Abstract
A better understanding of the effects that oxidative lesions have on RNA is of importance to understand their role in the development/progression of disease. 8-oxo-7,8-dihydroguanine was incorporated into RNA to understand its structural and functional impact on RNA:RNA and RNA:DNA duplexes, hairpins and pseudoknots. One to three modifications were incorporated into dodecamers of RNA [AAGAGGGAUGAC] resulting in thermal destabilization (ΔTm – 10°C per lesion). Hairpins with tetraloops c-UUCG*-g* (8-10), a-ACCG-g* (11-12), c-UUG*G*-g* (13-16) and c-ACG*G*-g* (17-20) were modified and used to determine thermal stabilities, concluding that: (i) modifying the stem leads to destabilization unless adenosine is the opposing basepair of 8-oxoGua; (ii) modification at the loop is position- and sequence-dependent and varies from slight stabilization to large destabilization, in some cases leading to formation of other secondary structures (hairpin→duplex). Functional effects were established using the aptamer for preQ1 as model. Modification at G5 disrupted the stem P1 and inhibited recognition of the target molecule 7-methylamino-7-deazaguanine (preQ1). Modifying G11 results in increased thermal stability, albeit with a Kd 4-fold larger than its canonical analog. These studies show the capability of 8-oxoG to affect structure and function of RNA, resulting in distinct outcomes as a function of number and position of the lesion.
INTRODUCTION
Oxidative damage has been an area of intense study on DNA, due in part to its mutagenic implications (1). Although it is known that oxidative stress can lead to 14–25 times more damage in RNA (2,3) than its 2΄-deoxy-analog, oxidation of RNA has not been as studied. Interestingly, this phenomenon has been proposed to play a role in the development/progression of disease (4–6) thus making this topic of wide interest, relevance, and with potential for new discoveries (4,7–9). The focus of this study is on exploring the structural and functional effects of arguably one of the most important oxidative lesions, 8-oxo-7,8-dihydroguanine, in RNA using sequences that arrange into different structural motifs, i.e. RNA:RNA duplex, RNA:DNA heteroduplex, hairpins and pseudoknots, as models.
Structure of 8-oxo-7,8-dihydroguanine
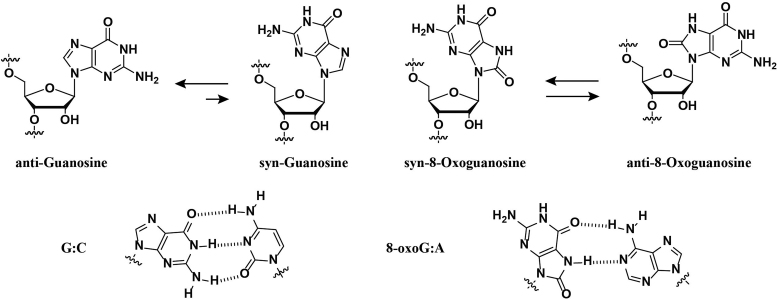
Among the four canonical nucleobases, guanosine has the lowest redox potential [1.29 V versus normal hydrogen electrode (NHE), pH 7] and is therefore a site prone for oxidation in DNA and RNA (10,11). One of the main products that forms is 8-oxo-7,8-dihydroguanine (8-oxoGua) and is generated from different pathways that can be initiated via the addition of reactive oxygen species (ROS) such as hydroxyl radical, superoxide or hydroperoxy radical onto the C8-position of guanosine (12,13). Structurally, it is well established that the keto-enol equilibrium lies toward the former, with two ionization values at (pKa, N1-H 8.6 and N7-H 11.7) (14). Importantly, functionalization at the C8-position of guanosine induces a conformational change around the glycosidic bond to its syn-isomer that may result in stable 8-oxoGua:A base-pair mismatches. The rotation around the glycosidic bond is, in part, the source of differences in structure and reactivity (vide infra, Scheme 1) (15,16).
Scheme 1.
Rotation around the glycosidic bond leads to the syn-conformation, that in turn, stabilizes the 8-oxoG:A basepair mismatch.
The redox activity of this lesion has been explored in Torula yeast RNA (17) and its presence has been shown in rRNA, which provides a site for redox active iron that in turn promotes its formation (18). 8-oxoGua has also been reported to form in mRNA in vivo via treatment of human cultured cells with paraquat (used as source to generate ROS) (19). This lesion has in turn a lower redox potential (0.74 V versus NHE, pH 7) (11) and may serve as an intermediate precursor to further oxidation products such as spiroiminodihydantoin and 5-guanidinohydantoin (20) and may also play a role in the generation of abasic sites in vitro (21).
Relationship to disease and biochemical pathways
The link between oxidative modifications in RNA and disease has been reviewed recently and was suggested as a contributing factor in the development/progression of diabetes, hemochromatosis, heart failure and β-cell destruction among others (6). Particular examples that illustrate the presence of 8-oxoGua include instances where its formation (i) was monitored via immunocytochemistry and related to increases in amyloid β in different regions of post-mortem brains of patients that suffered from Alzheimer's disease (22); (ii) was found on urine samples of patients with hereditary hemochromatosis in larger amounts than in DNA (23); and (iii) was linked to patients with muscle atrophy related to ageing or disuse (24).
Regarding functional aspects in mRNA, the presence of 8-oxoGua can stall the elongation phase of translation and inhibit protein synthesis in a position independent manner with a rate reduction greater than three orders of magnitude (25). This is consistent with another report, where peptide synthesis was greatly inhibited and led to the formation of truncated products (26). Structurally, the anti→syn conformational change induced by 8-oxoGua results in H-bond destabilization of canonical Watson Crick basepair, G:C, in addition to a basepair mismatching capability with adenine (Ade). This variation has been reported as the root of changes in function as follow:
- Basepairing properties of 8-oxoGua have been reported using synthetic oligonucleotides (ONs) of RNA. Thermal denaturation experiments demonstrated that the presence of a single lesion results in destabilization of an RNA:RNA duplex structure. Sequences containing C or U as the opposing basepair resulted in destabilization, while stabilization was observed in the cases of A or G (27), with respect to their canonical analogs. Interestingly, a different pattern was observed on RNA:DNA heteroduplex structures, where the basepairing preference of 8-oxoG was dC > T > dA > dG. A trend that was corroborated using reverse transcriptases [human immunodeficiency virus (HIV-RT), Rous associated virus 2 (RAV2-RT) and Moloney murine leukemia virus (MMLV)] in the synthesis of cDNA (28,29).
- 8-OxoGTP was efficiently incorporated into RNA opposite C and A by bacterial and human polymerases, Escherichia coli RNA pol and RNA Pol II, displaying similar kinetic parameters (30). These observations suggest that the presence of 8-oxoGTP in the nucleotide pool may result in its incorporation into RNA.
- The mispairing capability was also demonstrated on nick sealing (3΄-OH → 5΄-PO4) by Deinococcus radiodurans RNA ligase, noting that ONs containing 8-oxoGua were equally nicked when the opposing base was C or A. In this same work, 8-oxoGua inflicted a 3-fold decrease in the nicking rate with respect to its canonical analog (31).
- 8-oxoGua has also been incorporated into ONs of RNA to study their chemical stability toward strand scission in the presence of aniline. The lesion rendered a higher stability and resistance compared to its DNA analogs (using piperidine), however, it remains to be seen if 8-oxoGua may survive for longer periods of time in cellular environments (32).
Mechanisms that cope with oxidative stress and its resulting adducts (such as 8-oxoGua) have been proposed, e.g. elimination of 8-oxo-GTP from the nucleotide pool as an important process in protein synthesis (33); and selective protein recognition of RNA containing 8-oxoG (34–37). Despite all these studies, the thermal stability and effects of this lesion has not been tested in the context of RNA structure. To address this need, we set out to establish the effects and thermal stability of strands of RNA containing 8-oxoGua with sequences designed to fold into structural motifs that are ubiquitous in nature such as duplexes (multiple lesions), hairpins or pseudoknots (38–40). We chose a system with established behavior to test for function–structure relationships with the aptamer for pre-Q1 as model (41) for functional outcomes. This aptamer is a relatively well-understood system that, in contrast to other riboswitches, is small in size (33-nucleotide long) and with available crystal structures that contain its cognate bound ligand (preQ1) (42).
MATERIALS AND METHODS
General methods
1H NMR, 13C nuclear magnetic resonance (NMR) and 31P NMR spectra were recorded at 300, 75 and 121.5 MHz respectively, using a standard broadband multinuclear probe on a 300MHz Avance III platform from Bruker. IR spectra were recorded on a diamond attenuated total reflectance sampler using powders of pure materials. Pyridine, methylene chloride, dimethyl sulfoxide, N,N-dimethylformamide and Hunig's base were distilled over calcium hydride. Tetrahydrofuran was distilled over sodium and benzophenone. All other reagents were used as purchased without further purification. All intermediates and compounds analyzed for high-resolution mass spectrometry were carried out via electrospray ionization/atmospheric pressure chemical ionization.
The oxidative lesion 8-oxoGua was incorporated into ONs of RNA via standard solid-phase synthesis using its corresponding phosphoramidite. The synthesis was achieved by using an adapted procedure from those previously reported (26,27). We found that keeping the C8-benzyloxy group prior to tritylation led to increased yields using standard procedures. Attempts to install the silane protecting groups without the benzyloxy group failed to work in our hands or resulted in diminished yields. The resulting phosphoramidite was obtained in good yields and led to good coupling efficiencies (>97%, see Supplementary Data pp. S21–23).
It is worth noting that the hybridization of all ONs was carried out in the presence of MgCl2 (5 mM) and heat to 90°C with slow cooling to rt (over ca. 3 h), and with no detectable degradation. Since it has been reported that divalent Mg2+ catalyzes the cleavage of RNA in a temperature-, pH- and buffer-dependent manner (43,44) we further tested the described conditions. The lack of non-enzymatic hydrolysis was measured via mobility shift assays (no degradation products observed), or mass spectrometry (MALDI-TOF showed no degradation in samples before/after heating to 90°C in the described buffer system). These observations indicate the stability of the ONs used herein in the presence of 5 mM MgCl2, 10 mM NaCl, 1 mM sodium phosphate at pH 7.3 upon heating. Longer RNAs are known to be unstable under physiological MgCl2 concentrations in vitro (45), which suggests that the length of the strands used in this work (12–34 nt long) play an important role in this respect. A contributing role may be that larger strands can fold into structurally more complex structures that create micro-environments with distinct properties (46).
RNA synthesis
ONs 1–8, 10–13 and 15–23 were synthesized on a 394 ABI DNA/RNA synthesizer using controlled pore glass (CPG) supports and 2΄-O-t-butyldimethylsilyl-phosphoramidites (purchased from Glen Research). 0.25M 5-Ethylthio-1H-Tetrazole in Acetonitrile was used as the coupling reagent, 3% dichloroacetic acid in dichloromethane was used for de-tritylation, a 2,6-lutidine/acetic anhydride solution was used for capping and an I2/tetrahydrofuran/pyridine solution was used in the oxidation step. Coupling times of 10 min were used. ONs were deacetylated/debenzoylated and cleaved from the CPG support in the presence of 1:1 aq. methylamine (40%) and aq. ammonia (40%) with heat (60°C, 1.5 h). A mixture of N-methylpyrrolidinone/triethylamine/hydrogen fluoride (3:2:1) was used for deprotection of the tert-butyldimethylsilyl groups (60°C, 1 h) followed by purification via electrophoresis [20% denaturing polyacrylamide gel electrophoresis (PAGE)]. C18-Sep-Pak cartridges were obtained from Waters and used to desalt the purified oligomers using 5 mM NH4OAc as the elution buffer. ONs were redissolved in H2O and used as obtained for subsequent experiments. ONs 9 and 14 were purchased from IDT-DNA and, following quantification via UV-vis, used without further purification.
RNA characterization (MALDI-TOF)
Mass spectra (matrix assisted laser desorption-time of flight, MALDI-TOF MS) for all the ONs were prepared using a slightly changed procedure from that reported previously (47). C18 Zip Tip pipette tips were used to desalt and spot each ON as follows: (i) wash tip with 50% acetonitrile (10 μl × 2); (ii) equilibrate tip with 0.1% trifluoroacetic acid (TFA) (10 μl × 2); (iii) load tip with sample (typically 100–150 picomol); (iv) wash tip with 0.1% TFA (10 μl × 2); (v) wash tip with water (10 μl × 2); (vi) elute sample into matrix (10 μl of 25 mM-2,4,6-trihydroxyacetophenone monohydrate, 10 mM ammonium citrate, 300 mM ammonium fluoride in 50% acetonitrile); (vii) spot directly onto MALDI plate. All analyses were carried out on an ABI 4800 Plus MALDI-TOF/TOF mass spectrometer in positive mode.
UV-vis spectroscopy
Concentrations of ONs for which a secondary structure was not detected were obtained via UV-vis using a 1 mm pathlength with 1 μl volumes. All other were calculated at 65°C using a cuvette with a 1 cm pathlength. Extinction coefficients for monomers were obtained by preparing stock solutions (1–5 mM) of guanosine or 8-oxo-7,8-dihydroguanosine in water, followed by sequential dilutions to yield concentrations in the 100–500 μM range prior to measurement. Spectra corresponding to single stranded samples of 9–17 were carried out by preparing solutions of each ON (30 μM) in 5 mM MgCl2, 10 mM NaCl, 1 mM sodium phosphate adjusted to pH 7.3. Spectra corresponding to duplex samples were obtained by mixing both strands of interest in a 1:1 ratio followed by heat to 90°C with slow cooling to room temperature. Origin 9.1 was used to plot and normalize spectra of monomers and ONs for comparison.
CD spectroscopy
Circular dichroism (CD) spectra were recorded at various temperatures (PTC-348W1 peltier thermostat) using Quartz cuvettes with a 1 cm path length. Spectra were averaged over three scans (350–200 nm, 0.5 nm intervals, 1 nm bandwidth, 1 s response time) and background corrected with the appropriate buffer or solvent. Solutions containing the RNA strands had the following composition: 2.5 μM RNA, 5 mM MgCl2, 10 mM NaCl, 1 mM sodium phosphate-pH 7.3. All solutions prepared to record melting temperatures were hybridized prior to recording spectra by heating to 90°C followed by slow cooling to room temperature. Melting temperatures were recorded at 210 nm with a ramp of 1°/min and step size of 0.2 with temperature ranges from 4°C to 95°C. A thin layer of mineral oil was added on top of each solution to keep concentrations constant at higher temperatures. Origin 9.1 was used to determine all Tm values and to normalize CD spectra of ss-RNA and ds-ONs for both RNA:RNA duplexesand RNA:DNA heteroduplexes.
Oligonucleotide labeling
T4 polynucleotide kinase (PNK) and γ-32P-ATP-5΄-triphosphate were obtained from Perkin Elmer. ONs were labeled by mixing PNK, PNK buffer, ATP, RNA and water (final volume = 50 μl) according to manufacturer's procedure followed by heating to 37°C for 45 min. Radiolabeled materials were passed through a G-25 sephadex column followed by purification via electrophoresis (20% denaturing PAGE). The bands of interest (slowest) were cut and eluted over a phosphate saline buffer solution (10 mM NaCl, 10 mM Na2P2O7, pH 7.2) for 6 h at 37°C. The remaining solution was filtered and concentrated to dryness under reduced pressure followed by precipitation over NaOAc and ethanol. Supernatant was removed and the remaining ON was dried and re-dissolved in water. Activity was assessed using a Beckmann LS 6500 scintillation counter.
Electrophoretic mobility shift assays
Radiolabeled ONs were mixed in buffers under the desired conditions and all samples were heated to 90°C with slow cooling to room temperature before loading. All samples were electrophoresed using 20% non-denaturing PAGE. Samples were typically mixed in a 1:1 mixture with 75% glycerol buffer or 90% formamide buffer. Samples treated with formamide buffer were also heated to 75°C for 2 min with rapid cooling to rt to ensure complete denaturation. Quantification of radiolabeled ONs was carried out using a Molecular Dynamics Phosphorimager 840 equipped with ImageQuant Version 5.1 software.
Isothermal calorimetry
Measurements were acquired on a MicroCal iTC200 microcalorimeter (Microcal, Inc.) at 25°C with an RNA concentration of 10 μM and small molecule concentration of 100 μM. Both the ligand and small molecule were dissolved in a buffer consisting of 50 mM HEPES, 100 mM NaCl, and 6 mM MgCl2. PreQ1 was titrated into the sample cell in 2.5 μl injections, with a reference power of 10 μcal s-1, initial delay of 600 s, 180 s spacing, and a stirring speed of 750 rpm. Analysis of the data was performed using Origin 7.0 ITC software (Microcal Software Inc) via fitting to a single-site binding model.
RESULTS
RNA:RNA/RNA:DNA duplex studies
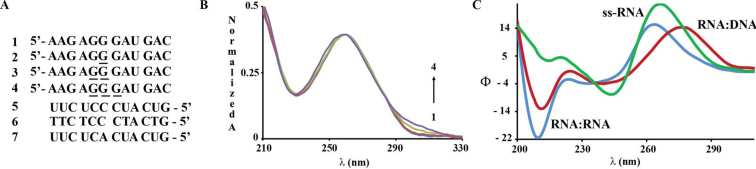
The oxidative modification (8-oxoGua) was incorporated into dodecamers of RNA designed to form a duplex with its complementary strand and without the possibility of forming other thermodynamically stable secondary structures, e.g. hairpin, self-duplex or internal loop. Structural parameters and thermal stabilities were obtained via CD and UV-vis spectroscopy by recording the change in ellipticity or absorbance at 210 and/or 270 nm as a function of increasing temperature. Phosphate buffered solutions (pH 7.2) containing magnesium chloride were used in all experiments. Furthermore, recording the change in absorbance or ellipticity at ca. 270 nm showed no change in the observed trends and only small differences in the ca. Tm (<1°C) values were observed with respect to those obtained at 210 nm (Table 1). The sequence that was used to establish the effects of the oxidative lesion on double stranded samples is shown on Figure 1 and its design is such that 1–3 modifications were tested, where all three sites are placed in a consecutive manner. UV-vis spectra showed a trend displaying higher hypochromicity in the band at ca. 290 nm as a function of increasing number of modifications, thus confirming the presence of the lesion (s) (Figure 1B and Supplementary Data, Supplementary Figure S49 for UV-vis spectra comparison of of 8-oxo-7,8-dihydroguanosine and guanosine). CD spectra obtained for the single strands (1-4) at room temperature did not display any differences among them and displayed the expected band with positive ellipticity at ca. 260 nm (Figure 1C). Following hybridization, all samples displayed A-form geometries, evidenced from their spectral main features, i.e. two bands with positive and negative ellipticity at ca. 270 and 210 nm respectively. There were no measurable differences in the spectra between ON duplexes 1:5 – 4:5 and all melting transitions displayed a monophasic behavior with thermal denaturation temperatures (Tm) in the 30–80°C range.
Table 1. Tm (o C) values of RNA:RNA duplexes of canonical and modified RNAs.
| RNA:RNA | Tm (210 nm) | ΔTm | Tm (270 nm) | ΔTm | OD | ΔTm |
|---|---|---|---|---|---|---|
| 1:5 | 72.6 ± 0.3 | 71.5 ± 0.4 | 70.7 ± 0.5 | |||
| 2:5 | 62.4 ± 0.6 | −10.2 ± 0.5 | 62.1 ± 0.4 | −9.4 ± 0.3 | 62.6 ± 0.7 | −8.1 ± 0.7 |
| 3:5 | 53.4 ± 0.3 | −19.2 ± 0.2 | 52.3 ± 0.6 | −19.2 ± 0.5 | 52.8 ± 0.9 | −17.9 ± 1.1 |
| 4:5 | 42.4 ± 0.3 | −30.2 ± 0.2 | 39.2 ± 0.7 | −32.3 ± 0.7 | 39.9 ± 0.2* | −30.8 ± 0.3 |
| mismatch | ||||||
| 1:7 | 55.9 ± 0.2 | −16.4 ± 0.1 | 56.4 ± 0.5 | −15.1 ± 0.4 | 56.8 ± 1 | −13.9 ± 1.3 |
| 2:7 | 60.3 ± 0.9 | −12.3 ± 0.9 | 59.6 ± 0.8 | −11.9 ± 0.8 | 59.5 ± 0.6 | −11.2 ± 0.6 |
| RNA:DNA | ||||||
| 1:6 | 63.5 ± 0.1 | 62.1 ± 0.4* | 64.9 ± 0.9* | |||
| 2:6 | 52.1 ± 0.6 | −11.5 ± 0.4 | 50.5 ± 0.4* | −10.2 ± 0.5 | 53.8 ± 1.4* | −11.1 ± 2.8 |
| 3:6 | 43.5 ± 0.1 | −20.1 ± 0.1 | 39.4 ± 0.9* | −19.2 ± 0.2 | 38.4 ± 1* | −26.5 ± 1.8 |
| 4:6 | 29.6 ± 0.8 | −33.9 ± 0.7 | 30.1 ± 0.8* | −30.2 ± 0.2 | nd | |
ΔTm (o C) indicates the difference when compared to the canonical analog. All Tm values were obtained by recording the CD signal at 210 or 270 nm. The error represents that of experiments carried out in triplicate or duplicate (*).
Figure 1.
(A) Sequences used with underlined Gs indicating positions where 8-oxoG was incorporated; (B) UV-vis spectra of strands 1–4 (1-blue, 2-red, 3-green, 4-purple) displaying a hyperchromic shift at ca. 310 nm as a function of number of modifications; and (C) spectra representing RNA:RNA and RNA:DNA heteroduplexes (2:5/2:6, 2 μM) and single stranded RNA (2, 7 μM).
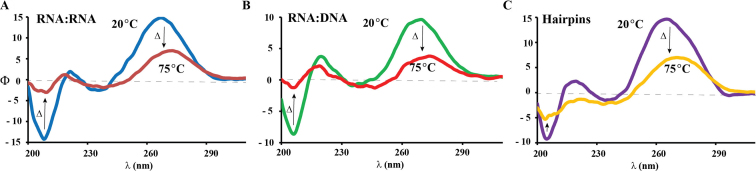
All thermal denaturation experiments were carried out using CD (48–50) or absorbance [Optical Density, (51,52)] by following the bands that can be unambiguously assigned to formation of an A-form duplex with negative ellipticity at 210 nm or positive ellipticity at 270 nm (Figure 2). Interestingly, each modification impacted the thermal stability with decreasing Tm values of ca. 10 ± 1°C per oxidative lesion (Table 1). The same experiments were conducted on RNA:DNA heteroduplex samples and displayed signals corresponding to an A-form geometry. Generally the obtained melting temperatures were ca. 10°C lower than the RNA:RNA duplex analogs, explained by the changes in the geometrical parameters (53), containing features of both A- and B-form helices (54). As in the case of the RNA:RNA duplex structures, increasing the number of modifications resulted in thermal destabilization, corroborated via decreases in the melting temperatures that ranged between 8.5–13.9°C per modification (Table 1, third column), as observed in the RNA:RNA duplex samples.
Figure 2.
Representative CD spectra of (A) RNA:RNA duplex (2:5), (B) RNA:DNA heteroduplex (2:6) and (C) hairpin (8) obtained at 20°and 75°C. Spectra for other oligonucleotides had the same features.
Furthermore, we carried out the same analysis on ONs containing a single basepair mismatch (G:A, 1:7) in the middle of the strand and, not surprisingly, significant destabilization was observed (ca. ΔTm, 16.4°C). On the other hand, basepairing between 8-oxoG and A resulted in added stability (RNA 2:7) with respect to canonical ON 1:7. These observations support that rotation around the glycosidic bond, yielding the syn-conformation, is facilitated in the case of 8-oxoG and that a basepair between 8-oxoG and A is thermodynamically more stable than its 8-oxoG:C analog.
The trends outlined in Table 1 follow the patterns reported previously on modified ONs at a single position, where 8-oxoGua results in decreased thermal stability on 8-oxoGua:C basepairs (ΔTm = −7.2°C on purine rich 13-mers) and increased stabilization on 8-oxoG:A basepairs (ΔTm = + 7.5°C) (27). As well as on RNA:DNA heteroduplex samples, where 8-oxoG results in destabilization when pairing dC and slight stabilization when pairing dA (the results by Kim et al. (29), show a greater affinity toward T). This is also in agreement with previous measurements carried out on DNA:DNA duplexes displaying depressed Tm values in the 1.6–13.8°C range on decamers, dodecamers, and 13-mers of DNA containing 8-oxodGua:C basepairs (55,56).
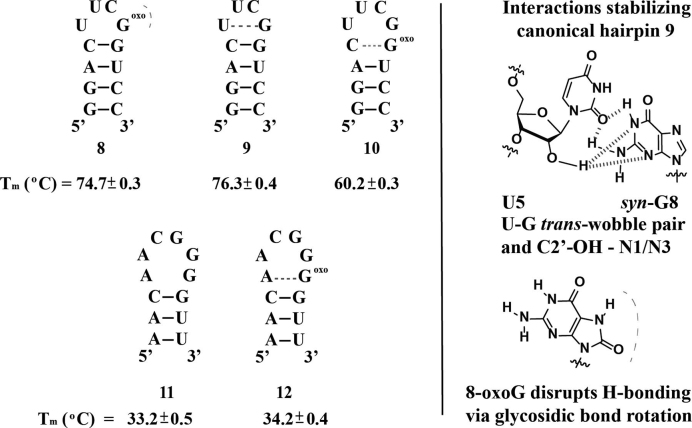
We then decided to test the effect of 8-oxoGua on hairpins of RNA with tetraloops containing the lesion at the closing basepair within the stem or at varying positions on the loop. For our initial studies, we chose a dodecamer containing tetraloop—(UUCG)—that is known to exist as a hairpin [Figure 3, (57)] under our experimental conditions (58, 59). Formation of the hairpin was corroborated via CD, which displayed a band with negative ellipticity at 210 nm and thermal denaturation values that were independent of concentration of RNA. The Tm value that was obtained for the canonical hairpin (ON 9) is ca. 4° higher than that obtained previously by Williams et al., which can be attributed to differences in the buffer system, i.e. added magnesium (5 mM), sodium phosphate (1mM) and sodium chloride (10 mM) in the case presented herein. As in the case of the duplex samples, modification at the closing basepair (ON 10) led to a destabilization that was reflected in a drop in the Tm of ca. 16°C. This observation can be explained by the disruption in the Hydrogen-bond network through the presence of the C-8 carbonyl moiety and is in agreement with calculated (60,61) values for a hairpin with a hexaloop and stem (a:u, g:c, g:c) of 60.3°C. Similarly, modification at the loop (ON 8) led to destabilization, reflected with a ΔTm of −1.6 ± 0.5°C. This observation was initially viewed as a surprising result if one considers that 8-oxoadenosine has been reported to stabilize a hairpin with tetraloop C-(UUCA*)-G (62). However, analysis of the solution NMR structure reported for hairpin 9 shows that stabilization of the hairpin is imparted by the presence of a U:G trans-wobble basepair (63,64), with position G8 existing in the syn-conformation (also observed via Raman spectroscopy) (65). Given the stabilization that is imparted by the various H-bonding interactions (Figure 2, right), it is reasonable to conclude that conformational changes arising from the presence of 8-oxoGua imparts the observed structural destabilization.
Figure 3.
Sequence and hairpins formed for structure 8–12 (left) and H-bonding network previously reported to stabilize the –UUCG– loop (57), determined via NMR and Raman (right). Blue and red dotted lines represent H-bonding that provides stabilization or destabilization respectively.
ONs 11 and 12 were synthesized to explore the basepair mismatch capabilities of 8-oxo-Gua with A in the context of hairpins at the first mismatch within the stem. Both ONs displayed features of hairpin structures and a modest stabilization (ca. ΔTm 1± 0.6) of the hairpin when 8-oxo-Gua basepairs A within the stem was observed. This difference is smaller than that obtained in the case of the RNA:RNA duplex samples (ONs 1:7/2:7, 4.3 ± 0.9), suggesting that formation of an 8-oxoG:A basepair mismatch plays a smaller role on this structural motif.
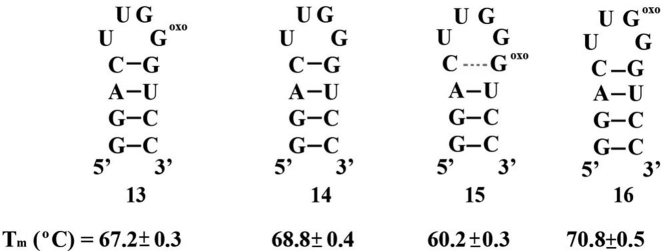
We then decided to test the effect of a modification at a different position in the tetraloop. To this end we synthesized ONs 13–16, where, position C7 of model hairpin 9 at the loop was replaced with a corresponding G (Figure 4). As with systems 8–12, all thermal denaturation measurements were independent of ON concentration. Tm measured for ON 14 was lower than the analogous canonical hairpin 9 by 7.5 ± 0.6°C, in agreement with previous observations, where replacement of this position with U decreases the stability by ca. 5.9°C (66). Contrasting the destabilization observed upon substitution at the loop on ONs 9 and 8, ON 13 displayed only a slight destabilization on the hairpin structure. This suggests that the U:G wobble pair has a lesser impact on structure stability of this hairpin and that it is more stable toward the presence of an oxidative lesion at this position. Furthermore, substitution at the basepair mismatch within the stem (ON 15) led to the expected destabilization and displayed the same thermal denaturation as that observed for ON 10, not surprising given the nature of the stems in both ONs 10 and 15. Interestingly, substitution at G7 (ON 16) had a stabilizing impact on structure that resulted in a ΔTm of ca. 2 ± 0.6°C with respect to canonical hairpin 14. This is a significant change that falls in a trend induced from substitution at this position: C (9, 76.3°C) to 8-oxoGua (16, 70.8°C) to G (14, 68.8°C) to U [64°C, (67)]; and is not an unreasonable result if one takes into consideration the conformational changes expected from 8-oxoG and the strong dependence of stability with sequence on the loop (68). To confirm the formation of a hairpin structure we carried out two control experiments; (i) concentration independent Tm values; and (ii) gel mobility shift assays; with both methods consistent with a unimolecular arrangement.
Figure 4.
Sequence and hairpins formed for structure 13–16 along with their corresponding Tm values.
To further explore the effect of a modification in the loop and its possible interaction with an opposing A, we synthesized hairpins 17–20, modeled after HP 11, containing an –ACGG– tetraloop and a stem that is fully complementary. Thermal denaturation temperatures measured for these ONs were independent of concentration and showed a monophasic transition, thus confirming a unimolecular, distinct structure. In agreement with hairpin formation, calculated values for ON 18, displayed Tm values that matched closely (ca. 51.8°C) (61) with our experimental results (Figure 5). As would be expected from destabilization of the duplex region, modification at the stem (ON 19) displayed a significant destabilization of the hairpin, reflected by a drop in the Tm of 25 ± 0.9°C. This result is in good agreement with calculated values for a hexaloop containing the au/au/cg stem with hexaloop –CACCGA– (ca. Tm = 27.4°C).
Figure 5.
Sequence and hairpins formed for structure 17–19 along with the corresponding 17-17΄ transformation under high salt concentrations and 8-OMe modified ON 17-OMe. Tm values were obtained via CD by measuring the ellipticity change at 270 nm.
In contrast to the observed destabilizing effect when comparing hairpins 8 and 9, substitution at position-8 in the loop with 8-oxoGua (ON 17) led to increased stabilization (ΔTm = + 9.3 ± 0.1°C). Although the result can be explained via the facilitated 8-oxoGua:A basepair formation, we reasoned that this value was larger than what we initially expected and therefore, decided to perform more experiments under different conditions. At first we tested a possible dependence on thermal denaturation temperatures as a function of [RNA]-(0.5-100 M) and [NaCl]-(1 mM–1M). Slight changes in the Tm with increased [NaCl] along with small increments in the ellipticity of both of the main dichroic bands (hypochromic at 210 nm and hypsochromic at 270 nm) were observed, suggestive of a quasi-palindromic structure (ON 17΄) stabilized by the presence of two 8-oxoGua:A basepair mismatches. To confirm our hypothesis we carried out electrophoretic mobility shift assays, which showed formation of a slower moving band (duplex) upon increasing [Na+] (see Supplementary Data, Supplementary Figure S47). This behavior is not uncommon and has been observed on hairpins containing internal basepair mismatches (69) or upon methylation of guanosine at different positions within other tetraloops (70). The same observations were obtained in the absence of magnesium, where structure 17΄ was favored under high salt [Na+] concentrations. Interestingly, neither ONs 18, 19, nor 20 displayed this behavior under any of the described conditions, pointing toward the importance of position G8 in the hairpin→duplex transformation. As a last control in probing the stability of 8-oxoGua pairing with A within the loop, we introduced a substitution containing a methoxy group at the C8 position (ON 17-OMe). We hypothesized that the presence of this group would not have an effect on the thermal stability of the hairpin for two reasons: (i) as with the 8-oxoGua lesion, this modification is known to favor the syn-conformation; and (ii) due to the lack of the N7-H, no added stability is expected from possible H-bonding interactions with A5. First, we determined that ON 17-OMe folded into its corresponding hairpin (concentration independent Tm) as the most thermodynamically stable structure. In agreement with our hypothesis, the presence of this functional group did not affect the structure significantly, with a Tm slightly lower than its canonical analog 18 (ΔTm = 0.8 ± 0.4°C) but significantly lower than the 8-oxoG:A stabilized hairpin 17 (ΔTm = 10.1 ± 0.4°C). In addition, this ON did not display the structural transformation to its quasi-palindromic sequence under the conditions tested (up to 1 M NaCl), confirming the significance of an 8-oxoGua lesion in this position.
Lastly we explored the effect at position-7 in the loop with ON 20, which displayed a ΔTm = 4.1 ± 0.5°C higher than its canonical analog but lower than the loop-modified ON 17 (ΔTm = 5.2 ± 0.5°C). This follows the trend observed between ONs 14 and16, albeit with a larger stabilizing effect that is possibly due to basepair mismatch interactions between A5 and 8-oxoG7.
Functional studies using the pre-Q1 aptamer as a model
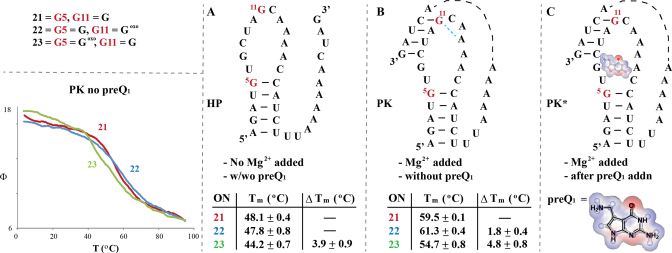
With the acquired information, we then tested the functional impacts of this lesion and opted to explore a system with a well-established behavior. We decided to use the aptamer of the preQ1 class I riboswitch, based on its size (34 nt long), degree of understanding available on this system and the presence of guanosine at various positions to test the effect of 8-oxoGua. The 34-nt RNA portion from Fusobacterium nucleatum specifically recognizes 7-aminomethyl-7-deazaguanine via a pre-organized pseudoknot in the presence of MgCl2 (71) (PK, Figure 6). These riboswitches regulate the biosynthesis of the hypermodified nucleotide queuosine, present at the wobble position of tRNAs with an anticodon loop of GUN (72). In our experiments, we chose to carry out substitutions at positions G5 and G11 (ONs 23 and 22) to probe formation of the hairpin→pseudoknot as well as the recognition of the small molecule. While both of these guanosine units are necessary, given their role in providing structure stability via base-pairing interactions, we expected for position G5 to have a larger impact on the overall structure. The small molecule pre-Q1 was synthesized and purified according to established procedures (73,74). The mechanism for recognition of preQ1 is known to occur via a conformational selection mechanism (75) that uses pre-organized pseudoknot PK to target the small molecule and form complex PK* (Figure 6).
Figure 6.
Sequence corresponding to the aptamer of a preQ1-I riboswitch (A) and the proposed structural transformation in the presence of Mg2+, (B) and the small molecule preQ1 (C), before/after 8-oxoG modification at positions G5 and G11 (labeled in red, ONs 21–23). The structure of preQ1 is shown with its charge density representation in the background. Provided tables indicate the measured Tm values for each ON, recorded via CD at 270 nm (Tm measurements for PK are displayed in the inset, lower left corner).
Since the addition of preQ1 is known to induce a measurable change via CD (76), we decided to probe for variations at different structural stages in this manner using ONs 21–23 (Figure 6). We hypothesized that substitution at G5 would prevent pre-organization while substitution at G11 would still enable formation of a pseudoknot, albeit thermodynamically less stable than its canonical analog 21. We decided to use a buffer system that would be compatible with calorimetry studies, vide infra (50 mM HEPES, 5 mM MgCl2, 100 mM NaCl) at the expense of increased background signal below 220 nm (bands of interest were at ca. 270 and 240 nm). To ensure that the same concentration was used in all cases, UV-vis and CD were recorded at 75°C before sample preparation. Importantly, we relied on the obtained ΔTm values as no differences in the CD spectra were observed under any of the conditions described in this section. First we probed the effect of 8-oxoG on hairpin (HP) stability, where, thermal denaturation temperatures were obtained in the absence of MgCl2. Knowing of the dependence of pseudoknot (PK) formation with [MgCl2] we took these values as ONs forming hairpins HP 21–23 exclusively. Gratifyingly, Tm values measured under these experimental conditions displayed a trend consistent with what one would predict from the behavior of hairpins containing 8-oxoGua at the stem or at the loop. That is, HP 23 with a modification at the stem (G5) displayed a decrease in the Tm that is consistent with destabilization associated with the presence of 8-oxoG positioned at the closing base-pair (Figure 6A). Furthermore, modification at the loop (position G11, HP 22) did not affect the thermal denaturation temperature of the hairpin given its remote location with respect to the stem. These results were independent of the presence of preQ1 (5 molar equivalents added in control experiments). The noteworthy result in this series is the fact that HPs 21 and 22 have unchanged Tm (different from results in the presence of MgCl2, vide infra), suggesting that the obtained ΔTm with HP 23 is indicative of destabilization of the hairpin structure.
Taking into account that the presence of Mg2+ ions are known to stabilize formation of secondary structures, we then carried out experiments in the presence of MgCl2 (5 mM) without the small molecule. Given the likelihood that pre-organization to pseudoknot PK occurs with the addition of this divalent cation, our goal was to gain information on the effect of 8-oxoGua on a different structural motif. As indicated on Figure 6B, the ΔTm between canonical PK 21 and G5-modified PK 23 was within error of strands in the absence of Mg2+, which is indicative of the destabilization arising from formation of HP. The main difference in this series arose with the G11 modified PK 22, where increased stabilization was observed with respect to the canonical analog PK 21 and a ΔTm of ca. + 1.8°C. This observation (in addition to the unchanged CD spectra) suggests that a similar degree of pre-organization or duplex character is present in these ONs (hairpin or pseudoknot) and that the presence of G11 adds stability that may be associated with the A-rich sequence and the capability of 8-oxoG to form basepair mismatches with A. The H-bonding illustrated with A14 is highlighted on Figure 6B since this is an interaction that has been reported [in crystal structures (77) and models (78)], however, networks with other adenosine units in the strand cannot be ruled out. In order to measure the effect of preQ1 we carried out the thermal denaturation experiments after the addition of the small molecule (3 molar equivalents) to observe the same trend (Tm, 22 > 21 > 23) as that observed without preQ1 or Mg2+ (Figure 6C). Tm measurements in the presence of Mg2+ and preQ1 yielded values that were within error of those obtained in the absence of the small molecule. Examples of aptamers where the difference in the structural thermal stability before and after addition of the target molecule is not significant or slightly lower have been reported (79).
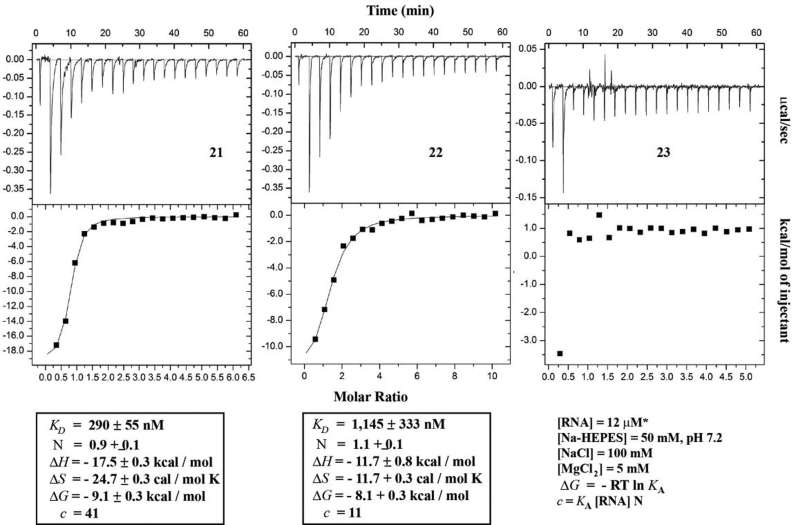
We next set out to establish the recognition properties of all ONs via isothermal titration calorimetry (ITC) as described previously (80). ITC analysis carried out on control aptamer 21 displayed a dissociation constant (Kd) within error of that previously found via fluorescence [same sequence; (41)] and ca. 20-fold higher than that previously reported via ITC, possibly due to variations in the sequence (81). On the other hand, G11-modified aptamer 22 displayed binding to preQ1 with a 5-fold decrease in the dissociation constant and a decrease in the binding affinity (ΔΔG = + 1 kcal/mol) (Figure 7). Furthermore, consistent with the disruption of pseudoknot PK formation, binding to preQ1 was inhibited in the case of G5-modified ON 23.
Figure 7.
ITC data for the recognition of ONs 21–23 toward preQ1 is shown. ITC for ON 22 was taken with [RNA] = 5.5 μM in the case shown.
DISCUSSION
The structural and functional effects that 7,8-dihydro-8-oxoguanine (8-oxoGua) has on ONs of RNA folding into different structural motifs is reported. It was found that each lesion destabilized duplex structure by ΔTm of ca. 10°C per lesion (1:5, 2:5, 3:5, 4:5). The additive value can be seen as a measure of how a carbonyl group destabilizes the overall structure while retaining its Watson–Crick base pairing capabilities with C. This is in agreement with a recent report in which the presence of a methoxy group at the C8-position amounts to a thermal destabilization of ca. 6.9°C and a larger benzyloxy group follows this trend to ca. 12.2°C (82). Furthermore positioning of an 8-oxoGua lesion opposing A stabilized the duplex structure by ca. 4.4°C, consistent with the previous observation that 8-oxoGua basepairs with A upon glydosidic bond rotation to its syn-conformational isomer (ONs 1:7, 2:7). The same destabilization trend was observed on RNA:DNA heteroduplex samples (ONs 1:6, 2:6, 3:6, 4:6), indicating that the carbonyl group exerts a similar destabilization effect on either RNA:RNA or RNA:DNA structures. It is noteworthy that Tm measurements obtained at 210 and 270 nm are in close agreement of each other, a result that has been corroborated by our group recently [using modified ONs of RNA (83)] and that highlights the added versatility of using CD for this purpose. Furthermore, the thermal destabilization induced by 8-oxoGua on dodecamers of RNA is within range of previous studies that used DNA:DNA or RNA:RNA duplexes (55,56). We are in the process of examining the effects of the presence of this lesion on duplexes with different sequence contexts and/or position of the lesion.
We then set out to establish how the presence of the oxidative lesion affects structure of hairpins with tetraloops c-UUCG*-g* (8-10), a-ACCG-g* (11-12), c-UUG*G*-g* (13-16) and c-ACG*G*-g* (17-20), where an asterisk (*) represents a site of modification, and rationalized the results as follow:
Modification at the closing base-pair on the stem resulted in destabilization of the hairpin in all cases (ONs 10, 15, 19), evidenced from depressed Tm values, consistent with disruption of H-bonding at the closing basepair and resulting in a hexaloop with three base-pairs at the stem. Interestingly, while modification of a duplex structure (ON 2:5) retains pairing interactions, the impact that 8-oxoG has on the loop is larger as the Watson–Crick bonding is not present. On the other hand, a slight stabilization was observed upon changing of the base to adenosine (ON 11). A direct correlation between this position and structural stability has been reported with other modifications such as 8-oxoadenosine (62, destabilizing), pseudouridine (84, stabilizing) and canonical basepairs (85, varies) and thus, it is reasonable that 8-oxoGua destabilizes structure unless it has an A as an opposing basepair.
Modification at position 4 of the tetraloop had various effects that depended on the position and sequence of the hairpin, and that resulted in stabilization, destabilization or formation of other secondary structures. Hairpin 8 (-UUCG*-) led to a large destabilization, presumably due to disruption of the U-G trans- wobble pair known to stabilize this hairpin. On the other hand, modified hairpin 13 resulted in a small degree of destabilization (Δ\Tm ca. + 1°C) while hairpin 17 stabilized the hairpin (ΔTm ca. + 9.3°C) via an 8-oxoGua:Ade mismatch. Furthermore, the effect on this position in ON 17 was unique in the family of hairpins studied herein in that a hairpin→duplex transformation was observed as a function of [NaCl], also facilitated via a quasi-palindromic sequence (ON 17΄) induced by two 8-oxoGua:Ade basepair mismatches. These observations indicate that this position is highly sensitive to modifications and thus conformational changes that are reflected in the overall structure.
Modification at position 3 of the tetraloop (ON 16 & 20) resulted in stabilization of the structure that was reflected in higher ΔTm of 2 and 4.1°C, with a higher stabilization in the latter occurring from a possible 8-oxoGua:Ade intraloop basepair. Clearly more examples are necessary to establish on whether this is a general trend, however, at least in these sequences, the stabilization is significant and may have an impact on function.
Tetraloops are common secondary structures that have been identified to carry out a variety of biological roles in silencing (86), and as precursors in RNA folding via RNA–RNA and RNA–protein binding interactions (87) among others and thus the importance of maintaining their integrity. While structure destabilization, following oxidative damage, may be a desirable path that leads to degradation of the strand, formation of thermodynamically more stable structures will result in a longer-lived damaged strand that may result in unwanted behavior, e.g. errors in protein synthesis, alterations in RNA–protein interactions, or affected binding affinities (as reported herein).
To put this information in the context of function, we modified the aptamer for preQ1 at positions G5 and G11 (ONs 21–23) and found no change in the CD bands before addition of the small molecule, suggesting a similar degree of secondary structure present, e.g. double-stranded regions. Thermal denaturation measurements complemented this information with the trend (Tm 22 > 21 > 23), assigned to the destabilization of hairpin HP in the case of G5 and added H-bonding interactions in the case of ON 22. In agreement with this statement, ITC analysis showed that ON 23 does not recognize the small molecule while modified aptamer at G11 selects preQ1 as target, albeit with a dissociation constant 4-fold higher than the canonical analog. These binding affinity differences are in agreement with magnitudes observed previously for mutation studies on other aptamers (88). While the effect at position G5 has been addressed (via destabilization at the stem region), that at G11 may be more complex. Since the Tm shows an increase in the thermal stability, it is possible that 8-oxoGua contributes to the stability of the pseudoknot via other H-bonding interactions (PK 22) while sacrificing binding affinity. The lower change in entropy obtained for unmodified aptamer 21 (Figure 7, ΔS) suggests that it has a more rigid structure, a phenomenon that has been observed in other DNA aptamers (89).
In conclusion, we have established that the effects of 8-oxoguanine may vary from destabilization or stabilization of secondary structures, with some cases shifting to a different structural motif, in a position dependent manner. Four small hairpins were used as models that proved to be useful in explaining the function of an oxidatively damaged aptamer. In addition, the position at which a guanosine is oxidized does have an impact on the functional capabilities of the RNA of interest. As new discoveries are made in the field of oxidative damage of RNA, the described behavior should prove useful in addressing function and potential outcomes of the effects of oxidation on RNA and its relationship to other pathways such as maturation, degradation or protein binding amongst others.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the University of Colorado Denver for support in the purchase of the CD spectrometer. Mass Spectrometry of ONs was carried out at the Mass spectrometry core facilities, University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences, Anschutz Medical Campus. We would like to acknowledge Shaun Bevers for advice on the ITC measurements, which were carried out at the Biophysics core facilities, Structural Biology and Biochemistry, University of Colorado Anschutz Medical Campus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Colorado Denver start-up funds; Research and Creative Activities Award (RaCAS, CU Denver) (to K.S.G., Y.J.C.). Funding for open access charge: University of Colorado Denver.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cadet J., Douki J.L., Ravanat J.L.. Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc. Chem. Res. 2008; 41:1075–1083. [DOI] [PubMed] [Google Scholar]
- 2.Hofer T., Badouard C., Edyta B., Ravanat J.-L., Mattsson Å., Cotgreave I.A.. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 2005; 386:333–337. [DOI] [PubMed] [Google Scholar]
- 3.von Sonntag C. Schreck S. DNA and Double-stranded oligonucleotides. Free-Radical-Induced DNA Damage and its Repair. 2006; Berlin Heidelberg: Springer-Verlag; 371–377. [Google Scholar]
- 4.Poulsen H.E., Specht E., Broedbaek K., Henriksen T., Ellervik C., Mandrup-Poulsen T., Tonnesen M., Nielsen P.E., Andersen H.U., Weimann A.. RNA modifications by oxidation: a novel disease mechanism?. Free Rad. Biol. Med. 2012; 52:1353–1361. [DOI] [PubMed] [Google Scholar]
- 5.Castellani R.J., Nunomura A., Rolston R.K., Moreira P.I., Takeda A., Perry G., Smith M.A.. Sublethal RNA oxidation as a mechanism for neurodegenerative disease. Int. J. Mol. Sci. 2008; 9:789–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreira P.I., Nunomura A., Nakamura M., Takeda A., Shenk J.C., Smith M.A., Perry G.. Nucleic acid oxidation in alzheimer disease. Free Rad. Biol. Med. 2008; 44:1493–1505. [DOI] [PubMed] [Google Scholar]
- 7.Küpfer P.A., Leumann C.J.. Erdmann VA, Markiewicz WT, Barciszewski J. Oxidative damage on RNA nucleobases. Chemical Biology of Nucleic Acids, RNA Technologies. 2014; Berlin Heidelberg: Springer-Verlag; 76–89. [Google Scholar]
- 8.Wurtmann E.J., Wolin S.L.. RNA under attack: cellular handling of RNA damage. Crit. Rev. Biochem. Mol. 2009; 44:34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feyzi E., Sundheim O., Westbye M.P., Aas P.A., Vågbø C.B., Otterlei M., Slupphaug G., Krokan H.E.. RNA base damage and repair. Curr. Pharm. Biotechnol. 2007; 8:326–331. [DOI] [PubMed] [Google Scholar]
- 10.Steenken S., Jovanovic S.V.. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997; 119:617–618. [Google Scholar]
- 11.Steenken S., Jovanovic S.V., Bietti M., Bernhard K.. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2΄-deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J. Am. Chem. Soc. 2000; 122:2373–2374. [Google Scholar]
- 12.Singh T.A., Rao B.S.M., O'Neill P.. Radical chemistry of 8-oxo-7,8-dihydro-2΄-deoxyadenosine and 8-oxo-7,8-dihydro-2΄-deoxyguanosine: apulse radiolysis study. J. Phys. Chem. B. 2010; 114:16611–16617. [DOI] [PubMed] [Google Scholar]
- 13.Steenken S. Purine bases, nucleosides, and nucleotides: aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts. Chem. Rev. 1989; 89:503–520. [Google Scholar]
- 14.Cho B.P., Kadlubar F.F., Culp S.J., Evans F.E.. 15N Nuclear magnetic resonance studies on the tautomerism of 8-hydroxy-2΄-deoxyguanosine, 8-hydroxyguanosine, and other C8-substituted guanine nucleosides. Chem. Res. Toxicol. 1990; 3:445–452. [DOI] [PubMed] [Google Scholar]
- 15.Thiviyanathan V., Somasunderam A., Hazra T.K., Mitra S., Gorenstein D.G.. Solution structure of a DNA duplex containing 8-hydroxy-2΄-deoxyguanosine opposite deoxyguanosine. J. Mol. Biol. 2003; 325:433–442. [DOI] [PubMed] [Google Scholar]
- 16.McAuley-Hecht K.E., Leonard G.A., Gibson N.J., Thomson J.B., Watson W.P., Hunter W.N., Brown T.. Crystal structure of a DNA duplex containing 8-hydroxyguanine-adenine base pairs. Biochemistry. 1994; 33:10266–10270. [DOI] [PubMed] [Google Scholar]
- 17.Yanagawa H., Ogawa Y., Ueno M.. Redox ribonucleosides. J. Biol. Chem. 1992; 267:13320–13326. [PubMed] [Google Scholar]
- 18.Honda K., Smith M.A., Zhu X., Baus D., Merrick W.C., Tartakoff A.M., Hattier T., Harris P.L., Siedlak S.L., Fujioka H. et al. Ribosomal RNA in alzheimer disease is oxidized by bound redox-active iron. J. Biol. Chem. 2005; 280:20978–20986. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka M., Chock P.B., Stadtman E.R.. Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alshykhly O.R., Fleming A.M., Burrows C.J.. 5-Carboxamido-5-formamido-2-iminohydantoin, in addition to 8-oxo-7,8-dihydroguanine, is the pamor product of the iron-Fenton or X-ray radiation-induced oxidation of guanine under aerobic reducing conditions in nucleoside and DNA contexts. J. Org. Chem. 2015; 80:6996–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka M., Song H., Küpfer P.A., Leumann C.J., Sonntag W.E.. An assay for RNA oxidation induced abasic sites using the aldehyde reactive probe. Free Rad. Res. 2011; 45:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nunomura A., Tamaoki T., Tanaka K., Motohashi N., Nakamura M., Hayashi T., Yamaguchi H., Shimohama S., Lee H.-G., Zhu X. et al. Intraneuronal amyloid β accumulation and oxidative damage to nucleic acids in Alzheimer disease. Neurobiol. Dis. 2010; 37:731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broedbaek K., Poulsen H.E., Weimann A., Kom G.D., Schwedhelm E., Nielsen P., Böger R.H.. Urinary excretion of biomarkers of oxidatively damaged DNA and RNA in hereditary hemochromatosis. Free Rad. Biol. Med. 2009; 47:1230–1233. [DOI] [PubMed] [Google Scholar]
- 24.Hofer T., Marzetti E., Xu J., Seo A.Y., Gulec S., Knutson M.D., Leeuwenburgh C., Dupont-Versteegden E.E.. Increased iron content and RNA oxidative damage in skeletal muscle with aging and disuse atrophy. Exp. Gerontol. 2008; 43:563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S.. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014; 9:1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calabretta A., Küpfer P.A., Leumann C.J.. The effect of RNA base lesions on mRNA translation. Nucleic Acids Res. 2015; 43:4713–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koga Y., Taniguchi Y., Sadaki S.. Synthesis of the oligoribonucleotides incorporating 8-oxo-guanosine and evaluation of their base pairing properties. Nucleosides Nucleotides Nucleic Acids. 2013; 32:124–136. [DOI] [PubMed] [Google Scholar]
- 28.Kim S.K., Yokoyama S., Takaku H., Moon B.J.. Oligoribonucleotides containing 8-oxo-7,8-dihydroguanoisne and 8-oxo-7,8-dihydro-2΄-O-methylguanosine: Synthesis and base pairing properties. Bioorg. Med. Chem. Lett. 1998; 8:939–944. [DOI] [PubMed] [Google Scholar]
- 29.Kim S.K., Lee S.H., Kwon O.-S., Moon B.J.. DNA•RNA Heteroduplex containing 8-oxo-7,8-dihydroguanosine: base pairing, structures, and thermodynamic stability. J. Biochem. Mol. Biol. 2004; 37:657–662. [DOI] [PubMed] [Google Scholar]
- 30.Kamiya H., Suzuki A., Yamaguchi Y., Handa H., Harashima H.. Incorporation of 8-hydroxyguanosine (8-oxo-7,8-dihydroguanosine) 5΄-triphosphate by bacterial and human RNA polymerases. Free Rad. Biol. Med. 2009; 46:1703–1707. [DOI] [PubMed] [Google Scholar]
- 31.Schmier B.J., Shuman S.. Effects of 3΄-OH and 5΄-PO4 base mispairs and damaged base lesions on the fidelity of nick sealing by Deinococcus radiodurans RNA ligase. J. Bacteriol. 2014; 196:1704–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming A.M., Alshykhly O., Zhu J., Muller J.G., Burrows C.J.. Rates of chemical cleavage of DNA and RNA olibomers containing guanine oxidation products. Chem. Res. Toxicol. 2015; 28:1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishibashi T., Hayakawa H., Ito R., Miyazawa M., Yamagata Y., Sekiguchi M.. Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Res. 2005; 33:3779–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J., Li Z.. Human polynucleotide phosphorylase reduces oxidative RNA damage and ptrotects HeLa cell against oxidatieve stress. Biochem. Biophys. Res. Commun. 2008; 372:288–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayakawa H., Sekiguchi M.. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry. 2006; 45:6749–6755. [DOI] [PubMed] [Google Scholar]
- 36.Hayakawa H., Uchiumi T., Fukuda T., Ashizuka M., Kohno K., Kuwano M., Sekiguchi M.. Binding capacity of human YB-1 protein for RNA containing 8-oxoguanine. Biochemistry. 2002; 41:12739–12744. [DOI] [PubMed] [Google Scholar]
- 37.Hayakawa H., Kuwano M., Sekiguchi M.. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry. 2001; 40:9977–9982. [DOI] [PubMed] [Google Scholar]
- 38.Deng N.-J., Cieplak P.. Free energy profile of RNA hairpins: a molecular dynamics simulation study. Biophys. J. 2010; 98:627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma H., Proctor D.J., Kierzek E., Kierzek R., Bevilacqua P.C., Gruebele M.. Exploring the energy landscape of a small RNA hairpin. J. Am. Chem. Soc. 2006; 128:1523–1530. [DOI] [PubMed] [Google Scholar]
- 40.Gareiss P.C., Sobczak K., McNaughton B.R., Palde P.B., Thornton C.A., Miller B.L.. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA – MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1). J. Am. Chem. Soc. 2008; 130:16254–16261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santner T., Rieder U., Kreutz C., Micura R.. Pseudoknot preorganization of the preQ1 class I riboswitch. J. Am. Chem. Soc. 2012; 134:11928–11931. [DOI] [PubMed] [Google Scholar]
- 42.Spitale R.C., Torelli A.T., Krucinska J., Bandarian V., Wedekind J.E.. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J. Biol. Chem. 2009; 284:11012–11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerard G.G., Collins S., Smith M.D.. Excess dNTPs minimize RNA hydrolysis during reverse transcription. Biotechniques. 2002; 33:984–990. [DOI] [PubMed] [Google Scholar]
- 44.AbouHaidar M.G., Ivanov I.G.. Non-enzymatic RNA promoted by the combined catalytic activity of buffers and magnesium ions. Naturforsch. 1999; 54c:542–548. [DOI] [PubMed] [Google Scholar]
- 45.Kilburn D., Behrouzi R., Lee H.-T., Sarkar K., Briber R.M., Woodson S.A.. Entropic stabilization of folded RNA in crowded solutions measured by SAXS. Nucleic Acids Res. 2016; doi:10.1093/nar/gkw597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pullman A., Pullman B.. Molecular electrostatic potential of the nucleic acids. Q. Rev. Biophys. 1978; 14:289–380. [DOI] [PubMed] [Google Scholar]
- 47.Chapman E.J., DeRose V.J.. Enzymatic processing of platinated RNAs. J. Am. Chem. Soc. 2010; 132:1946–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mergny J.-L., Lacroix L.. Analysis of thermal melting curves. Oligonucleotides. 2003; 13:515–537. [DOI] [PubMed] [Google Scholar]
- 49.Jin R., Breslauer K.J., Jones R.A., Gaffney B.L.. Tetraplex formation of a guanine-containing nonamerid DNA fragment. Science. 1990; 250:543–546. [DOI] [PubMed] [Google Scholar]
- 50.Guo Q., Lu M., Kallenbach N.R.. Adenine affects the structure and stability of telomeric sequences. J. Biol. Chem. 1992; 267:15293–15300. [PubMed] [Google Scholar]
- 51.Barbas C.F. III, Burton D.R., Silverman G.J.. Quantitation of DNA and RNA. CSH Protoc. 2007; doi:10.1101/pdb.ip47. [DOI] [PubMed] [Google Scholar]
- 52.Keel A.Y., Jha B.K., Kieft J.S.. Structural architecture of an RNA that competitively inhibits RNase L. RNA. 2012; 18:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheatham T.E. III, Kollman P.A.. Molecular dynamics simulations highlight the structural differences among DNA:DNA, RNA:RNA, DNA:RNA hybrid duplexes. J. Am. Chem. Soc. 1997; 119:4805–4825. [Google Scholar]
- 54.Anosova I., Kowal E.A., Dunn M.R., Chaput J.C., Van Horn W.D., Egli M.. The structural diversity of artificial genetic polymers. Nucleic Acids Res. 2016; 44:1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh S.K., Szulik M.W., Ganguly M., Khutsishvili I., Stone M.P., Marky L.A., Gold B.. Characterization of DNA with an 8-oxoguanine modification. Nucleic Acids Res. 2011; 39:6789–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plum G.E., Grollman A.P., Johnson F., Breslauer K.J.. Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry. 1995; 34:16148–16160. [DOI] [PubMed] [Google Scholar]
- 57.Allain R.H.-T., Varani G.. Structure of the P1 helix group I self-splicing introns. J. Mol. Biol. 1995; 250:333–353. [DOI] [PubMed] [Google Scholar]
- 58.Kanyo J.E., Duhamel J., Lu P.. Secondary structure of the r(CUUCGGG) tetraloop. Nucleic Acids Res. 1996; 24:4015–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams D.J., Hall K.B.. Experimental and computational studies of the G[UUCG]C RNA tetraloop. J. Mol. Biol. 2000; 297:1045–1061. [DOI] [PubMed] [Google Scholar]
- 60.Markham N.R., Zuker M.. Keith JM. UNAFold: software for nucleic acid folding and hybridization. Bioinformatics, Volume II. Structure, Function and Applications, number 453 in Methods in Molecular Biology. 2008; Totowa: Humana Press; 3–31. [DOI] [PubMed] [Google Scholar]
- 61.Markham N.R., Zuker M.. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005; 33:W577–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chauca-Diaz A.M., Choi Y.J., Resendiz M.J.E.. Synthesis and biophysical properties of oligonucleotides of RNA containing 7,8-dihydro-8-hydroxyadenosine. Biopolymers. 2015; 3:167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nozinovic S., Fürtig B., Jonker H.R.A., Richter C., Schwalbe H.. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010; 38:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duchart E., Schwalbe H.. Residue specific ribose and nucleobase dynamics of the cUUCGg RNA tetraloop motif by NMR 13C relaxation. J. Biomol. NMR. 2005; 32:295–308. [DOI] [PubMed] [Google Scholar]
- 65.Baumruk V., Gouyette C., Huynh-Dinh T., Sun J.-S., Ghomi M.. Comparison between CUUG and UUCG tetraloops: thermodynamic stability and structural features analyzed by UV absorption and vibrational spectroscopy. Nucleic Acids Res. 2001; 29:4089–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Antao V.P., Tinoco I. Jr. Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992; 20:819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antao V.P., Lai S.Y., Tinoco I. Jr. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991; 19:5901–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varani G. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 1995; 24:379–404. [DOI] [PubMed] [Google Scholar]
- 69.Meroueh M., Chow C.S.. Thermodynamics of RNA hairpins containing single internal mismatches. Nucleic Acids Res. 1999; 27:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Micura R., Pils W., Höbartner C., Grubmayr K., Evert M.-O., Jaun B.. Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic Acids Res. 2001; 29:3997–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rieder U., Kreutz C., Micura R.. Folding of a transcriptionally acting PreQ1 riboswitch. Proc. Nat. Ac. Sci. U.S.A. 2010; 107:10804–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eichhorn C.D., Kang M., Feigon J.. Structure and function of preQ1 riboswitches. Biochem. Biophys. Acta. 2014; 1839:939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gerber H.-D., Klebe G.. Concise and efficient syntheses of preQ1 base, Q base, and (ent)-Q base. Org. Biomol. Chem. 2012; 10:8660–8668. [DOI] [PubMed] [Google Scholar]
- 74.Klepper F., Polborn K., Carell T.. Robust synthesis and crystal-structure analysis of 7-cyano-7-deazaguanine (preQ0 base) and 7-(aminomethyl)-7-deazaguanine (preQ1 base). Helv. Chim. Acta. 2005; 88:2610–2616. [Google Scholar]
- 75.Moschen T., Wunderlich C.H., Spitzer R., Levic J., Micura R., Tollinger M., Kreutz C.. Ligand-detected relaxation dispersion NMR spectroscopy: Dinamics of preQ1-RNA binding. Angew. Chem. Int. Ed. 2015; 54:56–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Resendiz M.J.E., Schön A., Freire E., Greenberg M.M.. Photochemical control of RNA structure by disrupting π-stacking. J. Am. Chem. Soc. 2012; 134:12478–12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jenkins J.L., Krucinska J., McCarty R.M., Bandarian V., Wedekind J.E.. Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. J. Biol. Chem. 2011; 286:24626–24637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Banáš P., Sklenovský P., Wedekind J.E., Šponer J., Otyepka M.. Molecular mechanism of preQ1 riboswitch action: a molecular dynamics study. J. Phys. Chem. B. 2012; 116:12721–12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wacker A., Buck J., Mathieu D., Richter C., Wöhnert J., Schwalbe H.. Structure and dynamics of the deoxyguanosine-sensing riboswitch studied by NMR-spectroscopy. Nucleic Acids Res. 2011; 39:6802–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liberman J.A., Bogue J.T., Jenkins J.L., Salim M., Wedekind J.E.. ITC analysis of ligand binding to preQ1 riboswitches. Method Enzymol. 2014; 549:435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suddala K.C., Rinaldi A.J., Feng J., Mustoe A.M., Eidhhorn C.D., Liberman J.A., Wedekind J.E., Al-Hashimi H.M., Brooks C.L. III, Walter N.G.. Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Res. 2013; 41:10462–10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baranowski C.S., Kotkowiak W., Kierzek R., Pasternak A.. Hybridization properties of RNA containing 8-methoxyguanosine and 8-benzyloxyguanosine. PLOS One. 2015; 10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen C.J., Dzowo Y.K., Wolfbrandt C., Townsend J.S., Kukatin S., Wang H., Resendiz M.J.E.. Synthesis, thermal stability, biophysical properties, and molecular modeling of oligonucleotides of RNA containing 2-O-2-thiophenylmethyl groups. J. Org. Chem. 2016; doi:10.1021/acs.joc.6b01615. [DOI] [PubMed] [Google Scholar]
- 84.Sumita M., Desaulniers J.-P., Chang Y.-C., Chui H.M.-P., Closs L. II, Chow C.S.. Effects of nucleotide substitution and modification on the stability and structure of helix 69 from 28S rRNA. RNA. 2005; 11:1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Serra M.J., Lyttle M.H., Axenson T.J., Schadt C.A., Turner D.H.. RNA hairpin loop stability depends on closing base pair. Nucleic Acids Res. 1993; 21:3845–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duszczyk M.M., Wutz A., Rybin V., Sattler M.. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA. 2011; 17:1973–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thapar R., Denmon A.P., Nikonowicz E.P.. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. WIREs RNA. 2014; 5:49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gilbert S.D., Love C.E., Edwards A.L., Batey R.T.. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007; 46:13297–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pagano B., Martino L., Randazzo A., Giancola C.. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008; 94:562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.