a nucleosome-free gap in the middle of the array (Figure 6A, position #7, 13-CN). Randomly positioned and centrally localized single nucleosome-free gaps increase the experimentally observed rate of EPC in chromatin to nearly the same extent (Figure 6B and C) but with some enhancement for the gap in the center of the array (Figure 6D). The presence of the gap increases the distance between chain ends (∼23% more on average in simulated constructs missing the central nucleosome; Supplementary Figure S3A) and broadens the distribution of distances between chain ends. Although the gapped segment straightens on average, the added flexibility in the center of the construct makes it easier for proteins bound at the chain ends to come into direct contact (Figure 3C and Supplementary Figure S3B). Moreover, the bending fluctuations of the gap site bring the regulatory proteins into closest contact when the gap is located in the middle of the chain. This behavior follows from the geometry of the system. The gapped chromatin arrays are analogous to a series of lines of constant total length broken in two at various points by a fixed bend. Like the gapped arrays, the distance between the ends of the broken lines is shortest when the bend lies at the midpoint. Thus, a fiber with a gap at one end more closely resembles the intact fiber than one with the gap in the middle. The computations, however, do not capture as much of the enhancement in long-range communication as found with the transcription assay.
Figure 6.
Single nucleosome-free gaps strongly facilitate EPC on the 13-nucleosome array. (A) Computationally determined likelihood of enhancer-promoter interactions (normalized to the corresponding values for histone-free DNA) on 601207×13 arrays having a single nucleosome-free gap at every possible position. The array with the gap in the center (13-CN) is predicted to have a greater stimulating effect than any other gapped site on the rate of long-range communication compared to that on the saturated array without nucleosome-free regions (13). (B) Schematic diagram of the saturated 601207×13 nucleosomal array and the 601207×(13-1CN) array missing the central nucleosome. (C) Transcription of the fully saturated array (13) compared to arrays missing the central nucleosome (13-1CN) or one nucleosome in random positions (13-1R array). Analysis of labeled transcripts by denaturing PAGE. Removal of a single nucleosome from random positions in the array leads to more efficient EPC. The rates of EPC were measured as described in Figure 3A. (D) Quantitative analysis of the specific transcripts shown in Figure 6C. Error bars indicate standard deviations based on four independent measurements using two different reconstitutes (see Supplementary Table S2 for the results of t-test).
These numerical discrepancies may be related to the sequence and length of DNA in the nucleosome-depleted gaps. Consideration of the sequence-dependent features of DNA has little effect on the simulated end-to-end properties of short chains (44), i.e. a few turns of double helix comparable in length to the DNA linkers in chromatin arrays with 172 and 177-bp nucleosome repeats. The differences become more pronounced with increase in chain length (45). DNA treated, as here, as an inextensible, naturally straight chain with bending and twisting properties that are independent of base sequence tends to be more extended and globally stiffer on average than a model that takes account of the small sequence-dependent differences in the spatial arrangements of successive base pairs found in high-resolution structures (46). The subtle differences between the two models build up with chain length and can have profound effects on the predicted configurations of chains of the length of a single nucleosome-depleted gap. For example, specific sequences may bend in preferred directions that could enhance long-range contacts. Alternatively, the computations may underestimate the interactions of proteins on either side of the gap site. The assumed strength of nucleosome–nucleosome interactions has a profound effect on the simulated configurations of dinucleosomes (47). Elucidation of a mechanism that could account for the current differences between the predicted and observed levels of EP interactions in nucleosome arrays bearing a nucleosome-free gap is beyond the scope of the present work.
Taken together, the observed and modeled data suggest that a single nucleosome-free gap may increase or decrease the rate of EPC in chromatin arrays depending upon the NRL. A single nucleosome-free gap present in the middle of a 13-mer nucleosome array has a slightly stronger, stimulating effect on EPC than the single gaps present in other locations along the array.
Presence of multiple nucleosome-free gaps decreases the rate of EPC
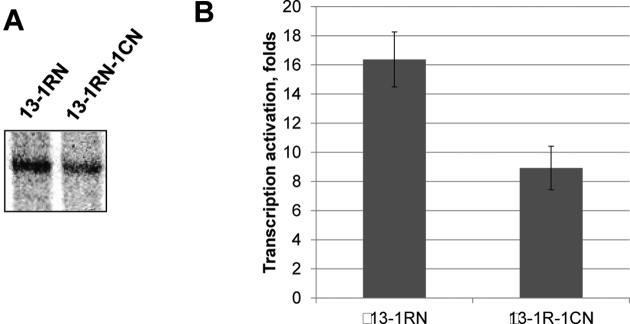
DNA is a less efficient communication device than chromatin (19). Although single nucleosome-free DNA regions facilitate EPC on longer (≥7 nucleosomes) arrays (Figures 4 and 6), the effect of multiple nucleosome-free gaps on EPC is likely to be less stimulatory. To evaluate this possibility, a 601207×(13-1CN-1RN) chromatin array was assembled using the 601207×(13-1CN) template under conditions when, in addition to the central nucleosome-free gap, other randomly positioned single nucleosome-free gaps were introduced (Figure 5A, right panel). As a result, two nucleosome-free gaps were present on nearly every array in the mix: one in the central position and one in a different, randomly chosen position within the array. EPC on the array missing up to two nucleosomes was 1.6 times less efficient than on the array missing one nucleosome (Figure 7A and B) and ∼30% less efficient than on the saturated array (compare with Figure 6C and D).
Figure 7.
The presence of multiple nucleosome-free gaps decreases the rate of EPC. (A) Transcription of the (13-1R) and (13-1R-1C) nucleosomal arrays. Analysis of labeled transcripts by denaturing PAGE. Whereas the omission of one nucleosome in the 601207×13 array increases the efficiency of EPC, two missing nucleosomes decrease EPC. The rates of EPC were measured as described in Figure 3A. (B) Quantitative analysis of the specific transcripts shown in Figure 7A. Error bars indicate standard deviations based on four independent measurements using two different reconstitutes (see Supplementary Table S2 for the results of t-test).
Taken together, the data show that single nucleosome-free gaps facilitate EPC on arrays containing more than seven nucleosomes. Consistently, the presence of additional nucleosome-free gap(s) on the longer arrays impairs EPC, indicating that when the density of nucleosome-free regions in an array is higher than one gap per seven nucleosomes, the gaps become inhibitory for EPC.
DISCUSSION
This work makes use of an in vitro experimental system that provides quantitative information on in-cis, enhancer–promoter communication (EPC) under physiologically relevant conditions (18) and a mesoscale model of chromatin that relates the effects of nucleosomal fine structure and linker DNA on long-range polymeric interactions (48). Nucleosome arrays with different spacings and different levels of saturation were assembled (Figures 1, 2 and 5). Using this system we have shown that: (i) increasing internucleosomal DNA linker length from 25 to 60 bp leads to more efficient EPC (Figure 3) while modulating the overall structure and flexibility of the nucleosome arrays (Supplementary Figures S2, S3); (ii) nucleosome-free gaps are essential for efficient EPC on nucleosomal arrays with 177-bp internucleosomal spacing and more than seven nucleosomes, but inhibit EPC on shorter nucleosomal arrays with the same spacing (Figure 4); (iii) single nucleosome-free gaps in different random positions within 13-mer nucleosomal arrays with longer spacing have positive effects on EPC (Figure 6); (iv) removal of a second nucleosome in random positions further decreases the rate of EPC on some of these arrays (Figure 7). Taken together, nucleosome-free gaps present within chromatin between an enhancer and a promoter can strongly affect the rate of EPC and thus could potentially participate in the regulation of gene expression.
Our previous work established that chromatin per se is an efficient device supporting high EPC rates (19). In contrast to EPC on linear, histone-free DNA, the rate of EPC in chromatin is nearly independent of distance over the range from ∼700 to 5000 bp (16); in the same experimental system histone-free DNA does not support EPC over distances more than ∼1 kb (16,49). The high rate of EPC on chromatin arrays is a consequence of the compact, yet dynamic structure of chromatin formed through internucleosomal interactions involving the histone tails (16).
Our current data show that both the length of the internucleosomal DNA linkers and the presence of single nucleosome-free gaps can influence the rate of long-distance EPC on nucleosome arrays. These histone-free DNA regions may serve as points of higher mobility on the chromatin fiber that facilitate EPC. At the same time single nucleosome-free gaps can increase or decrease the rate of EPC in chromatin arrays depending upon the number and spacing of nucleosomes. Furthermore, the introduction of a second nucleosome-free gap on 601207×13 arrays impairs the rate of EPC, possibly by decreasing the likelihood of forming configurations of the chromatin fiber that support efficient EPC. Taken together, the data suggest that the density of nucleosome-free gaps in native chromatin (∼one gap per 8–12 nucleosomes (50)) is important for efficient EPC.
Chromatin fibers in vivo do not have regular structures and consist of regions with irregular nucleosome spacing and nucleosome-free gaps. Nucleosome-free gaps are highly enriched in active chromatin, particularly in DNA regions that support transcription factor binding (51,52). The presence of nucleosome-free gaps facilitates formation of more dynamic chromatin (11), making the fiber more flexible and potentially a better device to support efficient EPC. Overall, nucleosome-free gaps could play differential roles in EPC and thus could affect the level of gene expression both positively and negatively. Furthermore, chromatin in living cells is associated with linker histones and non-histone proteins (53) that may also affect long-range communication and thus merit investigation in future studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr S. Grigoryev for providing us with the plasmid containing the 601207×12 sequence.
Footnotes
Present address: Yury S. Polikanov, Department of Biological Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
USPHS research grant [GM-34809 to W.K.O.]; NSF grant [MCB-1050470 to V.M.S.] and Fox Chase Cancer Center start-up funds (to V.M.S.); Russian Science Foundation grant (14-24-00031). Funding for open access charge: Fox Chase Cancer Center start-up funds (to V.M.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1. de Laat W., Duboule D.. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013; 502:499–506. [DOI] [PubMed] [Google Scholar]
- 2. Gibcus J.H., Dekker J.. The hierarchy of the 3D genome. Mol. Cell. 2013; 49:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krivega I., Dean A.. Enhancer and promoter interactions-long distance calls. Curr. Opin. Genet. Dev. 2012; 22:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harmston N., Lenhard B.. Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 2013; 41:7185–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noordermeer D., Branco M.R., Splinter E., Klous P., van Ijcken W., Swagemakers S., Koutsourakis M., van der Spek P., Pombo A., de Laat W.. Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet. 2008; 4:e1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilbert N., Boyle S., Fiegler H., Woodfine K., Carter N.P., Bickmore W.A.. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004; 118:555–566. [DOI] [PubMed] [Google Scholar]
- 7. Ghirlando R., Felsenfeld G.. Hydrodynamic studies on defined heterochromatin fragments support a 30-nm fiber having six nucleosomes per turn. J. Mol. Biol. 2008; 376:1417–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Platani M., Goldberg I., Lamond A.I., Swedlow J.R.. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell. Biol. 2002; 4:502–508. [DOI] [PubMed] [Google Scholar]
- 9. Gartenberg M.R., Neumann F.R., Laroche T., Blaszczyk M., Gasser S.M.. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 2004; 119:955–967. [DOI] [PubMed] [Google Scholar]
- 10. Boedicker J.Q., Garcia H.G., Johnson S., Phillips R.. DNA sequence-dependent mechanics and protein-assisted bending in repressor-mediated loop formation. Phys Biol. 2013; 10:066005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diesinger P.M., Kunkel S., Langowski J., Heermann D.W.. Histone depletion facilitates chromatin loops on the kilobasepair scale. Biophys. J. 2010; 99:2995–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taberlay P.C., Statham A.L., Kelly T.K., Clark S.J., Jones P.A.. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 2014; 24:1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collepardo-Guevara R., Schlick T.. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc. Natl. Acad. Sci U.S.A. 2014; 111:8061–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Holde K.E. Chromatin. 1989; Springer-Verlag. [Google Scholar]
- 15. Polikanov Y.S., Rubtsov M.A., Studitsky V.M.. Biochemical analysis of enhancer-promoter communication in chromatin. Methods. 2007; 41:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kulaeva O.I., Zheng G., Polikanov Y.S., Colasanti A.V., Clauvelin N., Mukhopadhyay S., Sengupta A.M., Studitsky V.M., Olson W.K.. Internucleosomal interactions mediated by histone tails allow distant communication in chromatin. J. Biol. Chem. 2012; 287:20248–20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walter W., Studitsky V.M.. Construction, analysis, and transcription of model nucleosomal templates. Methods. 2004; 33:18–24. [DOI] [PubMed] [Google Scholar]
- 18. Polikanov Y.S., Studitsky V.M.. Analysis of distant communication on defined chromatin templates in vitro. Methods Mol. Biol. 2009; 543:563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rubtsov M.A., Polikanov Y.S., Bondarenko V.A., Wang Y.H., Studitsky V.M.. Chromatin structure can strongly facilitate enhancer action over a distance. Proc. Natl. Acad. Sci U.S.A. 2006; 103:17690–17695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dorigo B., Schalch T., Bystricky K., Richmond T.J.. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003; 327:85–96. [DOI] [PubMed] [Google Scholar]
- 21. Clauvelin N., Lo P., Kulaeva O.I., Nizovtseva E.V., Diaz-Montes J., Zola J., Parashar M., Studitsky V.M., Olson W.K.. Nucleosome positioning and composition modulate in silico chromatin flexibility. J. Phys. Condens. Matter. 2015; 27:064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arents G., Burlingame R.W., Wang B.C., Love W.E., Moudrianakis E.N.. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci U.S.A. 1991; 88:10148–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 24. Tolstorukov M.Y., Colasanti A.V., McCandlish D., Olson W.K., Zhurkin V.B.. A novel ‘roll-and-slide’ mechanism of DNA folding in chromatin. Implications for nucleosome positioning. J. Mol. Biol. 2007; 371:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J.. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002; 319:1097–1113. [DOI] [PubMed] [Google Scholar]
- 26. Harp J.M., Hanson B.L., Timm D.E., Bunick G.J.. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr. D Biol. Crystallogr. 2000; 56:1513–1534. [DOI] [PubMed] [Google Scholar]
- 27. Czapla L., Swigon D., Olson W.K.. Sequence-dependent effects in the cyclization of short DNA. J. Chem. Theory Comput. 2006; 2:685–695. [DOI] [PubMed] [Google Scholar]
- 28. Dickerson R.E., Bansal M., Calladine C.R., Diekmann S., Hunter W.N., Kennard O., von Kitzing E., Lavery R., Nelson H.C.M., Olson W.K. et al. Definitions and nomenclature of nucleic acid structure parameters. J. Mol. Biol. 1989; 208:787–791. [Google Scholar]
- 29. Lu X.-J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding, and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thastrom A., Lowary P.T., Widlund H.R., Cao H., Kubista M., Widom J.. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 1999; 288:213–229. [DOI] [PubMed] [Google Scholar]
- 31. Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R.R., Richmond T.J.. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004; 306:1571–1573. [DOI] [PubMed] [Google Scholar]
- 32. Routh A., Sandin S., Rhodes D.. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci U.S.A. 2008; 105:8872–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ninfa A.J., Magasanik B.. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1986; 83:5909–5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popham D.L., Szeto D., Keener J., Kustu S.. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989; 243:629–635. [DOI] [PubMed] [Google Scholar]
- 35. Buck M., Cannon W.. Activator-independent formation of a closed complex between sigma 54- holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol. Microbiol. 1992; 6:1625–1630. [DOI] [PubMed] [Google Scholar]
- 36. Su W., Porter S., Kustu S., Echols H.. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:5504–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y., Bondarenko V., Ninfa A., Studitsky V.M.. DNA supercoiling allows enhancer action over a large distance. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:14883–14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Polikanov Y.S., Bondarenko V.A., Tchernaenko V., Jiang Y.I., Lutter L.C., Vologodskii A., Studitsky V.M.. Probability of the site juxtaposition determines the rate of protein-mediated DNA looping. Biophys. J. 2007; 93:2726–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Correll S.J., Schubert M.H., Grigoryev S.A.. Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J. 2012; 31:2416–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schlick T., Hayes J., Grigoryev S.. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J. Biol. Chem. 2012; 287:5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Norouzi D., Zhurkin V.B.. Topological polymorphism of the two-start chromatin fiber. Biophys. J. 2015; 108:2591–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Osipova T.N., Karpova E.V., Vorob'ev V.I.. Chromatin higher-order structure: two-start double superhelix formed by zig-zag shaped nucleosome chain with folded linker DNA. J. Biomol. Struct. Dyn. 1990; 8:11–22. [DOI] [PubMed] [Google Scholar]
- 43. Woodcock C.L., Frado L.L., Rattner J.B.. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J. Cell Biol. 1984; 99:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng G., Czapla L., Srinivasan A.R., Olson W.K.. How stiff is DNA. Phys. Chem. Chem. Phys. 2010; 12:1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Olson W.K., Colasanti A.V., Czapla L., Zheng G.. Voth GA. Coarse-Graining of Condensed Phase and Biomolecular Systems. 2008; Taylor and Francis Group, LLC; 205–223. [Google Scholar]
- 46. Olson W.K., Gorin A.A., Lu X.-J., Hock L.M., Zhurkin V.B.. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci., U.S.A. 1998; 95:11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Teif V.B., Kepper N., Yserentant K., Wedemann G., Rippe K.. Affinity, stoichiometry and cooperativity of heterochromatin protein 1 (HP1) binding to nucleosomal arrays. J. Phys. Condens. Matter. 2015; 27:064110. [DOI] [PubMed] [Google Scholar]
- 48. Olson W.K., Clauvelin N., Colasanti A.V., Singh G., Zheng G.. Insights into gene expression and packaging from computer simulations. Biophys. Rev. 2012; 4:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bondarenko V., Liu Y., Ninfa A., Studitsky V.M.. Action of prokaryotic enhancer over a distance does not require continued presence of promoter-bound sigma54 subunit. Nucleic Acids Res. 2002; 30:636–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ricci M.A., Manzo C., Garcia-Parajo M.F., Lakadamyali M., Cosma M.P.. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015; 160:1145–1158. [DOI] [PubMed] [Google Scholar]
- 51. Ranjan A., Mizuguchi G., FitzGerald P.C., Wei D., Wang F., Huang Y., Luk E., Woodcock C.L., Wu C.. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013; 154:1232–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Muller F., Tora L.. Chromatin and DNA sequences in defining promoters for transcription initiation. Biochim. Biophys. Acta. 2014; 1839:118–128. [DOI] [PubMed] [Google Scholar]
- 53. Ozer G., Luque A., Schlick T.. The chromatin fiber: multiscale problems and approaches. Curr. Opin. Struct. Biol. 2015; 31:124–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.